Abstract
Transcription factors of the achaete-scute and atonal bHLH proneural gene family play important roles in neuronal differentiation. They are also involved in neuronal subtype specification through collaboration with homeodomain (HD) transcription factors. However, concerted regulation of these genes and in turn progenitor fate toward distinct lineages within the developing vertebrate brain is not well understood. Fezf2 is an evolutionarily conserved zinc finger protein important for monoaminergic neuronal development in zebrafish. Here, we show that Fezf2 is also critical for GABAergic neuronal fate and investigate how a single transcription factor regulates the identity of multiple neuronal lineages in the developing ventral forebrain. First, our genetic analyses reveal the requirement of the achaete-scute-like genes ascl1a and 1b in serotonergic and GABAergic neuron development, but they are dispensable for the specification of dopaminergic neurons, which is dependent on the atonal-like gene neurog1. Second, the expression of fezf2, ascl1a/1b, and neurog1 demarcates distinct progenitor subpopulations, where fezf2 is required for activating but not maintaining the expression of bHLH genes. Third, Fezf2 is required to activate HD genes otpb and dlx2, which are involved in dopaminergic and GABAergic neuronal development, respectively. Finally, we uncover that Fezf2 is sufficient to increase dopaminergic neuronal numbers but not serotonergic or GABAergic lineages. Together, these findings reveal new mechanisms by which multilineage differentiation is coordinately regulated by a single transcription factor in the vertebrate ventral forebrain.
Introduction
The development of multicellular organisms involves the production of diverse cell types that are of specific identity. The need for both diversity and specificity is particularly prominent in the vertebrate CNS, which initially unfolds from a simple monolayer of germinal neuroepithelia that have the potential to give rise to a vast repertoire of cell types (Jessell, 2000; Livesey and Cepko, 2001; Temple, 2001; Agathocleous and Harris, 2009; Kriegstein and Alvarez-Buylla, 2009). The generation of both proper numbers and types of differentiated cells over a prolonged period necessitates an orchestrated control of progenitor fate. Transcription factors encoded by the basic helix-loop-helix (bHLH)-containing proneural genes, originally discovered through classical genetic studies in Drosophila (Ghysen and Dambly-Chaudiere, 1989; Jan and Jan, 1994), have been shown to play important roles in vertebrate neurogenesis (Anderson and Jan, 1997; Bertrand et al., 2002). Cell cycle exit and neurogenesis are initiated in progenitor cells by upregulation of atonal-like neurogenin (Ma et al., 1996) and achaete-scute-like ascl genes (Guillemot et al., 1993). In addition to promoting the expression of neuronal differentiation markers, proneural genes are also critical for determining subtype-specific neuronal identity, through collaboration with transcription factors of the homeodomain (HD) class (Jessell, 2000; Shirasaki and Pfaff, 2002).
Fezf2 (also known as Fezl, ZNF312, or Zfp312) is an evolutionarily conserved forebrain transcription regulator (Shimizu and Hibi, 2009). In zebrafish embryos, fezf2 is required for proper development of both dopaminergic (DA) and serotonergic (5-HT) neurons (Guo et al., 1999a; Levkowitz et al., 2003). The bHLH proneural gene neurog1 is required for DA neuron development and is regulated by Fezf2 (Jeong et al., 2006). However, it remains to be understood the nature of this regulation, and moreover, how Fezf2 regulates the development of 5-HT neurons is an open question.
In this study, we uncover a new and essential role of fezf2 in GABAergic neuron development and address how fezf2 regulates the cell fate of multiple neuronal lineages in the developing zebrafish ventral forebrain. Our combined molecular genetic studies identify another family of bHLH genes, ascl1a and ascl1b, as essential regulators of 5-HT and GABAergic neuron development and show that Fezf2 activates distinct bHLH genes in stereotypically positioned progenitor subpopulations. Additionally, Fezf2 can also activate HD genes in these lineages, thereby directing them toward specific neuronal fate.
Materials and Methods
Zebrafish strains.
Wild-type embryos (of either sex) were obtained from natural spawning of AB adults of both sexes and raised as described previously (Kimmel et al., 1995). The following mutants and transgenic lines (of either sex) were used: tofm808 (Guo et al., 1999a), neurog1hi1059Tg (Golling et al., 2002), piat25214 (Pogoda et al., 2006), Tg(Hsp70:: Gal4) (Scheer and Camnos-Ortega, 1999), and Tg(UAS::Fezf2) (Jeong et al., 2007).
In situ hybridization, immunostaining, and 5-ethynyl-2′-deoxyuridine assay.
RNA in situ hybridization and immunohistochemistry were performed as described previously (Guo et al., 1999b). For dual fluorescent in situ hybridization (FISH), the antisense RNA probe for fezf2 was labeled with digoxigenin (DIG) (DIG RNA labeling mix; Roche), and the anti-DIG-peroxidase (POD) secondary antibody (Roche; 1:400) was used, followed by color development using Alexa Fluor 555 tyramide (Invitrogen). All other probes were labeled with dinitrophenyl (DNP) (DNP RNA labeling mix; NTP set from Fermentas; DNP-11-UTP from PerkinElmer), and the anti-DNP-POD secondary antibody (1:200; PerkinElmer) was used, followed by color development using Alexa Fluor 488 tyramide (Invitrogen). The procedure was performed as previously described (Filippi et al., 2007). For 5-ethynyl-2′-deoxyuridine (EdU) labeling, EdU was injected into the yolk of embryos at different developmental stages. Embryos were fixed at 48 h postfertilization (hpf) and stained according to the manufacturer's protocol (Invitrogen). For quantification of DA, 5-HT, or GABAergic neurons in the posterior tuberculum (PT)/hypothalamus (H) region, in situ or antibody-labeled cells were visualized under a compound differential interference contrast or fluorescent microscope, and counted manually. This was feasible because of the small number of these neurons in the PT/H region. At least five embryos were quantified per genotype per condition.
Morpholino antisense oligonucleotide injection.
Morpholinos (Gene Tools) were resuspended in nuclease-free water and stored at −80°C. The following gene-specific morpholinos were used in this study: ascl1b MO (Amoyel et al., 2005) and fezf2 MO (Jeong et al., 2007); and 3–4 nl of 0.25 mm were used per embryo.
Results
Fezf2 is essential for the development of GABAergic neurons in the zebrafish ventral forebrain
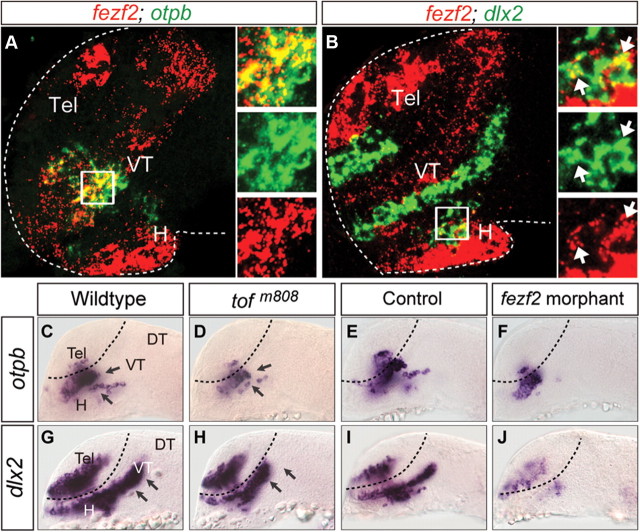
We previously isolated the zebrafish mutant for fezf2, too few (tofm808), which displays defects in the development of DA and 5-HT neurons (Guo et al., 1999a; Levkowitz et al., 2003) in the developing ventral forebrain, which is composed of the dorsal thalamus (DT), ventral thalamus (VT), PT, and H. To determine whether fezf2 regulates the development of additional neuronal types, we examined GABAergic neurons using in situ hybridization for Gad67, a key enzyme involved in GABA synthesis. A loss of gad67 expression was detected prominently in PT and H in the 48 hpf tofm808 mutant (Fig. 1B), whereas those in the telencephalon (Tel), DT, VT (Fig. 1B), and hindbrain (Fig. 1D) were largely unaffected. We have previously also developed morpholino antisense oligonucleotides targeting fezf2, and shown that fezf2 morphants have a severer forebrain defect than the tofm808 mutant, such that the VT subdivision is defective (Jeong et al., 2007). Since VT is predominantly composed of GABAergic neurons, we examined fezf2 morphants for their GABAergic neuronal state and found a significant deficit in VT (Fig. 1F), whereas those in the Tel, midbrain/hindbrain regions remained unaffected (Fig. 1F,H). Together, these results reveal that fezf2 is essential for proper development of GABAergic neurons in the zebrafish ventral forebrain.
Figure 1.
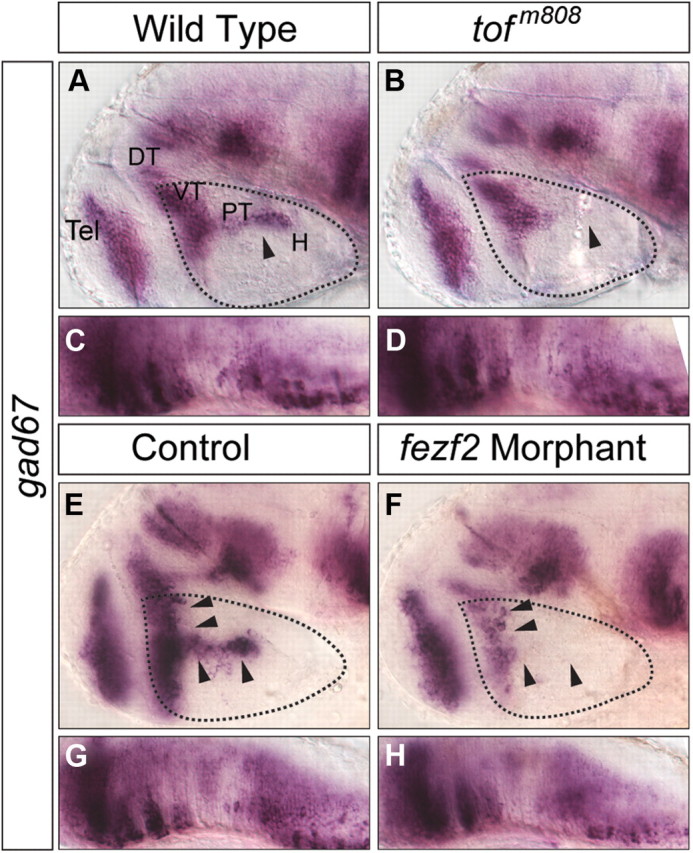
GABAergic neurons marked by gad67 transcript expression are reduced in the ventral forebrain of tofm808 mutant embryos (A–D) and fezf2 morphants (E–H). The solid arrowheads point to the differences between wild-type, mutant embryos and fezf2 morphants. Expression in the hindbrain was shown as controls (C, D, G, H). DT, Dorsal thalamus; H, hypothalamus; PT, posterior tuberculum; VT, ventral thalamus; Tel, telencephalon.
The development of DA versus 5-HT or GABAergic neurons depends on distinct bHLH proneural genes
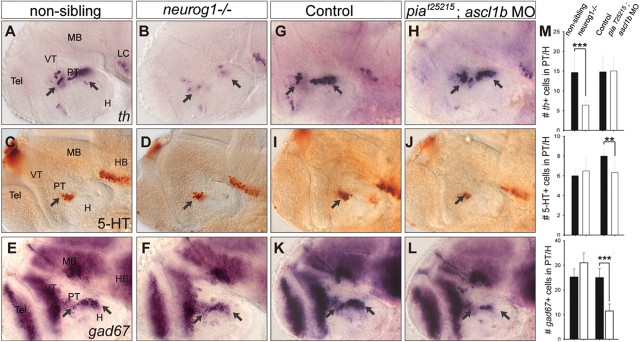
How does Fezf2 direct multilineage differentiation of neuronal progenitors toward DA, 5-HT, or GABAergic fate? Since our previous study uncovered an essential role of the proneural gene neurog1 for DA neuron development (Jeong et al., 2006), we asked whether neurog1 or other members of the proneural gene family are involved in 5-HT or GABAergic neuron specification, by analyzing these neuronal phenotypes in zebrafish mutants for neurog1 (neurog1hi1059) (Golling et al., 2002) and ascl1a (known as piat25215) (Pogoda et al., 2006). In agreement with the analysis of neurog1 morphants (Jeong et al., 2006), DA neurons were reduced in the neurog1−/− mutant (Fig. 2A,B,M), but 5-HT and GABAergic neurons were unaffected (Fig. 2C–F,M). After observing that all three neuronal types were normal in the ascl1a/piat25215 mutant, we analyzed them in the ascl1a/piat25215 mutant injected with a morpholino antisense oligonucleotide targeting ascl1b and found that, while DA neurons were normal (Fig. 2G,H,M), 5-HT (Fig. 2I,J,M) and GABAergic (Fig. 2K–M) neurons in the PT and H were reduced. The hindbrain raphe 5-HT neurons were also reduced in ascl1a and ascl1b double-deficient embryos, consistent with a known role of ascl1 family of genes in vertebrate raphe 5-HT neuronal development (Pattyn et al., 2004). Together, these results indicate that different bHLH genes differentially govern DA, 5-HT, and GABAergic neuronal development in the ventral forebrain.
Figure 2.
GABAergic, DA, and 5-HT neuron development in neurog1- or ascl1a/b-deficient embryos. DA, 5-HT, and GABAergic neurons in the control (A, C, E, G, I, K), neurog1−/− mutant (B, D, F, M), and ascl1b morpholino-injected ascl1a−/− mutant (piat25214) (H, J, L, M) embryos as revealed by whole-mount in situ hybridization with antisense RNA probes directed against gad67, th, or immunostaining with anti-5-HT antibody. DT, Dorsal thalamus; H, hypothalamus; PT, posterior tuberculum; VT, ventral thalamus; Tel, telencephalon. Please refer to Materials and Methods for quantification of these neurons in the PT/H region. Error bars indicate SEM. **P < 0.01; ***p < 0.001.
The expression of fezf2 and bHLH proneural genes implies regulatory hierarchy and defines distinct progenitor subpopulations
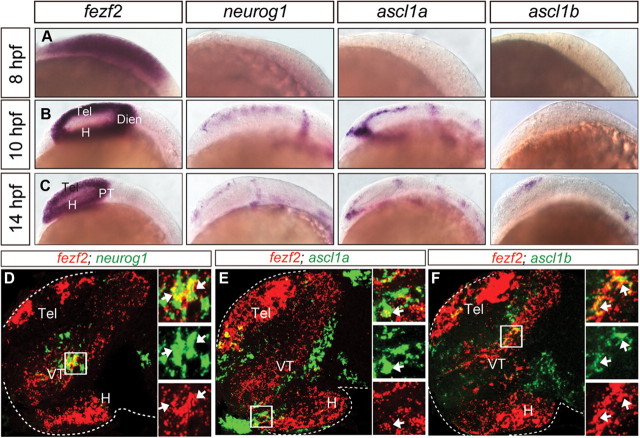
To discern the relationship between fezf2 and proneural genes in neuronal specification, we analyzed their spatial and temporal expression patterns. Through whole-mount in situ hybridization, we found that fezf2 was expressed in the forebrain primordium of the early gastrula embryo, preceding that of all three bHLH genes (Fig. 3A). As development progressed, proneural gene expression became detectable in subpopulations of forebrain progenitors (Fig. 3B,C). Dual FISH at ∼24 hpf provided a higher resolution view of their expression patterns through confocal microscopy. Surprisingly, only a small population of fezf2+neurog1+ cells was detected in the central domain of VT, among those that are singly positive for either fezf2 or neurog1 (Fig. 3D). An even smaller overlap was detected between the expression of fezf2 and ascl1a (Fig. 3E) or ascl1b (Fig. 3F), in anterior H and dorsal VT, respectively. These results reveal distinct progenitor subpopulations in the ventral forebrain demarcated by their expression profiles of fezf2 and bHLH genes.
Figure 3.
Expression patterns of fezf2 and proneuronal genes. A–C, fezf2 expression precedes that of bHLH genes during early development. Images of whole-mount ISH with antisense RNA probes directed against fezf2, neurog1, ascl1a, ascl1b in wild-type embryos at 8 hpf (A), 12 hpf (B), and 14 hpf (C). D–F, Lateral views (∼5 μm confocal projection, which is smaller than an average cell diameter of ∼10 μm) of the 20 hpf embryonic brains show the expression of neurog1 (D), ascl1a/b (E, F) (green), and fezf2 (red), revealed by FISH. H, Hypothalamus; Dien, diencephalon; PT, prethalamus; VT, ventral thalamus; Tel, telencephalon.
Fezf2 is required to activate bHLH genes in distinct progenitor subpopulations
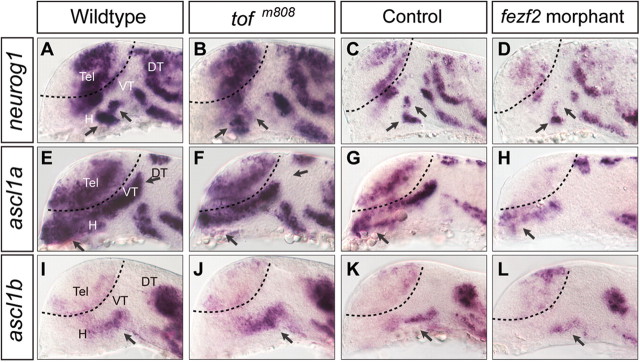
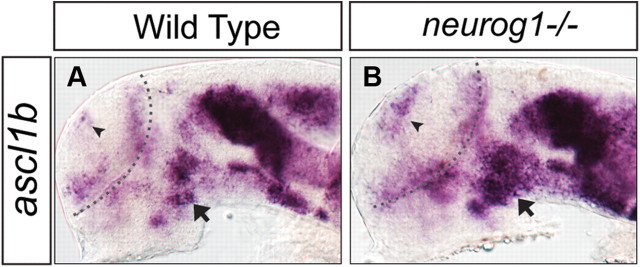
As previously reported (Jeong et al., 2006), we observed a transient but highly reproducible reduction of neurog1 expression in the ventral forebrain (Fig. 4B) of the ∼26-somite stage (∼22 hpf) tofm808 mutant, and this reduction appeared exacerbated in the fezf2 morphant (Fig. 4D). Similar reduction of gene expression was observed for ascl1a (Fig. 4E–H). Unexpectedly, however, ascl1b expression was slightly increased in the ventral forebrain of the tofm808 mutant (Fig. 4I,J) but appeared reduced in the fezf2 morphant (Fig. 4K,L). This increased ascl1b expression could be due to the loss of neurog1 in the tofm808 mutant, as neurog1 was previously shown to repress ascl1 expression in the mouse brain (Fode et al., 2000). We examined ascl1b expression in the neurog1−/− mutant, and indeed an increased ascl1b expression was observed in the ventral forebrain of the neurog1−/− mutant (Fig. 5), supporting the notion that increased ascl1b expression in the tofm808 mutant may be indirectly due to the loss of neurog1-mediated repression, whereas a further reduction of fezf2 activity in the morphant uncovers Fezf2-dependent activation of ascl1b. Together, these results provide genetic evidence to support that Fezf2 is an upstream activator of bHLH genes in the ventral forebrain.
Figure 4.
Proneural gene expression in fezf2-deficient embryos. Images of whole-mount in situ hybridization with antisense RNA probes directed against neurog1, ascl1a, and ascl1b in wild-type (A, E, I), tofm808 mutant embryos (B, F, J), control morpholino- (C, G, K), and fezf2 morpholino-injected embryos (D, H, L) at ∼20 hpf. All images are lateral views of anterior brain regions. Anterior is to the left, and dorsal is up. H, Hypothalamus; PT, posterior tuberculum; Tel, telencephalon.
Figure 5.
Ectopic ascl1b expression in neurog1-deficient embryos. The arrowhead indicates the increased expression in the dorsal telencephalon; the arrows indicate the increased ascl1b expression in the diencephalic region. All images are lateral views of anterior brain regions. Anterior is to the left, and dorsal is up.
Fezf2 is required to activate the HD genes otpb and dlx2 in distinct ventral forebrain progenitor subpopulations
The bHLH proneural genes specify neuronal subtype identity through collaboration with HD genes (Scardigli et al., 2001; Lee and Pfaff, 2003). We therefore examined whether Fezf2 regulated HD genes in addition to proneural genes. The HD gene otpb is expressed in the ventral forebrain (Fig. 6A) and is required for the specification of DA neurons (Blechman et al., 2007; Ryu et al., 2007). Another HD gene, dlx2, which is known to regulate GABAergic neuronal development in mammals (Anderson et al., 1997), is also expressed in the region (Fig. 6B). In the fezf2 mutant and morphant, the expression of these genes was defective in distinct subpopulations of progenitors (Fig. 6C--J). Thus, these results suggest that Fezf2 can activate HD genes in addition to bHLH genes.
Figure 6.
Fezf2 regulates the expression of HD genes otpb and dlx2. A, B, Small subsets of fezf2+ progenitor cells express optb or dlx2 as revealed by FISH. Lateral views of the 20 hpf embryonic brains show that the expression of otpb (A), dlx2 (B) (green) displays small overlap with that of fezf2 (red). C–J, Images of whole-mount in situ hybridization with antisense RNA probes directed against otpb and dlx2 in wild-type (C, G), tofm808 mutant embryos (D, H), control morpholino- (E, I), and fezf2 morpholino-injected embryos (F, J) at ∼20 hpf. All images are lateral views of anterior brain regions. Anterior is to the left, and dorsal is up. DT, Dorsal thalamus; H, hypothalamus; VT, ventral thalamus; Tel, telencephalon.
Fezf2 is sufficient to increase DA but not 5-HT and GABAergic neurons
Given that fezf2 is required for the development of DA, 5-HT, and GABAergic neurons, we next asked whether it is sufficient to generate more of these neurons when overexpressed. Since direct delivery of fezf2 mRNA into one-cell stage embryos causes severe deformity, we examined the consequence of fezf2 overexpression using the heat shock promoter and Gal4-UAS system (Scheer and Camnos-Ortega, 1999; Jeong et al., 2007). To determine when is the appropriate time for heat shock, we performed in vivo birth-dating analysis for the small and quantifiable numbers of DA, 5-HT, and GABAergic neurons in the PT/H region, which were also most severely affected in the tofm808 mutant (Fig. 7). By injecting EdU into wild-type embryos at different developmental stages and analyzing cells that are double-labeled with EdU and neuron-specific markers at 2 d postfertilization (dpf) (Fig. 7A), we found that DA, 5-HT, and GABAergic neurons in PT/H were born in a highly intermingled fashion, with >60% being generated during a common developmental window from 10 to 18 hpf (Fig. 7B–F).
Figure 7.
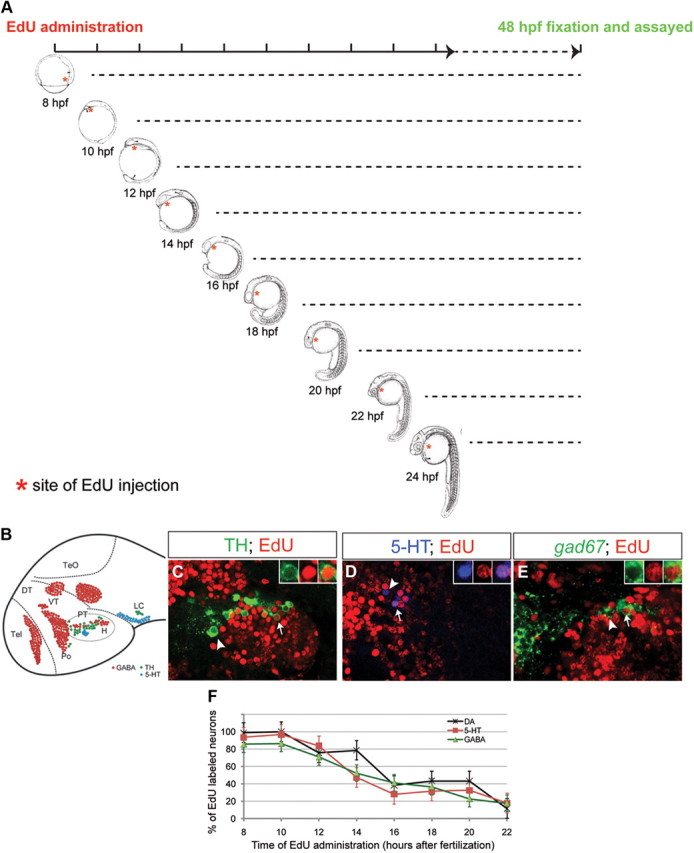
Birth-dating analysis of DA, 5-HT, and GABAergic neurons in the ventral forebrain. A, EdU was injected at different embryonic stages as indicated. At 48 hpf, embryos were collected and subjected to EdU staining together with immunofluorescence or FISH to mark specific neuronal subtypes. B, Schematic representations of the examined neuronal types in a 2 dpf wild-type embryo. C–E, Single confocal Z section (∼1 μm thick) of wild-type zebrafish embryos at 48 hpf. Proliferating DA (C), 5-HT (D), and GABAergic (E) progenitors were labeled by EdU at 14 hpf, and visualized by EdU staining together with immunofluorescence with antibodies against tyrosine hydroxylase (TH) and 5-HT or whole-mount FISH with antisense RNA probe directed against gad67. The arrows indicate EdU+ neurons, whereas the arrowheads indicate neurons with no EdU staining. F, Plot of the percentage of proliferating DA, 5-HT, and GABAergic progenitor cells as a function of time in wild-type embryos (n = 3–5 for individual neuronal type), showing that they are born in a similar developmental window. All images are lateral views of anterior brain regions. Anterior is to the left, and dorsal is up. Please refer to Materials and Methods for quantification of these neurons in the PT/H region.
We therefore performed heat shock at multiple developmental stages from 6-somite to 14-somite, and analyzed the state of DA, 5-HT, and GABAergic neurons at 48 hpf. Induction of fezf2 at 6-, 8-, and 10-somite stages led to significantly increased production of DA neurons in the ventral forebrain, and some increase of hindbrain locus ceruleus noradrenergic neurons were also observed (Fig. 8A,D). fezf2 overexpression, however, did not increase GABAergic or 5-HT neurons at any stages during which heat shock was performed (Fig. 8B–D).
Figure 8.
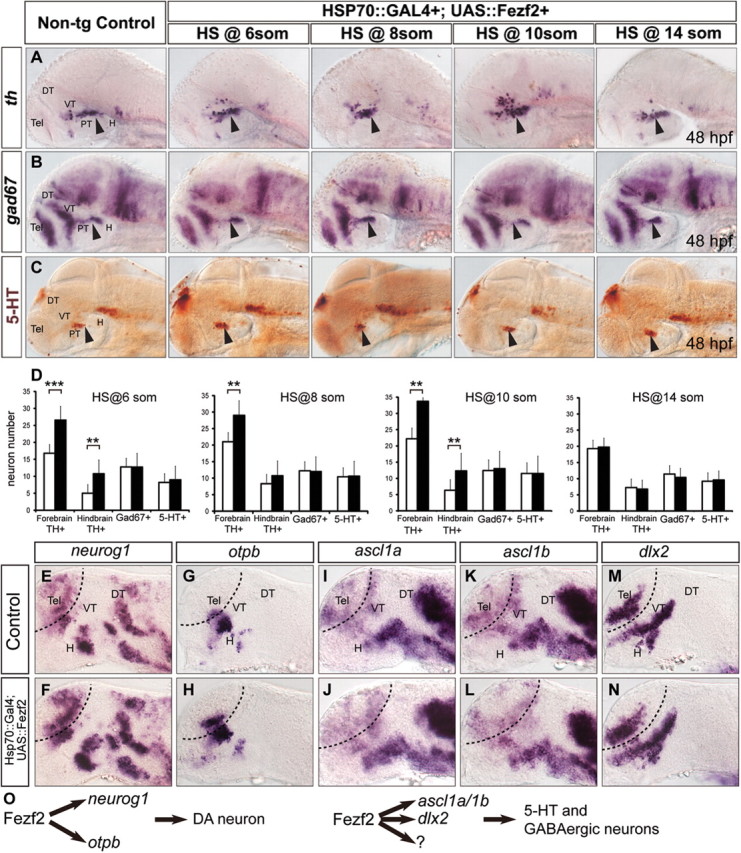
Neurogenesis in the ventral forebrain of fezf2-overexpressing embryos. DA (A), GABAergic (B), and 5-HT (C) neurons in embryos with forced fezf2 expression at different stages during embryonic development. D, Quantifications of DA, GABAergic, and 5-HT neurons in forebrain and hindbrain region of nontransgenic control and fezf2 overexpressing embryos. n ≥ 5 per genotype. ***p < 0.001; **p < 0.01. Error bars indicate SEM. E–N, Embryos derived from a cross between Hsp70::Gal4 and UAS::Fezf2 transgenic fish are heat shocked at 14 hpf and stained with neurog1 (E, F), otpb (G, H), ascl1a (I, J), ascl1b (K, L), and dlx2 (M, N)-directed probes at 20 hpf. O, A schematic model. Fezf2 regulates the generation of DA neurons through activating neurog1 and otpb. Meanwhile, Fezf2 regulates the production of 5-HT and GABAergic neurons through activating ascl1a/1b, dlx2, respectively, and possibly other undetermined mechanisms. DT, Dorsal thalamus; H, hypothalamus; PT, posterior tuberculum; VT, ventral thalamus; Tel, telencephalon. Please refer to Materials and Methods for quantification of these neurons in the PT/H region.
To further explore how fezf2 overexpression increases DA but not 5-HT or GABAergic neurons, we asked whether fezf2 is sufficient to differentially activate proneural and HD genes. Heat shock was performed at the 10-somite stage, and embryos were processed for in situ hybridization at 20 hpf. Fezf2 overexpression was sufficient to increase the expression of neurog1 (Fig. 8E,F) and otpb (Fig. 8G,H), but not that of ascl1a (Fig. 8I,J), ascl1b (Fig. 8K,L), or dlx2 (Fig. 8M,N). It is worth pointing out that the increase in neurog1 and otpb expression appeared as an expansion of the endogenous expression domains rather than a formation of new domains at ectopic locations. These results suggest that Fezf2 is a rate-limiting factor in the induction of neurog1 and otpb expression in the ventral forebrain.
Discussion
How multipotent progenitors give rise to multiple neuronal lineages during brain development is not well understood. Through combined molecular genetic loss- and gain-of-function approaches, we show that Fezf2 guides ventral forebrain neural progenitors toward DA, 5-HT, or GABAergic neuronal fate through activating achaete-scute and atonal type of bHLH proneural as well as HD genes in stereotypically positioned progenitor subpopulations (Fig. 8O, a schematic model). This finding provides new insights into the government of multilineage differentiation of neuronal progenitor cells, with potential implications for improving specific reprogramming of pluripotent stem or somatic cells into appropriate neuronal lineages.
Our findings also prompt a new question: How does Fezf2 activate different bHLH genes in distinct subsets of progenitors? The highly stereotyped patterns of organization of DA, 5-HT, and GABAergic progenitor domains and neurons in the ventral forebrain argue against a model of stochastic induction. Thus, fezf2-expressing ventral forebrain progenitors must represent a heterogeneous population of cells, either due to extrinsic or intrinsic programming. Future studies at single-cell resolution and at biochemical levels are necessary to further parse out the complex gene regulatory network underlying multilineage differentiation.
Does Fezf2 directly activate proneural genes in the zebrafish ventral forebrain? It is worth noting that Fezf2 contains a conserved engrailed homology 1 (EH1) repressor motif. The EH1 type of motif is known to interact with Groucho or Transducin-like enhancer of split (TLE) type transcriptional corepressors (Shimizu and Hibi, 2009). A study in the mouse dorsal telencephalon shows that Fezf2 together with its paralog Fezf1 directly represses the expression of Hes5, thereby indirectly activating Neurogenin 2 (Shimizu et al., 2010). Interestingly, however, many Groucho or TLE-interacting proteins, for example, TCF/LEF family of proteins involved in the Wnt signaling pathway (Bienz, 1998), are known to function as both activators and repressors, depending on intramolecular and intermolecular interactions in distinct cellular contexts. Thus, it remains a possibility that Fezf2 can directly activate proneural genes in the zebrafish ventral forebrain. Indeed, we have observed that Fezf2 appears to have distinct functions in the zebrafish dorsal Tel versus in the ventral forebrain (N. Yang, unpublished data). It would therefore be of interest in the future to analyze the role of Fezf2 in zebrafish telencephalic neurogenesis, as well as determining the biochemical nature of Fezf2 regulation of proneural genes.
Expression analyses as well as genetic studies support a priming rather than a maintenance role of Fezf2 in regulating the expression of bHLH and HD genes. First, at the mRNA level, a rather minimal overlap between the expression of fezf2 and bHLH or HD genes is detected, suggesting that fezf2 expression is downregulated shortly after the induction of bHLH and HD genes in a temporally specific manner. Second, impairment of fezf2 activity reduces rather than abolishes bHLH and HD gene expression. While incomplete inactivation of fezf2 is a possible cause, the data also suggest that other factors in the region may be able to activate these genes in the absence of fezf2. Consistent with this idea, the loss of bHLH gene expression in the ventral forebrain is only transiently detectable in ∼22 hpf fezf2-deficient embryos. Nevertheless, such defect has detrimental effects on neuronal lineage differentiation. Together, we propose that fezf2 plays a temporally critical role in promoting the differentiation of neuronal progenitors into early-born neurons, through priming but not maintaining the activation of bHLH and HD genes.
Our gain-of-function experiments reveal that overexpression of Fezf2 is sufficient to increase DA but not 5-HT or GABAergic neurons in the ventral forebrain. This is in part due to the ability of Fezf2 to increase the expression of neurog1 and otpb but not that of ascl1a, 1b, and dlx2. These results reveal a rate-limiting effect of Fezf2 in DA neuron development and suggest that Fexf2 exerts a different mode of regulation in different neuronal lineages. What accounts for this difference is not clear. One possibility is that fezf2 may be expressed at different levels in distinct progenitor populations, thereby exerting a concentration-dependent effect on their differentiation. It will be of interest to examine whether and how fezf2 may be differentially expressed in progenitor cells, and what impact it might have on progenitor fate choice.
Footnotes
This work was supported by NIH Grant NS042626. We thank the late J. Campos-Ortega for the hsp-gal4 and uas-NICD transgenic lines, A. Amsterdam and N. Hopkins for the neurogenin 1 mutant, M. Hammerschmidt for the pia mutant, and various other laboratories for their generous sharing of plasmids. We also thank Dr. B. Lu and Guo Laboratory members for comments on this manuscript and helpful discussion.
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Amoyel M, Cheng YC, Jiang YJ, Wilkinson DG. Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development. 2005;132:775–785. doi: 10.1242/dev.01616. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Jan YN. The determination of the neuronal phenotype. In: Cowan WM, editor. Molecular and cellular approaches to neural development. New York: Oxford UP; 1997. pp. 26–63. [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bienz M. TCF: transcriptional activator or repressor? Curr Opin Cell Biol. 1998;10:366–372. doi: 10.1016/s0955-0674(98)80013-6. [DOI] [PubMed] [Google Scholar]
- Blechman J, Borodovsky N, Eisenberg M, Nabel-Rosen H, Grimm J, Levkowitz G. Specification of hypothalamic neurons by dual regulation of the homeodomain protein Orthopedia. Development. 2007;134:4417–4426. doi: 10.1242/dev.011262. [DOI] [PubMed] [Google Scholar]
- Filippi A, Dürr K, Ryu S, Willaredt M, Holzschuh J, Driever W. Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development. BMC Dev Biol. 2007;7:135. doi: 10.1186/1471-213X-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. Genesis of the Drosophila peripheral nervous system. Trends Genet. 1989;5:251–255. doi: 10.1016/0168-9525(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev Biol. 1999a;208:473–487. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein Soulless/Phox2a. Neuron. 1999b;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Neuronal cell fate specification in Drosophila. Curr Opin Neurobiol. 1994;4:8–13. doi: 10.1016/0959-4388(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Jeong JY, Einhorn Z, Mercurio S, Lee S, Lau B, Mione M, Wilson SW, Guo S. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc Natl Acad Sci U S A. 2006;103:5143–5148. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JY, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, Guo S. Patterning the zebrafish diencephalon by the conserved zinc finger protein Fezl. Development. 2007;134:127–136. doi: 10.1242/dev.02705. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Zeller J, Sirotkin HI, French D, Schilbach S, Hashimoto H, Hibi M, Talbot WS, Rosenthal A. Zinc finger protein too few controls the development of monoaminergic neurons. Nat Neurosci. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Asci/Mash1 is required for the development of central serotonergic neurons. Nat Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, von der Hardt S, Herzog W, Kramer C, Schwarz H, Hammerschmidt M. The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis. Development. 2006;133:1079–1089. doi: 10.1242/dev.02296. [DOI] [PubMed] [Google Scholar]
- Ryu S, Mahler J, Acampora D, Holzschuh J, Erhardt S, Omodei D, Simeone A, Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr Biol. 2007;17:873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev Growth Differ. 2009;51:221–231. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakazawa M, Kani S, Bae YK, Kageyama R, Hibi M. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]