A CBFA2T3-GLIS2 fusion gene was identified in 31% of non–Down syndrome AMKL.
Abstract
Acute megakaryoblastic leukemia (AMKL) is a heterogeneous disease generally associated with poor prognosis. Gene expression profiles indicate the existence of distinct molecular subgroups, and several genetic alterations have been characterized in the past years, including the t(1;22)(p13;q13) and the trisomy 21 associated with GATA1 mutations. However, the majority of patients do not present with known mutations, and the limited access to primary patient leukemic cells impedes the efficient development of novel therapeutic strategies. In this study, using a xenotransplantation approach, we have modeled human pediatric AMKL in immunodeficient mice. Analysis of high-throughput RNA sequencing identified recurrent fusion genes defining new molecular subgroups. One subgroup of patients presented with MLL or NUP98 fusion genes leading to up-regulation of the HOX A cluster genes. A novel CBFA2T3-GLIS2 fusion gene resulting from a cryptic inversion of chromosome 16 was identified in another subgroup of 31% of non–Down syndrome AMKL and strongly associated with a gene expression signature of Hedgehog pathway activation. These molecular data provide useful markers for the diagnosis and follow up of patients. Finally, we show that AMKL xenograft models constitute a relevant in vivo preclinical screening platform to validate the efficacy of novel therapies such as Aurora A kinase inhibitors.
Acute megakaryoblastic leukemia (AMKL) is a heterogeneous subtype of acute myeloid leukemia (AML) and is more frequent in children than in adults (Lion et al., 1992; Dastugue et al., 2002; Paredes-Aguilera et al., 2003). The clinical features of AMKL, including rare occurrence, low blast counts, myelofibrosis, and the young age of patients have rendered difficult the molecular characterization of genetic alterations and establishment of models using primary patient cells. In adults, AMKL leukemic blasts often present a complex karyotype and frequently occur upon leukemic transformation of chronic myeloproliferative syndromes, including polycythemia vera, essential thrombocythemia, and primary myelofibrosis, which are associated with activating mutations in JAK2 or MPL (James et al., 2005; Pikman et al., 2006). In pediatric AMKL, two molecular subtypes have been characterized. The first group is represented by Down syndrome (DS) patients and presents with acquired GATA1 mutations leading to the expression of a GATA1-short (GATA1s) isoform lacking the wild-type transactivation domain (Wechsler et al., 2002; Roy et al., 2009). In non-DS AMKL, one third of infants present with the t(1;22)(p13;q13) chromosomal translocation, resulting in expression of the OTT-MAL fusion protein (Ma et al., 2001; Mercher et al., 2001, 2002). To date, only few point mutations in genes known to be involved in hematopoietic malignancies have been reported. Among them, the relevance of mutations in members of pathways involved in proliferation or survival is highlighted by the demonstration of activating point mutations in JAK2, JAK3 (Jelinek et al., 2005; Mercher et al., 2006; Walters et al., 2006), and MPL (Malinge et al., 2008) in AMKL patients and by the observation that mouse models of Gata-1s or Ott-MAL expression alone do not develop full-blown malignancy (Li et al., 2005; Mercher et al., 2009), whereas those with coexpression of Ott-MAL or Gata-1s with a mutant MPLW515L do (Mercher et al., 2009; Malinge et al., 2012). Together, the genetic basis of at least 50% of non-DS AMKL remains elusive. A recent study indicates that pediatric AMKL presents a high number of structural alterations with 9.33 copy-number alterations compared with 2.38 copy-number alterations on average for other subtypes of pediatric AML (Radtke et al., 2009). These observations suggest that structural genomic aberrations represent the major genetic basis in non-DS AMKL pathogenesis and that additional alterations remain to be identified and characterized at the molecular level.
Our limited understanding of the molecular basis for non-DS AMKL also affects the current treatment options. Indeed, although DS AMKL responds well to current therapies, non-DS AMKL patients have a poor prognosis with the majority of patients relapsing within 1 yr (Malinge et al., 2009). The development of accurate models of AMKL with primary patient leukemic cells is therefore needed to aid the development of novel therapies. In this study, we have developed xenotransplantation models in which human AMKL cells expanded and recapitulated the human disease, giving the opportunity to perform molecular analyses and assess the efficacy of a novel differentiation therapeutic strategy in vivo.
RESULTS
Xenotransplantation of AMKL primary patient cells models human disease
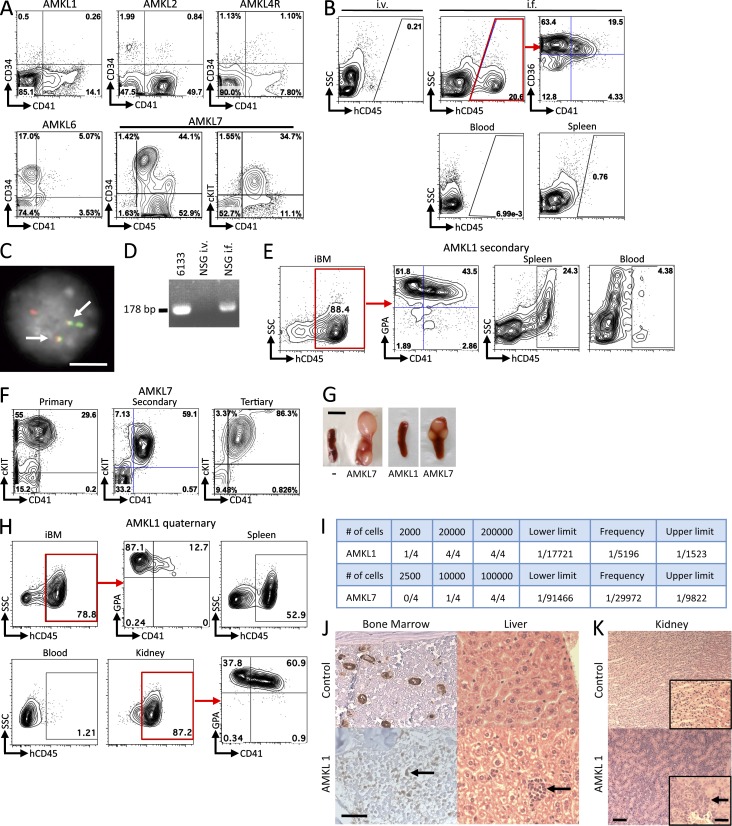
We first assessed whether xenotransplantation in immunodeficient mice is a suitable approach to model pediatric non-DS AMKL. Blast cells from the blood or BM of seven AMKL patients were immunophenotyped (Fig. 1 A and not depicted) and injected (1–2 × 106 cells/mouse) into sublethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjll/SzJ (NSG) mice by either i.v. or intrafemoral (i.f.) injection. Because of the small number of AMKL blasts, we generally used most of the patient sample to inject three to five recipients and retained <5 × 105 cells for molecular validation. For one patient, we transplanted both diagnosis (AMKL4) and relapse (AMKL4R) samples. Assessment of the engraftment was performed 6 wk after injection through blood and BM sampling followed by flow cytometry analysis for human surface markers. Six out of eight injected samples gave rise to a significant engraftment, and some cryopreserved samples were able to engraft (Table 1). Of note, the two samples that did not engraft either with i.v. of i.f. injections were from adult AMKL patients. Two representative examples of successful engraftments, with cells from AMKL1 (harboring an OTT-MAL fusion) and AMKL7 patients (lacking known molecular alteration), are detailed (Fig. 1). Importantly, recipients of i.v. injections did not harbor significant hCD45+ leukemic cells in the BM as opposed to recipients of i.f. injections (Fig. 1 B). Using fluorescent in situ hybridization (FISH) and RT-PCR on cells derived from AMKL1 recipient animals, we could detect fusion signals and an OTT-MAL fusion transcript (Fig. 1, C and D, respectively), confirming that engrafted cells were derived from the injected AMKL1 patient leukemic blasts with the OTT-MAL fusion. Additional flow cytometry analysis showed that human cells often coexpressed megakaryocytic and erythroid markers as well as immature hematopoietic markers such as c-KIT (Fig. 1, A, B, and E). Human leukemic cells obtained from successfully engrafted primary recipients were flow-sorted to obtain DNA and RNA to perform molecular analyses, and remaining cells were serially transplanted into secondary animals (0.5 × 106 cells/recipient). The phenotype of the leukemic cells was stable in secondary and subsequent transplantations (Fig. 1, E, F, and H). To further characterize the AMKL1 and AMKL7 samples, we investigated the frequency of leukemia-propagating cells by transplanting serial dilutions of hCD45+ cells purified by flow cytometry from secondary to tertiary recipients. Tertiary recipients presenting >0.1% of human cells in the BM at 4 wk after transplantation were considered positive for leukemia engraftment and propagation. The results indicate a frequency of leukemia-propagating cells of 1 in 5,196 for AMKL1 and 1 in 29,972 for AMKL7 (Fig. 1 I and not depicted).
Figure 1.
Engraftment of human AMKL samples into NSG recipients. (A) Immunophenotype of patient cells at diagnosis. (B) Flow cytometry analysis of blood and BM samples from recipients 6 wk after injection of 106 AMKL1 patient cells. Cells were injected either i.v. or i.f. An antibody against the human CD45 marker was used to specifically detect human cells in BM (top), blood, and spleen from recipients. (C) FISH analysis of leukemic cells collected from primary NSG recipients engrafted with AMKL1 patient cells using probes overlapping the OTT (green) and MAL (red) loci. Note the leukemic cells present: one normal OTT signal, one normal MAL signal, and two split signals indicative of a balanced translocation. Arrows point to fusion signals. (D) RT-PCR analysis of RNA from BM cells collected from primary NSG recipients engrafted with AMKL1 patient cells to detect an OTT-MAL fusion transcript. RNA from 6133, a cell line expressing the OTT-MAL fusion, was used as a positive control. (E) Immunophenotype of AMKL1 secondary NSG recipients 6 wk after injection with 0.5 × 106 patient cells. (F) Immunophenotype of AMKL7 patient cells from primary, secondary, and tertiary NSG recipients 6 wk after injection. (G) Consistent splenomegaly with nodular infiltration in AMKL7 recipients is not observed with other AMKL samples including AMKL1. “−“ is a noninjected control NSG mouse. (H) 0.5 × 106 cells from tertiary NSG recipients (AMKL1) were injected i.f. into quaternary recipients. Immunophenotype of injected BM (iBM), spleen, blood, and kidney cells at end point. (I) Human CD45+ cells were purified by flow cytometry from AMKL1 and AMKL7 secondary NSG recipients and injected at different doses in NSG recipients. After 4 wk, BM sampling and flow cytometry were performed. Recipients presenting a percentage of human cells (hCD45+) >0.1% were considered positive. The frequency of leukemia-propagating cells and the lower and upper limits were calculated using the L-Calc software with a 95% confidence interval. (J) Histopathological analysis of BM (left: von Willebrand immunohistochemistry; arrow points to an immature blast showing a low staining with von Willebrand marker) and liver (right: hematoxylin and eosin staining; arrow points to a focal infiltration of blasts) from AMKL1 recipients (bottom) and noninjected control NSG mouse (top). (K) Histopathological analysis of kidney from control and AMKL1 quaternary recipients (arrow points to infiltrated blasts). Insets show a higher magnification of the same section. Bars: (C) 10 µm; (G) 5 mm; (K) 200 µm; (J and K [insets]) 100 µm.
Table 1.
Summary of patient samples xenotransplanted in NSG immunodeficient animals
| Patient | Age | Gender | Sample | % of blasts | Karyotype | Engraftment | Mean % |
| AMKL1 | 6 mo | F | Fresh | 20 | 46,XX, t(1;22)(p13;q13) [17]/46, idem, ider(11) add(11)(p14) t(11;12)(q11;q23) [2]/46,XX [1] | 2/4 | 89.7 |
| AMKL2 | 21 mo | M | Frozen | 55 | 55, XY, +6, +9, t(9;11)(p22;q23), +10, +13, +19, +20, +21, +22, +22 [18] / 46, XY [2] | 1/3 | 42.7 |
| AMKL3 | 53 yr | M | Fresh | 70 | ND | 0/4 | NA |
| AMKL4 | 15 mo | M | Frozen | 20 | 51-52 XY, t(2;7)(q23;q21), t(2;11)(p11;p11-12), +4, +6, +8, +9, +/− 10, del(13)(q12-q13.14), +21 [17]/ 52-53, idem, + 13 [2]/ 46, XY [1] | 4/4 | 60.7 |
| AMKL4R | 26 mo | M | Fresh | 75 | ND | 3/3 | 32.3 |
| AMKL5 | 70 yr | F | Frozen | 80 | ND | 0/4 | NA |
| AMKL6 | 72 yr | M | Fresh | 87 | ND | 1/4 | 16 |
| AMKL7 | 42 mo | M | Frozen | 80 | ND | 2/5 | 66.6 |
F, female; M, male; NA, not applicable; R, relapse. Age at diagnosis is indicated in months or years. “Mean %” indicates the mean percentage of human cells.
We then performed histopathological analysis to confirm that although AMKL7 primary recipients consistently presented striking nodular infiltration of the spleen (Fig. 1 G), the majority of AMKL recipients did not show massive invasion of the spleen or blood with leukemic blasts (Fig. 1, B and G; and not depicted). Recipient animals also exhibited BM engraftment as revealed by positive staining of immature blasts for the von Willebrand marker using immunohistochemistry on BM sections (Fig. 1 J, left). Focal infiltrations of the liver sinusoids were also observed (Fig. 1 J, right). Interestingly, quaternary but not primary recipients of AMKL1 cells presented infiltration of spleen and a severe infiltration of the kidney (Fig. 1, H and K; and not depicted) that was previously described in another mouse knockin model of AMKL with Ott-MAL expression (Mercher et al., 2009).
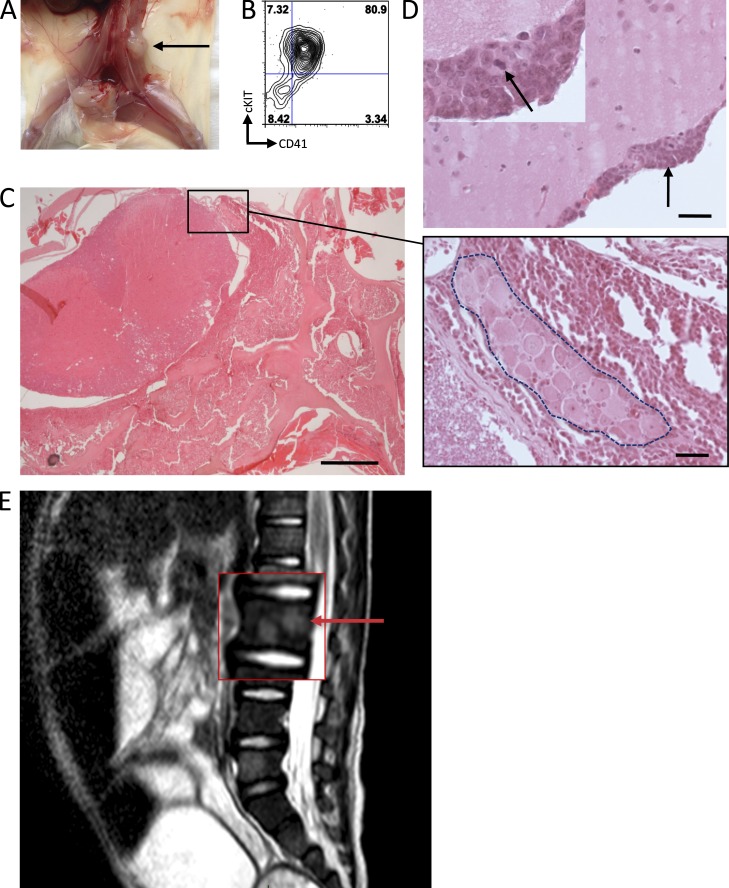
In one case (AMKL7), we observed consistent hind leg paralysis in all primary, secondary, and tertiary recipients. In some animals, a macroscopic tumor was observed near the spine, and analysis of the tumor by flow cytometry indicated that they were leukemic cells with a megakaryoblastic phenotype (Fig. 2, A and B). Detailed histopathological analysis of other animals indicated marked infiltration of the spinal cord leptomeninges and of the vertebra BM with leukemic cells compressing the spinal cord (Fig. 2 C). In addition, the cerebral leptomeninges presented multifocal infiltrations with proliferating leukemic cells (Fig. 2 D). Interestingly, this observation paralleled the clinical record of patient AMKL7 at advanced stages of the disease as indicated by abnormal signals upon magnetic resonance imaging (MRI) analysis indicative of spinal cord infiltration within several vertebral bodies (Fig. 2 E).
Figure 2.
CNS involvement in AMKL. (A) Recipient from AMKL7 cells showing spinal cord–localized tumor indicated by an arrow. (B) FACS analysis of the spinal cord tumor shown in A indicates that it is constituted of megakaryoblastic cells. (C) Histopathological analysis of the spinal cord. Close-up shows a spinal ganglion (inside dashed area) surrounded by leukemic cells. (D) Histopathological analysis of the brain reveals infiltration of the leptomeninges with leukemic cells. Arrow in the main image points to an infiltration of leukemic cells also shown in the top left inset; arrow in the inset points to a cell in mitosis. Bars: (C, left) 5,000 µm; (C [right] and D) 50 µm. (E) MRI image of the AMKL7 patient showing abnormal signals in several spine areas, suggesting leukemia infiltration of the nervous system. Arrow points to an abnormal signal.
Together, our data indicate that xenotransplantation of non-DS primary AMKL patient cells in immunodeficient NSG recipient mice enables engraftment of leukemic cells. These models recapitulate features of the human disease, including low blast counts in the blood, organomegaly, and in some instances central nervous system (CNS) infiltration, confirming that they can therefore be considered as faithful models of human AMKL.
High-throughput RNA-seq identifies novel recurrent fusion genes in AMKL
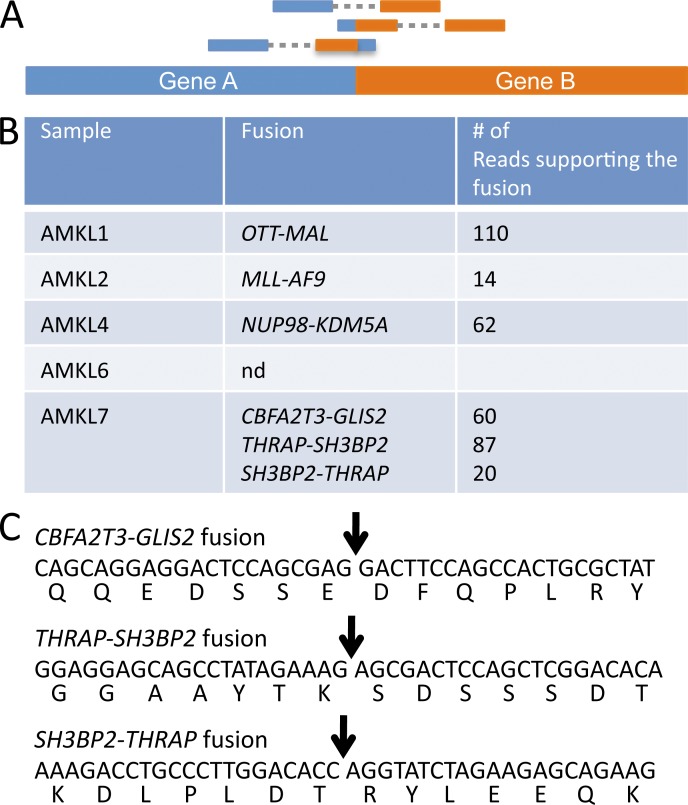
To investigate the molecular basis of non-DS AMKL, we performed high-throughput sequencing of RNA (RNA-seq) obtained from FACS-sorted primary recipient leukemic cells. Based on the observation that structural alteration may represent a major type of alteration in AMKL, we searched for fusion transcripts that could result from chromosomal translocations, inversions, or deletions (Fig. 3 A). To this end, we used the TopHat2 and DeFuse software and tuned the program parameters using results from the AMKL1 sample, which was known to express the OTT-MAL fusion oncogene. Interestingly, fusion genes were detected in all samples (summarized in Fig. 3 B), except AMKL6 which was obtained from an adult patient. AMKL2 harbored an MLL-AF9 fusion, consistent with the detection of a t(9;11)(p22;q23) translocation upon subsequent karyotype analysis (Table 1). AMKL4 presented a NUP98-KDM5A fusion that was previously identified in AML (van Zutven et al., 2006). AMKL7 had three novel in-frame fusion transcripts: a CBFA2T3-GLIS2 and a reciprocal fusion, THRAP3-SH3BP2 and SH3BP2-THRAP3 (Figs. 3 C and 4).
Figure 3.
High-throughput RNA-seq identifies novel recurrent fusion genes. (A) Scheme of the high-throughput sequencing reads supporting the presence of a fusion gene. (B) Summary of the fusion genes detected in the different AMKL samples engrafted into immunodeficient animals. (C) Sequence of the novel fusion transcripts found in AMKL and predicted sequence of the fusion proteins. Arrows indicate the fusion point. nd, none detected.
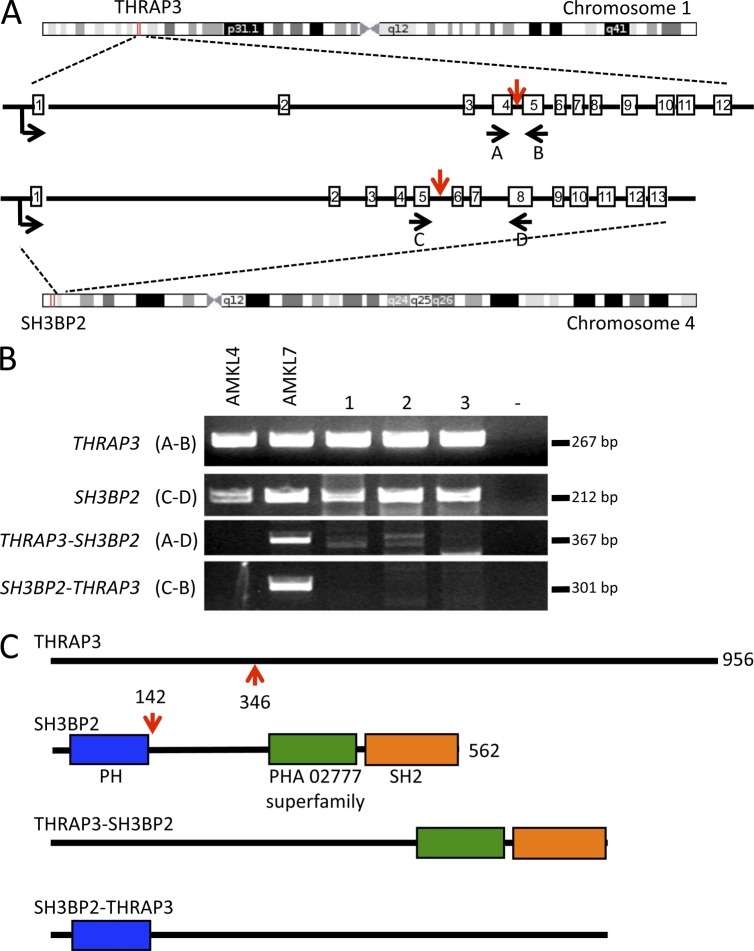
Figure 4.
The THRAP3-SH3BP2 fusion. (A) Schematic representation of the fusion between THRAP3 (located on chromosome 1) and SH3BP2 (located on chromosome 4). Localization on chromosome and exon–intron gene structure are indicated. Red vertical arrows indicate the targeted introns. Horizontal black arrows indicate localization of the primers used for RT-PCR analysis described in B to detect the fusion transcripts. (B) RT-PCR analysis on a validation cohort of AMKL patients using the primers described in A. Bands appearing in samples 1, 2, and 3 for the THRAP-SH3BP2 fusion were not confirmed by direct sequencing and therefore represent nonspecific amplification. (C) Schematic representation of the THRAP3, SH3BP2, THRAP3-SH3BP2, and SH3BP2-THRAP3 predicted proteins. Red arrows indicate fusion points on each protein.
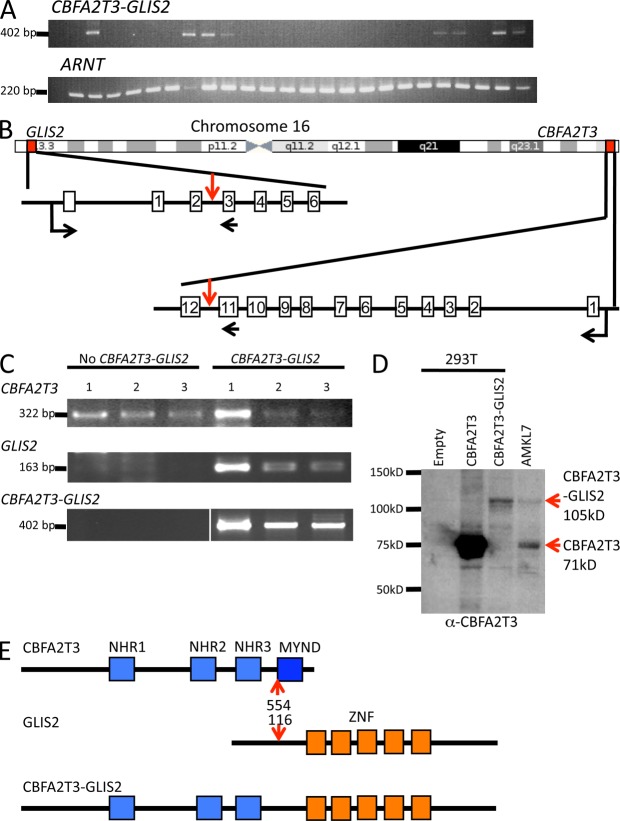
To assess the recurrence of these fusions, we performed RT-PCR analysis on an independent validation cohort of 22 non-DS pediatric AMKL, 9 DS AMKL, and 8 adult AMKL samples (Bourquin et al., 2006). The NUP98-KDM5A was found in one non-DS pediatric AMKL. The CBFA2T3-GLIS2 fusion was detected in seven non-DS pediatric AMKL samples (31%) but not in DS AMKL or adult AMKL (Figs. 3 B and 5 A), whereas the THRAP3-SH3BP2 and SH3BP2-THRAP3 fusions were not detected in the validation cohort. These fusions genes were mutually exclusive with the OTT-MAL fusion. Together, these results indicate that the NUP98-KDM5A fusion is a recurrent event in non-DS AMKL and that a new CBFA2T3-GLIS2 fusion characterizes the molecular basis of a novel major molecular subgroup of non-DS AMKL.
Figure 5.
The CBFA2T3-GLIS2 fusion is recurrent in AMKL. (A) RT-PCR analysis on a validation cohort of AMKL patients using primers located on CBFA2T3 exon 11 and GLIS2 exon 3 (top). Detection of the ARNT transcript was used as an RNA quality control. (B) Schematic representation of the chromosome 16 chromosomal inversion leading to the fusion between CBFA2T3 and GLIS2. Localizations on chromosome and exon–intron gene structures are indicated. Red vertical arrows indicate the introns targeted by the inversion. Horizontal black arrows indicate localization of the primers used for RT-PCR analysis described in C to detect the fusion transcript. (C) RT-PCR analysis of CBFA2T3 and GLIS2 expression in patients with or without the CBFA2T3-GLIS2 fusion. White line indicates that intervening lanes have been spliced out. (D) Western blot analysis of AMKL7 cells using an anti-CBFA2T3 antibody. AMKL7 cell lysates were prepared from fresh cells obtained from tertiary recipients. For controls, 293T cells were transfected with empty or CBFA2T3- or CBFA2T3-GLIS2–encoding vectors. (E) Schematic representation of the CBFA2T3, GLIS2, and CBFA2T3-GLIS2 predicted proteins. Red arrows indicate fusion points on each protein. ZNF, Krüppel-like zinc finger domain.
The CBFA2T3 and GLIS2 genes are both localized, in inverted orientation, on each arm of chromosome 16 close to the telomeres, in 16q24.3 and 16p13.3, respectively. Therefore, the CBFA2T3-GLIS2 fusion results from a chromosomal inversion of chromosome 16 (Fig. 5 B), leading to a fusion between exon 11 of CBFA2T3 and exon 3 of GLIS2. The GLIS2-CBFA2T3 reciprocal fusion was not detected by RNA-seq, suggesting that GLIS2 is not expressed in megakaryoblastic cells. RT-PCR analyses to detect expression of endogenous CBFA2T3 and GLIS2 in AMKL samples with and without the CBFA2T3-GLIS2 fusion confirmed this hypothesis. Indeed, CBFA2T3 was expressed in all AMKL samples, whereas GLIS2 expression was only detected in AMKL samples presenting the CBFA2T3-GLIS2 fusion (Fig. 5 C), indicating that the fusion with CBFA2T3 allows for ectopic expression of GLIS2 in megakaryoblastic cells. Accordingly, both the CBFA2T3 and the CBFA2T3-GLIS2 proteins could be detected in AMKL7 cells (Fig. 5 D).
CBFA2T3, also known as ETO2 or MTG16, is a member of the ETO/CBFA2T1/MTG8 family containing three members related to the Drosophila melanogaster nervy gene. Of note, both CBFA2T1 and CBFA2T3 are fused to RUNX1 (also known as AML1) in the t(8;21)(q22;q22) translocation of AML and t(16;21)(q24;q22) of therapy-related AML, respectively (Yamagata et al., 2005). However, the structures of the fusions are different, suggesting that the mechanisms of transformation are unrelated. The four NHRs (nervy homology regions) of CBFA2T3 present amino acid identity of ∼90% with those of ETO (Fig. 5 E), and both proteins have been found in HDAC (histone deacetylase corepressor) complexes (Davis et al., 2003). As a result of the fusion to GLIS2, the last NHR domain of CBFA2T3, which includes an MYND (myeloid, nervy, and Deaf-1 domain) class of zinc finger domain reported to interact with the N-CoR repressor complex, is lost. GLIS2 (GLI-similar 2) is a member of the Krüppel-like zinc finger transcription factors that are closely related to the GLI family of proteins mediating the transcriptional response to the Hedgehog pathway activation (Kang et al., 2010; Lichti-Kaiser et al., 2012). In the CBFA2T3-GLIS2 fusion, all of the GLIS2 zinc finger domains responsible for interaction with DNA and transactivation (Vasanth et al., 2011) are present (Fig. 5 E). These data suggested that this fusion between two transcriptional regulators results in aberrant expression of genes controlled either by CBFA2T3 or GLIS2.
CBFA2T3-GLIS2 AMKL exhibits a distinct expression signature
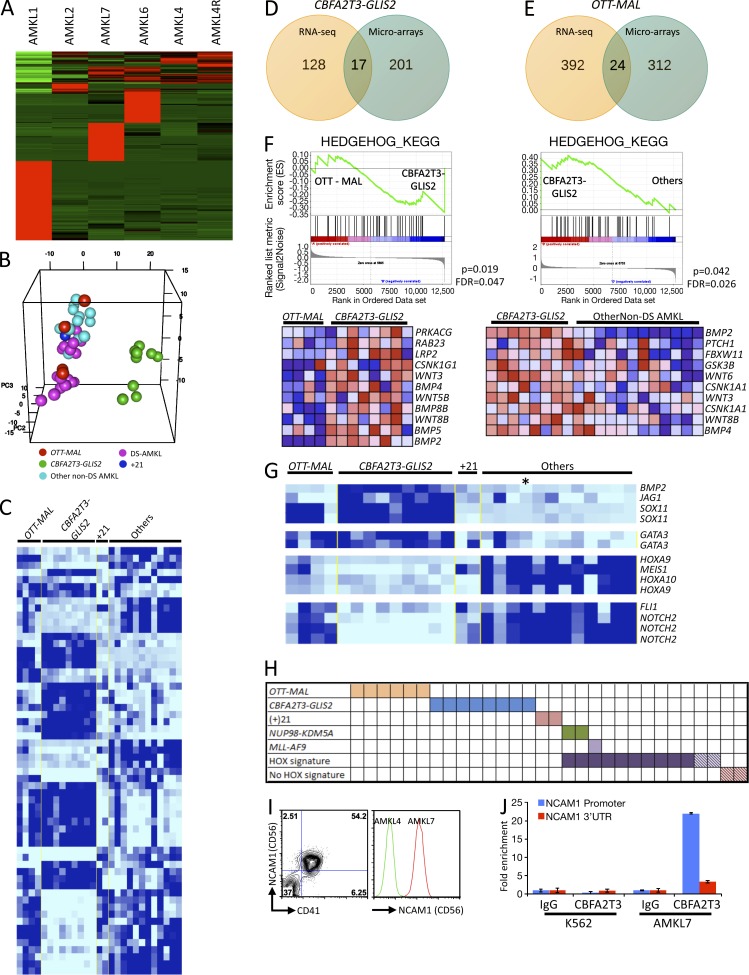
To characterize the molecular consequences of CBFA2T3-GLIS2 expression, we first analyzed the gene expression profile of each AMKL sample by RNA-seq and observed that AMKL1 (OTT-MAL fusion) and AMKL7 (CBFA2T3-GLIS2 fusion) presented specific molecular signatures (Fig. 6 A). Principal component analysis of the differentially regulated genes allowed homogeneous clustering of CBFA2T3-GLIS2 samples apart from other samples (Fig. 6 B). To confirm these observations, we performed meta-analysis of global expression data on the independent cohort in which we detected the additional patients with the CBFA2T3-GLIS2 fusion. We observed that the CBFA2T3-GLIS2 samples also presented a molecular signature different from OTT-MAL and other non-DS AMKL samples (Fig. 6 C and Table S1). Therefore, detection of the CBFA2T3-GLIS2 fusion now allows the separation of these patients from the OTT-MAL patients that were originally in the same consensus cluster (Fig. 6, B and C; Bourquin et al., 2006). To identify the most robustly deregulated gene signature using both the RNA-seq data containing few samples and the microarray data containing more samples, we focused on the intersection between the RNA-seq and microarray signatures (Fig. 6, D and E; and Table S2). For CBFA2T3-GLIS2 samples, all the genes of the intersection were up-regulated compared with other samples, supporting the idea that CBFA2T3-GLIS2 is a transcriptional activator. Among these genes were BMP2 and GATA3, two known target genes of the Hedgehog signaling pathway (Craven et al., 2004; Lan and Jiang, 2009), as well as IGFBP7, CCND2, NCAM1, and the homeobox HOPX (Table S2). Of note, the sample size of the RNA-seq dataset and the low overlap between the RNA-seq and microarray signature likely reflect the fact that RNA-seq data are based on analysis of one sample in each molecular subgroup and the corresponding low statistical power. To further investigate the alteration of the Hedgehog pathway, we performed gene set enrichment analysis (GSEA) on microarray data using a list of genes involved in the Hedgehog pathway obtained from the KEGG database to compare OTT-MAL and CBFA2T3-GLIS2 or CBFA2T3-GLIS2 and other non-DS AMKL samples. In both comparisons, GSEA results showed a significant enrichment of this list in CBFA2T3-GLIS2 samples (Fig. 6 F, top), with leading edge genes including BMP2, BMP4, WNT3, and WNT8B (Fig. 6 F, bottom).
Figure 6.
CBFA2T3-GLIS2 AMKLs exhibit a distinct expression signature. (A) Expression analysis of RNA-seq data showing molecular signature of each sample versus all of the other samples. (B) Genes implicated in the signatures indicated in A were used to perform principal component analysis. (C) Class comparison of AMKL samples presenting the OTT-MAL fusion, CBFA2T3-GLIS2 fusion, acquired trisomy 21, and other alterations. (D) Venn diagram representing common genes between RNA-seq and microarray signatures of AMKL samples presenting CBFA2T3-GLIS2 fusion. (E) Venn diagram representing common genes between RNA-seq and microarray signatures of AMKL samples presenting OTT-MAL fusion. (F) GSEA using a Hedgehog gene list comparing OTT-MAL with CBFA2T3-GLIS2 patients (left) and CBFA2T3-GLIS2 with other non-DS AMKL patients (right). The leading edge genes for each comparison are represented in the bottom panels. FDR, false discovery rate. (G) Selected genes from molecular signatures of the different AMKL subgroups. Asterisk indicates the patient presenting the NUP98-KDM5A fusion. (H) Schematic representation of all fusion genes found in AMKL samples. Hatched squares represent patients with no available material for detection of the novel fusions by bispecific PCR but for which global expression data are available. (I) Flow cytometry analysis of CD56 expression on AMKL7 (CBFA2T3-GLIS2) leukemic blasts (right). Comparison of the CD56 expression between AMKL4 (NUP98-KDM5A) and AMKL7. (J) ChIP analysis using a CBFA2T3-specific antibody or a nonspecific antibody (IgG) on AMKL7 cells or control K562 cells. Quantitative PCR was then performed on immunoprecipitated DNA using primer pairs located in the proximal NCAM1 promoter or in the 3′ UTR. Error bars indicate mean ± SEM.
To assess the molecular signature of NUP98-KDM5A, we searched the genes differentially expressed in other non-DS AMKL samples by comparison with samples with OTT-MAL, CBFA2T3-GLIS2, and acquired trisomy 21 (+21). Most of the samples in this group, including the NUP98-KDM5A sample, presented a strong up-regulation of HOXA9, HOXA10, and MEIS1 compared with OTT-MAL, CBFA2T3-GLIS2, and +21 samples (Fig. 6 G), confirming that most of the remaining non-DS AMKL samples have a deregulation of HOX genes (Fig. 6 H; Bourquin et al., 2006; Wang et al., 2009).
Surface markers differentially up-regulated in CBFA2T3-GLIS2 samples represent potentially interesting molecular markers for the diagnosis and follow up of AMKL patients with this chromosomal alteration. The surface marker NCAM1 (CD56) presented a mean differential expression of 35-fold by microarray and >200-fold by RNA-seq. To validate expression results at the protein level, we compared the expression of CD56 between AMKL7 (CBFA2T3-GLIS2) and AMKL4 (NUP98-KDM5A) cells. Using flow cytometry, we observed that AMKL7 blasts with the CBFA2T3-GLIS2 fusion are indeed CD41+CD56+ and that CD56 was drastically more expressed than on NUP98-KDM5A–expressing AMKL4 leukemic cells (Fig. 6 I). To assess whether CD56 could be a direct transcriptional target of the CBFA2T3-GLIS2 fusion, we performed chromatin immunoprecipitation (ChIP) analysis using a CBFA2T3-specific antibody or a nonspecific antibody (IgG) on AMKL7 cells or control K562 cells. Quantitative PCR was then performed on immunoprecipitated DNA using several primer pairs located in the proximal NCAM1 promoter or in the 3′ untranslated region (UTR). Specific enrichment at the proximal promoter but not the 3′ UTR was observed (Fig. 6 J and not depicted) with the CBFA2T3 antibody in AMKL7 cells that express the CBFA2T3-GLIS2 fusion. In contrast, enrichment was not observed in the K562 cell line expressing the endogenous CBFA2T3 protein. These data suggest that the CBFA2T3-GLIS2 fusion protein binds to the proximal promoter of the NCAM1 gene.
Together, high-throughput sequencing of RNA from AMKL blasts revealed several novel cryptic fusion genes, including a CBFA2T3-GLIS2 fusion present in 1/7 non-DS AMKL sample in our RNA-seq cohort and 7/22 in a validation cohort. Samples showing this fusion exhibit a distinct molecular signature in global gene expression analysis compared with other non-DS AMKL samples, including up-regulation of NCAM1 and of several target genes of the Hedgehog signaling pathway.
Aurora A kinase (AURKA) inhibitors induce differentiation and inhibit proliferation of AMKL patient blasts in vitro and in vivo
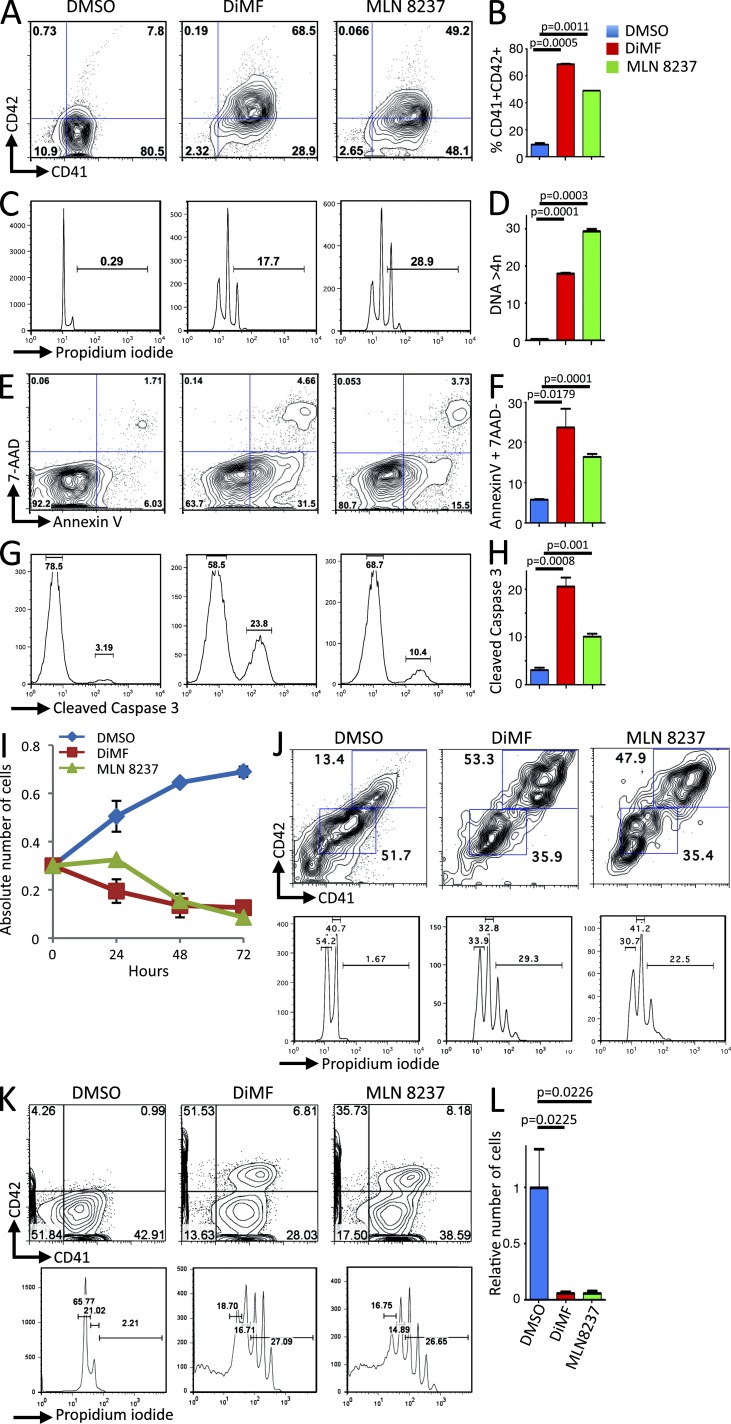
Finally, we investigated whether these novel models of human AMKL are suitable for the screening of novel therapeutic approaches. Dimethylfasudil (DiMF) was recently identified as a small-molecule inhibitor of AMKL cells through a screening strategy searching for inducers of megakaryocyte polyploidization that would inhibit proliferation and engage terminal differentiation of megakaryoblastic leukemic cells (Wen et al., 2012). Functional analysis indicated that an important target of DiMF is AURKA. To assess the efficacy of DiMF and AURKA inhibition on the proliferation of human AMKL cells with various genetic alterations, we cultured AMKL leukemic blasts purified by FACS from immunodeficient recipients with DiMF and MLN8237, another AURKA inhibitor currently being tested in clinical trials for other malignancies. We first treated in vitro leukemic cells recovered from AMKL7 (CBFA2T3-GLIS2) recipient animals with the two drugs and monitored their proliferation, differentiation, and ploidy over 72 h. Both molecules induced a significant enhancement of the expression of the maturing megakaryocyte-specific marker CD42 (Fig. 7, A and B) paralleled with a significant increase in ploidy content (Fig. 7, C and D). This differentiated phenotype was associated with an induction of apoptosis detected by increased Annexin V (Fig. 7, E and F) and cleaved caspase 3 (Fig. 7, G and H) and with a drastic inhibition of cell proliferation (Fig. 7 I). Similar results were obtained with other AMKL blasts, including AMKL1 (OTT-MAL) and AMKL4 (NUP98-KDM5A; Fig. 7, J–L).
Figure 7.
Aurora kinase inhibitors inhibit proliferation of AMKL patient blasts in vitro. (A) AMKL7 cells purified from immunodeficient recipients were cultured in vitro in the presence of DMSO (control), 5 µM DiMF, or 250 nM MLN8237 for 72 h. A representative flow cytometry analysis of CD41 and CD42 megakaryocyte–specific markers is shown. (B) Histogram representation of CD41+CD42+ maturing megakaryocyte elements shown in A. (C) Representative ploidy analysis of cells treated as in A. (D) Histogram representation of cells with a ploidy > 4n shown in C. (E) Representative apoptosis analysis of cells treated as in A. (F) Histogram representation of apoptotic cells (AnnexinV+7AAD− cells) shown in E. (G) Representative flow cytometry analysis of cleaved caspase 3 in cells treated as in A. (H) Histogram representation of cleaved caspase 3–positive cells shown in G. (B, D, F, and H) Mean value ± SEM of duplicate (B and D) or triplicate (F and H) experiments is shown. (I) Proliferation of AMKL7 leukemic cells upon treatment with aurora kinase inhibitors. Mean ± SEM of the number of viable cells (trypan blue exclusion, duplicate experiments) is shown. (J) Effect of 5 µM DiMF and 250 nM MLN8237 treatment on AMKL1 cells. Flow cytometry analysis of ploidy and differentiation after 6 d of in vitro treatment. (K) Effect of 5 µM DiMF and 250 nM MLN8237 treatment on AMKL4 cells. Flow cytometry analysis of ploidy and differentiation after 4 d of in vitro treatment. (L) Viable number of AMKL4 cells after 4 d of treatment as in K. Means ± SEM of duplicate experiments are shown.
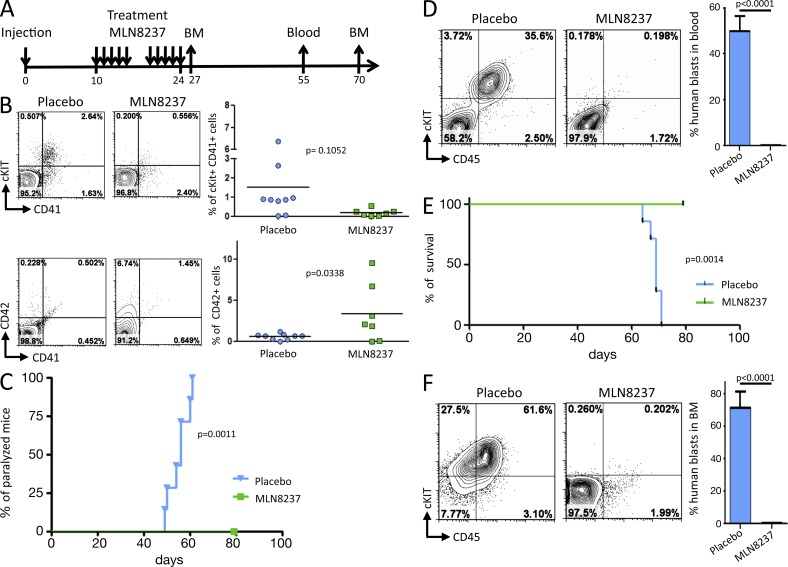
We then investigated whether treatment with an AURKA inhibitor affected disease development in vivo. 2 × 106 AMKL7 leukemic blasts obtained from tertiary recipients were injected per animal into a cohort of quaternary recipients that were treated with either 15 mg/kg MLN8237 or a placebo (Fig. 8 A). 10 d after injection, recipients were randomized to the placebo or MLN8237 groups and treated twice daily by oral gavage for 10 d. Although MLN8237 was well tolerated by the majority of the mice, 2/7 recipients lost >20% of their weight and died soon after the treatment period, but importantly, without signs of significant leukemic infiltration. The remaining 5/7 MLN8237-treated recipients displayed no visible signs of toxicity and presented with weight loss <10% on average and could therefore be further analyzed. BM sampling 3 d after treatment revealed that MLN8237 led to a significant increase in the fraction of human CD42+ cells compared with placebo-treated animals (Fig. 8 B), which supports an induction of terminal differentiation by MLN8237 in vivo. Placebo-treated recipients presented a median time to the first signs of hind leg paralysis of 56 d, whereas MLN8237-treated recipients did not present signs of paralysis for up to 80 d (Fig. 8 C). Blood samples collected 30 d after treatment indicated a striking decrease in the number of human blasts in MLN8237-treated compared with placebo-treated animals (Fig. 8 D). Placebo-treated recipients presented a median survival of 70 d, whereas none of the MLN8237 animals succumbed to disease up to 80 d (Fig. 8 E). Analysis of the BM from placebo- and MLN8237-treated animals at day 70 confirmed the low infiltration in MLN8237-treated recipients, whereas placebo-treated recipients presented massive infiltration (Fig. 8 F). Together, these results indicate that treatment of xenograft models of human AMKL with MLN8237, a clinically available AURKA inhibitor, efficiently inhibits proliferation of AMKL leukemic blasts, reduces disease burden, and increases survival.
Figure 8.
MLN8237 efficiently reduces disease burden and prolongs survival of a xenograft model of human AMKL expressing the CBFA2T3-GLIS2 fusion. (A) Schematic representation of the in vivo drug trial with MLN8237. 2 × 106 AMKL7 cells were injected per recipient. 10 d later, two groups of seven recipients were treated either with placebo or 15 mg/kg MLN8237. BM and blood were sampled at the indicated time points for analysis. (B) BM cells collected at day 27 were analyzed by flow cytometry. (left) A representative flow cytometry analysis of h–c-KIT, hCD41, and hCD42 markers is shown. (right) Scatter plots indicate the result for all recipients. Horizontal lines indicate mean. (C) Representation of the onset of hind leg paralysis (Placebo: n = 7, MLN8237: n = 5). (D) Representative analysis (left) and histogram representation (right) of the blood samples collected at day 55. (Placebo: n = 7, MLN8237: n = 5). (E) Kaplan-Meier survival curves of placebo-treated (n = 7) and MLN8237-treated (n = 5) animals. (F) BM cells collected at day 70 were analyzed by flow cytometry. (left) A representative flow cytometry analysis of cKIT, CD41, and CD42 markers is shown. (right) Histogram representation of the results (Placebo: n = 2, MLN8237: n = 5). (D and F) Error bars indicate mean ± SEM.
DISCUSSION
AMKL is a heterogeneous subtype of AML generally associated with poor prognosis. Challenges in understanding the molecular basis of this disease derive in part from the difficulties in obtaining a sufficient amount of material, which together with the lack of experimental model limits the development of novel therapeutic strategies. Using a xenotransplantation approach with primary human AMKL patient samples into immunodeficient mice, we have established novel models of AMKL recapitulating features of the human disease. The leukemic blasts engrafted in immunodeficient recipient mice allowed the identification and characterization of novel genetic lesions and were relevant models to test the efficacy of AURKA inhibitors as a potential therapeutic strategy for the treatment of AMKL with various genetic alterations.
Xenotransplantation has been widely used as a strategy to propagate human leukemic cells in immunodeficient mice. To date however, no specific study of AMKL has been reported. In this study, we have shown that primary AMKL patient leukemic cells engraft NSG recipients. Although engraftment efficiency was higher with fresh samples, viable cells frozen in DMSO could also successfully engraft. We observed that the immunophenotype of leukemic cells that grow in immunodeficient recipients were heterogeneous, especially based on expression of the megakaryocytic marker CD41 that could be present on all leukemic cells or only on a small fraction. Recipient animals presented several features of human disease, including low blast count in the blood, massive infiltration of BM with immature megakaryoblastic elements, and variable infiltration of the liver and spleen. Strikingly, analysis of recipients injected with AMKL cells carrying the novel CBFA2T3-GLIS2 fusion oncogene showed consistent infiltration of the spinal cord, which was also detected by MRI analysis of the patient at late stages of the disease. Although, engraftment of additional samples will be required to assess whether this phenotype is associated with the CBFA2T3-GLIS2 fusion, several observations suggest its relevance for pathology. For example, AMKL, like other subtypes of leukemia can present with CNS infiltrations, which are generally associated with poor prognosis. In addition, the CBFA2T3-GLIS2 patients present a homogenous gene expression signature including strong expression of CD56, a marker proposed to correlate with CNS infiltration in acute lymphoblastic leukemia (Ravandi et al., 2002). Together, these models may constitute useful tools to further investigate the cellular and molecular basis of CNS infiltration in leukemia.
Our observations also provided important novel insights into the genetic basis of non-DS AMKL. Indeed, high-throughput sequencing of the RNA obtained from leukemic blasts amplified in immunodeficient recipients identified several previously undetected fusion genes. First, we characterized two patients with a NUP98-KDM5A fusion identical to a previously identified chromosomal translocation invisible by classical cytogenetics in AML (van Zutven et al., 2006) and an MLL-AF9 fusion in another non-DS AMKL patient. Although these fusions are rare in non-DS AMKL (8% and 4%, respectively in our cohort), MLL and NUP98 alterations were previously reported in AMKL (Borkhardt et al., 1995; Allen et al., 1998; Morerio et al., 2005; van Zutven et al., 2006; Takita et al., 2009; Marques-Salles et al., 2010). Both mutations result in deregulation of HOX A cluster genes, respectively, through recruitment of the histone H3K79 methyltransferase DOT1L by several MLL fusions (Nguyen and Zhang, 2011) and through recruitment of the histone acetyltransferase CBP/p300 and prevention of differentiation-associated removal of H3K4me3 by NUP98 fusions (Wang et al., 2009). Importantly, a larger subgroup of non-DS AMKLs show a HOX cluster signature with aberrant expression of HOXA9, HOXA10, and MEIS1 (Bourquin et al., 2006), suggesting that although individual MLL or NUP98 fusions are rare in AMKL, up-regulation of HOX genes may constitute the common mechanism within this subgroup of non-DS AMKL. Therefore, additional efforts to characterize other MLL and NUP98 fusions are warranted and may lead to identification of other genetic alterations resulting in HOX gene activation.
Second, we identified a new recurrent CBFA2T3-GLIS2 fusion resulting from a cytogenetically cryptic inversion of chromosome 16. The CBFA2T3-GLIS2 fusion was detected in 31% of pediatric non-DS AMKL, was not detected in adult AMKL or DS-AMKL, and was mutually exclusive with the OTT-MAL fusion (Gruber et al. 2012. American Association for Cancer Research. Abst. 4867). CBFA2T3 is a member of the ETO family involved in the NCoR/mSin3a/HDAC corepressor complex and is expressed during normal hematopoiesis. GLIS2 is a Krüppel-like zinc finger DNA-binding transcription factor related to GLI involved in the response to the Hedgehog pathway activation and is not expressed in megakaryoblastic cells. Of note, GLIS2 inactivating mutations have been found in nephronophthisis, an autosomal recessive renal disease, and the inactivation of Glis2 in the mouse results in progressive renal atrophy without reported hematopoietic phenotype (Attanasio et al., 2007; Kim et al., 2008). Interestingly, patients with the CBFA2T3-GLIS2 fusion present a clear expression signature clustering them apart from other non-DS AMKL. Among these differentially regulated genes are several known targets of the Hedgehog pathway including BMP2, BMP4, GATA3, and CCND2 (Craven et al., 2004; Lan and Jiang, 2009). These observations support the hypothesis that the fusion with CBFA2T3 leads to ectopic expression of the GLIS2 transcription factor, which in turn induces up-regulation of several Hedgehog target genes during megakaryopoiesis. Interestingly, Hedgehog pathway stimulation was shown to increase megakaryocyte differentiation of CD34+ hematopoietic cells (Martínez et al., 2006), and BMP4 was reported to be involved in TPO signaling and to play an important role in megakaryopoiesis (Jeanpierre et al., 2008), suggesting that aberrant activation of Hedgehog target genes by the CBFA2T3-GLIS2 fusion participates in the association between this fusion gene and megakaryoblastic leukemia. Together with previous studies (Borkhardt et al., 1995; Allen et al., 1998; Morerio et al., 2005; Bourquin et al., 2006; van Zutven et al., 2006; Takita et al., 2009; Marques-Salles et al., 2010), our genetic data indicate that non-DS AMKL can be divided into at least four molecular subgroups including patients with the OTT-MAL fusion, CBFA2T3-GLIS2 fusion, acquired trisomy 21 with a GATA1 mutation, and patients presenting a molecular signature of HOX A gene cluster activation including MLL or NUP98 alterations.
These results may also have direct clinical impact. Indeed, identification and characterization of these novel genetic lesions constitute additional molecular markers for the diagnosis and may serve for the detection of the minimal residual disease during follow up of AMKL patients as described previously with the OTT-MAL fusion (Ballerini et al., 2003). Of note, although the prognostic value of the CBFA2T3-GLIS2 fusion will require additional investigation, this fusion was associated with treatment-refractory disease in the patient analyzed here.
Finally, to experimentally address whether these animal models may serve as platforms to assess the relevance of novel therapeutic strategies for the treatment of AMKL, we investigated the effect of DiMF, an inhibitor of AURKA and a potent ploidy inducer of various megakaryoblastic leukemia cell lines and primary cells on growth of AMKL cells in vivo (Wen et al., 2012). Using AMKL patient cells harboring various genetic lesions, including OTT-MAL, CBFA2T3-GLIS2, NUP98-KDM5A, and MLL-AF9 fusions, we show that DiMF efficiently induces differentiation and polyploidization of leukemic blasts and drastically inhibits proliferation in vitro for all cases. Similar results were obtained with MLN8237, an AURKA inhibitor in clinical trials for other tumors. Most importantly, in vivo treatment of mice xenografted with human AMKL cells bearing the CBFA2T3-GLIS2 fusion with MLN8237 significantly reduced disease burden and prolonged survival of recipient mice. Although treatment of additional patient samples is required to precisely assess the relative sensitivity of each molecular subgroup of AMKL to AURKA inhibitors, these observations highlight that these compounds are promising candidates for the treatment of human AMKL independently of their genetic lesions. Together, we have demonstrated that modeling AMKL using a xenotransplantation approach provides an integrated platform to gain insights into the molecular genetics of AMKL and test novel therapeutic approaches. Our results support the idea that CBFA2T3-GLIS2 activates Hedgehog pathway genes. Because Hedgehog signaling is not required for self-renewal of normal adult hematopoietic progenitors (Gao et al., 2009; Hofmann et al., 2009), it will be interesting to test whether this molecular subgroup of AMKL is sensitive to Hedgehog pathway inhibitors targeting GLI factors.
MATERIALS AND METHODS
Patients.
Human AMKL patient blood or BM samples were obtained with the informed consent of the patient in accordance with national ethics rules. Cells were subjected to Ficoll gradient, immunophenotyped, and used directly or frozen in FBS supplemented with 10% DMSO.
Animal experiments.
106 AMKL cells were xenotransplanted in toto to sublethally irradiated (2–3 Gy) NSG immunodeficient mice (The Jackson Laboratory) by i.f. or by i.v. injections. Animals were maintained in pathogen-free conditions. Engraftment was monitored by blood and BM sampling at 6-wk after injection and performed under isoflurane anesthesia. Secondary recipients were injected with 0.5 × 106 cells per animal. For histological analyses, sample organs were routinely processed into paraffin section and stained with hematoxylin and eosin. Immunohistochemistry was performed using the von Willebrand factor antibody (Abcam). For the in vivo drug trial, 2 × 106 AMKL7 leukemic blasts obtained from tertiary recipients were injected per animal into a cohort of recipients that were used for treatment with either 15 mg/kg MLN8237 or a placebo. MLN8237 was dissolved in 100 mg/ml of an arginine solution. Placebo was comprised of the arginine solution alone. 10 d after injection, recipients were randomly assigned to the placebo and MLN8237 groups. Recipients were treated twice daily by oral gavage for 10 d (5 d of treatment, 2 d off, 5 d of treatment). Weight was monitored daily during the treatment period. BM samples were analyzed 3 d after the end of the treatment, blood samples were analyzed 1 mo after the end of the treatment, and final BM analyses were performed 49 d after the end of the treatment. Animal experiments were approved by the Commissariat à l’Énergie Atomique and Institut Gustave Roussy animal care and use committees.
Cell culture.
AMKL cells obtained directly from immunodeficient animals or after purification by flow cytometry were cultured in RPMI 1640 medium supplemented with 20% FBS (Gibco), penicillin-streptomycin (Gibco), and 10 ng/ml each of human SCF, IL3, IL6, IL11, TPO, and EPO (PeproTech).
Flow cytometry.
Cells analyzed by flow cytometry were stained in 1× PBS supplemented with 2% FBS. Antibodies including CD41, CD42, GPA, CD36, CD34, CD14, and cKIT were purchased from BD except otherwise mentioned. Whole BM cells from mice were collected, subjected to red blood cell lysis, and stained for FACS analysis. Cells were analyzed on a FACSCantoII or BDLSRII flow cytometer (BD). To obtain a pure population of AMKL blasts, cells were stained with cKIT, CD41, and CD45 and purified on FACSAriaIII (BD) BD Influx or MoFlo (Beckman Coulter) cell sorters. Postacquisition analyses were performed using FlowJo software (Tree Star).
FISH analysis.
The OTT-MAL fusion was detected by standard FISH methods using the BAC probes OTT (green: RP11-142E15 and RP11-948I5) and MAL (red: RP11-597P1 and RP11-229A8).
Sequencing and expression analysis.
RNA-seq was performed on an Illumina Hi-seq 2000 using paired-end sequencing of 150–250-bp inserts and 100-bp reads (Fasteris). 52–83 × 106 reads were obtained per sample. The quality of the reads was first evaluated using FastQC. Then we used TopHat (Trapnell et al., 2009), which is a fast splice junction mapper for RNA-Seq, to map the reads on the human genome. Bam files were transformed into sam files and sorted by read name using the samtools utilities. Fusion transcripts were detected using the TopHat 2 (Trapnell et al., 2009; Kim and Salzberg, 2011) and DeFuse (McPherson et al., 2011) programs. The known presence of an OTT-MAL fusion transcript in AMKL1 allowed calibration of the following parameters. TopHat2: mate-inner-dist: 200, fusion-anchor-length: 13, num-fusion-reads: 1, num-fusion-pairs: 1. DeFuse: max_insert_size: 200, split_min_anchor: 13, split_count_treshold: 5, span_count_treshold: 3. Fusions common to the two algorithms were considered for technical validation. For expression analysis, we counted the number of reads per gene using the HTSeq python package (Anders and Huber, 2010), using the gtf annotation file from UCSC (hg19). To test for differential expression, we used the R package DESeq with negative binomial distribution and a shrinkage estimator for the distribution’s variance. P-values < 0.05 (adjusted by Benjamini and Hochberg procedure) were considered significant. Analysis of gene expression in AMKL patients was performed using the set of Affymetrix U133A array data deposited at GEO (under accession no. GSE4119; Bourquin et al., 2006).
ChIP analysis and Western blotting.
ChIP analyses were performed according to the MagnaChIP kit (Millipore). In brief, 10 × 106 AMKL7 or K562 cells were fixed with 0.75% formaldehyde, lysed, and sonicated (10-min cycle on Covaris apparatus; KBioscience). Sheared chromatin was immunoprecipitated overnight using the following antibodies: anti-CBFA2T3 (Abcam), normal rabbit IgG (Santa Cruz Biotechnology, Inc.), and anti–histone H3 (Millipore). 1/10 of the sheared chromatin (Input) was used as a reference for the quantitative PCR analysis using the primers proximal NCAM1 promoter (forward, 5′-GCATCTGCCTCCCTGTCTCT-3′; and reverse, 5′-CTCGCAACTCGGAGATCCTT-3′) and 3′ UTR (forward, 5′-GCAGCTTTGGAGGTGGAACT-3′; and reverse, 5′-GAGAGAAGGCCCCCGAAGTA-3′) on a 7500 Fast Real-Time PCR system (Applied Biosystems). AMKL7 and 293T cells lysates were prepared as follows: (a) cells were lysed in 20 mM Tris-HCl, pH 7.4, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, 1% Triton X-100, and 10% glycerol supplemented with protease inhibitors, (b) cells were sonicated, and (c) NaCl was added for a final concentration of 150 mM. Western blotting was then performed using standard procedures, and the anti-CBFA2T3 antibody was incubated overnight.
Statistical analysis.
Data are depicted as means ± SEM or SD as denoted. P-values were calculated using either the Mann–Whitney test or the two-tailed unpaired Student’s t test (Prism version 4; GraphPad Software). P-values for survival curves were calculated using a Log-rank (Mantel-Cox) test (Prism version 4).
Online supplemental material.
Table S1 shows class comparison between OTT-MAL, CBFA2T3-GLIS2, and other non-DS AMKLs. Table S2 shows the intersection between RNA-seq and microarray expression analyses. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121343/DC1.
Supplementary Material
Acknowledgments
We thank Julien Tillet and Christophe Joubert from the Commissariat à l’Énergie Atomique (CEA) animal facility and Patrick Gonin, Justine Deniau, and Aurélie Gasnier from Institut Gustave Roussy animal facilities for excellent mouse care. We also thank Olivia Bawa for histological sections and stainings and Philippe Rameau from the Institut Gustave Roussy Flow Cytometry Core Facility. We thank Paule Zanardo and Rahma Benaicha for excellent administrative assistance.
Our laboratories were supported by grants from Institut National de la Santé et de la Recherche Médicale, CEA, Institut National du Cancer (INCA), Agence Nationale Recherche (ANR-09-BLAN-0275-02-MEGON), Région Ile de France, Association pour la Recherche sur le Cancer (to T. Mercher and W. Vainchenker), Ligue Nationale Contre le Cancer (to F. Pflumio and O.A. Barnard), Association Laurette Fugain (to F. Pflumio), and the Samuel Waxman Cancer Research Foundation (to J.D. Crispino). C. Thiollier was supported by a PhD fellowship from the Fondation pour la Recherche Médicale. B. Gerby was supported by a PhD fellowship from Ligue Nationale Contre le Cancer and Société Francaise d’Hématologie. C. Ignacimouttou and S. Poglio were supported by fellowships from INCA. T. Mercher was supported by fellowships from the Fondation Gustave Roussy and José Carreras Leukemia Foundation–European Hematology Association (DJCLS EHA 2009 F 09/02).
The authors declare that they have no relevant competing financial interests.
Footnotes
Abbreviations used:
- AMKL
- acute megakaryoblastic leukemia
- AML
- acute myeloid leukemia
- AURKA
- Aurora A kinase
- ChIP
- chromatin immunoprecipitation
- CNS
- central nervous system
- DiMF
- dimethylfasudil
- DS
- Down syndrome
- FISH
- fluorescent in situ hybridization
- GSEA
- gene set enrichment analysis
- i.f.
- intrafemoral
- MRI
- magnetic resonance imaging
- UTR
- untranslated region
References
- Allen R.J., Smith S.D., Moldwin R.L., Lu M.M., Giordano L., Vignon C., Suto Y., Harden A., Tomek R., Veldman T., et al. 1998. Establishment and characterization of a megakaryoblast cell line with amplification of MLL. Leukemia. 12:1119–1127 10.1038/sj.leu.2401002 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio M., Uhlenhaut N.H., Sousa V.H., O’Toole J.F., Otto E., Anlag K., Klugmann C., Treier A.C., Helou J., Sayer J.A., et al. 2007. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 39:1018–1024 10.1038/ng2072 [DOI] [PubMed] [Google Scholar]
- Ballerini P., Blaise A., Mercher T., Pellegrino B., Perot C., van den Akker J., Gatbois E., Adam M., Douay L., Berger R., et al. 2003. A novel real-time RT-PCR assay for quantification of OTT-MAL fusion transcript reliable for diagnosis of t(1;22) and minimal residual disease (MRD) detection. Leukemia. 17:1193–1196 10.1038/sj.leu.2402914 [DOI] [PubMed] [Google Scholar]
- Borkhardt A., Haas O.A., Strobl W., Repp R., Mann G., Gadner H., Lampert F. 1995. A novel type of MLL/AF10 fusion transcript in a child with acute megakaryocytic leukemia (AML-M7). Leukemia. 9:1796–1797 [PubMed] [Google Scholar]
- Bourquin J.P., Subramanian A., Langebrake C., Reinhardt D., Bernard O., Ballerini P., Baruchel A., Cavé H., Dastugue N., Hasle H., et al. 2006. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc. Natl. Acad. Sci. USA. 103:3339–3344 10.1073/pnas.0511150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven S.E., Lim K.C., Ye W., Engel J.D., de Sauvage F., Rosenthal A. 2004. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 131:1165–1173 10.1242/dev.01024 [DOI] [PubMed] [Google Scholar]
- Dastugue N., Lafage-Pochitaloff M., Pagès M.P., Radford I., Bastard C., Talmant P., Mozziconacci M.J., Léonard C., Bilhou-Nabéra C., Cabrol C., et al. ; Groupe Français d’Hematologie Cellulaire 2002. Cytogenetic profile of childhood and adult megakaryoblastic leukemia (M7): a study of the Groupe Français de Cytogénétique Hématologique (GFCH). Blood. 100:618–626 10.1182/blood-2001-12-0241 [DOI] [PubMed] [Google Scholar]
- Davis J.N., McGhee L., Meyers S. 2003. The ETO (MTG8) gene family. Gene. 303:1–10 10.1016/S0378-1119(02)01172-1 [DOI] [PubMed] [Google Scholar]
- Gao J., Graves S., Koch U., Liu S., Jankovic V., Buonamici S., El Andaloussi A., Nimer S.D., Kee B.L., Taichman R., et al. 2009. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell. 4:548–558 10.1016/j.stem.2009.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I., Stover E.H., Cullen D.E., Mao J., Morgan K.J., Lee B.H., Kharas M.G., Miller P.G., Cornejo M.G., Okabe R., et al. 2009. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell. 4:559–567 10.1016/j.stem.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C., Ugo V., Le Couédic J.P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., et al. 2005. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 434:1144–1148 10.1038/nature03546 [DOI] [PubMed] [Google Scholar]
- Jeanpierre S., Nicolini F.E., Kaniewski B., Dumontet C., Rimokh R., Puisieux A., Maguer-Satta V. 2008. BMP4 regulation of human megakaryocytic differentiation is involved in thrombopoietin signaling. Blood. 112:3154–3163 10.1182/blood-2008-03-145326 [DOI] [PubMed] [Google Scholar]
- Jelinek J., Oki Y., Gharibyan V., Bueso-Ramos C., Prchal J.T., Verstovsek S., Beran M., Estey E., Kantarjian H.M., Issa J.P. 2005. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood. 106:3370–3373 10.1182/blood-2005-05-1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., ZeRuth G., Lichti-Kaiser K., Vasanth S., Yin Z., Kim Y.S., Jetten A.M. 2010. Gli-similar (Glis) Krüppel-like zinc finger proteins: insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histol. Histopathol. 25:1481–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Salzberg S.L. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12:R72 10.1186/gb-2011-12-8-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kang H.S., Herbert R., Beak J.Y., Collins J.B., Grissom S.F., Jetten A.M. 2008. Kruppel-like zinc finger protein Glis2 is essential for the maintenance of normal renal functions. Mol. Cell. Biol. 28:2358–2367 10.1128/MCB.01722-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Jiang R. 2009. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 136:1387–1396 10.1242/dev.028167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Godinho F.J., Klusmann J.H., Garriga-Canut M., Yu C., Orkin S.H. 2005. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat. Genet. 37:613–619 10.1038/ng1566 [DOI] [PubMed] [Google Scholar]
- Lichti-Kaiser K., ZeRuth G., Kang H.S., Vasanth S., Jetten A.M. 2012. Gli-similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam. Horm. 88:141–171 10.1016/B978-0-12-394622-5.00007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion T., Haas O.A., Harbott J., Bannier E., Ritterbach J., Jankovic M., Fink F.M., Stojimirovic A., Herrmann J., Riehm H.J., et al. 1992. The translocation t(1;22)(p13;q13) is a nonrandom marker specifically associated with acute megakaryocytic leukemia in young children. Blood. 79:3325–3330 [PubMed] [Google Scholar]
- Ma Z., Morris S.W., Valentine V., Li M., Herbrick J.A., Cui X., Bouman D., Li Y., Mehta P.K., Nizetic D., et al. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28:220–221 10.1038/90054 [DOI] [PubMed] [Google Scholar]
- Malinge S., Ragu C., Della-Valle V., Pisani D., Constantinescu S.N., Perez C., Villeval J.L., Reinhardt D., Landman-Parker J., Michaux L., et al. 2008. Activating mutations in human acute megakaryoblastic leukemia. Blood. 112:4220–4226 10.1182/blood-2008-01-136366 [DOI] [PubMed] [Google Scholar]
- Malinge S., Izraeli S., Crispino J.D. 2009. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood. 113:2619–2628 10.1182/blood-2008-11-163501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S., Bliss-Moreau M., Kirsammer G., Diebold L., Chlon T., Gurbuxani S., Crispino J.D. 2012. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J. Clin. Invest. 122:948–962 10.1172/JCI60455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Salles Tde.J., Mkrtchyan H., Leite E.P., Soares-Ventura E.M., Muniz M.T., Silva E.F., Liehr T., Silva M.L., Santos N. 2010. Complex karyotype defined by molecular cytogenetic FISH and M-FISH in an infant with acute megakaryoblastic leukemia and neurofibromatosis. Cancer Genet. Cytogenet. 200:167–169 10.1016/j.cancergencyto.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Martínez M.C., Larbret F., Zobairi F., Coulombe J., Debili N., Vainchenker W., Ruat M., Freyssinet J.M. 2006. Transfer of differentiation signal by membrane microvesicles harboring hedgehog morphogens. Blood. 108:3012–3020 10.1182/blood-2006-04-019109 [DOI] [PubMed] [Google Scholar]
- McPherson A., Hormozdiari F., Zayed A., Giuliany R., Ha G., Sun M.G., Griffith M., Heravi Moussavi A., Senz J., Melnyk N., et al. 2011. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLOS Comput. Biol. 7:e1001138 10.1371/journal.pcbi.1001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T., Coniat M.B., Monni R., Mauchauffe M., Nguyen Khac F., Gressin L., Mugneret F., Leblanc T., Dastugue N., Berger R., Bernard O.A. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA. 98:5776–5779 10.1073/pnas.101001498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T., Busson-Le Coniat M., Nguyen Khac F., Ballerini P., Mauchauffé M., Bui H., Pellegrino B., Radford I., Valensi F., Mugneret F., et al. 2002. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes Chromosomes Cancer. 33:22–28 10.1002/gcc.1208 [DOI] [PubMed] [Google Scholar]
- Mercher T., Wernig G., Moore S.A., Levine R.L., Gu T.L., Fröhling S., Cullen D., Polakiewicz R.D., Bernard O.A., Boggon T.J., et al. 2006. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood. 108:2770–2779 10.1182/blood-2006-04-014712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T., Raffel G.D., Moore S.A., Cornejo M.G., Baudry-Bluteau D., Cagnard N., Jesneck J.L., Pikman Y., Cullen D., Williams I.R., et al. 2009. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J. Clin. Invest. 119:852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morerio C., Rapella A., Tassano E., Rosanda C., Panarello C. 2005. MLL-MLLT10 fusion gene in pediatric acute megakaryoblastic leukemia. Leuk. Res. 29:1223–1226 10.1016/j.leukres.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Nguyen A.T., Zhang Y. 2011. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25:1345–1358 10.1101/gad.2057811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Aguilera R., Romero-Guzman L., Lopez-Santiago N., Trejo R.A. 2003. Biology, clinical, and hematologic features of acute megakaryoblastic leukemia in children. Am. J. Hematol. 73:71–80 10.1002/ajh.10320 [DOI] [PubMed] [Google Scholar]
- Pikman Y., Lee B.H., Mercher T., McDowell E., Ebert B.L., Gozo M., Cuker A., Wernig G., Moore S., Galinsky I., et al. 2006. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 3:e270 10.1371/journal.pmed.0030270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke I., Mullighan C.G., Ishii M., Su X., Cheng J., Ma J., Ganti R., Cai Z., Goorha S., Pounds S.B., et al. 2009. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 106:12944–12949 10.1073/pnas.0903142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi F., Cortes J., Estrov Z., Thomas D., Giles F.J., Huh Y.O., Pierce S., O’Brien S., Faderl S., Kantarjian H.M. 2002. CD56 expression predicts occurrence of CNS disease in acute lymphoblastic leukemia. Leuk. Res. 26:643–649 10.1016/S0145-2126(01)00188-6 [DOI] [PubMed] [Google Scholar]
- Roy A., Roberts I., Norton A., Vyas P. 2009. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multi-step model of myeloid leukaemogenesis. Br. J. Haematol. 147:3–12 10.1111/j.1365-2141.2009.07789.x [DOI] [PubMed] [Google Scholar]
- Takita J., Motomura A., Koh K., Ida K., Taki T., Hayashi Y., Igarashi T. 2009. Acute megakaryoblastic leukemia in a child with the MLL-AF4 fusion gene. Eur. J. Haematol. 83:149–153 10.1111/j.1600-0609.2009.01275.x [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutven L.J., Onen E., Velthuizen S.C., van Drunen E., von Bergh A.R., van den Heuvel-Eibrink M.M., Veronese A., Mecucci C., Negrini M., de Greef G.E., Beverloo H.B. 2006. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 45:437–446 10.1002/gcc.20308 [DOI] [PubMed] [Google Scholar]
- Vasanth S., ZeRuth G., Kang H.S., Jetten A.M. 2011. Identification of nuclear localization, DNA binding, and transactivating mechanisms of Krüppel-like zinc finger protein Gli-similar 2 (Glis2). J. Biol. Chem. 286:4749–4759 10.1074/jbc.M110.165951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.K., Mercher T., Gu T.L., O’Hare T., Tyner J.W., Loriaux M., Goss V.L., Lee K.A., Eide C.A., Wong M.J., et al. 2006. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 10:65–75 10.1016/j.ccr.2006.06.002 [DOI] [PubMed] [Google Scholar]
- Wang G.G., Song J., Wang Z., Dormann H.L., Casadio F., Li H., Luo J.L., Patel D.J., Allis C.D. 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 459:847–851 10.1038/nature08036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J., Greene M., McDevitt M.A., Anastasi J., Karp J.E., Le Beau M.M., Crispino J.D. 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32:148–152 10.1038/ng955 [DOI] [PubMed] [Google Scholar]
- Wen Q., Goldenson B., Silver S.J., Schenone M., Dancik V., Huang Z., Wang L.Z., Lewis T.A., An W.F., Li X., et al. 2012. Identification of regulators of polyploidization presents therapeutic targets for treatment of AMKL. Cell. 150:575–589 10.1016/j.cell.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T., Maki K., Mitani K. 2005. Runx1/AML1 in normal and abnormal hematopoiesis. Int. J. Hematol. 82:1–8 10.1532/IJH97.05075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.