Abstract
The ability to regulate the expression of a gene greatly aids the process of uncovering its functions. The fission yeast Schizosaccharomyces pombe has so far lacked a system for rapidly controlling the expression of chromosomal genes, hindering its full potential as a model organism. Although the widely used nmt1 promoter displays a wide dynamic range of activity, it takes >14–15 h to de-repress. The urg1 promoter also shows a large dynamic range and can be induced quickly (<2 h), but its implementation requires laborious strain construction and it cannot be used to study meiosis. To overcome these limitations we constructed a tetracycline-regulated system for inducible expression of chromosomal genes in fission yeast, which is easily established and implemented. In this system the promoter of a gene is replaced by simple one-step substitution techniques with a tetracycline-regulated promoter cassette (tetO7-TATACYC1) in cells where TetR/TetR′-based transcription activators/repressors are also produced. Using top1 and nse6 as reporter genes, we show that Top1 and Nse6 appear after just 30 min of activating tetO7-TATACYC1 and plateau after ~4–6 h. The amount of synthesised protein is comparable to that produced from the attenuated nmt1 promoter Pnmt8, which should be closer to wild type levels for most genes than those generated from excessively strong promoters and can be controlled by changing the concentration of the effector antibiotic. This system also works efficiently during meiosis, thus making it a useful addition to the toolkit of the fission yeast community.
Keywords: Schizosaccharomyces pombe, fission yeast, tetracycline, gene expression, inducible promoter
Introduction
Being able to control the expression of a gene is an extremely powerful approach to uncovering its functions. An ideal controllable gene expression system should allow the user to turn on, or off, the transcription of a gene in a rapid, tight and reversible manner. It should also be quick and easy to implement. The unicellular eukaryote Schizosaccharomyces pombe, also known as fission yeast, is a very valuable model organism because in some respects it shows a greater degree of conservation of key molecular pathways with higher eukaryotes than those found in the distantly related, but more widely employed, budding yeast Saccharomyces cerevisiae (Moreno, et al., 1991; Nurse, 2000). Although a few systems for regulating gene expression have been developed for fission yeast they display several limitations. For instance, the commonly used nmt1 promoter (Pnmt1) is induced ~70-fold upon removal of thiamine from the growth medium and it can be easily used to replace the endogenous promoter of a gene of interest by routine one-step PCR integration protocols (Bahler, et al., 1998; Maundrell, 1993). Attenuated versions of this very strong promoter, called Pnmt4 and Pnmt8, have also been created, which allow the user to control the levels of gene transcription (Basi, et al., 1993). Yet, Pnmt1/4/8 take at least 14–15 h to induce (Kumar and Singh, 2006). The basal levels of gene expression under non-induced conditions are also relatively high, especially in the case of the full-strength Pnmt1, and to a lesser extent for Pnmt4. In addition, when cells are cultured in liquid medium, inducing Pnmt1/4/8 requires extensive washes to ensure that thiamine is completely removed from the medium, which is time-consuming and can induce cellular stress responses. Similarly, the recently developed urg1 promoter system, Purg1, allows quick (< 2 h) induction of gene expression, but its implementation involves relatively complicated strain construction: the gene whose expression has to be controlled must be integrated, by Cre-loxP site-specific recombination, at the urg1 locus in place of urg1+ itself (Watson, et al., 2011). Although this system is certainly useful for controlling the expression of a xenogene, it poses complications when used to control the transcription of an endogenous fission yeast gene. For example, if the copy of the gene inserted at the urg1 locus is to be the sole source of its expression, which is often the case, then the one found at the endogenous locus must be removed. Lastly, Purg1 cannot be used for studying meiosis because it is induced upon nitrogen starvation, which is a pre-requisite for triggering mating and meiosis in fission yeast.
A general drawback of using endogenous promoters to alter the transcription of a gene is that controlling them often requires changes in the growth medium, e.g. removal of thiamine and uracil for Pnmt and Purg1 respectively, which can interfere with cellular metabolism. A solution to this problem is to use heterologous promoters in combination with synthetic transcription factors, such as tetracycline-regulated promoters, where addition of the effector antibiotic tetracycline can either induce (Tet-On) or repress (Tet-Off) gene expression. This system relies on the ability of the Escherichia coli tetracycline repressor TetR, or its “cousin” TetR′, to bind to the tetO sequence only under specific conditions, i.e. either in the presence or absence of tetracycline, respectively (Gossen and Bujard, 2002). Sophisticated Tet-On and Tet-Off systems, which allow for tight and fast regulation of gene expression, have been developed by fusing TetR/TetR′ to certain transcription activation/repression domains and substituting the promoter of a desired gene with tetO arrays (Belli, et al., 1998a; Belli, et al., 1998b; Yen, et al., 2003). Tetracycline-regulated expression systems have been developed for fission yeast but they show limitations. The approach developed by Faryar and Gatz (1992) can be used to control the expression of plasmid-borne genes only. The same applies to the system devised by Erler et al. (2006), with the difference that their plasmids can be genomically integrated and the selection marker they contain can be recycled by Cre-loxP site-specific recombination. It follows that the endogenous copy of the gene whose expression has to be controlled must be knocked out to ensure that the tetracycline-regulated one is the sole source of its transcription. This requires complicated and time-consuming strain manipulation that could be avoided. Thus, we set out to develop a tetracycline-regulated system to control the expression of chromosomal genes in fission yeast in a rapid, tight and reversible manner, which is both easy and quick to implement.
Materials and Methods
Strains and growth conditions
Standard fission yeast culture and handling were used as described by Moreno et al. (1991). All strains are ura4-D18 leu1-32. Table 1 contains the list of strains used to prepare this report.
Table 1.
A list of the yeast strains used in this study. All strains are ura4-D18 leu1-32.
| Strain # | Genotype | Source |
|---|---|---|
| NBY780 | h+ | |
| NBY781 | h− | |
| NBY3770 | tetO7-TATACYC1-FLAG3-top1:hphMX6, h− | This study |
| NBY4461 | tetO7-TATACYC1-FLAG3-top1:hphMX6, pDM291-tetR:ura4+, h− | This study |
| NBY3771 | tetO7-TATACYC1-FLAG3-top1:hphMX6, pDM291-tetR-tup11Δ70:ura4+, h− | This study |
| NBY2553 | Pnmt4-FLAG3-top1:hphMX6, h+ | Heideker et al. (2011) |
| NBY2554 | Pnmt8-FLAG3-top1:hphMX6, h+ | Heideker et al. (2011) |
| NBY670 | top1::kanMX6, h+ | Heideker et al. (2011) |
| NBY3992 | tetO7-TATACYC1-FLAG3-nse6, h+ | This study |
| NBY3912 | tetO7-TATACYC1-FLAG3-nse6:hphMX6, pDM291-tetR-tup11Δ70:ura4+, h− | This study |
| NBY871 | nse6::kanMX6, h− | Pebernard et al. (2006) |
| NBY835 | nse6::kanMX6, h+ | Pebernard et al. (2006) |
| NBY4209 | tetO7-TATACYC1-FLAG3-nse6:hphMX6, pDM291-tetR-tup11Δ70:ura4+, h+ | This study |
| NBY4756 | tetO7-TATACYC1-FLAG3-top1:hphMX6, pDM291-tetR′-tup11Δ70:ura4+, h− | This study |
PCR amplification of integrative cassettes, transformation and verification of integration
Integrative cassettes for promoter replacement were generated by PCR with Ex Taq DNA polymerase (Clontech, Mountain View, CA) following the manufacturer’s instruction. Fission yeast cells were transformed essentially as described by Suga and Hatakeyama (2001). Integration of gene targeting cassettes or linearised plasmids at a specific genomic locus was confirmed by PCR directly from isolated yeast colonies.
Spot assay
For spot assays, cells were propagated in YES medium at 30 °C to logarithmic phase, spotted on YES agar supplemented with the relevant drug in five-fold dilutions from a starting OD600 of 0.5, and then grown at 30 °C for 2–3 days. Irradiation with UV light at 365 nm was carried out in a Stratalinker-1800 crosslinker instrument (Stratagene, La Jolla, CA).
Preparation of whole-cell extracts and western blotting
For western blotting, ~1–3 108 cells were lysed by beating four times at 5.0 m s−1 for 20 s in a FastPrep-24 instrument (MP Biochemicals, Solon, OH) in 150 μL of 8 M urea, 50 mM Tris-HCl pH 6.8, 1× Complete Protease Inhibitor Cocktail, 2 mM PMSF supplied with 100 μL of silica-zirconia beads (BioSpec Product, Bartlesville, OK). The lysates were cleared by centrifugation at 16,000 g for 10 min at 4 °C. The protein concentration of the cleared lysates was measured using Bio-Rad’s protein assay (Bio-Rad, Hercules, CA) and equalized to 3 μg μL−1. These samples were supplemented with LDS Sample Buffer (Life Technologies, Carlsbad, CA) and DTT to a final concentration of 1× and 50 mM, respectively, and denatured at 70 °C for 15 min. 20 μg of protein was resolved on 4–20% Tris-Glycine gels (Expedeon, San Diego, CA or Life Technologies, Carlsbad, CA) and transferred to a nitrocellulose membrane using an iBlot Dry Blotting Transfer System (Life Technologies, Carlsbad, CA). The membrane was blocked in 5% w/v non-fat milk in Tris-buffered saline solution with 0.1% v/v Tween-20 before being blotted with antibodies against FLAG or tubulin (Sigma-Aldrich, Saint Louis, MO). After incubation with a horseradish peroxidase- or IRDye-conjugated secondary antibody (Pierce, Rockford, IL or Li-Cor, Lincoln, NE, respectively), the membrane was subjected to enhanced chemiluminescence (Thermo Scientific, Waltham, MA) or imaged on an ODYSSEY scanner (Li-Cor, Lincoln, NE), respectively.
Construction of plasmids
To create fission yeast-compatible pFA6a-tetO7-TATACYC1 plasmids the tetO7-TATACYC1 cassette was amplified by PCR from pCM325 (Yen, et al., 2003) with oNZ3 and oNZ4 (Table 2) and used to replace Pnmt1/4/8 from existing pFA6a vectors (Bahler, et al., 1998; Noguchi, et al., 2008), using BglII and PacI.
Table 2.
A list of the oligonucleotides used in this study.
| Primer # | Sequence |
|---|---|
| oNZ3 | 5′-CGCAGATCTGGGTAATATAGATCAATTCCTCGA-3′ |
| oNZ4 | 5′-CGCATTTTAATTAACCTCCAGGATGATAAACGGATCCCCCG-3′ |
| oNZ5 | 5′-GGAGGTACCTACGAAAAAGATATCAACAAGTTG-3′ |
| oNZ6 | 5′-CGTGGTACCTCAAGGAGATGCAGGGTC-3′ |
| oNZ14 | 5′-CCGGGCGCCCGCGCCACTTCTAAATAAGCG-3′ |
| oNZ15 | 5′-CAATTTTAAAACCTATTTGCACCCCCTAGCGGATCTGCCGGTAG-3′ |
| oNZ16 | 5′-GGTGCAAATAGGTTTTAAAATTG-3′ |
| oNZ17 | 5′-ATGCATCCAACCCACCTAAATGGCCCGGGCATGGTACGATATATAATTAG-3′ |
| oNZ33 | 5′-CCATACCCGGGGGTCGAGGAGCTTGGCCC-3′ |
| oNZ27 | 5′-CCGGCCAGCTGGGGTAAAAGGAATGTCTCCC-3′ |
| oNZ28 | 5′-GAATGCTTTCATGTATAAATCAAGGAGCTCGCATTACTAATAGAAAG-3′ |
| oNZ29 | 5′-CTTGATTTATACATGAAAGCATTC-3′ |
| oNZ30 | 5′-GGCCGCAGCTGCATGGTAATATTGATACAGAACG-3′ |
To construct the pDM291-(tetR/tetR′)-tup11Δ70 plasmids, a DNA fragment containing the tup11 open reading frame, missing the region encoding for its first 70 amino acids, was generated from wild type genomic DNA using oNZ5 and oNZ6, and inserted into pCM217 or pCM223 (Belli, et al., 1998b) with KpnI. Next, a DNA fragment containing the adh1 terminator (Tadh1) was amplified from pFA6a-hphMX6-P3nmt1-FLAG3 with oNZ14 and oNZ15 and fused by PCR to a DNA molecule encompassing a region located 3′ of the ura4 locus (oNZ16 and oNZ17). The resulting product was cloned into pCM217-tup11Δ70 with KasI and BstXI. pCM217-tetR′-tup11Δ70 was created by extracting a fragment comprising part of the CMV promoter and tetR′-tup11Δ70 from pCM223-tup11Δ70 with NcoI and BglII and cloning it into pCM217-tup11Δ70. The DNA molecule encompassing the (tetR/tetR′)-tup11Δ70 cassette was generated from pCM217-tup11Δ70 or pCM217-tetR′-tup11Δ70 by PCR with oNZ33 and oNZ17, digested with SmaI and ligated into a pDM291 plasmid (Grimm, et al., 1988) that had been cut with HpaI and whose BamHI site had been disrupted. In this construct the (tetR/tetR′)-tup11Δ70 fusion is transcribed in the opposite direction from the ura4+ marker.
The pUG6SP-(tTA/tTa′) vectors are derivatives of pUG6-tTA (Yen, et al., 2003). pUG6-tTA′ was created by replacing tetR in pUG6-tTA with a DNA fragment containing tetR′ from pCM251 (Belli, et al., 1998b), which had been digested with SphI and BbsI. The nmt1 terminator was generated by PCR from pREP1 with oNZ27 and oNZ28 and fused to a ~600-bp region that is located between the arg3 and arg11 loci that was amplified from genomic DNA with oNZ29 and oNZ30. The resulting DNA fragment was inserted into pUG6-tTA/tTA′ with PvuII.
Analysis of meiosis
For meiotic studies, ~1–2 107 cells of each relevant haploid strain were mixed in 10 μL of water and plated on sporulation SSA medium lacking or supplied with anhydrotetracycline. After 2 days at 25 °C, the cell mixture was observed on an Eclipse E800 microscope (Nikon Metrology, Brighton, MI) and images were acquired with a Quantix camera (Photometrics, Tucson, AZ). Staining with 4′,6-diamidino-2-phenylindole (DAPI) and the measurement of spore viability were performed as described in Wehrkamp-Richter et al. (2012).
Results
In order to engineer a tetracycline-inducible promoter for fission yeast we built upon the systems developed by Belli et al. (1998b) and Yen et al. (2003) for budding yeast, because of their versatility, ease of use and how quickly they can be activated. In such systems the endogenous promoter of a gene is replaced, by one-step PCR integration, with a cassette that contains an array of two or seven tetO sequences (tetO2 or tetO7) immediately upstream of the TATA and leader regions of the CYC1 gene from S. cerevisiae (TATACYC1). Co-producing fusions of TetR and TetR′ to specific transcription activation (e.g. VP16) and repression (e.g. Tup1) modules in the same cell allows for low basal transcription under non-induced conditions and good expression upon induction. In order to adapt and improve this system for S. pombe, we engineered various pFA6a-family vectors that are commonly used to substitute the endogenous promoter of a gene with Pnmt (Bahler, et al., 1998; Noguchi, et al., 2008) to contain the tetO2/tetO7-TATACYC1 cassette in place of Pnmt itself (Figure 1A). We also constructed fission yeast-compatible vectors for the constitutive production of fusions of TetR/TerR′ to either the transcription activation domain of the herpes simplex virus protein VP16 or a truncation of the transcription co-repressor Tup11, Tup11Δ70. This construct lacks the first 70 amino acids found in the full-length Tup11, which is the region that in the budding yeast orthologue Tup1 mediates interactions with Ssn6 (Figures 2A and 2B). We used Tup11Δ70 because Tup11 does not need to interact with Ssn6 to repress gene expression. In fact in the Ssn6-Tup1 repressor complex, it is Tup1 that executes the silencing role while Ssn6 acts as a bridge for the promoter-specific DNA-binding subunit (Tzamarias and Struhl, 1994; Tzamarias and Struhl, 1995). The chimeras we generated include a λ cI linker between their constituent moieties to increase flexibility. Both sets of plasmids contain a recyclable selection marker, hisG-ura4+-hisG or loxP-KanMX6-loxP, which can be removed by either plating cells on 5-fluoro-orotic acid-containing medium or by producing the Cre recombinase, respectively. The vectors we constructed also contain a region that is homologous to a specific locus in the fission yeast genome, which allows them to be chromosomally integrated following digestion with the relevant restriction enzyme. In all of the strains presented in this report, TetR/TetR′, or their derivatives, are produced from a chromosomally integrated form of the relevant expression vector. Having created fusions of Tup11Δ70 and VP16 to both TetR and TetR′ means that, depending on which set of chimeras are introduced in a cell, tetracycline can be used either to turn on gene expression or to turn it off.
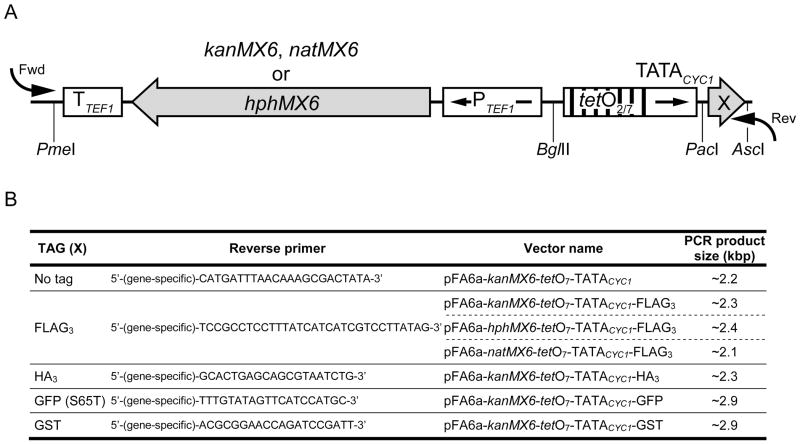
Figure 1.
The pFA6a-tetO7-TATACYC1 module. (A) Schematic representation of the pFA6a-tetO7-TATACYC1 module employed to generate gene-targeting cassettes for promoter replacement/N-terminal tagging. The position of the restriction sites used for cloning is shown. Arrows inside boxes show the direction of transcription. Arrows outside boxes indicate the position where the forward (Fwd) and reverse (Rev) PCR primers anneal. X = Tag. (B) Reverse primers for amplifying the promoter replacement/tagging cassettes from pFA6a-tetO7-TATACYC1 vectors. The forward primer is common to all vectors and should conform to the following sequence 5′-(gene-specific sequence)-GAATTCGAGCTCGTTTAAAC-3′. The reverse primer is specific to each unique template. The gene-specific part of the primer typically corresponded to 80 bp of sequences upstream (Fwd) and downstream (Rev) of the start codon of the relevant gene. The expected sizes of the PCR products are shown.
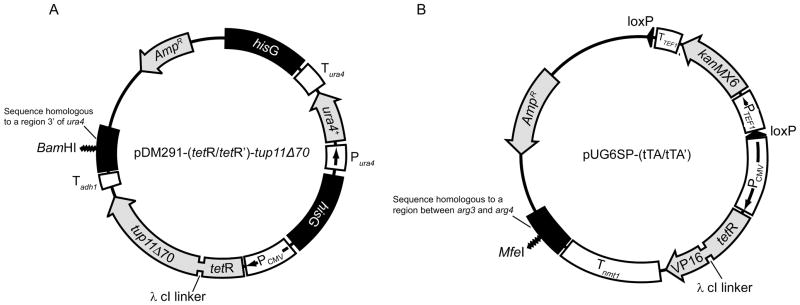
Figure 2.
The pDM291-(tetR/tetR′)-tup11Δ70 and pUG6SP-(tTA/tTA′) vectors. Schematic representations of (A) pDM291-(tetR/tetR′)-tup11Δ70 and (B) pUG6SP-(tTA/tTa′). The position of the restriction sites, (A) BamHI and (B) MfeI, which can be used to linearise the plasmids prior to transformation into yeast cells is shown. Arrows inside boxes show the direction of transcription. pDM291-tetR′-tup11Δ70 and pUG6SP-tTA′ are identical to pDM291-tetR-tup11Δ70 and pUG6SP-tTA except for the fact they contain tetR′ instead of tetR.
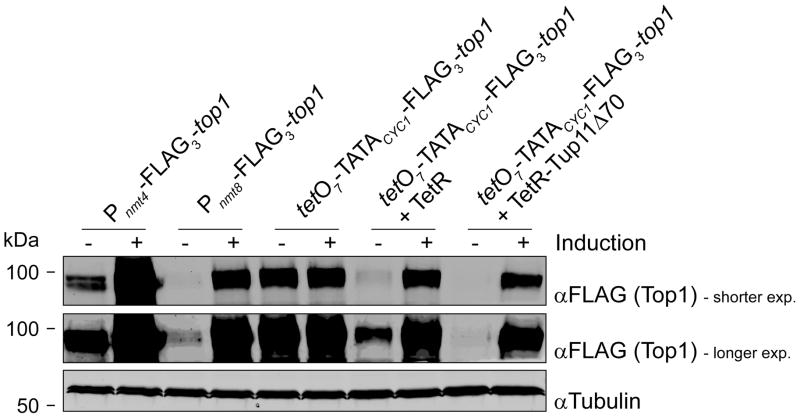
Initially we explored whether tetO7-TATACYC1 can be used to effectively control gene expression. We replaced the endogenous promoter of top1+, which codes for topoisomerase I, with the tetO7-TATACYC1-FLAG3 cassette. Western blotting analysis of extracts from tetO7-TATACYC1-FLAG3-top1 cells showed the presence of an anti-FLAG antibody-reactive band that migrated as the FLAG3-tagged Top1 produced from previously validated Pnmt4/8-FLAG3-top1 strains (Figure 3; Heideker, et al., 2011). These data also showed that the amount of FLAG3-Top1 generated from an uninhibited tetO7-TATACYC1 was comparable to that produced from the induced Pnmt8. Such an extent of expression is likely to be closer to wild type levels for most genes than those driven by the overly powerful Pnmt4, Pnmt1 or Purg1. Thus, tetO7-TATACYC1 is an ideal promoter for all those experiments where “physiological” amounts of a protein need to be produced in a regulated manner. Unless the amount of required gene expression significantly exceeds the intrinsic activity of TATACYC1, it follows that producing the tetracycline-regulated activator TetR/TetR′-VP16 should not be necessary in most cases; for instance, see below. This is advantageous because it reduces the number of manipulations that are necessary to construct the relevant “tet” strains, i.e. integration pUG6-tTA/tTa′ and removal of its selection marker.
Figure 3.
On and off states of tetO7-TATACYC1 vs. Pnmt4 and Pnmt8. A comparison of top1 expression driven from Pnmt4, Pnmt8 and tetO7-TATACYC1 under repressed and induced conditions, as determined by analyzing FLAG3-Top1 protein levels by western blotting. Pnmt4/8 were induced by growing cells in medium lacking thiamine for at least 24 h. tetO7-TATACYC1 was de-repressed by propagating cells in 2.5 μg mL−1 ahTet for at least 6 h.
Constitutively producing tetR alone from an expression vector chromosomally integrated near the ura4 locus, in the absence of an effector antibiotic, was sufficient to reduce the amount of FLAG3-Top1 to very low, but not undetectable, levels in the tetO7-TATACYC1-FLAG3-top1 cells (Figure 3). Conversely, TetR-Tup11Δ70 reduced the amount of FLAG3-Top1 to imperceptible levels, which was reverted by addition to the growth medium of the tetracycline analogue anhydrotetracycline (ahTet). We used ahTet instead of doxycycline, which is more commonly employed to control Tet-On/Tet-Off systems, because we, as previously reported by Erler, et al. (2006), also found that the former antibiotic worked more efficiently than the latter in fission yeast (data not shown). Fusions of TetR to transcription repressors other than Tup11, such as fission yeast Ssn6 or Sir2, did not repress gene expression from tetO7-TATACYC1 as well as TetR-Tup11Δ70 did (data not shown), and therefore they were not used further in this study. Poor repression of top1 expression was also observed when tetO2-TATACYC1 was used instead of tetO7-TATACYC1 (data not shown). In conclusion, in the absence of ahTet TetR-Tup11Δ70 efficiently represses transcription from tetO7-TATACYC1, while allowing it to become active following addition of the effector antibiotic. The tetO7-TATACYC1-FLAG3-top1 + tetR-tup11Δ70 strain will be hereafter referred to as tetO7-top1 for simplicity.
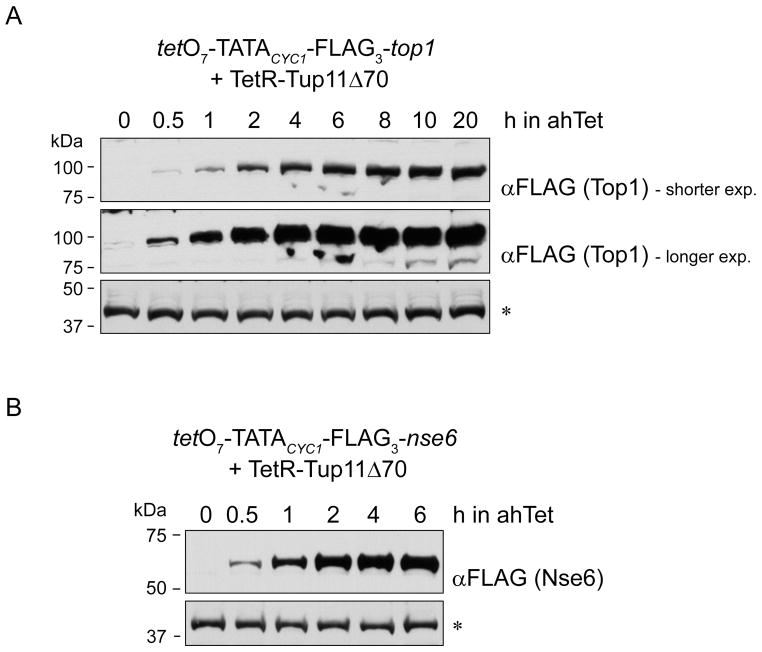
We also examined how quickly the expression of a gene could be turned on from tetO7-TATACYC1. We found that FLAG3-Top1 appeared after only 30 min of adding ahTet to the growth medium. The amount of this protein rose steadily thereafter and plateaued within 4–6 h (Figure 4A). This pattern of induction was not unique to Top1 but was also observed for FLAG3-Nse6 produced from a tetO7-nse6 strain (Figure 4B). Nse6 is a component of the structural maintenance of chromosome complex Smc5-6 (Pebernard, et al., 2006). Thus, tetO7-TATACYC1 can be used to rapidly express various genes in fission yeast.
Figure 4.
Kinetics of protein accumulation after induction of tetO7-TATACYC1. Time-course experiments showing the accumulation, by western blotting, of (A) FLAG3-Top1 -and (B) FLAG3-Nse6 following induction of tetO7-TATACYC1 by addition of 2.5 μg mL 1ahTet to the growth medium. A yeast protein of unknown identity that cross-reacted with the anti-FLAG tag antibody (*) is used as a loading control.
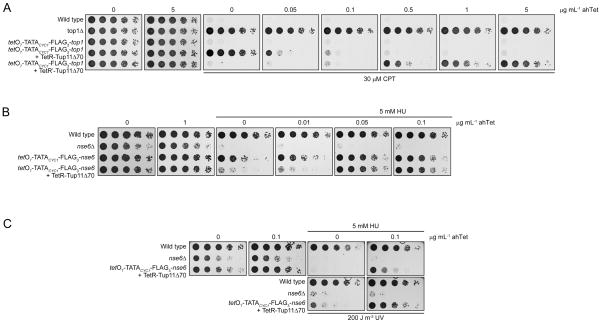
Next, we asked whether our tetracycline-regulated promoter could be used to investigate questions of biological relevance. To address this, we examined how the tetO7-top1 strain responded to the topoisomerase I inhibitor camptothecin (CPT) in the presence of different concentrations of ahTet vs. wild type and top1Δ cells (Figure 5A). We observed that, as expected, the wild type was unable to proliferate in the presence of 30 μM CPT. Instead, the top1Δ cells grew uninhibited. Uninhibited growth was also observed for the tetO7-top1 strain that was grown on medium lacking ahTet, that is, under conditions of gene repression, thus indicating that little or no Top1 was produced in these cells. In the presence of increasing amounts of ahTet, which leads to activation of transcription from tetO7-TATACYC1 in the presence of TetR-Tup11Δ70, the tetO7-top1 strain became progressively more sensitive to CPT, which implies that the concentrations of effector antibiotic determined the amount of Top1 produced by these cells. As expected producing TetR′-Tup11Δ70, instead of TetR-Tup11Δ70, in the tetO7-TATACYC1-FLAG3-top1 cells yielded a reverse growth pattern on CPT in the presence of increasing concentrations of ahTet to the one described above. These results show that both the tet-off and tet-on systems work equally well. They also demonstrate that the amount of ahTet added to the growth medium can be adjusted to control the extent of expression from tetO7-TATACYC1 to a desired level, which is a great advantage over the “all-or-none” promoters Pnmt and Purg1. Analogously, adding increasing amounts of ahTet to the growth medium of the tetO7-nse6 cells reverted their exquisite sensitivity towards the replication fork stalling agent hydroxyurea (HU) from an nse6Δ-like phenotype to a wild type-like one (Figure 5B).
Figure 5.
The phenotypes of the tetO7-top1 and tetO7-nse6 strains. The relevant yeast strains were serially diluted onto rich medium in the presence of the indicate drugs and grown at 30 °C.
For the tetO7-nse6 strain we observed that, in the absence of ahTet, its viability was greatly impaired following irradiation by UV light, too. However, this phenotype was not as penetrant as that observed for the nse6Δ mutant (Figure 5B, Pebernard, et al., 2006), which suggests that some, minimal, transcription of nse6 must occur from tetO7-TATACYC1 even under repressed conditions. This leakiness was more obvious when the cells were challenged by UV light, instead of HU, probably because the minimal amount of Nse6 required to counteract UV light-induced DNA damage, being an acute form of genotoxic stress, is likely to be less than that required to counteract HU, which was used as a chronic source of insults to DNA in Figure 5B.
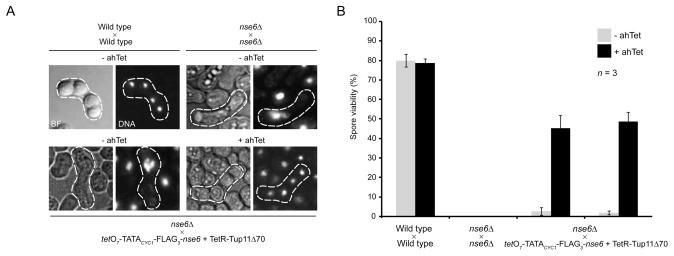
Having shown that, in the presence of TetR-Tup11Δ70, tetO7-TATACYC1 can be used to induce expression of a desired fission yeast gene from its endogenous locus very quickly, we asked whether this system could be used to induce gene expression during meiosis. This is an important question because although Purg1 can also be quickly activated it cannot be used to study meiosis: it is strongly activated by nitrogen starvation (Watt, et al., 2008), which is required to induce mating and meiosis in fission yeast. Endogenous promoters whose activation state changes upon entry into meiosis do exist, however their activation/repression cannot be controlled at will and is restricted to a specific stage of the meiotic cycle, e.g. the rec12 promoter is activated during pre-meiotic DNA synthesis (Farah, et al., 2009; Lin and Smith, 1994; Mata, et al., 2002). In order to address the above-mentioned question we used the tetO7-nse6 strain because nse6 is critical to carry out meiosis (Wehrkamp-Richter, et al., 2012). When nse6Δ cells are induced to undergo meiosis they produce morphologically aberrant asci and spores with very low viability (Figure 6, Wehrkamp-Richter, et al., 2012). When tetO7-nse6 cells were stimulated to undergo mating and sporulation with an nse6Δ strain in the absence or presence of ahTet we observed that, under conditions of gene repression, they produced mostly aberrant asci with inviable spores to an extent that was similar to, but not as penetrant as, that observed for the nse6Δ mutant (Figure 6). Again, this phenotype was reverted to a wild type-like behaviour by the addition of ahTet. Such results indicate that tetO7-TATACYC1 can be used to study biology associated with meiosis.
Figure 6.
tetO7-TATACYC1 can be induced during meiosis. Wild type and nse6Δ cells or a cross between the tetO7-nse6 and nse6Δ strains were stimulated to undergo meiosis in the presence or absence of 2.5 μg mL−1 ahTet. The resulting cell-ascus mixtures were: (A) imaged by microscopy to determine ascus morphology and the DNA content of spores by DAPI staining, and (B) processed to isolate individual spores whose viability was measured by plating them on rich medium. Relevant asci are highlighted by dashed white lines. Error bars indicate the standard error of the mean from three independent crosses. Crosses between the tetO7-nse6 and nse6Δ cells were carried out using both h+ (leftward set of bars in B) and h− (rightward set of bars in B) tetO7-nse6 strains and the matching nse6Δ mutants. BF = bright field, DNA = DAPI.
In conclusion, we report the construction and validation of a new system for rapidly controlling the expression of chromosomal genes in the fission yeast S. pombe, which relies on the use of a heterologous promoter, tetO7-TATACYC1, in combination with tetR-based synthetic transcription factors. This system is easy and rapid to establish because it can be implemented using ordinary gene targeting techniques. It also allows marker recycling, so that the number of genetic modifications to a strain is unrelated to the number of available markers. We have generated all the reagents necessary to establish both Tet-On and Tet-Off systems, including several pFA6a-tetO7-TATACYC1 plasmids that can be used to generate PCR fragments for promoter replacement and gene tagging.
Acknowledgments
M.N.B. is supported by a Scholar Award from the Leukemia & Lymphoma Society. This study was funded by NIH grants GM068608 and GM081840 awarded to M.N.B.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–51. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–6. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Belli G, Gari E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998a;14:1127–38. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1127::AID-YEA300>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998b;26:942–7. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler A, Maresca M, Fu J, Stewart AF. Recombineering reagents for improved inducible expression and selection marker re-use in Schizosaccharomyces pombe. Yeast. 2006;23:813–23. doi: 10.1002/yea.1396. [DOI] [PubMed] [Google Scholar]
- Farah JA, Cromie GA, Smith GR. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc Natl Acad Sci U S A. 2009;106:9356–61. doi: 10.1073/pnas.0902793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faryar K, Gatz C. Construction of a tetracycline-inducible promoter in Schizosaccharomyces pombe. Curr Genet. 1992;21:345–9. doi: 10.1007/BF00351693. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet. 2002;36:153–73. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–6. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Heideker J, Prudden J, Perry JJ, Tainer JA, Boddy MN. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1-mediated DNA damage and genome instability. PLoS Genet. 2011;7:e1001320. doi: 10.1371/journal.pgen.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Singh J. A truncated derivative of nmt1 promoter exhibits temperature-dependent induction of gene expression in Schizosaccharomyces pombe. Yeast. 2006;23:55–65. doi: 10.1002/yea.1343. [DOI] [PubMed] [Google Scholar]
- Lin Y, Smith GR. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–79. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–7. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–30. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Noguchi C, Garabedian MV, Malik M, Noguchi E. A vector system for genomic FLAG epitope-tagging in Schizosaccharomyces pombe. Biotechnol J. 2008;3:1280–5. doi: 10.1002/biot.200800140. [DOI] [PubMed] [Google Scholar]
- Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100:71–8. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, 3rd, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–30. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga M, Hatakeyama T. High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast. 2001;18:1015–21. doi: 10.1002/yea.753. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–61. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–31. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Watson AT, Werler P, Carr AM. Regulation of gene expression at the fission yeast Schizosaccharomyces pombe urg1 locus. Gene. 2011;484:75–85. doi: 10.1016/j.gene.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Watt S, Mata J, Lopez-Maury L, Marguerat S, Burns G, Bahler J. urg1: a uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One. 2008;3:e1428. doi: 10.1371/journal.pone.0001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrkamp-Richter S, Hyppa RW, Prudden J, Smith GR, Boddy MN. Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Gitsham P, Wishart J, Oliver SG, Zhang N. An improved tetO promoter replacement system for regulating the expression of yeast genes. Yeast. 2003;20:1255–62. doi: 10.1002/yea.1037. [DOI] [PubMed] [Google Scholar]