Transcriptional activator Oct2 and cofactor OBF-1 regulate B cell IL-6 to induce T cell production of IL-21, to support Tfh cell development in antiviral immunity.
Abstract
A strong humoral response to infection requires the collaboration of several hematopoietic cell types that communicate via antigen presentation, surface coreceptors and their ligands, and secreted factors. The proinflammatory cytokine IL-6 has been shown to promote the differentiation of activated CD4+ T cells into T follicular helper cells (TFH cells) during an immune response. TFH cells collaborate with B cells in the formation of germinal centers (GCs) during T cell–dependent antibody responses, in part through secretion of critical cytokines such as IL-21. In this study, we demonstrate that loss of either IL-6 or IL-21 has marginal effects on the generation of TFH cells and on the formation of GCs during the response to acute viral infection. However, mice lacking both IL-6 and IL-21 were unable to generate a robust TFH cell–dependent immune response. We found that IL-6 production in follicular B cells in the draining lymph node was an important early event during the antiviral response and that B cell–derived IL-6 was necessary and sufficient to induce IL-21 from CD4+ T cells in vitro and to support TFH cell development in vivo. Finally, the transcriptional activator Oct2 and its cofactor OBF-1 were identified as regulators of Il6 expression in B cells.
Protective long-term humoral immunity against pathogens depends on the generation of antibodies of high affinity that are capable of appropriate effector functions, a process which relies on the formation of germinal centers (GCs) in LNs or in the spleen during infection. GCs are essential but transient structures in which high affinity antibody-secreting cells and memory B cells are generated during a T cell–dependent (TD) antibody response. Although B cells constitute the majority of cells within a GC, macrophages, follicular DCs, and CD4+ T cells contribute to the defined architecture and the functionality of a GC during an immune response. These cells cooperate via antigen presentation, adhesion molecules, cell surface co-stimulatory molecules, and secreted factors to enable a robust GC reaction and an effective antibody response.
The formation and maintenance of GCs require a specialized subset of CD4+ T cells, T follicular helper cells (TFH cells; Yu and Vinuesa, 2010; Crotty, 2011; Nutt and Tarlinton, 2011). TFH cells that are induced during TD responses are characterized by the expression of several critical surface markers that interact with ligands on APCs such as DCs and B cells. These molecules include co-stimulatory molecules and their ligands (PD-1, ICOS, CD200, OX40, and CD40-ligand), adhesion mediators of the Slam/SAP family, and receptors for IL-6 and IL-21 (King et al., 2008; Nurieva et al., 2008; Ma et al., 2009; Yusuf et al., 2010).
The coordinated induction of the chemokine receptor CXCR5, and repression of CCR7, allows TFH to home to B cell follicles (Ansel et al., 1999; Haynes et al., 2007). CXCR5 induction depends on an OX40-mediated signal in TFH (Brocker et al., 1999). Antigen-presenting B cells meet their cognate TFH cells at the T–B border and engage in prolonged interactions, mediated by antigen and Slam/SAP proteins, to deliver signals that are essential for TFH maintenance and subsequent productive GC formation (Qi et al., 2008; Deenick et al., 2010). Once in the follicle, TFH cells provide help to activated B cells through the expression of molecules such as CD40-ligand and ICOS and through the secretion of cytokines, predominantly IL-4 and IL-21 (Chtanova et al., 2004; Reinhardt et al., 2009). IL-21, a pleiotropic cytokine, is a hallmark of TFH cells. It has been shown to induce proliferation and expression of Blimp1 and Bcl6 in B cells, thereby influencing their decision to differentiate into antibody-secreting cells or to continue to participate in the GC reaction (Ozaki et al., 2004; Arguni et al., 2006). Furthermore, IL-21 promotes switching to IgG1, IgG2a and IgG3 and inhibits IgE responses (Ozaki et al., 2002).
Recent studies have suggested that both IL-6 and IL-21 have pivotal roles in vivo in the generation of IL-21–secreting TFH cells and the formation of GCs (King et al., 2008; Nurieva et al., 2008; Suto et al., 2008). Differentiation of an activated CD4+ T cell into an IL-21–secreting TFH cell is dependent on the transcription factor Bcl6, which acts as a master regulator for CD4+ TFH cell differentiation (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). In vitro, IL-6 and IL-21 are able to stimulate Bcl6 and enhance Il21 expression in CD4+ T cells, consistent with these cytokines serving an inductive role for TFH (Suto et al., 2008). Nurieva et al. (2008) reported that mice deficient in IL-6 formed fewer GC B cells and have reduced TFH cell numbers after an immune challenge with sheep red blood cells. Similarly, other groups demonstrated a reduced frequency and size of GCs in IL-6–deficient mice (Kopf et al., 1998; Wu et al., 2009). In some of the aforementioned studies, the impaired formation of GCs in the IL-6–deficient mice was linked to a reduction of IL-21–producing TFH cells (Nurieva et al., 2008; Suto et al., 2008). IL-21 has also been implicated in the generation and maintenance of TFH cells and the formation of GCs in vivo (Nurieva et al., 2008; Vogelzang et al., 2008). Thus, it was proposed that IL-6 initially induces Bcl6 and Il21 expression in activated CD4+ T cells, and subsequently, IL-21 acts as a positive feedback loop to maintain Il21 and Bcl6 expression in the TFH (Nurieva et al., 2009; Linterman et al., 2010).
However, other studies have yielded conflicting results on the roles of IL-6 and IL-21 in TFH cell generation and GC formation. These studies indicate that IL-21 is not essential for the generation of TFH cells (Linterman et al., 2010; Zotos et al., 2010; Rankin et al., 2011) and that loss of IL-21R had little effect on initial GC development but was critical for GC maintenance during an immune response (Linterman et al., 2010; Zotos et al., 2010). Another study suggested that IL-6 was not required for the formation of TFH cells or the GC response (Poholek et al., 2010). Both IL-6 and IL-21 signal predominantly through the same intracellular signal transducer, Stat3 (Zeng et al., 2007; Nurieva et al., 2008; Eddahri et al., 2009). Given the conflicting data and potential redundancy caused by a shared signaling pathway, we wished to test whether IL-6 and IL-21 could be functionally redundant in the TD antibody response to infection. We show here that the loss of either IL-6 or IL-21 alone has little effect on the development of GCs and the formation of TFH cells in response to an acute viral infection. However, combined loss of both factors severely crippled the humoral immune response, including the development of GC B cells and the formation of TFH cells. We further show that IL-6 and IL-21 act with different kinetics during the GC response and that B cells, in an Oct2/OBF-1–dependent manner, can supply the IL-6 necessary for early induction of TFH development.
RESULTS
Combined loss of IL-6 and IL-21 compromises GC formation
Several studies examining the importance of either IL-6 or IL-21 in the formation and maintenance of GCs reported conflicting results (Kopf et al., 1998; Ozaki et al., 2002; Nurieva et al., 2008; Vogelzang et al., 2008; Wu et al., 2009; Poholek et al., 2010; Linterman et al., 2010; Zotos et al., 2010; Rankin et al., 2011). Many of these investigators variously used immunization with either hapten antigens or sheep red blood cells as the experimental model. We wished to assess the roles of IL-6 and IL-21, individually or in combination, in the formation of GCs and TFH cells in a physiological model of acute viral infection.
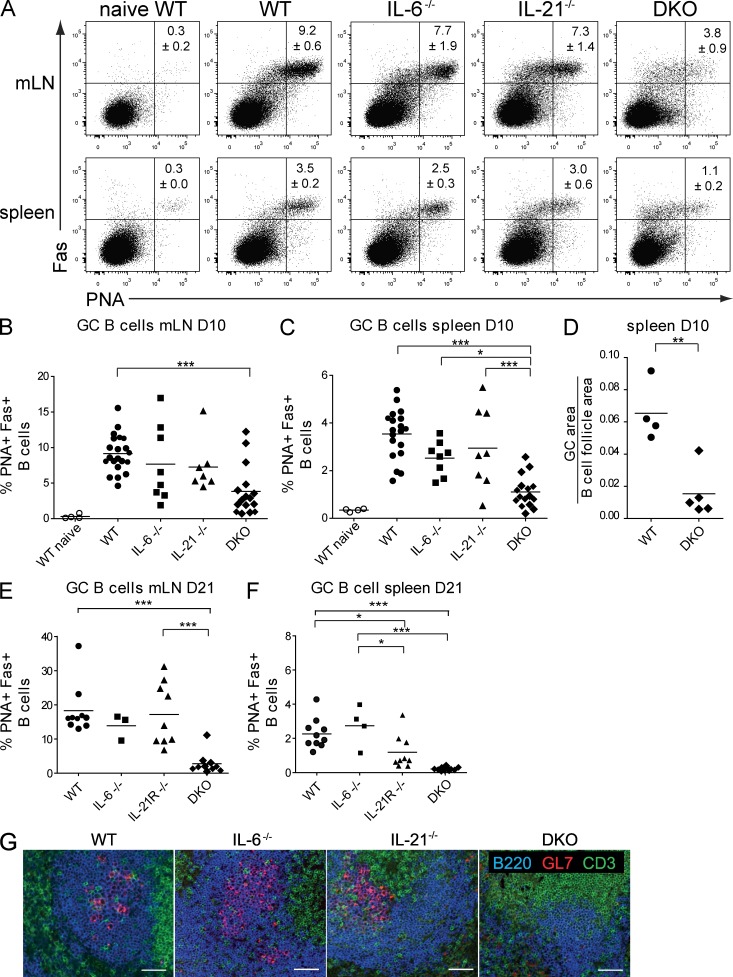
WT C57BL/6 or IL-6–, IL-21–, or IL-21R–deficient mice were infected intranasally with the HKx31 strain of influenza virus. GC B cells (B220+, Fas+, PNA+) in the lung-draining mediastinal LNs (mLNs) and the spleens of infected mice were assessed by flow cytometry at days 10 and 21 after infection. Individual loss of IL-6 or IL-21 had little impact on GC B cells in the draining LNs in this infection model (Fig. 1, A and B). In the spleen on day 10 after infection, neither IL-21– nor IL-6–deficient mice showed a significant difference of GC B cells compared with WT (Fig. 1 C). Similar results were observed when measuring GC B cells in mLNs and spleen on day 21 after infection in IL-6– or IL-21R–deficient mice (Fig. 1, E and F). Because IL-6 and IL-21 signal through a common transducer, Stat3, it was possible that they play redundant roles in the generation of GCs during an acute viral infection. To test this possibility, IL-6/IL-21 compound mutant mice (DKO) were generated and infected with influenza virus. 10 d after infection, GC formation was assessed and compared with WT and single mutant mice. IL-6/IL-21 DKO mice showed a significant reduction in GCs in both the draining LNs and the spleens on day 10, at the peak of the immune response (Fig. 1, A–D; Flynn et al., 1998), and this was even more pronounced at day 21 after infection (Fig. 1, E and F). The GC deficit in infected DKO mice was confirmed through immunohistochemical staining of spleen sections (Fig. 1, D and G), which showed significantly reduced GC B cell areas in the spleens of DKO mice compared with controls. These results indicate that IL-6 and IL-21 in combination play an essential role in the development of GC B cells in response to acute viral infection.
Figure 1.
Combined loss of IL-6 and IL-21 compromises GC formation in influenza infection. Analysis of GC B cells in C57BL/6 (WT), IL-6, IL-21, and IL-6/IL-21 double-deficient mice (DKO). Mice were analyzed on day 10 after influenza infection. Results shown are from three to six independent experiments, totaling 4 naive WT and 21 WT, 8 Il6−/−, 8 Il21−/−, and 16 DKO-infected mice, respectively. (A) Cells from the draining mLNs and from the spleen were stained for GC B cells with α-B220, α-Fas, and PNA, and the percentage of B220+ cells that were also PNA+/Fas+ is shown. (B and C) Frequency distribution of GC B cells in spleens and mLNs from WT and mutant mice analyzed on day 10 of infection. (D) Ratio of GC area to B cell follicle area in spleens of WT and DKO animals on day 10, as measured from histological sections. Each symbol represents an individual animal. (E and F) Frequency distribution of GC B cells from WT and mutant mice analyzed on day 21 of infection. Each symbol represents an individual animal. Statistical analyses used Tukey’s multiple comparison tests. ***, P < 0.001; **, P = 0.001–0.001; *, P = 0.01–0.05. Bars and numbers show mean percentage with ± SEM. Results are from three to six independent experiments. (G) Representative histological staining to detect GCs in spleens from control or mutant mice 10 d after influenza infection. Paraffin sections were stained with α-GL7, α-B220, and α-CD3. Bars, 50 µm.
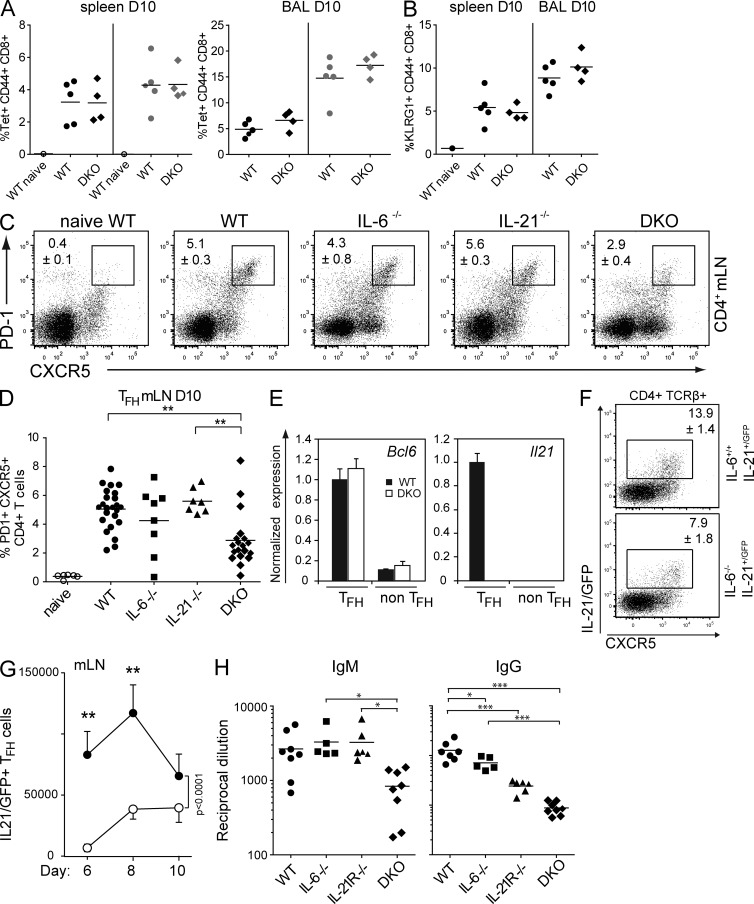
As IL-21 has been shown to contribute to CD8+ T cell responses (Casey and Mescher, 2007; Novy et al., 2011), we wanted to ensure that the defective GC development we observed in the double mutants was not influenced by a crippled CD8+ response to the influenza infection. We therefore measured virus-specific CD8+ T cell responses in WT and DKO mice during the peak of the immune response. There was no significant difference in the frequency of virus-specific CD8+ T cells between WT and mutant mice (Fig. 2 A). Furthermore, there was no difference in the percentage of mature effector KLRG1+CD44+CD8+ T cells in control and DKO mice (Fig. 2 B), demonstrating that, unlike the GC response, the antiviral CD8+ T cell response was unaffected by the loss of these two cytokines.
Figure 2.
Combined loss of IL-6 and IL-21 does not affect the virus-specific CD8 response but limits TFH formation and the antibody response in influenza infection. Analysis of anti-influenza CD8 responses in WT and DKO animals. Mice were analyzed on day 10 after infection. (A) Splenocytes and cells from the bronchoalveolar lavage (BAL) were stained with α-CD8, α-CD44, and either NP-tetramer or PA-tetramer. Frequency distribution of splenic, virus-specific CD8+ T cells (tetramer stains: NP, black symbols; PA, gray symbols) is shown in a representative of two independent experiments using three to five animals of each genotype. (B) Frequency distribution of KLRG1/CD44 double-positive CD8+ T cells in spleen and bronchoalveolar lavage. Each symbol represents an individual animal. Bars and numbers show mean percentage with ± SEM. (C) WT, IL-6– or IL-21–deficient, and DKO mice were infected with HKx31 influenza virus and analyzed on day 10 after infection. Cells from the mLNs were stained for TFH cells with α-CD4, α-CXCR5, and α–PD-1, and the percentage of PD-1/CXCR5 double-positive CD4+ T cells was measured. (D) Frequency of TFH from WT and mutant mice analyzed on day 10 after infection. Representative example shown of two to six independent experiments, totaling 6 naive WT, 23 WT, 8 Il6−/−, 8 Il21−/−, and 19 DKO-infected mice, respectively. (E) CD4+PD-1+CXCR5+ TFH cells and CD4+PD-1−CXCR5− T cells were sorted from spleen on day 14 of HKx31 influenza infection. Bcl6 and Il21 expression was measured by RT-qPCR. (As expected, Il21 is not expressed in the DKO mice. This is a control only.) Bars and numbers show relative gene expression normalized to the housekeeping gene, Hmbs, ± SEM (n = 3). (F) IL-21–GFP reporter mice, on a WT or Il6−/− background, were infected, and mLNs were harvested on days 6, 8, and 10 and stained for TFH cells (CD4, TCR-β, CXCR5, and PD-1). The dot plot shows IL-21–expressing cells in the CD4+/TCR-β+ gate on day 10. (G) Time course showing total numbers of IL-21–expressing TFH in the draining mLNs from days 6 to 10 of infection. Numbers show means ± SEM of five animals in each group. IL-21/GFP+ cells were also CD4+, TCR-β+, PD-1+, CXCR5+. (H) IgM and IgG HKx31-specific responses in WT, IL-6, IL-21R, and DKO mice. Serum IgM and IgG titers were measured by ELISA on day 14 of the influenza infection and are represented as the reciprocal of serum dilutions, giving an absorbance that was 50% of maximum value for the assay. Each symbol represents an individual animal. ***, P < 0.001; **, P = 0.001–0.001; *, P = 0.01–0.05. Bars and numbers show mean dilution ± SEM.
The combined actions of IL-6 and IL-21 control the early generation of TFH cells
The defective GC reaction in DKO mice raised the question of whether the relevant T helper cell response in these mice was impaired. We examined the draining LNs 10 d after influenza infection from WT, IL-6 and IL-21 singly deficient mice, and DKO mice for CD4+ T cells expressing the TFH markers CXCR5 and PD-1 (Vinuesa et al., 2005). Infected, but not naive, WT mice showed a distinct TFH cell population in the draining LNs. Loss of IL-6 or IL-21 alone did not cause a significant change in frequency of TFH cells (Fig. 2, C and D). However, there was a significant reduction of TFH cells at day 10 of the infection in DKO mice. Interestingly, by day 21, TFH frequencies were similar in all mice (not depicted), implying that IL-6 and IL-21 affect most strongly the early stages of TFH development.
Both IL-6 and IL-21 have been implicated in the induction of Bcl6 and Il21 expression (Nurieva et al., 2009; Linterman et al., 2010), hallmarks of TFH cells. We therefore assessed whether TFH cells that develop in the absence of IL-6 and IL-21 expressed Bcl6. To that end, we isolated CD4+PD-1+CXCR5+ TFH and CD4+PD-1−CXCR5− non-TFH cells from influenza-infected WT and DKO mice and measured Bcl6 and Il21 messenger RNA (mRNA) expression ex vivo by real-time quantitative PCR (qPCR). The combined loss of IL-6 and IL-21 did not alter Bcl6 expression in the TFH cells (Fig. 2 E). These results indicate that IL-6 and IL-21 play critical but redundant or complementary roles early during TFH cell generation or expansion, but TFH cells formed in their absence are normal.
Because in an earlier study (Zotos et al., 2010) we showed that loss of IL-21 or its receptor did not change the kinetics of TFH appearance after immunization, we focused on the influence of IL-6. To explore the rate of induction of TFH in vivo, we made use of our recently described IL-21–GFP knockin reporter mice (Lüthje et al., 2012). In these mice (which are heterozygous for a functional Il21 allele), IL-21–GFP+ CD4 cells can be clearly visualized as a subset of TFH cells that localize to the GCs during infection or immunization and express cytokine genes as well as the TFH hallmarks PD-1, CXCR5, and Bcl6. We enumerated CD4+/GFP+ cells in WT and Il6−/− mice bearing the IL-21–GFP allele (Fig. 2 F) during the early stages of the antiviral response (days 6–10; IL-21–GFP+ cells only appear after day 5 in this model; unpublished data) and found that TFH cells in Il6−/− mice were significantly delayed in their generation, but nearly matched WT numbers by day 10 in the mLNs (Fig. 2 G). As expected, GC B cells followed similar kinetics with Il6−/− mice, trailing their WT counterparts (not depicted). These data show that IL-6 strongly influences TFH induction or expansion early in the antiviral response.
Collectively, these data concur with previous studies showing that neither IL-6 nor IL-21 alone is required for the generation of GCs or TFH cells (Poholek et al., 2010; Linterman et al., 2010; Zotos et al., 2010; Rankin et al., 2011). However, we show here that the simultaneous loss of both cytokines strongly blunts both GC and TFH development. IL-6 deficiency significantly delays TFH induction in vivo. Thus, the highly interdependent TFH and GC B cell response to infection relies on the combined actions of IL-6 and IL-21
IL-6 and IL-21 are critical for an effective antibody response to acute viral infection
To assess the consequences of the impaired TFH and GC development in IL-6/IL-21 DKO mice on the humoral antiviral response, we measured antiviral IgM and IgG levels in the serum of WT, single mutant, and DKO mice on day 14 of the infection. Although the single loss of IL-6 or IL-21R alone (which phenocopies loss of IL-21 in the antibody response; Zotos et al., 2010) had no impact on the IgM response and only a modest impact on IgG titers, combined loss of IL-6 and IL-21 resulted in a significant (approximately threefold) reduction in virus-specific IgM (Fig. 2 H). IL-21R–deficient mice had an impaired IgG response (∼3-4-fold), but the combined absence of IL-21 and IL-6 magnified this effect, reducing IgG titers to ∼14-fold lower than in WT mice (Fig. 2 H), confirming that the combined actions of IL-6 and IL-21 are essential for a strong humoral response to acute viral infection.
IL-6 is induced early in B cells during an immune reaction
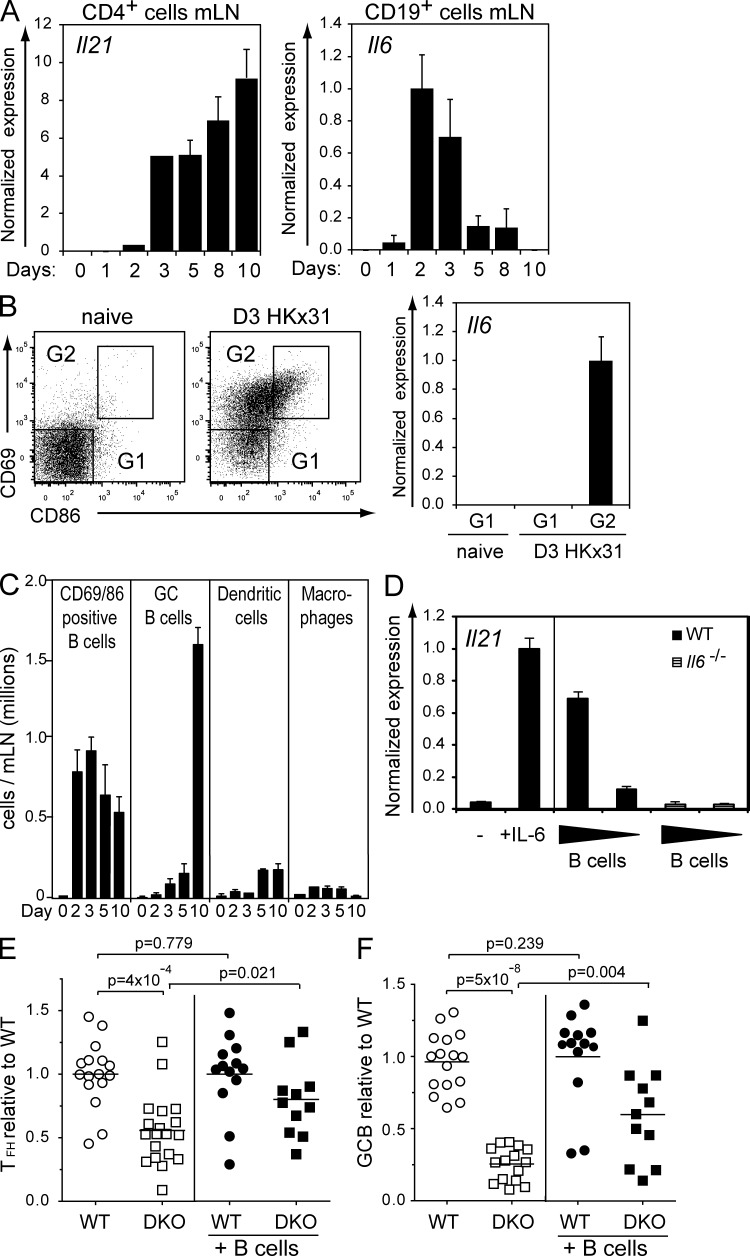
Although the combined loss of IL-6 and IL-21 impaired the formation of TFH cells and strongly reduced the GC response, the maintenance of TFH cells was independent of IL-6 and IL-21. We therefore reasoned that these cytokines play important roles early in a viral infection, during the initiation of TFH cell differentiation. To test whether the kinetics of expression of both cytokines was compatible with this prediction, we measured Il6 and Il21 mRNA levels in cells from the draining mLNs at different time points after influenza infection in WT mice. The expression of Il21 in CD4+ cells in the draining LNs stayed low early in the infection, reaching appreciable levels on days 3–5, which were sustained until at least day 10 (Fig. 3 A). In contrast, Il6 mRNA was transiently expressed in CD19+ B cells, rising sharply between days 1 and 2 of infection, peaking at days 2–3 and falling by day 5 (Fig. 3 A). Although APCs such as DCs and macrophages are thought to be a source of IL-6 in the early process of TFH priming (Kopf et al., 1998; Cucak et al., 2009), our data show that B cells also secrete IL-6 early after an acute viral infection (Fig. 3 A). To test whether IL-6 expression was restricted to newly activated cells, we isolated activated (CD86+CD69+) and resting (CD86−CD69−) B cells (CD19+CD11c−CD11b−) from the draining LNs of mice 3 d after infection and from naive controls. Among B cells, Il6 mRNA was restricted to the activated cell compartment in the infected mice (Fig. 3 B), indicating that activated B cells represent a rapid and abundant cellular source of IL-6 after infection. Furthermore, activated B cells increased rapidly and dramatically in number to become the most abundant APC in the draining mLNs between days 2 and 10 of infection (Fig. 3 C).
Figure 3.
IL-6 and IL-21 are expressed early during an influenza infection. (A) CD4+ and CD19+ cells were isolated from mLNs on the indicated days after influenza infection. Il21 or Il6 mRNA expression was measured by qPCR. (B) Cells from the mLNs of naive and infected animals were stained with α-CD19, α-CD11c, α-CD11b, α-CD69, and α-CD86. Resting (G1) and activated follicular B cells (G2) were sorted. Bars and numbers show relative Il6 expression normalized to Hmbs ± SEM (n = 3) in each sorted population. (C) Time course of total cell numbers of activated and GC B cells, DCs, and macrophages in mLNs of infected mice. Numbers are means ± SEM of five mice. (D) Co-culture of WT naive CD62L+/CD4+ T cells, stimulated with α-CD3/α-CD28, and different numbers of CpG-preactivated B cells from control or IL-6–deficient mice. The left panel shows CD3/CD28-activated T cells with medium alone or with recombinant IL-6. After 4 d of co-culture, the CD4+ T cells were sorted, and Il21 mRNA expression was determined. The results are representative of three independent experiments. (A and D) Error bars represent SDs of triplicate assays. (E and F) WT, congenic Ly5.1+ B cells were injected into WT and DKO animals 2 d before influenza infection. 10 d after infection, cells from the mLNs were stained for TFH and GC B cells as described for Figs. 2 C and 1 A, respectively. Figures show fold change of each animal’s TFH or GC B cells compared with the mean percentage of TFH or GC B cells from controls within each experiment. Results are from three to six independent experiments. (E) TFH ratio comparing WT and DKO animals without or with rescue by WT Ly5.1 B cells. (F) GC B cell ratio comparing WT and DKO animals without or with rescue by WT Ly5.1 B cells. Each symbol represents an individual animal. Statistical analyses were performed using the two-sample Wilcoxon test, and all p-values are two-sided. Bars and numbers show fold change ± SEM.
Next, we wished to determine whether IL-6 produced by activated B cells was sufficient to induce the generation of IL-21–producing CD4+ T cells in vitro. B cells were stimulated for 24 h with CpG1668, a known inducer of IL-6 production by B cells (Yi et al., 1996). They were then added, at different cell ratios, to cultures of α-CD3/α-CD28–stimulated naive CD4+ T cells. After 4 d, the CD4+ T cells were recovered and assayed for Il21 expression. In control cultures, and consistent with published results (Dienz et al., 2009), CD4+ T cell activation in the presence of soluble recombinant IL-6 induced marked Il21 expression (Fig. 3 D). Il21 was also strongly induced in activated T cells that were co-cultured with equal numbers of CpG-activated WT B cells, and the amount of Il21 mRNA expressed in the CD4+ T cells was proportional to the number of B cells added to the cultures. IL-6–deficient B cells failed to induce Il21 mRNA expression in co-cultured CD4+ T cells (Fig. 3 D), indicating that B cell–derived IL-6 is necessary and sufficient to induce IL-21 production by CD4+ T cells in this co-culture system.
Finally, we wished to determine whether B cell–derived IL-6 supported TFH formation in vivo. To this end, 1–2 × 107 IL-6–sufficient B cells from Ly5.1 congenic mice were injected on two consecutive days into either WT or DKO mice. The mice were then infected with influenza virus, and TFH generation and GC B cell formation in the draining LNs were analyzed. B cell transfer was relatively inefficient, but by 10 d after infection, a small proportion of donor-derived B cells (≤6% of total B cells) were evident in each experiment (not depicted). As shown (Figs. 2 and 3 E), loss of IL-6 and IL-21 leads to a clear reduction of TFH. However, transfer of IL-6–sufficient B cells led to a significant rescue of the TFH population in DKO mice (Fig. 3 E). In parallel, we observed a partial but significant rescue of GC formation in DKO mice that had received IL-6–sufficient B cells (Fig. 3 F). The rescue was notable considering the low ratio of IL-6–sufficient to –deficient B cells in the recipients. Collectively, these data show that IL-6 is expressed by activated follicular B cells in the draining LNs early after viral infection (days 2–3) and that IL-6 supplied by activated B cells is sufficient to drive IL-21 expression in CD4+ T cells in vitro and TFH cell development in vivo.
Oct2- and OBF-1–deficient B cells are impaired in IL-6 production
The factors that influence IL-6 production during TFH expansion and GC formation are largely unknown. We focused our attention on Oct2 (a DNA-binding POU/homeodomain transcriptional activator) and OBF-1 (OCA-B/Bob.1), a coactivator for Oct1 and Oct2 (Gstaiger et al., 1996; Lins et al., 2003), because our own studies on TLR signaling responses in B cell lines indicated that both Oct2- and OBF-1–deficient cells expressed less Il6 than controls (unpublished data), and a recent publication suggested that octamer-binding transcription factors are involved in the transcriptional regulation of the human IL6 gene (Smith et al., 2008).
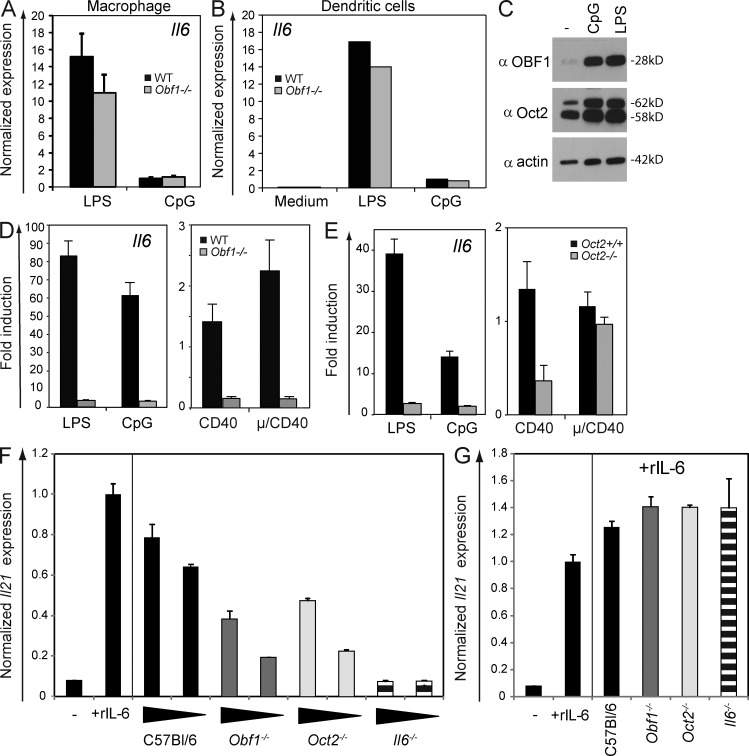
We therefore examined whether loss of OBF-1 or Oct2 has an effect on Il6 expression in primary B cells. OBF-1 expression is restricted to the lymphocyte compartment of the immune system. Consistent with the lack of OBF-1 expression in myeloid cells, OBF-1 loss had no impact on Il6 expression in macrophages (CD11b+/GR1+) or BM-derived DCs stimulated with LPS or CpG (Fig. 4, A and B). In contrast, Il6 expression in B cells was strongly influenced by the loss of OBF-1 or Oct2. B cells up-regulate Oct2 and OBF-1 upon activation (Fig. 4 C). Splenic B cells purified from WT and Oct2- and OBF-1–deficient mice were cultured with various mitogens, and Il6 expression was measured by qPCR. Although Il6 expression was induced under all conditions (and most strongly by TLR ligands) in WT B cells, its induction was very weak in both Oct2- and OBF-1–deficient B cells, especially in LPS or CpG cultures (Fig. 4, D and E). Consistent with these results, CpG-activated Oct2- or OBF-1–deficient B cells were strongly impaired in their capacity to induce Il21 transcription in co-cultured CD4+ T cells, most clearly seen when their numbers were limiting (Fig. 4 F). Addition of exogenous IL-6 to these co-cultures fully complemented the deficiencies of Oct2, OBF-1, or IL-6 mutant B cells, inducing robust Il21 mRNA expression in the responding CD4+ T cells (Fig. 4 G) and confirming that IL-6 is the dominant inductive cytokine in these cultures.
Figure 4.
Induction of IL-6 in activated B cells is dependent on Oct2 and OBF-1. (A and B) qPCR measurement of Il6 expression in sorted and LPS- or CpG-stimulated Gr1+, Mac1+ macrophages or BM-derived DCs from WT or Obf-1−/− mice. (C) Western blot analysis of Oct2 and OBF-1 in mature resting or CpG- or LPS-stimulated B cells. Blots were probed with α–OBF-1, α-Oct2, or α-actin. (D and E) Il6 expression in sorted and activated splenic B220+ B cells from OBF-1–deficient or WT mice and Oct+/+ or Oct−/− B cells from fetal liver reconstituted mice. Bars and numbers show relative gene expression normalized to Hmbs expression ± SEM (n = 3). (F) In vitro generation of IL-21–producing cells. Co-culture of WT naive CD4+, CD62L+ T cells, activated with α-CD3/α-CD28, with different numbers of CpG-preactivated B cells from WT, Obf-1−/−, Oct2−/−, or IL-6–deficient mice. After 4 d of co-culture, the CD4+ T cells were sorted, and Il21 expression was determined by qPCR. (A and F) Error bars represent SDs of triplicate assays. (G) T cells stimulated with medium alone, with recombinant IL-6, or with B cells and recombinant IL-6. Il21 expression was determined by qPCR and normalized as described for D–E. Bars and numbers show relative gene expression normalized to Hmbs expression ± SEM (n = 3).
Molecular regulation of the IL-6 locus by octamer-binding factors
To investigate whether octamer-binding factors directly interact with the murine Il6 locus control regions, we first analyzed the Il6 gene for consensus octamer-binding sites. The Il6 locus in mice spans a region of ∼7 kb on mouse chromosome 5. A previous study has identified binding sites for several transcription factors (AP-1, NF-κB, or CEBP family members) at a core promoter region ∼230 bp upstream of the transcription start site (Fig. 5 A; Baccam et al., 2003). Bioinformatics analysis using PROMO (Messeguer et al., 2002) revealed four consensus octamer-binding sites within the Il6 locus: octamer 1, ATTTGCAT −3309 to −3302; octamer 2, TTTTGCAT −1459 to −1452; octamer 3, ATTTGCAT 3793 to 3800; and octamer 4, ATTTGCAT 10615 to 10622 (Fig. 5 A).
Figure 5.
Functional octamer factor binding sites in the Il6 locus. (A) Organization of the Il6 locus and location of the core promoter region. Positions of four consensus octamer sites identified in silico are shown. AK039125 is an adjacent gene. (B) EMSA analysis on nuclear extracts from 24-h CpG-stimulated splenic B cells, performed using short fragments (160–207 bp) containing the consensus octamer site (ATGCAAT) from an Ig heavy chain promoter or octamer sequences identified in the Il6 locus (site 1, ATTTGCAT −3302; site 2, TTTTGCAT −1438; site 3, ATTTGCAT 3793; site 4, ATTTGCAT 10615). Specific complex formation was detected through supershifts using α-Oct2 or α–OBF-1 monoclonal antibodies, as indicated. Oct2–DNA complexes are indicated with asterisks. The results are representative of three independent experiments. (C and D) Immunoprecipitation (ChIP) of chromatin from purified splenic B cells from mice of the indicated genotypes, using preimmune and hyperimmune rabbit serum specific for Oct2. (C) Cd36 is a known Oct2 target gene (König et al., 1995). (D) ChIP on the same chromatin as in C, but examining the octamer-containing Il6 gene sequences identified in A and positive by EMSA (B). Values in all graphs are means ± SEM (n = 3).
To determine whether the putative octamer-binding sites in the Il6 gene are functional, nuclear extracts from CpG-stimulated B220+ B cells were used for electrophoretic mobility shift assays (EMSAs). Endogenous Oct2 bound to all four predicted octamer sites (Fig. 5 B). In accordance with earlier evidence and our unpublished data indicating that OBF-1 does not directly contact DNA but associates with DNA-bound Oct2 or Oct1 (Strubin et al., 1995), it was not possible to detect direct binding of OBF-1 to these octamer sites. Each of the sites, however, has the consensus sequence known to be necessary to recruit OBF-1 to the Oct–DNA complex (Gstaiger et al., 1996). Consistent with the EMSA results, chromatin immunoprecipitation (ChIP) revealed that Oct2 associates with all four sites in vivo (Fig. 5 D), as it does with the established Oct2 target gene, Cd36 (Fig. 5 C; König et al., 1995; Shore et al., 2002). These data show that several sites in the Il6 locus can be bound directly by Oct2 and suggest that OBF-1, in complex with Oct2, directly regulates Il6 expression in B cells. Together these experiments show that Il6 expression in B cells is strongly dependent on Oct2 and OBF-1.
Impaired TFH cell development in OBF-1–deficient mice
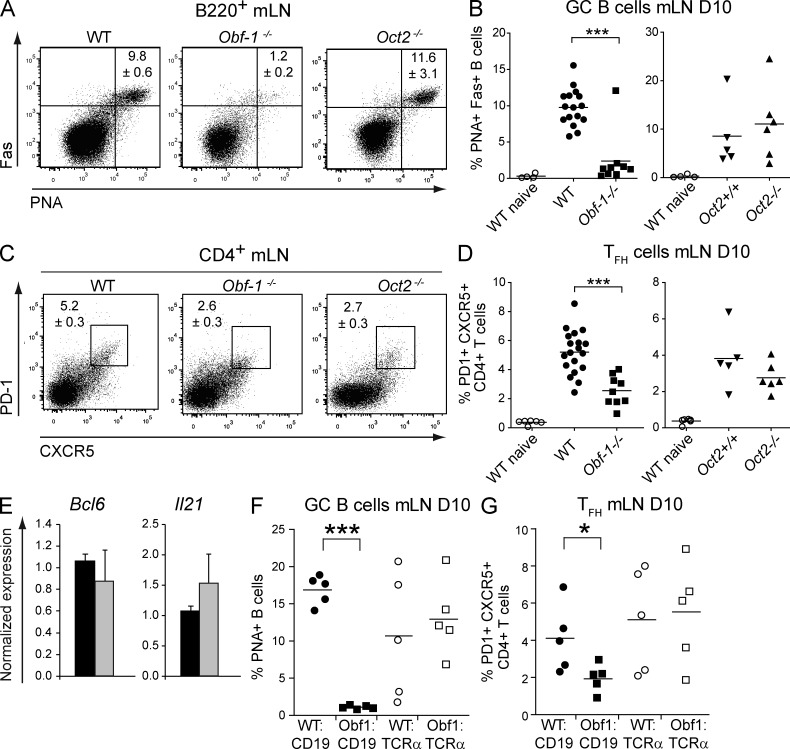
To determine whether OBF-1 or Oct2 is involved in GC and TFH development in response to viral infection, WT and OBF-1– or Oct2-deficient mice were infected with influenza virus. The formation of TFH cells and GC B cells was assessed on day 10 after infection. Although Oct2-deficient mice showed normal development of GC B cells in infected mice (in contrast to a study using hapten protein immunization; Schubart et al., 2001), GC B cells were severely reduced or absent in the lung-draining LNs of OBF-1–deficient mice compared with WT (Fig. 6, A and B). OBF-1 KO mice also showed a significantly reduced TFH cell compartment when compared with WT or Oct2−/− mice (Fig. 6, C and D). Consistent with these cellular deficiencies, virus-specific Ig was severely reduced in OBF-1–deficient mice (not depicted). Thus, optimal genesis of both GC B cells and TFH cells is dependent on OBF-1 but not Oct2.
Figure 6.
Loss of OBF-1 but not Oct2 results in loss of GCs and reduction of TFH during influenza infection. Analysis of GC B cells and TFH cells in control or OBF-1– or Oct2-deficient mice on day 10 of infection. (A and B) mLN cells were stained with α-B220, α-Fas, and PNA to detect GC B cells. Representative staining is shown in A, and summary of data for all mice is shown in B. (C and D) mLN cells were stained with α-CD4, α-CXCR5, and α–PD-1 to detect TFH cells. Representative staining is shown in C, and the frequency distribution for all mice is shown in D. (E) Analysis of Bcl6 and Il21 expression in CD4+, PD-1+, and CXCR5+ TFH sorted from spleens of WT and Obf-1−/− mice 10 d after influenza infection. Bars and numbers show mean normalized gene expression with ± SEM (n = 3). (F and G) TFH and GC B cells were analyzed in WT:CD19−/−, Obf-1−/−:CD19−/−, WT:TCRα−/−, and Obf-1−/−:TCRα−/− mixed BM chimeras 10 d after influenza infection. (F) Cells from the mLNs were stained for GC B cells as described in A and B. (G) Cells from the mLNs were stained for TFH cells, as described for E. Each symbol represents an individual animal. ***, P < 0.001; *, P = 0.01–0.05. Results are from three independent experiments.
OBF-1 has been previously implicated in the differentiation of helper T cells (Brunner et al., 2007). Thus we asked whether TFH cells of OBF-1–deficient mice show a normal functional phenotype in vivo and in vitro. First, we measured mRNA expression of the TFH cytokine Il21 and the TFH cell regulator Bcl6 in phenotypic TFH cells sorted from WT and OBF-1–deficient mice. Bcl6 and Il21 mRNA expression was normal in TFH cells from OBF-1 KO mice (Fig. 6 E). We then tested whether CD4+ T cells from OBF-1–deficient mice were able to differentiate into IL-21–producing cells T cells in vitro and found that WT and mutant T cells were equally capable of doing so (not depicted). Thus, once they are formed, TFH cells in the Obf1−/− mice have a normal phenotype. This strongly suggests that reduced numbers of TFH cells in the Obf1−/− mice are caused by a T cell–extrinsic defect. To test this, we generated mixed BM chimeras using BM from T cell–deficient or B cell–defective mice (TCRα−/− or Cd19−/−, respectively) and BM from OBF-1–deficient or control mice. 8 wk after reconstitution, the recipient mice were infected with influenza virus and analyzed 10 d later. In mice reconstituted with OBF-1–deficient T cells together with WT B cells, GC B and TFH cells formed normally (Fig. 6, F and G), indicating that OBF-1–deficient T cells are not impaired in their ability to differentiate to TFH and to provide sufficient help for GC B cell development in vivo. In contrast, mice with OBF-1–deficient B cells and WT T cells (Obf1−/−:CD19−/−) showed no GC and reduced TFH cell generation, demonstrating that defective GC formation and reduced TFH cell development are B cell intrinsic in Obf-1−/− mice.
DISCUSSION
Initiation of a TD antibody response
Very recently, the cellular, anatomical, and molecular events that occur during the earliest stages of a TD B cell response have been scrutinized by several groups (e.g., Crotty, 2011; Deenick et al., 2011; Vinuesa and Cyster, 2011). Among their findings are the critical importance of antigen and DCs as early inducers of TFH polarization in the T cell zone, the colocalization of TFH with antigen-specific B cells, and the need for prolonged contact between TFH precursors and cognate B cells (Garside et al., 1998; Haynes et al., 2007; Deenick et al., 2010; Poholek et al., 2010; Baumjohann et al., 2011; Choi et al., 2011; Kerfoot et al., 2011; Kitano et al., 2011). Prolonged B cell–T cell interaction is critical and is mediated by Slam/SAP family receptors, allowing the sustained antigen signal that apparently drives TFH cell differentiation and maintenance (Qi et al., 2008; Cannons et al., 2010). Once cognate T cells and B cells have been activated by antigen during an immune response to a pathogen, IL-6 is thought to drive TFH cell differentiation and IL-21 secretion (Fazilleau et al., 2009; King, 2009). Subsequently, during a B cell–T cell interaction in a GC, IL-21 can act on both the B cell, driving isotype switching and differentiation to an antibody-secreting plasma cell (Kwon et al., 2009; Linterman et al., 2010; Zotos et al., 2010), and in an autocrine fashion on the TFH cell, reinforcing signals that maintain the TFH phenotype (Nurieva et al., 2008; Vogelzang et al., 2008). Here we show that, in addition to antigen and other co-stimulatory surface molecule interactions, B cells release IL-6 to promote TFH in response to infection.
The role of IL-21 in GC formation and TFH development
Some uncertainty surrounds the role of IL-21 during TD B cell responses. Some studies described an impaired initial GC B cell response to various antigens in the absence of an IL-21 signal (Nurieva et al., 2008; Vogelzang et al., 2008; Bessa et al., 2010; Poholek et al., 2010; Eto et al., 2011; Rankin et al., 2011). In contrast, Ozaki et al. (2004) and Zotos et al. (2010) did not detect early GC abnormalities upon loss of IL-21 or IL-21R but saw impaired persistence of GCs. Here we also found normal GC formation in IL-21– or IL-21R–deficient mice on day 10 of an acute viral infection.
The influence of IL-21 on the formation and maintenance of TFH in vivo is also unclear. Some studies describe a reduction of TFH cells in the absence of IL-21 or IL-21R (Nurieva et al., 2008; Vogelzang et al., 2008), whereas others suggest that IL-21 is specifically required for TFH cell persistence but not formation (Linterman et al., 2010). Still others report no impact of IL-21 or IL-21R on the formation of TFH cells (Bessa et al., 2010; Poholek et al., 2010; Zotos et al., 2010; Eto et al., 2011; Rankin et al., 2011). We find here that loss of either IL-21 or IL-21R had little impact on TFH formation during influenza infection at any time point examined.
The role of IL-6 in GC formation and TFH development
Studies to define a role for the pleiotropic cytokine IL-6 in TD immune responses have also yielded conflicting results. Kopf et al. (1998) demonstrated that IL-6–deficient mice had reduced serum IgG2a and formed smaller GCs than controls upon DNP-OVA immunization, whereas others have demonstrated a more severe impact of IL-6 loss on the appearance of GC B cells (Nurieva et al., 2008; Wu et al., 2009). However, Poholek et al. (2010) and Eto et al. (2011) saw no significant reduction of GC B cells after LCMV infection in mice deficient for IL-6 or after IL-6 neutralization. We also find here that loss of IL-6 caused only minimal reduction of GC B cells at the peak of an influenza infection and had a mild effect on GC maintenance.
Although the activity of IL-6 as an inducer of IL-21 expression in CD4+ T cell cultures is well established (Suto et al., 2008; Dienz et al., 2009), its in vivo role in the generation of TFH is not. Nurieva et al. (2008) showed that IL-6 loss caused a strong reduction of TFH cells in response to sheep red blood cell immunization, conclusions which conflict with other studies (Poholek et al., 2010; Choi et al., 2011). Our kinetic data suggest that IL-6 acts primarily in the induction and/or expansion of TFH early in the immune response, as seen most clearly using the IL-21–GFP reporter system on an Il6−/− background, but that TFH cell numbers normalize as the response proceeds (Fig. 2).
One explanation for the conflicting data regarding the roles of IL-6 and IL-21 in GC and TFH responses is likely to be the variety of experimental systems used, including immunization with synthetic or nonreplicating antigens (hapten-coupled proteins or sheep red blood cells), or infectious agents, and the differing levels of inflammation (and so IL-6 production) that might result in each situation. Another may be that IL-6 and IL-21 act cooperatively, and loss of either factor alone can be compensated in vivo. The latter is consistent with the results presented here analyzing IL-6/IL-21 double-deficient mice, which clearly show that IL-6 and IL-21 act together on the formation, persistence, and function of GCs and TFH cells. Our findings disagree with aspects of a recent study (Eto et al., 2011), which showed that IL-6 neutralizing antibody had no additional impact on GC formation over IL-21 loss alone. It is possible that IL-6 neutralizing antibody cannot fully neutralize all IL-6 in vivo (particularly if the IL-6 is delivered within a tight junction from cognate B cell to TFH cell), leading to an underestimation of its contribution to the response. Nevertheless, we concur with the general conclusion of this paper, that IL-6 and IL-21 serve different functions in humoral immunity. However, IL-6 and IL-21 are not functionally redundant in the conventional sense; they may share signaling pathways, but they act at different times and on different cells during the response. During acute infection, IL-6 is produced by APCs, including B cells, as we show here. IL-6 acts on CD4+ T cells to initiate or reinforce their polarization toward TFH cells. IL-21 acts later on TFH polarized cells in an autocrine manner, through a positive feedback loop to reinforce TFH commitment (Suto et al., 2008), and on GC B cells to drive their differentiation. Finally, IL-6 and IL-21 are not the only cytokines initiating TD B cell immunity, as IL-4 and IL-27, another Stat3 signaling cytokine, have recently been implicated in TFH cell differentiation and GC responses (Batten et al., 2010; Vijayanand et al., 2012), and IL-12 has also been shown to be important in TFH development in humans (Ma et al., 2009).
A role for B cell–derived IL-6 in GC and TFH responses
We found that activated WT B cells can stimulate Il21 expression in CD4+ T cells in vitro and that IL-6 was necessary and sufficient for this effect. We also performed a detailed kinetic analysis of IL-6 and IL-21 expression in vivo and found that IL-6 was produced from B cells and myeloid cells in a transient fashion, peaking on day 2 to 3 after infection, then dropping over subsequent days. Although Il6 was expressed in myeloid cells isolated from influenza-infected mice, we found that viral antigens induced high Il6 expression in activated follicular B cells within the draining LN, by far the most abundant APC in the tissue at that time. Conversely, Il21 expression was restricted to CD4+ T cells, increasing from days 3 to 10. We therefore reasoned that IL-6 could play a critical early role in the GC response. Indeed, we were able to improve the weak TFH cell response of IL-6/IL-21 doubly deficient mice and the GC response through the provision of naive WT B cells to the animals just before infection. These data collectively support a role for paracrine secretion of IL-6 by B cells to CD4+ T cells as an important early step in TFH development or expansion. More recently, IL-6 was shown to play an essential late role in the clearance of a chronic viral infection, with follicular DCs supplying IL-6 to TFH, to drive GC formation and neutralizing antibody production (Harker et al., 2011). Collectively, these studies point to a need for provision of IL-6 both early and late for optimal TD antibody responses but suggest that the preferred cellular source of IL-6 changes as the response progresses and TFH cells interact with different cellular partners.
B cells require Oct2 and OBF-1 to produce IL-6
There is accumulating evidence for the direct role of these two transcription factors in the cytokine-mediated regulation of antibody responses (Corcoran et al., 2005; Emslie et al., 2008). Here we found that Il6 expression in B cells is dependent on Oct2 and OBF-1. We identified four consensus sites in the Il6 gene to which Oct2 bound in vitro and in vivo. It is known that OBF-1 is recruited, in an Oct factor–dependent way, to such consensus sequences (Cepek et al., 1996; Gstaiger et al., 1996; Shore et al., 2002). Therefore, we propose that in activated B cells, OBF-1 is able to bind with Oct2 to the consensus sites in the murine Il6 locus, activating the gene. This was specific to B cells, as IL-6 production was not affected in macrophages or DCs isolated from Obf-1−/− or Oct2−/− mice (Fig. 4 and not depicted). Consequently, both OBF-1– and Oct2-deficient B cells were weak in vitro inducers of Il21 expression in CD4+ T cells.
OBF-1 is essential for the formation of GC B cells (Schubart et al., 1996). However, the specific requirements for OBF-1 in the GCs are not fully understood. Impaired BCR signaling in OBF-1−/− B cells (Qin et al., 1998; Samardzic et al., 2002), possibly mediated through loss of SpiB expression (Bartholdy et al., 2006), is likely to be the dominant disability blocking GC development. Clearly, poor IL-6 production by B cells is not the sole defect underlying the total lack of GCs in Obf-1−/− mice, as Il6−/− and Oct2−/− mice can make GCs normally (Figs. 1 and 6). Here we observed a significant reduction of TFH cells in the draining LNs of influenza-infected OBF-1–deficient mice. As OBF-1 is expressed in activated T cells (Sauter and Matthias, 1997; Zwilling et al., 1997) and regulates essential T helper cytokines (Brunner et al., 2007), it was possible that TFH cells required OBF-1 intrinsically for their differentiation. However, analysis of mixed BM chimeras excluded a T cell–intrinsic role for OBF-1 in TFH formation. Conversely, WT T cells were impaired in their differentiation to TFH when Obf-1−/− B cells were present, indicating that the observed TFH and GC phenotypes in OBF-1–deficient mice are both B cell intrinsic.
A temporal model of TFH and GC generation
Bcl6 is the signature transcriptional regulator of both TFH and GC B cells, and Bcl6 reporter mice have recently revealed the in vivo dynamics of development of these cells during immune responses (Baumjohann et al., 2011; Kitano et al., 2011). Upon infection or immunization, antigen-presenting DCs prime CD4 T cells to rapidly but modestly induce Bcl6 expression and to initiate TFH cell differentiation. However, this signal provokes only incomplete TFH cell differentiation, as the nascent TFH cells do not express PD-1 or CXCR5 and cannot sustain Bcl6 expression or GC development. A second, stronger wave of Bcl6 expression in CD4+ cells, induced through contact with B cells, correlates with increased cell division and CXCR5 and PD-1 expression. These studies confirm that DCs are important for early TFH priming but that from approximately day 3.5 onwards, well before GCs are formed, B cells are required to sustain and reinforce Bcl6 expression and TFH expansion and to enable follicular entry (Haynes et al., 2007; Zaretsky et al., 2009; Deenick et al., 2010; Baumjohann et al., 2011; Goenka et al., 2011). In this paper, we demonstrate an important interplay between IL-6 and IL-21 in the formation of TFH and GCs. The kinetics of production and cellular sources of IL-6 and IL-21 documented here during acute viral infection are consistent with these factors being part of the critical communication between B cells and TFH that is required for GC formation.
Because IL-6 and IL-21 both signal through Stat3 (Zeng et al., 2007; Nurieva et al., 2008; Eddahri et al., 2009), it is possible that precursors of TFH need to exceed a certain Stat3 signaling threshold or signal duration to efficiently up-regulate and maintain the high Bcl6 levels required to commit fully to differentiation. Loss of one cytokine might be tolerated, but loss of both could drop the signal below this limit. Thus, IL-6 and IL-21 may cooperate to ensure that TH cells receive a sufficiently strong and durable signal to mature, enter the follicle, and support GC formation for a potent antibody response.
MATERIALS AND METHODS
Mice, immunization, and tissue recovery.
All mutant mice were >10 generations backcrossed onto the C57BL/6 background. IL-6–deficient (Kopf et al., 1998), IL-21–deficient (Parrish-Novak et al., 2000), IL-21R–deficient (Ozaki et al., 2002), IL-21–GFP reporter (Lüthje et al., 2012), Ly5.1 C57BL/6, recombination activating gene 1–deficient (Rag-1−/−; Spanopoulou et al., 1994), CD19-deficient (Engel et al., 1995), Oct2-deficient (Corcoran et al., 1993) and OBF-1–deficient (Schubart et al., 1996) mice were bred and maintained in the specific pathogen–free facilities of the Walter and Eliza Hall Institute of Medical Research. The Oct2+/+ and Oct2−/− mice used here were Rag1−/− mice reconstituted with fetal liver cells, as the Oct2 mutation is lethal when homozygous (Corcoran et al., 1993). TCR-α–deficient mice (Philpott et al., 1992) were maintained at the University of Melbourne. Reconstitution experiments used donor BM from TCRα−/−, Cd19−/−, Obf-1−/−, and C57BL/6 strains, mixed in equal ratios and injected into Rag-1−/− mice. At the indicated times after infections, mice were sacrificed, spleens and mLNs were removed, and single cell suspensions were prepared for analysis as previously described (Blink et al., 2005). All procedures were approved by the Animal Ethics Committee of the Walter and Eliza Hall Institute of Medical Research.
Viral infections.
Mice were inoculated intranasally with 104 pfu of the HKx31 (H3N2) influenza virus (Flynn et al., 1998; Belz et al., 2000). Virus stocks were grown in the allantoic cavity of 10 d embryonated hen’s eggs and stored in aliquots at −80°C.
Antibodies and flow cytometry.
Single cell suspensions of BM cells, splenocytes, or LNs were stained with fluorochrome or biotin-labeled antibodies. Cells were analyzed on an LSRII, FACSCalibur, or FACSCanto cytometer (BD) or were sorted using a MoFlo (Beckman Coulter) or FACSAria (BD) using a live lymphocyte gate (defined as negative for propidium iodide uptake). Data were analyzed with FlowJo (Tree Star) and Prism (GraphPad Software) software. Antibodies used were the following: α-CD4 (GK1.5; FITC; BD), α–PD-1 (RPMI-30; PE; BioLegend), α-CXCR5 (2G8; bio; BD), α-B220/CD45R (RA3-6B2; APC; BD), α-FAS/CD95 (DX2; PE; BD), CD86 (GL1; PE; BD), CD69 (H1.2F3; biotin; BD), PNA (FITC; Vector Laboratories), α-CD8 (53-67; FITC; BD), α-CD44 (IM7; APC; BD), α-KLRG1 (2F1; PECy7; BD), DbNP366 tetramer (PE; in house), DbPA224 tetramer (PE; in house), α-CD19 (ID3; APC; BD), α-IgM (331-12; ITC; in house), α-IgD (112GC; PE; in house), α-CD11c (H13; PE; BD), and α-Mac1/CD11b (M1/70; FITC; BD).
Immunofluorescence histology.
Splenic tissue samples were fixed in 4% paraformaldehyde. 7-µm sections were cut and stained with α-B220 (RA3-6B2; biotin; BD), α-CD3 (rabbit polyclonal; Thermo Fisher Scientific), and α-GL7 (supernatant; in house). Secondary antibodies used were streptavidin-Cy5 (BD), α–rabbit Ig Alexa Fluor 488 (goat polyclonal; Invitrogen), and α–rat Ig Alexa Fluor 555 (goat; Invitrogen). Multiple images from spleen sections were taken with an LSM 5 life microscope (Carl Zeiss), using the Mosaic module (Carl Zeiss) to stitch and align all images taken from one section. For analysis, images covering 1/4 to 1/2 of a spleen section were taken and processed from each sample. The images were analyzed and quantified with the AxioVision (Carl Zeiss) software.
Virus-specific ELISA.
Influenza-specific antibody titers were determined by ELISA (Sangster et al., 2000) using 96-well plates coated with 0.25 mg/well of purified, detergent-disrupted influenza HKx31. In brief, purified HKx31 influenza virus was disrupted in a 1/10 dilution of lysis buffer (0.05 M Tris, pH 7.5, 0.5% Triton X-100, and 0.6 M KCI) in PBS, pH 7.2. Protein concentration of HKx31 virus antigen was determined by Bradford assay. Bound antibody was detected with HRP (horseradish peroxidase)-conjugated goat α–mouse antibodies specific for IgM and HRP-conjugated rabbit α–mouse total IgG (SouthernBiotech) and visualized with ABTS substrate (2,2’-Azinobis (3-ethylbenzthiazoline Sulfonic Acid) Diammonium salt; A-1888; Sigma-Aldrich). Titers shown in Fig. 2 are those that gave 50% of the maximal response.
Cell preparation and culture.
Cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM glutamine, 100 mg/ml penicillin, 100 mg/ml streptomycin, and 50 µM 2-mercaptoethanol. Follicular B cells, DCs, and macrophages from the mLNs were isolated by cell sorting using α-CD19, α-CD11c, and α-Mac1 antibodies. Splenic B cells or B cells from the LNs were isolated using α-CD45R/B220- or α-CD19–coupled magnetic beads (Miltenyi Biotec).
For the preparation of BM-derived macrophages, BM was harvested from the femurs of 8–12-wk-old mice and cultured in bacterial-grade dishes for 6 d in RPMI medium supplemented with 10 ng/ml recombinant murine GM-CSF (PeproTech). On days 3 and 5 of the culture period, 70% of the culture supernatant containing nonadherent cells was removed and replaced with fresh media containing 10 ng/ml GM-CSF. On day 6, the loosely adherent and nonadherent cells were removed by vigorous washing. The remaining adherent macrophage population was harvested by incubating the cells for 5 min in PBS + 10 mM EDTA followed by gentle scraping with a rubber policeman (SARSTEDT).
For DC culture, BM cells were extracted and erythrocytes were removed by brief exposure to 0.168 M NH4Cl. Cells were cultured for 5 d at a density of 1.5 × 106 cells/ml in RPMI 1640 medium containing 100 ng/ml mouse Flt3L (PeproTech) at 37°C in 10% CO2 (Naik et al., 2005).
Splenic B cells, BM-derived macrophages, and BM-derived DCs were culture for 24–48 h in RPMI 1640. The cell cultures were stimulated with the following mitogens as indicated: 10 µg/ml LPS from Escherichia coli 0111:4B (Sigma-Aldrich), 1 µM oligonucleotide CpG 1668 (sequence 5′-TCCATGACGTTCCTGATGCT-3′, fully phosphothioated; GeneWorks). Anti-µ and anti-CD40 were used at 10 µg/ml as described previously (Corcoran and Karvelas, 1994).
For the T cell cultures, naive splenic CD4+CD62L+ T cells were isolated using α-CD4 FITC (GK1.5) and an α-FITC Multisort kit followed by α-CD62L–coupled magnetic beads (Miltenyi Biotech). Isolated naive T cells were cultured for 5 d in RPMI 1640 medium on plates coated with both 10 µg/ml α-CD3 (45-2C11) and in the presence of monoclonal 2 µg/ml α-CD28 (37.51), together with recombinant IL-6 (10% vol/vol, prepared in house) and 100 ng/ml recombinant IL-21 (PeproTech).
For B/T cell co-cultures, splenic B cells were isolated using α-CD19 or α-B220 beads (Miltenyi Biotech) and stimulated for 24 h with CpG as described above. Activated B cells were washed three times with PBS and co-cultured in different ratios (Fig. 3: 3 × 105 or 3 × 103 B cells to 3 × 105 T cells; Fig. 4: 2 × 105 or 6 × 104 B cells to 6 × 105 T cells) with naive C57BL/6 T cells in the conditions described above. After 5 d, the CD4+ cells were recovered by flow cytometric cell sorting.
B cell rescue.
1–2 × 107 splenic B cells were isolated from Ly5.1 congenic mice and injected i.v. on two subsequent days into C57BL/6 and IL-6/IL-21 DKO mice. On the third day, the host mice were infected with HKx31 influenza virus. Mice were sacrificed 10 d after infection, and GCs and TFH cells in mLNs were analyzed by flow cytometry. To adjust for technical variation between experiments that was not related to genotype, the percentages of TFH cells (of total CD4+ T cells) or GC B cells (of B220+ B cells) were normalized to the mean frequency of the WT control from each experimental cohort. We were thus able to determine the fold change of TFH or GC B cell percentage compared with each control group. We also compared TFH and GC B cell data from the transplanted mice with data from infected C57BL/6 and DKO mice that had not received B cells.
Western blotting.
OBF-1 and Oct2 expression was detected using our monoclonal rat α–mouse antibodies (clone 9A2, Corcoran et al. [2004]; clone 6E4, Corcoran et al. [2005]). Protein extracts corresponding to equal cell numbers were loaded onto the gel, with equal protein loading confirmed with Ponceau red stains of the membrane after protein transfer. A goat α-actin antiserum (Santa Cruz Biotechnology, Inc.) was used as a loading control.
Quantitative RT-PCR.
First-strand cDNA was transcribed (SuperScript III First-Strand Synthesis System; Invitrogen) from total RNA (RNeasy Micro kit; QIAGEN) using the manufacturers’ protocols. Real-time qPCR analysis was performed using a SYBR green system (Superarray), according to the manufacturer’s instructions. The expression data were analyzed on a sequence detection system (ABI Prism 7900HT; Applied Biosystems) and CFX384 real-time system (Bio-Rad Laboratories) using relative quantification of gene expression. Expression was normalized using hydroxymethylbilane synthase (Hmbs) as a housekeeping gene. Normalization of expression data was computed by the qGENE tool (Simon, 2003).
Primers used for cDNA amplification were as follows: Hmbs, (forward) 5′-GACCTGGTTGTTCACTCCCTGAAG-3′ and (reverse) 5′-GACAACAGCATCACAAGGGTTTTC-3′; Bcl6, (forward) 5′-GCCGGCTCAATAATCTCGTGAACAGGTCC-3′ and (reverse) 5′-CCAGCAGTATGGAGGCACATCTCTGTATGC-3′; Il21, (forward) 5′-TCAGCTCCACAAGATGTAAAGGG-3′ and (reverse) 5′-GGGCCACGAGGTCAATGAT-3′; and Il6, the QuantiTect Primer Assay for Mm_IL6 (QIAGEN).
EMSA.
Nuclear extracts were prepared (Schreiber et al., 1989) and EMSA was performed as previously described (Corcoran et al., 2004). Restriction fragment probes were labeled using [32P]dATP and Klenow DNA polymerase. Probes for Il6 locus were generated through PCR amplification using genomic C57BL/6 DNA as a template. 130-bp- to 350-bp-long PCR products were subsequently labeled using [32P]dATP and Klenow DNA polymerase.
Primer sequences for probes of Il6 locus are as follows: IL-6–P1, (forward) 5′-GGATACAATCAGCCCCATAC-3′ and (reverse) 5′-GTATGGGGCTGATTGTATCC-3′; IL-6–P2, (forward) 5′-ATCAACCGGCTTTTCATTTTA-3′ and (reverse) 5′-TGCTCCCATGTTTAATAGTTCAA-3′; IL-6–P3, (forward) 5′-CCAGTTGGAACATCTTCTGCG-3′ and (reverse) 5′-TGGGGTACAAAGCTAAACAAA-3′; and IL-6–P4, (forward) 5′-AGGTGAAATCTCAGGGTAGT-3′ and (reverse) 5′-TAAAACATGGGGTACAGAGT-3′.
ChIP.
ChIP was performed essentially as described previously (Emslie et al., 2008) except that Protein G Dynabeads (Invitrogen) were used to capture the protein–DNA immune complexes. The primers used to amplify the four putative Oct2-binding sites in the Il6 gene are listed in the previous section, and the Cd36 primers have been described previously (Emslie et al., 2008).
Acknowledgments
We are indebted to the facilities of our respective institutes, particularly those responsible for animal husbandry and flow cytometry. We also acknowledge the expert technical assistance of J. Jackson and M. Camilleri and are grateful for many helpful discussions with Dr. S.L. Nutt and other members of the B Cell Program at the Walter and Eliza Hall Institute for Medical Research. We also thank Dr. S. Tangye (Garvan Institute, Sydney, New South Wales, Australia) for critical input. Dr. T. Speed and P. Hickey provided invaluable advice on statistical analysis.
This work was supported by grants from the National Health and Medical Research Council (NHMRC) Australia (Independent Research Institute Infrastructure Support Scheme [IRIISS] grant #361646 and Program Grant #575500), the Sylvia and Charles Viertel Charitable Foundation, and the Victorian State Government Operational Infrastructure Support grant and was made possible through the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- GC
- germinal center
- mLN
- mediastinal LN
- mRNA
- messenger RNA
- qPCR
- quantitative PCR
- TD
- T cell dependent
References
- Ansel K.M., McHeyzer-Williams L.J., Ngo V.N., McHeyzer-Williams M.G., Cyster J.G. 1999. In vivo–activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190:1123–1134 10.1084/jem.190.8.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguni E., Arima M., Tsuruoka N., Sakamoto A., Hatano M., Tokuhisa T. 2006. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int. Immunol. 18:1079–1089 10.1093/intimm/dxl041 [DOI] [PubMed] [Google Scholar]
- Baccam M., Woo S.Y., Vinson C., Bishop G.A. 2003. CD40-mediated transcriptional regulation of the IL-6 gene in B lymphocytes: involvement of NF-kappa B, AP-1, and C/EBP. J. Immunol. 170:3099–3108 [DOI] [PubMed] [Google Scholar]
- Bartholdy B., Du Roure C., Bordon A., Emslie D., Corcoran L.M., Matthias P. 2006. The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc. Natl. Acad. Sci. USA. 103:11665–11670 10.1073/pnas.0509430103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten M., Ramamoorthi N., Kljavin N.M., Ma C.S., Cox J.H., Dengler H.S., Danilenko D.M., Caplazi P., Wong M., Fulcher D.A., et al. 2010. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J. Exp. Med. 207:2895–2906 10.1084/jem.20100064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Okada T., Ansel K.M. 2011. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187:2089–2092 10.4049/jimmunol.1101393 [DOI] [PubMed] [Google Scholar]
- Belz G.T., Stevenson P.G., Doherty P.C. 2000. Contemporary analysis of MHC-related immunodominance hierarchies in the CD8+ T cell response to influenza A viruses. J. Immunol. 165:2404–2409 [DOI] [PubMed] [Google Scholar]
- Bessa J., Kopf M., Bachmann M.F. 2010. Cutting edge: IL-21 and TLR signaling regulate germinal center responses in a B cell-intrinsic manner. J. Immunol. 184:4615–4619 10.4049/jimmunol.0903949 [DOI] [PubMed] [Google Scholar]
- Blink E.J., Light A., Kallies A., Nutt S.L., Hodgkin P.D., Tarlinton D.M. 2005. Early appearance of germinal center–derived memory B cells and plasma cells in blood after primary immunization. J. Exp. Med. 201:545–554 10.1084/jem.20042060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker T., Gulbranson-Judge A., Flynn S., Riedinger M., Raykundalia C., Lane P. 1999. CD4 T cell traffic control: in vivo evidence that ligation of OX40 on CD4 T cells by OX40-ligand expressed on dendritic cells leads to the accumulation of CD4 T cells in B follicles. Eur. J. Immunol. 29:1610–1616 [DOI] [PubMed] [Google Scholar]
- Brunner C., Sindrilaru A., Girkontaite I., Fischer K.D., Sunderkötter C., Wirth T. 2007. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J. 26:3191–3202 10.1038/sj.emboj.7601742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons J.L., Qi H., Lu K.T., Dutta M., Gomez-Rodriguez J., Cheng J., Wakeland E.K., Germain R.N., Schwartzberg P.L. 2010. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 32:253–265 10.1016/j.immuni.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey K.A., Mescher M.F. 2007. IL-21 promotes differentiation of naive CD8 T cells to a unique effector phenotype. J. Immunol. 178:7640–7648 [DOI] [PubMed] [Google Scholar]
- Cepek K.L., Chasman D.I., Sharp P.A. 1996. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev. 10:2079–2088 10.1101/gad.10.16.2079 [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T., Tangye S.G., Newton R., Frank N., Hodge M.R., Rolph M.S., Mackay C.R. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78 [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Karvelas M. 1994. Oct-2 is required early in T cell-independent B cell activation for G1 progression and for proliferation. Immunity. 1:635–645 10.1016/1074-7613(94)90035-3 [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Karvelas M., Nossal G.J., Ye Z.S., Jacks T., Baltimore D. 1993. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 7:570–582 10.1101/gad.7.4.570 [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Koentgen F., Dietrich W., Veale M., Humbert P.O. 2004. All known in vivo functions of the Oct-2 transcription factor require the C-terminal protein domain. J. Immunol. 172:2962–2969 [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Hasbold J., Dietrich W., Hawkins E., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D. 2005. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J. Exp. Med. 201:1385–1396 10.1084/jem.20042325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Cucak H., Yrlid U., Reizis B., Kalinke U., Johansson-Lindbom B. 2009. Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity. 31:491–501 10.1016/j.immuni.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Deenick E.K., Chan A., Ma C.S., Gatto D., Schwartzberg P.L., Brink R., Tangye S.G. 2010. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity. 33:241–253 10.1016/j.immuni.2010.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenick E.K., Ma C.S., Brink R., Tangye S.G. 2011. Regulation of T follicular helper cell formation and function by antigen presenting cells. Curr. Opin. Immunol. 23:111–118 10.1016/j.coi.2010.10.007 [DOI] [PubMed] [Google Scholar]
- Dienz O., Eaton S.M., Bond J.P., Neveu W., Moquin D., Noubade R., Briso E.M., Charland C., Leonard W.J., Ciliberto G., et al. 2009. The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J. Exp. Med. 206:69–78 10.1084/jem.20081571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddahri F., Denanglaire S., Bureau F., Spolski R., Leonard W.J., Leo O., Andris F. 2009. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 113:2426–2433 10.1182/blood-2008-04-154682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emslie D., D’Costa K., Hasbold J., Metcalf D., Takatsu K., Hodgkin P.O., Corcoran L.M. 2008. Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor α chain expression on activated B cells. J. Exp. Med. 205:409–421 10.1084/jem.20072049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P., Zhou L.-J., Ord D.C., Sato S., Koller B., Tedder T.F. 1995. Abnormal B lymphocyte development, activation, and differentiation in mice that lack or overexpress the CD19 signal transduction molecule. Immunity. 3:39–50 10.1016/1074-7613(95)90157-4 [DOI] [PubMed] [Google Scholar]
- Eto D., Lao C., DiToro D., Barnett B., Escobar T.C., Kageyama R., Yusuf I., Crotty S. 2011. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 6:e17739 10.1371/journal.pone.0017739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N., Mark L., McHeyzer-Williams L.J., McHeyzer-Williams M.G. 2009. Follicular helper T cells: lineage and location. Immunity. 30:324–335 10.1016/j.immuni.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn K.J., Belz G.T., Altman J.D., Ahmed R., Woodland D.L., Doherty P.C. 1998. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 8:683–691 10.1016/S1074-7613(00)80573-7 [DOI] [PubMed] [Google Scholar]
- Garside P., Ingulli E., Merica R.R., Johnson J.G., Noelle R.J., Jenkins M.K. 1998. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 281:96–99 10.1126/science.281.5373.96 [DOI] [PubMed] [Google Scholar]
- Goenka R., Barnett L.G., Silver J.S., O’Neill P.J., Hunter C.A., Cancro M.P., Laufer T.M. 2011. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 187:1091–1095 10.4049/jimmunol.1100853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M., Georgiev O., van Leeuwen H., van der Vliet P., Schaffner W. 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 15:2781–2790 [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., Zuniga E.I. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes N.M., Allen C.D., Lesley R., Ansel K.M., Killeen N., Cyster J.G. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099–5108 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfoot S.M., Yaari G., Patel J.R., Johnson K.L., Gonzalez D.G., Kleinstein S.H., Haberman A.M. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 34:947–960 10.1016/j.immuni.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Qin X.F., Gong S., Stevens S., Luo Y., Nussenzweig M., Roeder R.G. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 383:542–547 10.1038/383542a0 [DOI] [PubMed] [Google Scholar]
- King C. 2009. New insights into the differentiation and function of T follicular helper cells. Nat. Rev. Immunol. 9:757–766 10.1038/nri2644 [DOI] [PubMed] [Google Scholar]
- King C., Tangye S.G., Mackay C.R. 2008. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu. Rev. Immunol. 26:741–766 10.1146/annurev.immunol.26.021607.090344 [DOI] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- König H., Pfisterer P., Corcoran L.M., Wirth T. 1995. Identification of CD36 as the first gene dependent on the B-cell differentiation factor Oct-2. Genes Dev. 9:1598–1607 10.1101/gad.9.13.1598 [DOI] [PubMed] [Google Scholar]
- Kopf M., Herren S., Wiles M.V., Pepys M.B., Kosco-Vilbois M.H. 1998. Interleukin 6 influences germinal center development and antibody production via a contribution of C3 complement component. J. Exp. Med. 188:1895–1906 10.1084/jem.188.10.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Thierry-Mieg D., Thierry-Mieg J., Kim H.P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins K., Reményi A., Tomilin A., Massa S., Wilmanns M., Matthias P., Schöler H.R. 2003. OBF1 enhances transcriptional potential of Oct1. EMBO J. 22:2188–2198 10.1093/emboj/cdg199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Beaton L., Yu D., Ramiscal R.R., Srivastava M., Hogan J.J., Verma N.K., Smyth M.J., Rigby R.J., Vinuesa C.G. 2010. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207:353–363 10.1084/jem.20091738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje K., Kallies A., Shimohakamada Y., TBelz G.T., Light A., Tarlinton D.M., Nutt S.L. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 13:491–498 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- Ma C.S., Suryani S., Avery D.T., Chan A., Nanan R., Santner-Nanan B., Deenick E.K., Tangye S.G. 2009. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol. Cell Biol. 87:590–600 10.1038/icb.2009.64 [DOI] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M.M. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 18:333–334 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Corcoran L.M., Wu L. 2005. Development of murine plasmacytoid dendritic cell subsets. Immunol. Cell Biol. 83:563–570 10.1111/j.1440-1711.2005.01390.x [DOI] [PubMed] [Google Scholar]
- Novy P., Huang X., Leonard W.J., Yang Y. 2011. Intrinsic IL-21 signaling is critical for CD8 T cell survival and memory formation in response to vaccinia viral infection. J. Immunol. 186:2729–2738 10.4049/jimmunol.1003009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.-H., Watowich S.S., Jetten A.M., Tian Q., Dong C. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H., Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Tarlinton D.M. 2011. Germinal center B and follicular helper T cells: siblings, cousins or just good friends? Nat. Immunol. 12:472–477 10.1038/ni.2019 [DOI] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Feng C.G., Qi C.-F., Cheng J., Sher A., Morse H.C., III, Liu C., Schwartzberg P.L., Leonard W.J. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science. 298:1630–1634 10.1126/science.1077002 [DOI] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Ettinger R., Kim H.-P., Wang G., Qi C.-F., Hwu P., Shaffer D.J., Akilesh S., Roopenian D.C., et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361–5371 [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J., Dillon S.R., Nelson A., Hammond A., Sprecher C., Gross J.A., Johnston J., Madden K., Xu W., West J., et al. 2000. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 408:57–63 10.1038/35040504 [DOI] [PubMed] [Google Scholar]
- Philpott K.L., Viney J.L., Kay G., Rastan S., Gardiner E.M., Chae S., Hayday A.C., Owen M.J. 1992. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 256:1448–1452 10.1126/science.1604321 [DOI] [PubMed] [Google Scholar]
- Poholek A.C., Hansen K., Hernandez S.G., Eto D., Chandele A., Weinstein J.S., Dong X., Odegard J.M., Kaech S.M., Dent A.L., et al. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185:313–326 10.4049/jimmunol.0904023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Cannons J.L., Klauschen F., Schwartzberg P.L., Germain R.N. 2008. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 455:764–769 10.1038/nature07345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.F., Reichlin A., Luo Y., Roeder R.G., Nussenzweig M.C. 1998. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 17:5066–5075 10.1093/emboj/17.17.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin A.L., MacLeod H., Keegan S., Andreyeva T., Lowe L., Bloom L., Collins M., Nickerson-Nutter C., Young D., Guay H. 2011. IL-21 receptor is critical for the development of memory B cell responses. J. Immunol. 186:667–674 10.4049/jimmunol.0903207 [DOI] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.-E., Locksley R.M. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samardzic T., Gerlach J., Muller K., Marinkovic D., Hess J., Nitschke L., Wirth T. 2002. CD22 regulates early B cell development in BOB.1/OBF.1-deficient mice. Eur. J. Immunol. 32:2481–2489 [DOI] [PubMed] [Google Scholar]
- Sangster M.Y., Topham D.J., D’Costa S., Cardin R.D., Marion T.N., Myers L.K., Doherty P.C. 2000. Analysis of the virus-specific and nonspecific B cell response to a persistent B-lymphotropic gammaherpesvirus. J. Immunol. 164:1820–1828 [DOI] [PubMed] [Google Scholar]
- Sauter P., Matthias P. 1997. The B cell-specific coactivator OBF-1 (OCA-B, Bob-1) is inducible in T cells and its expression is dispensable for IL-2 gene induction. Immunobiology. 198:207–216 10.1016/S0171-2985(97)80041-1 [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M.M., Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart D.B., Rolink A., Kosco-Vilbois M.H., Botteri F., Matthias P. 1996. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 383:538–542 10.1038/383538a0 [DOI] [PubMed] [Google Scholar]
- Schubart K., Massa S., Schubart D., Corcoran L.M., Rolink A.G., Matthias P. 2001. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat. Immunol. 2:69–74 10.1038/83190 [DOI] [PubMed] [Google Scholar]
- Shore P., Dietrich W., Corcoran L.M. 2002. Oct-2 regulates CD36 gene expression via a consensus octamer, which excludes the co-activator OBF-1. Nucleic Acids Res. 30:1767–1773 10.1093/nar/30.8.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P. 2003. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 19:1439–1440 10.1093/bioinformatics/btg157 [DOI] [PubMed] [Google Scholar]
- Smith A.J.P., D’Aiuto F., Palmen J., Cooper J.A., Samuel J., Thompson S., Sanders J., Donos N., Nibali L., Brull D., et al. 2008. Association of serum interleukin-6 concentration with a functional IL6 -6331T>C polymorphism. Clin. Chem. 54:841–850 10.1373/clinchem.2007.098608 [DOI] [PubMed] [Google Scholar]
- Spanopoulou E., Roman C.A.J., Corcoran L.M., Schlissel M.S., Silver D.P., Nemazee D., Nussenzweig M.C., Shinton S.A., Hardy R.R., Baltimore D. 1994. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 8:1030–1042 10.1101/gad.8.9.1030 [DOI] [PubMed] [Google Scholar]
- Strubin M., Newell J.W., Matthias P. 1995. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 80:497–506 10.1016/0092-8674(95)90500-6 [DOI] [PubMed] [Google Scholar]
- Suto A., Kashiwakuma D., Kagami S.-i., Hirose K., Watanabe N., Yokote K., Saito Y., Nakayama T., Grusby M.J., Iwamoto I., Nakajima H. 2008. Development and characterization of IL-21–producing CD4+ T cells. J. Exp. Med. 205:1369–1379 10.1084/jem.20072057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayanand P., Seumois G., Simpson L.J., Abdul-Wajid S., Baumjohann D., Panduro M., Huang X., Interlandi J., Djuretic I.M., Brown D.R., et al. 2012. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 36:175–187 10.1016/j.immuni.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Cyster J.G. 2011. How T cells earn the follicular rite of passage. Immunity. 35:671–680 10.1016/j.immuni.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Vinuesa C.G., Cook M.C., Angelucci C., Athanasopoulos V., Rui L., Hill K.M., Yu D., Domaschenz H., Whittle B., Lambe T., et al. 2005. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 435:452–458 10.1038/nature03555 [DOI] [PubMed] [Google Scholar]
- Vogelzang A., McGuire H.M., Yu D., Sprent J., Mackay C.R., King C. 2008. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 29:127–137 10.1016/j.immuni.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Wu Y., El Shikh M.E., El Sayed R.M., Best A.M., Szakal A.K., Tew J.G. 2009. IL-6 produced by immune complex-activated follicular dendritic cells promotes germinal center reactions, IgG responses and somatic hypermutation. Int. Immunol. 21:745–756 10.1093/intimm/dxp041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi A.K., Chace J.H., Cowdery J.S., Krieg A.M. 1996. IFN-gamma promotes IL-6 and IgM secretion in response to CpG motifs in bacterial DNA and oligodeoxynucleotides. J. Immunol. 156:558–564 [PubMed] [Google Scholar]
- Yu D., Vinuesa C.G. 2010. The elusive identity of T follicular helper cells. Trends Immunol. 31:377–383 10.1016/j.it.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Yusuf I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., Crotty S. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185:190–202 10.4049/jimmunol.0903505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky A.G., Taylor J.J., King I.L., Marshall F.A., Mohrs M., Pearce E.J. 2009. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J. Exp. Med. 206:991–999 10.1084/jem.20090303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Spolski R., Casas E., Zhu W., Levy D.E., Leonard W.J. 2007. The molecular basis of IL-21-mediated proliferation. Blood. 109:4135–4142 10.1182/blood-2006-10-054973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotos D., Coquet J.M., Zhang Y., Light A., D’Costa K., Kallies A., Corcoran L.M., Godfrey D.I., Toellner K.-M., Smyth M.J., et al. 2010. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell–intrinsic mechanism. J. Exp. Med. 207:365–378 10.1084/jem.20091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S., Dieckmann A., Pfisterer P., Angel P., Wirth T. 1997. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 277:221–225 10.1126/science.277.5323.221 [DOI] [PubMed] [Google Scholar]