Abstract
Mitotic exit and cell division must be spatially and temporally integrated to facilitate equal division of genetic material between daughter cells. In the fission yeast, Schizosaccharomyces pombe, a spindle pole body (SPB) localized signaling cascade termed the septation initiation network (SIN) couples mitotic exit with cytokinesis. The SIN is controlled at many levels to ensure that cytokinesis is executed once per cell cycle and only after cells segregate their DNA. An interesting facet of the SIN is that its activity is asymmetric on the two SPBs during anaphase; however, how and why the SIN is asymmetric has remained elusive. Many key factors controlling SIN asymmetry have now been identified, shedding light on the significance of SIN asymmetry in regulating cytokinesis. In this review, we highlight recent advances in our understanding of SIN regulation, with an emphasis on how SIN asymmetry is achieved and how this aspect of SIN regulation fine-tunes cytokinesis.
Keywords: Septation initiation network (SIN), cytokinesis, mitosis, spindle pole body (SPB), S. pombe
Introduction
Generating two daughter cells with identical genetic content is the ultimate goal of cell division. In all eukaryotic organisms, cell division requires massive cytoskeletal rearrangements that allow for chromosome separation, recruitment of cytokinesis proteins to the division site, assembly of an actomyosin-based cytokinetic ring (CR) and CR constriction. Given the intricacy of the cell division process, it is not surprising that it demands a diverse cohort of proteins to orchestrate these events. These include (1) structural proteins to assemble into the mitotic spindle and CR and to provide spatial landmarks within the cell, (2) molecular motors to provide the kinetic requirements for chromosome separation and CR constriction and (3) signaling enzymes to provide temporal cues, typically by imparting activating or inhibitory messages onto their targets via post-translational modifications (Bohnert and Gould, 2011). Coordinating these assorted molecules to ultimately divide a cell in two is a complicated yet vital task to guarantee that the ensuing progenies do not inherit erroneous DNA content.
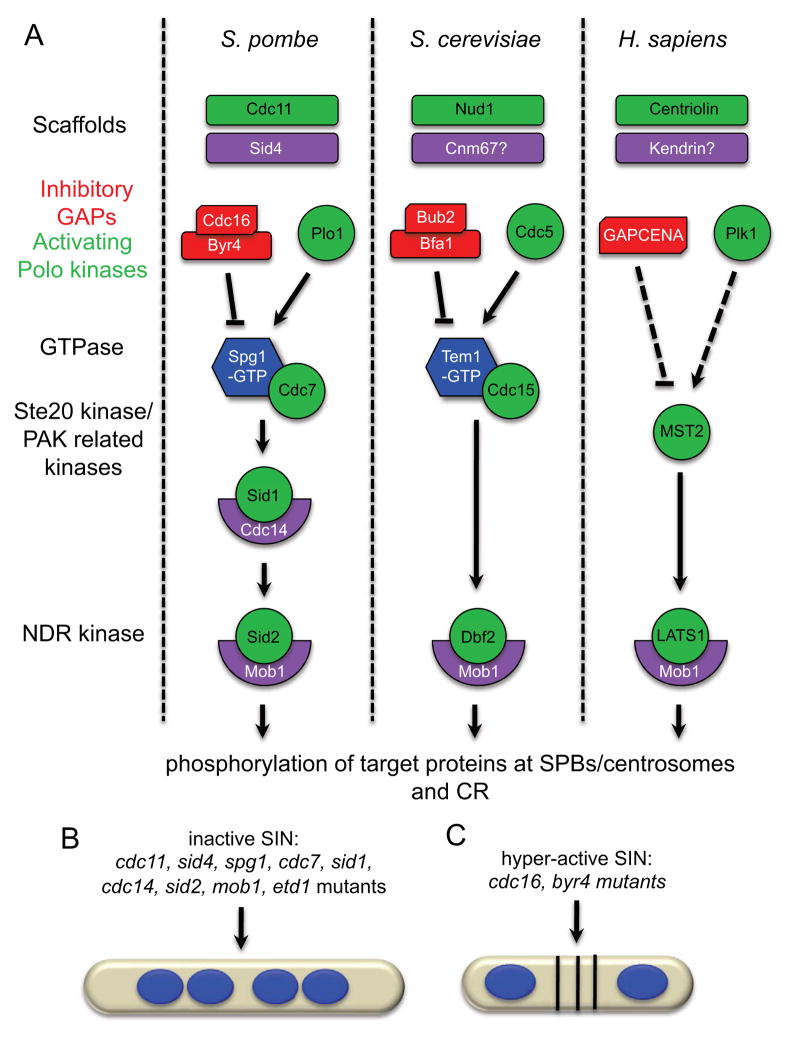
That mitosis (division of genetic material) precedes cytokinesis (division of cytoplasmic material) is critical to ensure the survival of each new cell and, thus, mitotic events must be intimately linked with cytokinetic events, such that they occur in an orderly fashion. Low CDK activity is a hallmark of mitotic exit and, therefore, many organisms respond to changes in CDK activity as a mechanism to couple mitosis with cytokinesis. The fission yeast Schizosaccharomyces pombe utilizes a conserved signaling pathway called the septation initiation network (SIN) that induces cytokinesis only when CDK activity drops in anaphase (Chang et al., 2001; Guertin et al., 2000), guaranteeing that cytokinesis occurs after chromosome segregation. A pathway homologous to the SIN, termed the mitotic exit network (MEN), exists in the budding yeast S. cerevisiae (Bardin and Amon, 2001; Seshan and Amon, 2004). Almost all SIN components have orthologs in the MEN and the pathways have similar organization (Figure 1A and Table 1). SIN/MEN orthologs also exist in metazoans (Figure 1A and Table 1), underscoring the conservation of these pathways; however, the functions of metazoan SIN/MEN pathways in cell division are less well characterized. Thus, understanding cytokinesis regulation by the yeast SIN/MEN should aid in our understanding of the metazoan pathways.
Figure 1.
A. The essential signaling components of the SIN/MEN pathways in S. pombe, S. cerevisiae and H. sapiens. In S. pombe, the SIN is anchored to SPBs via a bipartite scaffold complex, Cdc11-Sid4. During interphase, the Cdc16-Byr4 GAP complex inhibits the Spg1 GTPase to hold it in its GDP-bound form. Upon mitotic entry, Plo1 promotes Spg1 activation, perhaps through inhibition of Cdc16-Byr4 allowing Spg1 to switch to its active GTP-bound form. Spg1-GTP binds its effector kinase Cdc7 and elicits activation of the downstream SIN kinases Sid1-Cdc14 and Sid2-Mob1. Upon activation, Sid2-Mob1 translocates to the CR and presumably phopshorylates key substrates that promote CR assembly and constriction. Similar mechanisms of SIN/MEN activation also occur in S. cerevisiae and H. sapiens. B. Phenotype observed when the SIN is inactivated. Inactivating mutations in SIN activators cdc11, sid4, spg1, cdc7, sid1, cdc14, sid2, mob1, and etd1 produce multinucleate cells as a result of cytokinesis failure. C. Phenotype observed when the SIN is hyper-active. Inactivating mutations in the SIN inhibitors byr4 and cdc16 produce multi-septated cells.
Table 1.
List of S. pombe SIN proteins and their homologs in S. cerevisiae and H. sapiens.
| Core SIN components | |||
|---|---|---|---|
| S. pombe | S. cerevisiae | H. sapiens | gene product |
| Sid4 | Cnm67p? | Kendrin? | Scaffold |
| Cdc11 | Nud1p | Centriolin | Scaffold |
| Spg1 | Tem1p | ? | GTPase |
| Cdc7 | Cdc15p | MST2? | Ste20 family protein kinase |
| Sid1 | ? | MST2? | PAK-related protein kinase |
| Cdc14 | ? | ? | Sid1co-factor |
| Sid2 | Dbf2p | LATS1 | NDR family protein kinase |
| Mob1 | Mob1p | MOB1A | Sid2 co-factor |
| Byr4 | Bfa1p | ? | GAP scaffold |
| Cdc16 | Bub2p | GAPCENA | GAP |
| Etd1 | Lte1p | ? | GEF-like protein |
| SIN regulators | |||
| S. pombe | S. cerevisiae | H. sapiens | gene product |
| Plo1 | Cdc5p | PLK1 | Polo-like protein kinase |
| Dma1 | Dma1p/Dma2p | CHFR/RNF8 | E3 ubiquitin ligase |
| Clp1 | Cdc14p | CDC14 | protein phosphatase |
| Cdc2 | Cdc28p | CDK | protein kinase |
| Zfs1 | Tis11p/cth1p | ? | Zn finger protein |
| Par1 | Rts1p | ? | PP2A B′ subunit |
| Csc1 | Far10p | SLMAP | PP2A B‴ subunit |
The core SIN network is comprised of a GTPase, three protein kinases, an inhibitory GAP complex, and a scaffold complex that anchors the pathway to SPBs (Figures 1A and 2). The terminal kinase in the pathway, Sid2, transitions from the SPB to the cell division site in anaphase to drive cytokinesis (Sparks et al., 1999). How SIN proteins interact and transmit signals through the cascade to trigger cytokinesis is only beginning to be unraveled. Also, although it was observed many years ago that SIN signaling is asymmetric on the two anaphase SPBs, how and why the SIN becomes asymmetric has remained elusive. Recent studies have shed light on many of these issues and in this review we describe these findings and discuss their implications. We begin by describing what is known about the SIN’s role in initiating cytokinesis and how signals are propagated through the pathway. After discussing our current understanding of SIN regulation, with an emphasis on SIN asymmetry, we conclude with general remarks about outstanding questions in the field.
Figure 2.
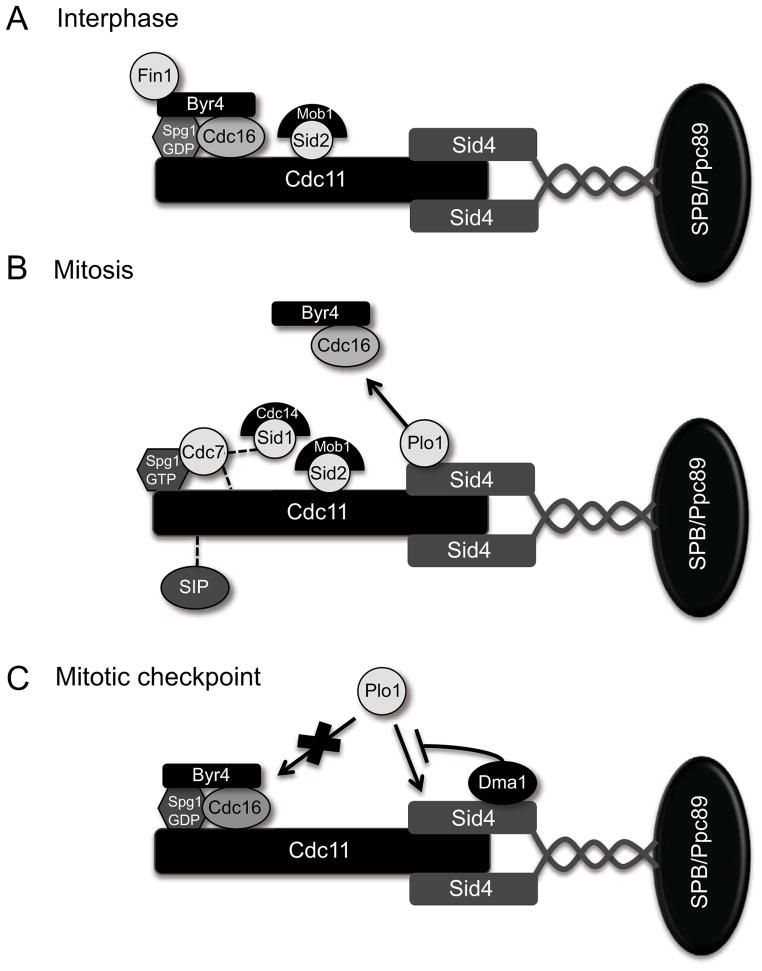
Organization of SIN components at SPBs during interphase (A). mitosis (B) and a mitotic checkpoint (C). Proteins in contact indicate interactions detected by two-hybrid or in vitro experiments and dashed lines indicate potential interactions based on epistatic experiments.
Identification of the SIN genes in S. pombe
Much of our current understanding of cell division derives from studying simpler model systems, such as Schizosaccharomyces pombe. This rod shaped unicellular organism grows primarily at its tips, undergoes a closed mitosis (no nuclear envelope breakdown), and divides via binary fission using an actomyosin-based CR. S. pombe is a useful model organism to study the cell cycle because its cell size is tightly coupled to its cell cycle stage, it is amenable to genetic and biochemical study, and a comprehensive collection of deletion and temperature-sensitive mutants are readily available (Goyal et al., 2011). Because many key genes required for S. pombe cytokinesis are conserved in metazoans, studying S. pombe cytokinesis has piloted many principal discoveries that have shaped our current understanding of cytokinesis in multiple organisms.
To better understand cytokinesis, several genetic screens were performed in S. pombe that enabled the identification of genes required specifically for division site specification, CR assembly, and CR constriction/septation (Balasubramanian et al., 1998; Chang et al., 1996; Minet et al., 1979; Nurse et al., 1976). One set of mutations impacting CR assembly, constriction, and septation displayed a number of genetic interactions with each other and were thus proposed to constitute a signal transduction cascade that initiated the final steps in cytokinesis (Marks et al., 1992). Subsequent biochemical characterization and epistatic analyses led to our current understanding of their functional integration in an ordered pathway that is now termed the septation initiation network (SIN) (Figure 1A).
Functions of the SIN in cytokinesis
SIN mutants generate one of two phenotypes: multi-nucleate cells or multi-septated cells that fail in cell cleavage (Figures 1B and 1C). The former phenotype is caused by SIN inactivation; the latter phenotype results from SIN hyper-activity. Both scenarios uncouple cell division from nuclear division; thus, the SIN coordinates cytokinesis with other cell cycle phases.
Detailed analyses of SIN mutant phenotypes indicate that the SIN is essential for CR assembly and constriction as well as septum formation. In S. pombe, the anillin-related Mid1 protein and the SIN drive CR assembly in early (pre-anaphase) and late mitosis (anaphase/telophase), respectively. In early mitosis, Mid1 localizes to cortical nodes near the site of division and recruits CR components (Motegi et al., 2004; Sohrmann et al., 1996; Wu et al., 2006). These nodes then coalesce into a ring-like structure, which matures into a continuous ring (Vavylonis et al., 2008; Wu et al., 2003). A CR can assemble in both mid1 (Sohrmann et al. 1996) and SIN mutants (Balasubramanian et al., 1998; Wu et al. 2003), suggesting that these two pathways are independent; however, distinct defects are observed in each case. mid1Δ mutants assemble ectopic rings in anaphase when the SIN becomes active, implying that the major function of Mid1 is to direct CR assembly to the correct location (Chang et al., 1996; Sohrmann et al., 1996). SIN mutants form a CR in early mitosis (presumably by the Mid1 pathway); however, it dissolves in anaphase suggesting that SIN signaling is required for CR maintenance/assembly in late mitosis (Balasubramanian et al., 1998). Disrupting both Mid1 and the SIN blocks CR assembly completely (Hachet and Simanis, 2008; Huang et al., 2008), indicating that each pathway makes important contributions to CR assembly. However, activating the SIN in interphase triggers CR assembly, demonstrating that the SIN is capable of driving CR assembly on its own (Schmidt et al. 1997). Because SIN mutants also fail to deposit septum material, the SIN might also promote the activity of enzymes involved in septum deposition, such as the glucan synthase Cps1 (Balasubramanian et al., 1998).
Although major progress has been made towards understanding Mid1-dependent CR assembly, the role of the SIN in CR assembly is less clear, particularly because the pertinent SIN substrates at the CR are unknown. The only SIN component that localizes to the CR is the terminal SIN kinase Sid2-Mob1 and, to date, the only reported Sid2 target at the CR is the Cdc14-like phosphatase Clp1 (Chen et al., 2008). During interphase, Clp1 is sequestered in the nucleolus and is released into the cytoplasm early in mitosis, such that it can localize to the CR ring and de-phosphorylate its substrates (Trautmann et al., 2001). Clp1 phosphorylation by Sid2 promotes binding of the 14-3-3 protein, Rad24, which maintains Clp1 in the cytoplasm during cytokinesis (Chen et al., 2008; Mishra et al., 2005). Without Sid2 phosphorylation, Clp1 returns prematurely to the nucleolus and cells exhibit cytokinesis defects. One direct Clp1 target is the PCH-family protein Cdc15, which is essential for CR assembly and must be de-phosphorylated to efficiently assemble the CR (Clifford et al., 2008; Roberts-Galbraith et al., 2010). Consistent with Sid2’s role in Clp1 regulation, Cdc15 at the CR is severely diminished when Sid2 function is compromised (Hachet and Simanis, 2008), most likely because Clp1 is not maintained in the cytoplasm to de-phosphorylate Cdc15. Thus, Sid2-dependent phosphorylation of Clp1 is important for the final steps in cytokinesis. However, other Sid2 substrates at the CR must exist, since Clp1 is non-essential, and identifying the essential Sid2 substrates will be important to completely understand how the SIN drives CR assembly and constriction.
Spindle pole bodies as a signaling hub for cytokinesis
Several studies indicate that SPBs provide an essential platform for SIN signaling. Specifically, ablating both mitotic SPBs results in cytokinesis failure (Magidson et al., 2006), indicating that cytokinesis requires signals emanating from SPBs. In accord with this observation, SIN components assemble at SPBs via a bipartite scaffold complex Sid4-Cdc11 (Figure 2A–C) (Chang and Gould, 2000; Krapp et al., 2001; Morrell et al. 2004; Tomlin et al., 2002). Sid4-Cdc11 localize to SPBs in all cell cycle phases and fluorescence recovery after photo-bleaching (FRAP) experiments indicate that association with the SPBs is stable (Feoktistova et al., 2012; Morrell et al., 2004). Another SPB protein, Ppc89, anchors the Cdc11-Sid4 scaffold to SPBs by directly binding the C-terminus of Sid4 (Rosenberg et al., 2006). Together, Sid4-Cdc11 establishes a signaling hub onto which SIN signaling components and their regulators assemble (Morrell et al., 2004).
In addition to providing a stable platform for SIN components, evidence suggests that post-translational modifications acquired by Cdc11-Sid4 modulate their scaffold functions and, thus, provide another level of SIN regulation (Figure 4B and table 2). Prior to CR assembly and constriction Cdc11 is hyper-phosphorylated, which enhances SIN activation by promoting recruitment of downstream SIN kinases (Feoktistova et al., 2012; Krapp et al., 2003). Sid4 is ubiquitinated during a mitotic checkpoint arrest, which inhibits recruitment of an essential SIN activator (Plo1) until the checkpoint has been satisfied (Johnson and Gould, 2011). In both cases, modifying Cdc11 and Sid4 alters their binding capacity for the signaling components with which they interact and, thereby, affects SIN signaling.
Figure 4.
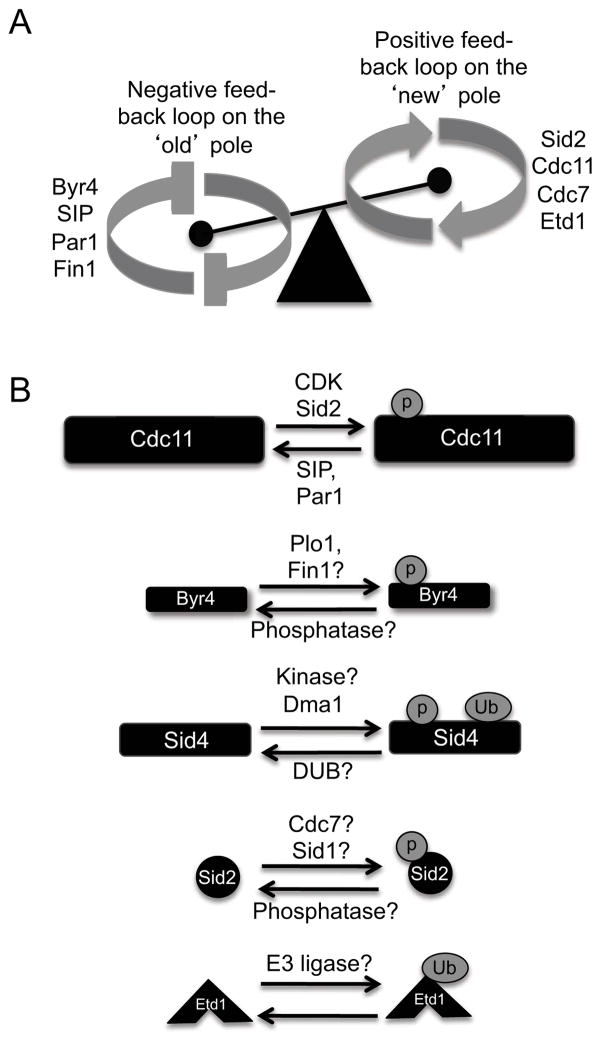
A. Model for potential positive and negative feedback loops that break SIN symmetry during anaphase. Cooperative efforts between Byr4, the Fin1 kinase and PP2A phosphatases inhibit the SIN on the ‘old’ pole, while Sid2, Cdc7, Cdc11 and Etd1 contribute to SIN hyper-activation on the ‘new’ pole. B. Post-translational modifications identified for the core SIN proteins and the validated or potential enzymes that promote or antagonize these modifications.
Table 2.
Post-translational modifications (PTMs) of SIN proteins and their homologs. Many targeted and genome-wide studies have identified PTMs on many SIN proteins and their counterparts in other organisms. Here, we list the yeast and human SIN/MEN homologs for which PTMs have been identified to date and provide references for each protein where more detailed information can be found.
| S. pombe | S. cerevisiae | H. sapiens | ||||||
|---|---|---|---|---|---|---|---|---|
| Protein | PTM(s) | references | Protein | PTM(s) | references | Protein | PTM(s) | references |
| Sid4 | Ub, P* | 1 | Cnm67p | P | 7–10 | Kendrin | P | 16–18 |
| Cdc11 | P | 2–4 | Nud1p | P | 7,9 | Centriolin | P | 19,20 |
| Byr4 | P | 1,2,5 | Bfa1p | P | 11,12 | ? | n/a | |
| Cdc16 | - | Bub2p | P | 13 | GAPCENA | P | 21 | |
| Spg1 | - | Tem1p | - | ? | n/a | |||
| Cdc7 | P | 5 | Cdc15p | P | 8,9,14 | MST2 | P | 17,20–25 |
| Sid1 | - | ? | n/a | MST2 | P | 17,20–25 | ||
| Cdc14 | - | ? | n/a | ? | n/a | |||
| Sid2 | P | 2,5,6 | Dbf2p | P | 8,9,14 | LATS1 | P | 21–23,26–30 |
| Mob1 | - | Mob1p | P | 8,9,15 | MOB1 | P | 21–23,31–33 | |
P-phosphorylation, Ub-ubiquitination, (−)-none detected, n/a-not applicable,
our unpublished results.
S. pombe references:
S. cerevisiae references:
H. sapien references:
Signaling through the SIN
SIN signaling progresses through sequential activation of the Ras super-family GTPase Spg1 and its downstream effectors. Spg1 localizes to SPBs constitutively by direct interaction with Cdc11 (Morrell et al., 2004) and can drive cytokinesis in any cell cycle stage when over-expressed (Schmidt et al., 1997). During interphase, Spg1 associates with a bipartite GAP, Byr4-Cdc16, which maintains Spg1 in its inactive state and is required for interphase SPB localization of Spg1 (Figure 2A) (Furge et al., 1999; Furge et al., 1998; Krapp et al., 2008; Song et al., 1996). Upon mitotic entry, Byr4-Cdc16 dissociates from SPBs, allowing Spg1 to switch to its GTP-bound active state (Figure 2B) (Li et al., 2000).
Spg1 activation subsequently triggers the activity of three protein kinases (Cdc7, Sid1 and Sid2) in a step-wise manner (Figure 1A). The Ste20 family protein kinase, Cdc7, preferentially binds the GTP-bound activated form of Spg1, and the two proteins depend on each other for SPB localization when Spg1 is in its active form (Fankhauser and Simanis, 1994; Krapp et al. 2008; Sohrmann et al., 1998). Because Cdc7 preferentially binds Spg1 is in its active form, its presence at the SPB can be used to monitor Spg1 activity in vivo. Cdc7 protein levels and kinase activity do not fluctuate throughout the cell cycle; thus, Cdc7 function is mainly regulated by its SPB recruitment (Sohrmann et al., 1998). Cdc7 localizes to both SPBs early in mitosis and as the spindle elongates, Cdc7 disappears from one SPB and accumulates at the opposite SPB (Sohrmann et al., 1998). Byr4-Cdc16 returns to the SPB in which Cdc7 disappears, inactivating and preventing further SIN signaling on this pole (Li et al., 2000). Thus far, a target of the Cdc7 protein kinase has not been identified, although by analogy to the budding yeast homologs (Mah et al. 2005), Sid2 is a likely Cdc7 target.
At anaphase onset, the protein kinase Sid1 and its binding partner Cdc14 localize to the SPB with active Spg1 (Fankhauser and Simanis, 1993; Guertin et al., 2000). Sid1 requires Sid4, Cdc11, Cdc14, Spg1 and Cdc7 for its SPB recruitment placing it downstream of Cdc7 recruitment (Guertin et al., 2000). Sid1 protein levels are not cell cycle dependent; however, Sid1 kinase activity peaks in late anaphase/telophase, coincident with its SPB localization (Guertin and McCollum, 2001). Sid1-Cdc14 SPB localization also depends on decreased CDK activity (Guertin et al., 2000), which normally occurs at anaphase onset, providing one mechanism to couple exit from mitosis with initiation of cytokinesis. Unfortunately, our knowledge of Sid1 targets is also lacking, but Sid2 and/or its binding partner Mob1 are potential substrates.
Sid2, a member of the NDR (nuclear Dbf2-related) family of kinases, and its partner Mob1 localize to SPBs constitutively and function downstream of Sid1-Cdc14 (Hou et al., 2004; Hou et al., 2000; Salimova et al., 2000; Sparks et al., 1999). Sid2 kinase activity requires Mob1 association and its activity peaks prior to septation (Hou et al., 2004; Sparks et al., 1999). Sid2-Mob1 also localize to the division site (Sparks et al., 1999), where Sid2 presumably phosphorylates its substrates to drive CR assembly and constriction. Sid2-Mob1 division site localization depends on an intact microtubule cytoskeleton (Sparks et al., 1999), but the mechanisms of Sid2-Mob1 re-localization are still unknown. Similar to other NDR family kinases, Sid2 phosphorylation is important for Mob1 association and, thus, for its catalytic activity (Hou et al., 2004). The kinase(s) that phosphorylate Sid2 are unknown, but the human Sid2 homolog, LATS1, is phosphorylated and activated by the Ste20 kinase MST2 (Chan et al., 2005), implicating Cdc7 or Sid1 as candidates. The phosphatase(s) for Sid2 are also unknown, but several studies implicate PP2A phosphatases in SIN inhibition (discussed later) and PP2A phosphatases have been reported to antagonize NDR kinases in mammalian cells (Millward et al., 1999). Because Sid2 is the terminal SIN kinase, regulating Sid2’s phosphostatus is likely to be an important aspect of SIN regulation.
The SIN in checkpoint signaling
Several studies show that the SIN is not only required for cytokinesis, but also plays an integral role in checkpoint pathways that ensure coordination of major mitotic events. When chromosomes are not properly attached to the mitotic spindle, SIN activity is inhibited to prevent the CR from cutting through unsegregated chromosomes. One protein that inhibits the SIN under these conditions is Dma1, an E3 ubiquitin ligase that contains an N-terminal phosphothreonine-binding FHA domain and a C-terminal RING domain (Guertin et al., 2002; Murone and Simanis 1996). Dma1 localizes symmetrically to SPBs and also to the cell division site (Guertin et al., 2002; Murone and Simanis, 1996). At the SPB, Dma1 ubiquitinates the SIN scaffold, Sid4, to impede recruitment of the SIN activator, Plo1 (Figure 2C) (Johnson and Gould, 2011). Dma1’s FHA domain is required for its SPB localization (Guertin et al., 2002) and Sid4 is a phosphoprotein in vivo (our unpublished results), suggesting that Sid4 phosphorylation might be important for this checkpoint pathway as well. However, signals connecting events at the kinetochore-microtubule interface to the SIN at the cytoplasmic face of the SPB are unknown.
The SIN also participates in a ‘cytokinesis checkpoint’ that monitors the integrity of the CR (Le Goff et al., 1999; Liu et al., 2000). This checkpoint pathway was first identified through characterization of the 1,3-β-glucan synthase cps1 (Liu et al., 2000; Liu et al., 1999), required for septum formation (Ishiguro et al., 1997). cps1 mutants arrest with a stable CR and two nuclei that each complete S-phase, but do not enter mitosis. This implies that a monitoring system prevents mitotic entrance if the previous cytokinesis fails and further demonstrates that septum formation and CR constriction are coupled. The CR itself is required for this cell cycle arrest, since CR disassembly by Latrunculin A treatment allowed cells to progress into mitosis (Liu et al., 2000). Inactivating the SIN bypasses the cps1 arrest, indicating that the SIN is required for this checkpoint, although the SIN’s role has not been defined yet (Le Goff et al., 1999; Liu et al., 1999). Clp1 is also required for this checkpoint pathway and given the relationship between Clp1 and the SIN, it is reasonable to think that Clp1 might be a major effecter of this checkpoint pathway (Cueille et al., 2001; Trautmann et al., 2001). This is supported by the observation that Clp1 is necessary for proper cytokinesis if the CR is perturbed (Mishra et al., 2004).
Asymmetry in SIN signaling
As mentioned previously, SIN signaling is asymmetric on the two SPBs during anaphase (Figure 3). By exploiting the slow folding nature of red fluorescent protein (RFP) to mark the ‘old’ SPB, it was discovered that the SIN is hyper-activated on the ‘new’ SPB (Grallert et al., 2004). MEN activity is also asymmetric on the two SPBs; however, in contrast to the SIN, the MEN is active on the ‘old’ SPB (Pereira et al., 2001).
Figure 3.
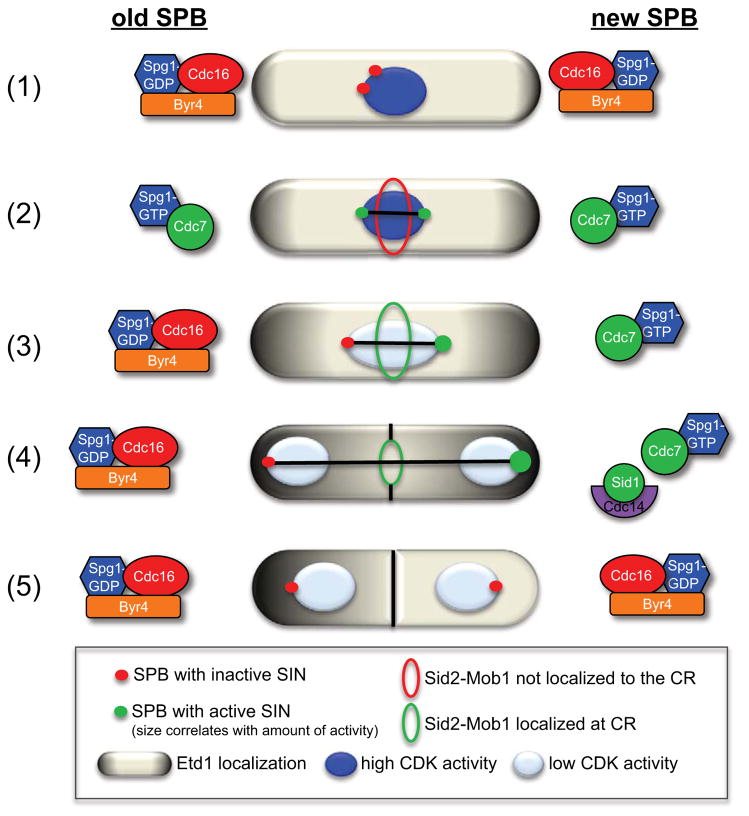
Localization patterns of the SIN signaling proteins at the ‘old’ and ‘new’ SPBs throughout mitosis. (1) In interphase and early pro-metaphase, Byr4-Cdc16 localize to both SPBs and maintains Spg1 in its GDP-bound inactive state. (2) Then, in late pro-metaphase to metaphase, Cdc7 localizes to both SPBs via interaction with Spg1-GTP. (3) As the spindle begins to elongate in anaphase, Cdc7 disappears from the ‘old’ pole and Byr4-Cdc16 returns to the ‘old’ pole to inactivate the SIN and establish SIN asymmetry. (4) Later in mitosis when CDK activity is low, Sid1-Cdc14 localizes to the ‘new’ SPB with active SIN signaling and as the SPBs reach the cell cortex, Etd1 contacts Spg1 and further activates the SIN on the ‘new’ pole. Since Spg1 is bound to the inhibitory GAP complex (Byr4-Cdc16) on the ‘old’ pole, Etd1 is probably prevented from contacting Spg1 on the ‘old’ pole. Once the spindle is fully elongated, Sid2-Mob1 translocates to the division site and induces CR constriction. (5) After septation, Etd1 disappears from the cell compartment with active SIN signaling and Byr4-Cdc16 returns to this SPB to terminate SIN signaling.
SIN asymmetry can be monitored by examining the localization of certain SIN proteins throughout mitosis. Cdc7 localizes to both SPBs in metaphase, but becomes asymmetric once the spindle elongates (Sohrmann et al., 1998). In wild-type cells, Sid1 localizes exclusively to one of the two SPBs during anaphase (Guertin et al., 2000) and presumably the SPB that retains Cdc7, since Sid1 requires Cdc7 for SPB localization (Guertin et al., 2000). In contrast, Byr4 localizes to the interphase SPB and disappears from the ‘new’ pole as the SPBs separate to opposite sides of the nucleus (Cerutti and Simanis, 1999). Byr4 asymmetry precedes that of Cdc7, and Byr4 and Cdc7 SPB localizations are always reciprocal (Cerutti and Simanis, 1999). Thus, Byr4-Cdc16 and Cdc7 localization dictate SIN asymmetry in anaphase B by inactivating Spg1 on the ‘old’ pole and promoting SIN activity on the ‘new’ pole, respectively.
Why is the SIN activated on the new SPB?
It is puzzling why the SIN is preferentially inactivated on the ‘old’ pole and hyper-activated on the ‘new’ pole during anaphase given that the essential scaffold proteins required for SPB localization of the SIN proteins appear to be identical on the two mitotic SPBs. One study made a compelling observation that when the ‘new’ SPB was ablated in anaphase cells, Cdc7 accumulated on the ‘old’ SPB (Magidson et al., 2006). Although Cdc7 interaction with the ‘old’ SPB was transient and its signal weaker than what is normally seen on the ‘new’ pole, the amount of Cdc7 recruited to the ‘old’ SPB was sufficient to drive cytokinesis. Thus, the ‘old’ SPB has the potential to scaffold SIN activity (Magidson et al., 2006).
So what prevents SIN activity on the ‘old’ SPB? One possibility is that inhibitory factor(s) on the ‘new’ SPB prevent recruitment of SIN proteins on the ‘old’ SPB (Magidson et al., 2006). Abolishing the ‘new’ SPB would silence the inhibitory signal(s) and permit recruitment of the SIN components on the ‘old’ SPB. Another possibility is that some SIN proteins have a stronger affinity for the ‘new’ SPB, which sequesters SIN proteins away from the ‘old’ pole. Thus, if the ‘new’ SPB is inaccessible, the ‘old’ pole no longer has to compete with the new SPB for SIN proteins. A potential factor that might confer varying SPB affinities is post-translational modifications of SPB proteins. This is supported by the findings that SPBs and centrosomes are modified over consecutive cell cycles (Grallert et al., 2004; Vorobjev and Chentsov Yu, 1982). Because many SPB proteins have slow turn-over rates, it is likely that proteins associated with the ‘old’ SPB will have encountered one cell cycle already and, therefore, might acquire post-translational modifications that are not incorporated on ‘new’ SPB proteins. Given that many of the proteins involved in SIN regulation are protein kinases and phosphatases, generating phospho-specific antibodies to localize specific phosphorylation events at the two SPBs should help clarify modifications that are particularly important in establishing SIN asymmetry.
SIN asymmetry in terminating cytokinesis
A key question is why do cells possess such elaborate mechanisms to generate asymmetric SIN signaling. Results from one study demonstrated that in wild-type cells, the SIN is inactivated precisely when the CR completes constriction and asymmetric SIN signaling is important to inactivate the SIN when cytokinesis is complete (Garcia-Cortes and McCollum, 2009). Using binucleate dikaryon cells (which can have either symmetric or asymmetric SIN signaling), this study also showed that cells with symmetric SIN activity were defective in terminating SIN signaling and formed additional rings and septa (Garcia-Cortes and McCollum, 2009). Consistent with this observation, inactivating a SIN inhibitory PP2A complex (SIP, discussed in the next section) results in 100% of anaphase cells with symmetric Cdc7 localization and also produces a few cells with premature and multiple septations (Singh et al. 2011). The relatively mild defect in terminating SIN signaling displayed by SIP mutants might be explained by the observation that the SIN eventually becomes asymmetric in these mutants just before septation completes (Singh et al., 2011), indicating that other factors contribute to SIN asymmetry. Collectively, these studies suggest that SIN asymmetry contributes to silencing the SIN after cytokinesis completes. The formation of additional rings and septa could also indicate that SIN asymmetry is important to initiate cytokinesis only once and the presence of two signaling hubs creates conflicting signals, resulting in multiple rounds of septation.
In addition to core SIN components, many SIN regulators distribute asymmetrically within the cell and aid in breaking SIN symmetry. The SIN activator, Etd1, identified in a screen for new cell cycle regulators, physically interacts with spg1 (Daga et al., 2005; Garcia-Cortes and McCollum, 2009; Jimenez and Oballe, 1994). Etd1 localizes at the cell cortex, rather than the SPB, such that Spg1 and Etd1 contact each other only when the mitotic spindle is fully elongated (Figure 3). This would allow Etd1 to influence SIN activity only after chromosomes have segregated completely, providing yet another mechanism for coupling chromosome segregation and septation. After cytokinesis, Etd1 is partitioned away from the cell compartment containing the SPB with active SIN signaling (Figure 3), which allows SIN inactivation on this pole (Garcia-Cortes and McCollum, 2009). The mechanism by which Etd1 is lost from one cell compartment remains to be determined, but given the interdependent relationship between Etd1 and the SIN, they might regulate each other in a positive feedback loop. Whether Etd1 or SIN activity tips the balance to initiate this feedback loop remains to be determined. A similar mechanism has been proposed for the S. cerevisiae MEN regulator, Lte1, which localizes to the bud cortex and is proposed to activate Tem1 when the spindle has elongated and entered the bud (Bardin et al., 2000; Pereira et al., 2000). Thus, Lte1 and Etd1 might have conserved roles in cytokinesis regulation.
Regulating SIN asymmetry
Return of Byr4-Cdc16 to the ‘old’ SPB in anaphase is a pivotal aspect of breaking SIN symmetry. However, how Byr4-Cdc16 specifically recognizes the ‘old’ pole during anaphase is still unclear. To date, factors required for SIN asymmetry include two protein kinases (Sid2-Mob1 and Fin1) (Feoktistova et al., 2012; Grallert et al., 2004) and two protein phosphatase type 2 (PP2A) complexes (PP2A-B′ and PP2A-B‴) (Jiang and Hallberg, 2001; Le Goff et al., 2001; Singh et al., 2011). Interestingly, the SIN scaffold protein Cdc11 is a common target for Sid2-Mob1 and both PP2A phosphatases and Cdc11 phospho-regulation affects the establishment of SIN asymmetry (Feoktistova et al., 2012; Krapp et al., 2003; Singh et al., 2011) (Figure 4A and 4B).
In a sid2 temperature-sensitive mutant (sid2-250), Cdc7-GFP localizes symmetrically throughout mitosis, indicating that Cdc7 asymmetry requires Sid2 function (Feoktistova et al., 2012). Furthermore, Sid2-mediated phosphorylation of Cdc11 promotes Cdc7-Cdc11 interaction. Since Sid2 activity requires Cdc7 activity (Sparks et al., 1999), this suggests that Sid2 provides positive feedback to promote its own activity and contributes to SIN activity on the ‘new’ pole. However, there are likely other Sid2-Mob1 targets involved in promoting Cdc7 asymmetry, since mutating the Sid2 target sites on Cdc11 does not phenocopy the symmetric Cdc7 localization pattern observed in the sid2-250 mutant.
As mentioned earlier, PP2A phosphatases have been implicated in regulating SIN asymmetry. PP2A phosphatases are present in all eukaryotic cells and function in almost every biological process, including mitotic exit and cytokinesis (Jiang, 2006; Wurzenberger and Gerlich, 2011). The PP2A holoenzyme complex contains a scaffold subunit (A), a catalytic subunit (C) and a regulatory subunit (B), of which there are 4 distinct classes: B′, B″, B‴ and B″″. The first PP2A complex genes implicated in SIN regulation were the B′-type regulatory subunits, par1 and par2 (Jiang and Hallberg, 2000; Jiang and Hallberg, 2001). Genetic analyses of par1 and par2 indicated that these genes negatively regulate SIN function upstream of cdc7 (Jiang and Hallberg, 2001; Le Goff et al., 2001). Although Par1 localizes to SPBs in a symmetrical fashion, par1 mutants display an increased number of anaphase cells with symmetric Cdc7 localization, implying that Par1 has a role in establishing and/or maintaining SIN asymmetry (Jiang and Hallberg, 2001).
Another PP2A complex (the SIP, mentioned above) consists of at least 6 subunits and forms a PP2A-B‴ complex required for SIN asy7mmetry (Singh et al., 2011). Similar to Byr4, SIP localizes to both SPBs in metaphase and to the ‘old’ SPB during anaphase. In the absence of SIP, Byr4 does not re-localize to the ‘old’ SPB and Cdc7 and Sid1 remain symmetric throughout anaphase (Singh et al., 2011). This implies that SIP acts directly on Byr4 or upstream of Byr4 to promote Byr4 re-localization on the ‘old’ pole, which subsequently breaks SIN symmetry. Cdc11 is also a potential target for both PP2A-Par1 and SIP, since Cdc11 phosphorylation at the end of mitosis in par1 and SIP mutant cells is increased compared to wild-type cells (Krapp et al., 2003; Singh et al., 2011). Thus, these PP2A phosphatase complexes might work in a concerted fashion to antagonize phosphorylation of Cdc11 and/or other targets on the ‘old’ SPB (Figure 4A and 4B).
Another protein implicated in breaking SIN symmetry is the NIMA-related kinase, Fin1 (Grallert and Hagan, 2002; Grallert et al., 2004). Although the mechanism is unclear, Fin1’s unusual localization pattern provides one clue; it localizes asymmetrically to SPBs in about half of anaphase cells (Grallert and Hagan, 2002; Grallert et al., 2004). Pedigree analyses demonstrated that Fin1’s SPB association depends on SPB age (Grallert et al., 2004); however, how Fin1 discerns SPB maturity remains unclear. Importantly, Fin1’s SPB localization requires SIN activity, suggesting that Fin1 and the SIN are involved in a negative feedback loop, possibly to inactivate SIN signaling on the ‘old’ SPB more robustly (Figure 4A). Finally, although Fin1 targets remain to be identified, Byr4 is a potential candidate because Fin1 interacts with Byr4 by two-hybrid analysis and Byr4 is hyper-phosphorylated in early mitosis (Grallert et al., 2004; Johnson and Gould, 2011). Thus, while a positive feedback loop driven by Sid2 contributes to SIN activation on the ‘new’ pole, a negative feedback loop driven by PP2A phosphatases and Fin1 contributes to SIN inactivation on the ‘old’ pole (Figure 4A). However, how these factors cooperate to ‘tip the balance’ and break SIN symmetry is not well understood yet.
SIN regulators
Other proteins, which are not considered part of the core SIN machinery, act peripherally to modulate SIN activity. A major positive SIN regulator is the Polo-like kinase Plo1. Plo1 has myriad functions in mitosis and cytokinesis, including formation of a bipolar spindle, CR assembly, division site selection, and septum formation (Mulvihill and Hyams, 2002; Ohkura et al., 1995). Early in mitosis Plo1 concentrates at SPBs, the mitotic spindle and the CR (Bahler et al. 1998). Plo1 directly interacts with the scaffold Sid4 and activates the SIN pathway when over-expressed (Figure 2B) (Morrell et al., 2004; Mulvihill et al., 1999; Ohkura et al., 1995; Tanaka et al., 2001). Thus, Plo1 has long been touted as an upstream activator of the SIN pathway; however, Plo1’s target at the SIN remains unknown. Phosphorylation of the S. cerevisiae Byr4 homolog, Bfa1, by the Polo-like kinase Cdc5 inhibits its GAP activity and promotes its dissociation from SPBs (Geymonat et al., 2003; Hu et al., 2001). It is unclear whether a similar mechanism occurs in S. pombe, but Plo1 can phosphorylate Byr4 in vitro and Byr4 residence at SPBs inversely correlates with Plo1 SPB residence (Johnson and Gould, 2011). Thus, it is plausible that Byr4 is the major Plo1 target in the SIN (Figure 2B). Other factors which positively impact SIN signaling include the protein kinase Lsk1 (Karagiannis et al., 2005) and the protein phosphatase calcineurin (Lu et al., 2002), although their associations with the SIN await further characterization.
Genes whose functions antagonize SIN signaling include the PP2A B type subunit pab1 (Lahoz et al., 2010) and the PP2A activator ypa2 (Goyal and Simanis, 2012). Deletion of pab1 suppresses the cytokinesis defects of etd1Δ cells, suggesting that Pab1 counteracts etd1 function (Lahoz et al. 2010). Pab1 interacts with SIN components Mob1, Sid1, Sid2 and Cdc11 in yeast two-hybrid experiments, suggesting that PP2A-Pab1 dephosphorylates a SIN component, but the target(s) remain unknown (Lahoz et al., 2010). The PP2A activator, ypa2, was identified in a genetic screen for cold-sensitive suppressors of the cdc7-24 mutant and genetic analyses suggests that Ypa2 inhibits the SIN (Goyal and Simanis, 2012). The PP2A complex that Ypa2 activates is unknown, but it is reasonable to predict that it impacts PP2A-Par1, SIP or Pab1, which as discussed above, inhibit the SIN. Other negative regulators include a putative RNA binding protein scw1 (Jin and McCollum, 2003; Karagiannis et al., 2002) and a zinc-finger protein zfs1 (Beltraminelli et al., 1999); however, their roles in SIN regulation are not understood.
The SIN in meiosis
Given its role in driving septum formation, it is not surprising that the SIN also operates during meiosis. Specifically, the SIN is activated during the second meiotic division and is required for fore-spore membrane assembly (Krapp et al., 2006). During meiosis, F-actin assembles into 4 ring structures, termed Meiotic actin rings (MeiAR). Constriction of the MeiARs is the final step in fore-spore membrane assembly and the SIN controls the rate of MeiAR constriction (Yan and Balasubramanian, 2012).
A Sid2-like kinase, Slk1, is expressed specifically during meiosis and requires the SIN for its localization to SPBs (Perez-Hidalgo et al., 2008; Yan et al., 2008). Deletion of slk1 results in decreased sporulation efficiency and deletion of both slk1 and sid2 prevents sporulation completely. Thus, Slk1 and Sid2 have some redundant roles in forespore membrane assembly, but meiotic substrates of Slk1 and Sid2 remain to be identified.
The SIN inhibitor, Dma1, is also implicated in regulating forespore membrane assembly (Krapp et al., 2010; Li et al., 2010). Consistent with a role in meiosis, Dma1 is upregulated during meiosis I and II, and similar to its mitotic localization pattern, Dma1 requires the SIN scaffold Sid4 for its SPB localization in meiotic cells (Li et al., 2010). How Dma1 impacts meiotic progression is not known, but genetic analyses suggest that it might influence Slk1 signaling.
SIN-like pathways in other organisms
Signaling networks homologous to the SIN exist in other organisms including the budding yeast Saccharomyces cerevisiae (Seshan and Amon, 2004), the filamentous fungi Aspergillus nidulans (Bruno et al., 2001; Kim et al., 2006; Kim et al., 2009) and basidiomycete Ustilago mayadis (Sandrock et al., 2006). In metazoans, many SIN homologs exist, suggesting that a similar pathway is present; however, molecular information for the human pathway is lacking (Figure 1A and Table 1).
The NDR protein kinase, LATS1/WARTS, shares many functional similarities with S. pombe Sid2. First identified in D. melanogaster as members of the hippo/salvador/warts pathway, LATS/WARTS kinases function in signaling pathways involved in cell proliferation and apoptosis (Halder and Johnson, 2011). Similar to Sid2, LATS1 functions in mitotic exit and cytokinesis and localizes to centrosomes constitutively and to the division site in late mitosis (Bothos et al., 2005; Hirota et al., 2000; Xia et al., 2002; Yang et al. 2001; Yang et al., 2004). Like other NDR family kinases, LATS1 associates with its co-factor/activator MOB1A, contributing to LATS1 activation (Chow et al., 2009; Hergovich et al., 2006). LATS1 and MOB1A are further activated by the Ste20 protein kinase, MST2, which shares homology with S. pombe Sid1 and Cdc7 (Chan et al., 2005; Hirabayashi et al., 2008). Many human Ste20 protein kinases also require association with GTPases for their activity (Dan et al., 2001); however, the GTPase(s) with which MST2 associates, if one exists, is still unknown.
The GAP protein, GAPCENA is homologous to S. pombe Cdc16 and localizes to centrosomes (Cuif et al., 1999). The GTPase that GAPCENA associates with at the centrosomes is not known, but identifying its centrosome-localized GTPase partner might reveal Spg1 homolog(s). Centriolin is a centrosome and mid-body localized protein that shares homology with the SIN scaffold protein Cdc11 and participates in cytokinesis (Gromley et al., 2005). Whether the signaling components for the proposed human “mitotic exit network” associate with Centriolin remains to be determined. Ascertaining whether signaling through the human network mirrors that of the yeast SIN/MEN pathways will entail detailed investigations of the protein-protein interactions between the human homologs and further characterization at a molecular level.
Concluding remarks
Significant progress has been made in learning how the SIN is organized and how it regulates cytokinesis. Presently, we view identifying substrates of SIN kinases, particularly Sid2 substrates at the CR, as the most pressing task to advance our understanding. By identifying Sid2 targets, we anticipate learning much about the mechanism of CR assembly and contraction. It will also be of interest to investigate in greater detail how signals within the SIN network are transduced. Phosphorylation of key SIN proteins appears to be important to propagate signals within the SIN network and teasing apart these mechanisms will expand our understanding of SIN regulation and especially how SIN asymmetry is established. Computational modeling may provide significant value in these endeavors. Lastly, identifying more SIN homologs in metazoans and characterizing their functions and interactions with each other will be pertinent to determine if the SIN, its targets, and its modes of regulating cytokinesis are conserved.
Acknowledgments
We thank members of the Gould lab, especially Janel McLean, Adam Bohnert and Anna Feoktistova for critically reading the manuscript and many helpful discussions. A.E.J. was supported by the Cellular, Biochemical and Molecular Sciences training program, NIH T32 GM08554, D.M. is supported by NIH grant GM058406 and K.L.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7(7):1389–96. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143(6):1603–16. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149(3):1265–75. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2(11):815–26. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102(1):21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltraminelli N, Murone M, Simanis V. The S. pombe zfs1 gene is required to prevent septation if mitotic progression is inhibited. J Cell Sci. 1999;112(Pt 18):3103–14. doi: 10.1242/jcs.112.18.3103. [DOI] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7(6):e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. On the cutting edge: post-translational modifications in cytokinesis. Trends Cell Biol. 2011;21(5):283–92. doi: 10.1016/j.tcb.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65(15):6568–75. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Morrell JL, Hamer JE, Staiger CJ. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol Microbiol. 2001;42(1):3–12. doi: 10.1046/j.1365-2958.2001.02605.x. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Simanis V. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J Cell Sci. 1999;112 (Pt 14):2313–21. doi: 10.1242/jcs.112.14.2313. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24(12):2076–86. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109 (Pt 1):131–42. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci U S A. 2000;97(10):5249–54. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Morrell JL, Feoktistova A, Gould KL. Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol Cell Biol. 2001;21(19):6681–94. doi: 10.1128/MCB.21.19.6681-6694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18(20):1594–9. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2193–8. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chow A, Hao Y, Yang X. Molecular characterization of human homologs of yeast MOB1. Int J Cancer. 2009;126(9):2079–89. doi: 10.1002/ijc.24878. [DOI] [PubMed] [Google Scholar]
- Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, 3rd, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181(1):79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueille N, Salimova E, Esteban V, Blanco M, Moreno S, Bueno A, Simanis V. Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J Cell Sci. 2001;114(Pt 14):2649–64. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18(7):1772–82. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Lahoz A, Munoz MJ, Moreno S, Jimenez J. Etd1p is a novel protein that links the SIN cascade with cytokinesis. EMBO J. 2005;24(13):2436–46. doi: 10.1038/sj.emboj.7600705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators ofMAP kinase cascades. Trends Cell Biol. 2001;11(5):220–30. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31(3):438–48. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Simanis V. The Schizosaccharomyces pombe cdc14 gene is required for septum formation and can also inhibit nuclear division. Mol Biol Cell. 1993;4(5):531–9. doi: 10.1091/mbc.4.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 1994;13(13):3011–9. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova A, Morrell-Falvey J, Chen JS, Singh NS, Balasubramanian MK, Gould KL. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol Biol Cell. 2012;23(9):1636–45. doi: 10.1091/mbc.E11-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Furge KA, Cheng QC, Jwa M, Shin S, Song K, Albright CF. Regions of Byr4, a regulator of septation in fission yeast, that bind Spg1 or Cdc16 and form a two-component GTPase-activating protein with Cdc16. J Biol Chem. 1999;274(16):11339–43. doi: 10.1074/jbc.274.16.11339. [DOI] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8(17):947–54. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Cortes JC, McCollum D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J Cell Biol. 2009;186(5):739–53. doi: 10.1083/jcb.200902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J Biol Chem. 2003;278(17):14591–4. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- Goyal A, Simanis V. Characterization of ypa1 and ypa2, the Schizosaccharomyces pombeorthologs of the peptidyl proyl isomerases that activate PP2A, reveals a role for Ypa2p in the regulation of cytokinesis. Genetics. 2012;190(4):1235–50. doi: 10.1534/genetics.111.138040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Takaine M, Simanis V, Nakano K. Dividing the spoils of growth and the cell cycle: The fission yeast as a model for the study of cytokinesis. Cytoskeleton (Hoboken) 2011;68(2):69–88. doi: 10.1002/cm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Hagan IM. Schizosaccharomyces pombe NIMA-related kinase, Fin1, regulates spindle formation and an affinity of Polo for the SPB. EMBO J. 2002;21(12):3096–107. doi: 10.1093/emboj/cdf294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 2004;18(9):1007–21. doi: 10.1101/gad.296204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123(1):75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4(3):310–27. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19(8):1803–15. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, McCollum D. Interaction between the noncatalytic region of Sid1p kinase and Cdc14p is required for full catalytic activity and localization of Sid1p. J Biol Chem. 2001;276(30):28185–9. doi: 10.1074/jbc.M103802200. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev Cell. 2002;3(6):779–90. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22(22):3205–16. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138(1):9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006;345(1):50–8. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- Hirabayashi S, Nakagawa K, Sumita K, Hidaka S, Kawai T, Ikeda M, Kawata A, Ohno K, Hata Y. Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene. 2008;27(31):4281–92. doi: 10.1038/onc.2008.66. [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149(5):1073–86. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol. 2004;24(8):3262–76. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10(10):619–22. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- Hu F, Elledge SJ. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle. 2002;1(5):351–5. [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107(5):655–65. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183(6):979–88. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert N, Navaratnam N, Augert A, Da Costa M, Martien S, Wang J, Martinez D, Abbadie C, Carling D, de Launoit Y, et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 2010;29(2):376–86. doi: 10.1038/emboj.2009.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro J, Saitou A, Duran A, Ribas JC. cps1+, a Schizosaccharomyces pombe gene homolog of Saccharomyces cerevisiae FKS genes whose mutation confers hypersensitivity to cyclosporin A and papulacandin B. J Bacteriol. 1997;179(24):7653–62. doi: 10.1128/jb.179.24.7653-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hallberg RL. Isolation and characterization of par1(+) and par2(+): two Schizosaccharomyces pombe genes encoding B’ subunits of protein phosphatase 2A. Genetics. 2000;154(3):1025–38. doi: 10.1093/genetics/154.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hallberg RL. Correct regulation of the septation initiation network in Schizosaccharomyces pombe requires the activities of par1 and par2. Genetics. 2001;158(4):1413–29. doi: 10.1093/genetics/158.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70(2):440–9. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Oballe J. Ethanol-hypersensitive and ethanol-dependent cdc- mutants in Schizosaccharomyces pombe. Mol Gen Genet. 1994;245(1):86–95. doi: 10.1007/BF00279754. [DOI] [PubMed] [Google Scholar]
- Jin QW, McCollum D. Scw1p antagonizes the septation initiation network to regulate septum formation and cell separation in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2003;2(3):510–20. doi: 10.1128/EC.2.3.510-520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J. 2011;30(2):341–54. doi: 10.1038/emboj.2010.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis J, Bimbo A, Rajagopalan S, Liu J, Balasubramanian MK. The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell. 2005;16(1):358–71. doi: 10.1091/mbc.E04-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis J, Oulton R, Young PG. The Scw1 RNA-binding domain protein regulates septation and cell-wall structure in fission yeast. Genetics. 2002;162(1):45–58. doi: 10.1093/genetics/162.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Shu S, Coppola MD, Kaneko S, Yuan ZQ, Cheng JQ. Regulation of proapoptotic mammalian ste20-like kinase MST2 by the IGF1-Akt pathway. PLoS One. 2010;5(3):e9616. doi: 10.1371/journal.pone.0009616. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim J, Luo G, Bahk YY, Song K. Cdc5-dependent asymmetriclocalization of bfa1 fine-tunes timely mitotic exit. PLoS Genet. 2012;8(1):e1002450. doi: 10.1371/journal.pgen.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Lu L, Shao R, Chin J, Liu B. Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics. 2006;173(2):685–96. doi: 10.1534/genetics.105.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Zeng CJ, Nayak T, Shao R, Huang AC, Oakley BR, Liu B. Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2009;20(12):2874–84. doi: 10.1091/mbc.E08-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Cano E, Simanis V. Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr Biol. 2003;13(2):168–72. doi: 10.1016/s0960-9822(02)01417-3. [DOI] [PubMed] [Google Scholar]
- Krapp A, Collin P, Cano Del Rosario E, Simanis V. Homoeostasis between the GTPase Spg1p and its GAP in the regulation of cytokinesis in S. pombe. J Cell Sci. 2008;121(Pt 5):601–8. doi: 10.1242/jcs.022772. [DOI] [PubMed] [Google Scholar]
- Krapp A, Collin P, Cokoja A, Dischinger S, Cano E, Simanis V. The Schizosaccharomyces pombe septation initiation network (SIN) is required for spore formation in meiosis. J Cell Sci. 2006;119(Pt 14):2882–91. doi: 10.1242/jcs.03025. [DOI] [PubMed] [Google Scholar]
- Krapp A, Del Rosario EC, Simanis V. The role of Schizosaccharomyces pombe dma1 in spore formation during meiosis. J Cell Sci. 2010;123(Pt 19):3284–93. doi: 10.1242/jcs.069112. [DOI] [PubMed] [Google Scholar]
- Krapp A, Schmidt S, Cano E, Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol. 2001;11(20):1559–68. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Lahoz A, Alcaide-Gavilan M, Daga RR, Jimenez J. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics. 2010;186(4):1261–70. doi: 10.1534/genetics.110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X, Buvelot S, Salimova E, Guerry F, Schmidt S, Cueille N, Cano E, Simanis V. The protein phosphatase 2A B′-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 2001;508(1):136–42. doi: 10.1016/s0014-5793(01)03047-2. [DOI] [PubMed] [Google Scholar]
- Le Goff X, Woollard A, Simanis V. Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol Gen Genet. 1999;262(1):163–72. doi: 10.1007/s004380051071. [DOI] [PubMed] [Google Scholar]
- Li C, Furge KA, Cheng QC, Albright CF. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J Biol Chem. 2000;275(19):14381–7. doi: 10.1074/jbc.275.19.14381. [DOI] [PubMed] [Google Scholar]
- Li WZ, Yu ZY, Ma PF, Wang Y, Jin QW. A novel role of Dma1 in regulating forespore membrane assembly and sporulation in fission yeast. Mol Biol Cell. 2010;21(24):4349–60. doi: 10.1091/mbc.E10-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6(3):1190–7. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Balasubramanian MK. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci. 2000;113 (Pt 7):1223–30. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, McCollum D, Balasubramanian MK. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153(3):1193–203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sugiura R, Yada T, Cheng H, Sio SO, Shuntoh H, Kuno T. Calcineurin is implicated in the regulation of the septation initiation network in fission yeast. Genes Cells. 2002;7(10):1009–19. doi: 10.1046/j.1365-2443.2002.00582.x. [DOI] [PubMed] [Google Scholar]
- Magidson V, Chang F, Khodjakov A. Regulation of cytokinesis by spindle-pole bodies. Nat Cell Biol. 2006;8(8):891–3. doi: 10.1038/ncb1449. [DOI] [PubMed] [Google Scholar]
- Mah AS, Elia AE, Devgan G, Ptacek J, Schutkowski M, Snyder M, Yaffe MB, Deshaies RJ. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J, Fankhauser C, Simanis V. Genetic interactions in the control of septation in Schizosaccharomyces pombe. J Cell Sci. 1992;101 (Pt 4):801–8. doi: 10.1242/jcs.101.4.801. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals systemwide modulation of protein-protein interactions. Sci Signal. 2009;2(84):ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- Millward TA, Hess D, Hemmings BA. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem. 1999;274(48):33847–50. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison JM. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1979;137(1):440–6. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Karagiannis J, Sevugan M, Singh P, Balasubramanian MK. The 14–3–3 protein rad24p modulates functionof the cdc14p family phosphatase clp1p/flp1p in fission yeast. Curr Biol. 2005;15(15):1376–83. doi: 10.1016/j.cub.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Mishra M, Karagiannis J, Trautmann S, Wang H, McCollum D, Balasubramanian MK. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minorperturbation of the cell division machinery in Schizosaccharomyces pombe. J Cell Sci. 2004;117(Pt 17):3897–910. doi: 10.1242/jcs.01244. [DOI] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2199–204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell JL, Tomlin GC, Rajagopalan S, Venkatram S, Feoktistova AS, Tasto JJ, Mehta S, Jennings JL, Link A, Balasubramanian MK, et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr Biol. 2004;14(7):579–84. doi: 10.1016/j.cub.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165(5):685–95. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill DP, Hyams JS. Cytokinetic actomyosin ring formation and septation in fission yeast are dependent on the full recruitment of the polo-like kinase Plo1 to the spindle pole body and a functional spindle assembly checkpoint. J Cell Sci. 2002;115(Pt 18):3575–86. doi: 10.1242/jcs.00031. [DOI] [PubMed] [Google Scholar]
- Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol Biol Cell. 1999;10(8):2771–85. doi: 10.1091/mbc.10.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 1996;15(23):6605–16. [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103(14):5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146(2):167–78. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9(9):1059–73. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8(7):1751–64. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6(1):1–10. [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20(22):6359–70. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Hidalgo L, Rozalen AE, Martin-Castellanos C, Moreno S. Slk1 is a meiosis-specific Sid2-related kinase that coordinates meiotic nuclear division with growth of the forespore membrane. J Cell Sci. 2008;121(Pt 9):1383–92. doi: 10.1242/jcs.023812. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39(1):86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res. 2010;70(3):1195–203. doi: 10.1158/0008-5472.CAN-09-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JA, Tomlin GC, McDonald WH, Snydsman BE, Muller EG, Yates JR, 3rd, Gould KL. Ppc89 links multiple proteins, including the septation initiation network, to the core of the fission yeast spindle-pole body. Mol Biol Cell. 2006;17(9):3793–805. doi: 10.1091/mbc.E06-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J Cell Sci. 2000;113 (Pt 10):1695–704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Sandrock B, Bohmer C, Bolker M. Dual function of the germinal centre kinase Don3 during mitosis and cytokinesis in Ustilago maydis. Mol Microbiol. 2006;62(3):655–66. doi: 10.1111/j.1365-2958.2006.05405.x. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11(12):1519–34. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Seshan A, Amon A. Linked for life: temporal and spatial coordination of late mitotic events. Curr Opin Cell Biol. 2004;16(1):41–8. doi: 10.1016/j.ceb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Singh NS, Shao N, McLean JR, Sevugan M, Ren L, Chew TG, Bimbo A, Sharma R, Tang X, Gould KL, et al. SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr Biol. 2011;21(23):1968–78. doi: 10.1016/j.cub.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104(25):10364–9. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10(21):2707–19. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12(1):84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Mach KE, Chen CY, Reynolds T, Albright CF. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J Cell Biol. 1996;133(6):1307–19. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146(4):777–90. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 2001;20(6):1259–70. doi: 10.1093/emboj/20.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin GC, Morrell JL, Gould KL. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol Biol Cell. 2002;13(4):1203–14. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol. 2001;11(12):931–40. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319(5859):97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93(3):938–49. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Malik R, Nigg EA, Korner R. Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal Chem. 2008;80(24):9526–33. doi: 10.1021/ac801708p. [DOI] [PubMed] [Google Scholar]
- Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7(3):1088–97. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5(5):723–34. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174(3):391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12(8):469–82. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- Xia H, Qi H, Li Y, Pei J, Barton J, Blackstad M, Xu T, Tao W. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21(8):1233–41. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- Yan H, Balasubramanian MK. A Meiotic Actin Ring (MeiAR) Essential for Proper Sporulation in Fission Yeast. J Cell Sci. 2012 doi: 10.1242/jcs.091561. [DOI] [PubMed] [Google Scholar]
- Yan H, Ge W, Chew TG, Chow JY, McCollum D, Neiman AM, Balasubramanian MK. The meiosis-specific Sid2p-related protein Slk1p regulates forespore membrane assembly in fission yeast. Mol Biol Cell. 2008;19(9):3676–90. doi: 10.1091/mbc.E07-10-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li DM, Chen W, Xu T. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene. 2001;20(45):6516–23. doi: 10.1038/sj.onc.1204817. [DOI] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2004;6(7):609–17. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- Yu LR, Zhu Z, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6(11):4150–62. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]