Abstract
We have developed GFam, a platform for automatic annotation of gene/protein families. GFam provides a framework for genome initiatives and model organism resources to build domain-based families, derive meaningful functional labels and offers a seamless approach to propagate functional annotation across periodic genome updates. GFam is a hybrid approach that uses a greedy algorithm to chain component domains from InterPro annotation provided by its 12 member resources followed by a sequence-based connected component analysis of un-annotated sequence regions to derive consensus domain architecture for each sequence and subsequently generate families based on common architectures. Our integrated approach increases sequence coverage by 7.2 percentage points and residue coverage by 14.6 percentage points higher than the coverage relative to the best single-constituent database within InterPro for the proteome of Arabidopsis. The true power of GFam lies in maximizing annotation provided by the different InterPro data sources that offer resource-specific coverage for different regions of a sequence. GFam’s capability to capture higher sequence and residue coverage can be useful for genome annotation, comparative genomics and functional studies. GFam is a general-purpose software and can be used for any collection of protein sequences. The software is open source and can be obtained from http://www.paccanarolab.org/software/gfam/.
INTRODUCTION
An important post-sequencing aim for a new genome project or model organism database is to use computational methods to annotate protein function (1,2,3). Comparisons of protein-coding sequences from complete genomes revealed that gene duplication, divergence and rearrangement are predominant mechanisms that drive the expansion of a species' protein complement during evolution (4–7). This means that proteins can be grouped into families, where members are likely to perform similar functions. While providing important clues on the molecular and cellular role of a protein, elucidating gene families in a genome is also crucial for understanding the evolutionary forces that shape genomes and drive speciation (8–10). Currently, there are two broad approaches to group individual proteins into families. (i) Domain-based approaches: resources such as Pfam (11), SUPERFAMILY (12), SMART (13), Gene3D (14) and HMMTIGR (15) consider domains as a structural and/or functional unit of evolution of a protein. Typically, these resources use a collection of hidden Markov models (HMMs) where each HMM represents a domain family that are used for annotating sequences without experimentally determined function(s). The domains described by these resources can comprise the entire protein or a portion of the sequence; however, in multi-cellular eukaryotes, most often, a protein is most frequently composed of more than one domain (16). It is important to note that Pfam comprises Pfam-A, which are curated families, and Pfam-B, which are automated families built from homologous sequence clusters. At this time, InterPro (17) integrates only Pfam-A families. For the rest of the article, we refer to domains derived from Pfam-A assignments as Pfam. InterPro (17) is a widely used meta-resource that provides such domain annotation and integrates protein signatures, repeats, families and patterns from 12 different resources—these include GENE3D (14), HAMAP (18), PANTHER (19), PIRSF (20), PRINTS (21), PROSITE patterns (22), PROSITE profiles (22), Pfam (11), ProDom (23), SMART (13), SUPERFAMILY (12) and TIGRFAMs (15). InterPro integrates different overlapping signatures that match the same set of proteins in the same region on the sequence, by placing them into a single entry. This grouping of equivalent signatures from different sources together provides a consistent way of looking at protein signatures. Although InterPro annotation can provide useful information about component domains and their functions, it falls short of providing consensus domain architecture for a protein sequence given a linear list of domain assignments from all these 12 resources. In other words, InterPro provides redundant information on component domain annotation for proteins. The inherent power of InterPro as a resource is that member databases have individual strengths and offer specific advantages in sequence annotation. This means that there will be sequence regions that are uniquely covered by specific resources. To our knowledge, there is no external database or stand-alone software that provides consensus domain architectures given InterPro annotation. In addition, there can also be gaps in sequence coverage outside InterPro domain coverage where proteins can have large regions without any annotation. Although this is necessarily not an area of immediate focus for InterPro or its component databases, we believe this need can be addressed if one wants consensus domain architecture. (ii) Sequence clustering approaches: several methods have been proposed to group sequences into protein families, which are based on clustering a graph in which nodes are the proteins and links between them are weighted with a measure related to the sequence similarity between the proteins. Clustering methods such as ProDom (23), ProClust (24), SYSTERS (25), TribeMCL (26), SCPS (27, 28), FORCE (29) and CluSTr (30) group proteins into families based on a set threshold of a sequence similarity measure [for instance, BLAST (31,32) E value or percent identity]; two protein sequences are considered potentially homologous if their similarity is above the chosen threshold. Although clustering-based methods consider whole sequences to generate families, they are not as sensitive as HMM-based methods in identifying distant evolutionary relationships. The choice of distance metric used in the clustering is a major limiting factor for detecting remote homologous relationships among proteins. Further, the multi-domain nature of proteins compounded by fold irregularities (33) such as domain insertion (34,35), circular permutation (36) and other non-contiguous sequence arrangement can complicate accurate family assignment. It is important to realize that these two approaches are complementary and offer specific advantages that be combined algorithmically to improve sequence annotation (11,37). The two approaches are complementary in the following respects. Although existing Pfam-A HMM models can be used to identify remote homologues, most of the new families added to the Pfam database during each release basically come from one of three sources: (i) a family seeded by a structure deposited in the Protein Data Bank—wwPDB that was not covered by the previous Pfam release. (ii) Pfam-B families that were used as a starting point for building Pfam-A, focusing particularly on Pfam-B clusters without a corresponding annotated family in InterPro. Pfam-B families are automatically derived and built from homologous sequence clusters using the Automatic Domain Decomposition Algorithm (ADDA) algorithm (37). (iii) In addition to these two sources, many curated Pfam-A families have been contributed by suggestions from the community. Several novel domains that subsequently became part of Pfam have been identified using homologous sequence searching and sequence analysis procedures that involve a clustering step to identify and describe novel protein families (38). For instance, the Pfam-A domain NYN (PF01936) was first identified and described by Anantharaman and Aravind (39) in 2006. Clustering approaches are adopted to provide an initial set of homologous sequences; however, to identify more distant homologues, these close homologues typically serve as seed sequences can then be used to build a HMM model that are then used to collect remote homologous sequences. Thus, the resolution at which these approaches operate are different and offer complementary advantages that can be combined to improve sequence annotation.
We have developed a hybrid method that combines the power of these two approaches to group proteins and provides a unified way to look at protein families. Specifically, we have developed GFam, a meta-tool that chains protein domain annotation provided by InterPro in a non-redundant manner with the power of sequence clustering methods to identify novel domains. GFam then uses this information to build consensus domain architecture for each sequence and subsequently classifies families based on common domain architecture. By consensus domain architecture, we refer to a unified domain arrangement derived from the different data sources in a way that avoids representing the same real domain with domain assignments from two or more data sources at the same time. When more than one data source classifies a fragment of the sequence as belonging to a specific domain, GFam resolves such overlaps and select one data source to be included in the consensus.
Although building families is important, it is equally desirable to describe a family’s function through a meaningful label and a system for database resources to propagate such annotation across genome releases. These labels contain information on the function of a protein in a highly condensed form; furthermore, in the absence of more specific protein names, these labels are frequently used in the definition line of a sequence file, providing a convenient way to quickly analyse and group gene sets resulting from large-scale transcriptomics, proteomics or metabolomics experiments, as well as serve as a source of function data for annotation of new genomes. In the following sections, we describe in detail, the implementation of GFam to derive family assignment for the proteome of Arabidopsis, the coverage we obtain from GFam assignment, the functional labels we curated for these families and a workflow to transfer such annotation seamlessly across genome updates.
MATERIALS AND METHODS
Implementation of GFam: steps of the GFam pipeline
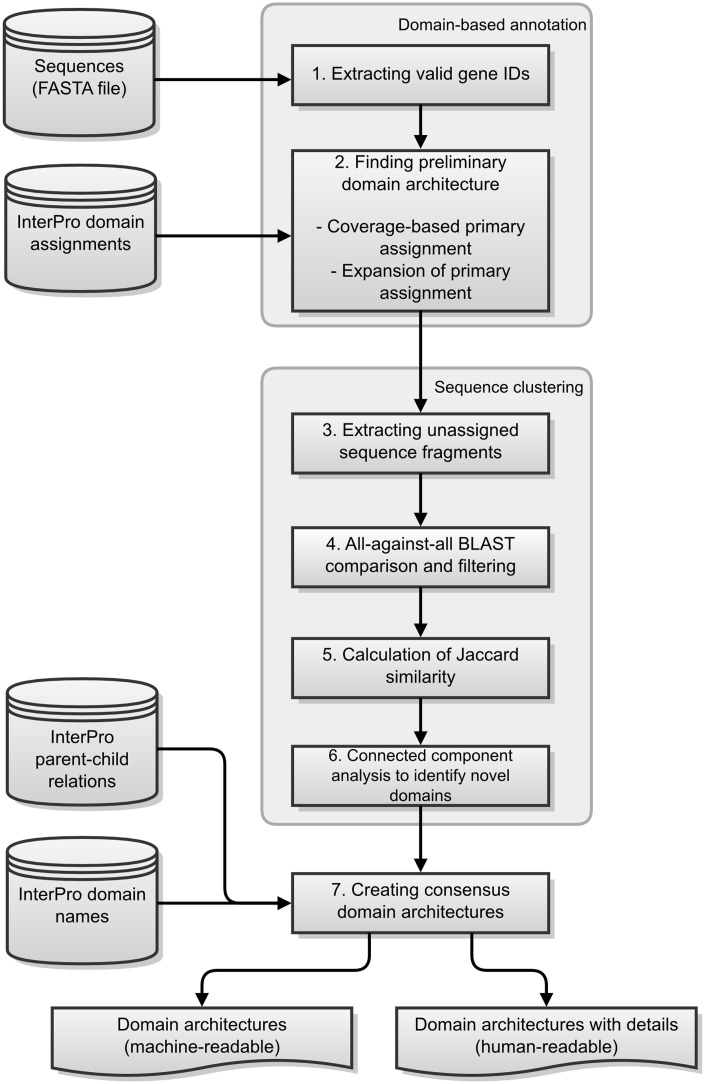
The GFam pipeline consists of multiple steps. In this section, we will describe the input files on which the GFam pipeline operates, the order in which the steps are executed and the output files produced. First, a short overview of the whole process will be given, followed by a more detailed description of each step. Figure 1 provides a schematic of the various steps of the GFam pipeline.
Figure 1.
Schematic of the various steps in the GFam pipeline.
Overview of a GFam analysis
GFam infers annotations for sequences by first finding consensus domain architecture for each sequence. The calculation of the consensus domain architecture is complex as GFam has to account not only for the known domain assignments by integrating the various InterPro resources but also for the possible existence of novel, previously uncharacterized domains. The whole pipeline can be broken down into seven steps as follows:
Extract valid protein identifiers from the sequence file.
Determine preliminary domain architecture for each sequence by considering known domains from the domain assignment file. The domain assignment file is a raw output of an InterProScan (40) run on the sequences of interest. InterProScan is a tool that combines the protein function recognition methods of InterPro member databases into one application.
Find unassigned regions of each sequence; i.e. the regions that are not assigned to any domain in the preliminary domain architecture.
Run an all-against-all BLAST comparison of the unassigned sequence fragments and filter BLAST results to determine which fragments may correspond to the same novel domain. Such filtering is based primarily on E value and alignment length. At this point, we obtain a graph where each fragment is a node, and two fragments are connected if they passed the BLAST filter.
Calculate a similarity score for every pair of valid fragment connected by an edge and remove those connections with a low similarity.
Find connected components of the remaining graph. Each connected component will correspond to a putative novel domain.
Calculate consensus domain architecture by merging the preliminary domain architecture with the newly detected novel domains.
These steps and the required input files will be described more in detail in the next few subsections.
Step 1—extracting valid protein identifiers
In this step, the input sequence file is read, and the protein identifiers are extracted from the definition line of FASTA-formatted sequences. The protein identifier is assumed to be the first word of the definition line. If the definition lines in the original FASTA file follow some other format, one can supply a regular expression in the configuration file that can be used to extract the actual Identifier (ID) from the definition line.
Step 2—preliminary domain architecture
This step processes the output file with domain assignments from the raw output of an InterProScan run and determines preliminary domain architecture for each sequence. This step will include only domains from the InterProScan output. Domain architectures for each sequence are determined in isolation, so the domain architecture of one sequence has no effect on another.
For each sequence, we first collect the set of domain assignments from the domain assignment file. Each assignment has a data source (e.g. HMMPfam, SUPERFAMILY and HMMSmart), a domain ID according to the schema of the source, the starting and ending indices of the domain in the amino acid chain, an InterPro ID to which the domain ID is mapped and an E value. First, the list is filtered based on E values, where one might apply different E value thresholds for different data sources. This leads to a list of trusted domain assignments that are unlikely to be artefacts. After that, GFam performs multiple passes on the list of trusted domain assignments, starting with a subset focused on more informative data sources. Less informative data sources are used in the later stages, and it is possible that some data sources are not considered at all. Data sources are classified as ‘more informative’ and ‘less informative’ based on expert input; for instance, to obtain final GFam assignment for Arabidopsis, we ignored assignments from Seg, Coil, HAMAP, FPrintscan and PatternScan as they are not informative for various reasons. We explain these reasons in the next section. Because such a classification of ‘more informative’ and ‘less informative’ data sources can be different for different types of applications, users have the option to take a look at all possible data sources and decide which ones should be used at which stages of the algorithm.
During the first pass, one single data source that gives the highest coverage for the sequence on its own is selected. This data source will be referred to as the primary data source, and the domains of the primary data source will be called the primary assignment. After the first pass, the primary assignment will be extended by domains from other data sources in a greedy manner using the following rules. These rules were derived to provide an optimum solution by taking into account several factors, most importantly coverage and uniform annotation.
Larger domains from other data sources will be considered first. (In other words, the remaining assignments not included already in the primary assignment are sorted by length in descending order.)
Domains are considered one at a time for addition to the primary assignment.
If a domain is the exact duplicate of some other domain already added (in the sense that it starts and ends at the same amino acid index), the domain is excluded from further consideration.
If a domain to be added overlaps with an already added domain from another data source, the domain is excluded from further consideration. Herein, a domain from another data source may refer either to a domain that was already selected from the primary data source or a domain that was selected during the secondary (extension) phase. In one stage of the extension phase, we process domains in decreasing length from all ‘enabled’ data sources, and it may happen that a domain that was selected in the extension phase already occupies a place where GFam tries to add a shorter domain from another data source. Herein, ‘enabled’ data sources refer to InterPro data sources that are considered for inclusion in a GFam run by the user.
If a domain to be added is contained completely within another domain from the same data source, it is added to the primary assignment and the process continues with the next domain from Step 2. Note that the opposite cannot happen, as we consider domains in decreasing order of their sizes.
If a domain to be added overlaps partially with an already added domain from the same data source, the size of the overlap determines how it will be resolved. If the overlap size is smaller than a given threshold, the domain will be added, and the process continues from Step 2. Otherwise, the new domain is excluded from further consideration, and the process continues from Step 2 until there are no more domains left in the current stage.
We call this six-step procedure the expansion of a primary assignment. We recall that GFam works in multiple stages: the first stage creates the primary assignment with a limited set of trusted data sources, and the second stage expands the primary assignment with an extended set of data sources. Following stages may be necessary with even more extended sets of data sources. For Arabidopsis, we found the following strategy to be successful:
Assignments from HAMAP, PatternScan, FPrintScan (41), Seg (low-complex regions) (42) and Coil (coiled-coil prediction) (43) were discarded for the following reasons: (i) HAMAP may not be a suitable resource for eukaryotic family annotation as it is geared towards completely sequenced microbial proteome sets and provides manually curated microbial protein families in UniProtKB/Swiss-Prot (44). For Arabidopsis, there were only 133 domains annotated by HAMAP, and all domains had E values larger than 0.001. (ii) PatternScan and FPrintScan are resources for identifying motifs in a sequence and are more limited in use for understanding larger evolutionary units or domains. The match size ranged between 3 and 103 amino acids for PatternScan and between 4 and 30 amino acids for FPrintScan and, hence, is too short. (iii) Seg and Coil were ignored as these define regions of low-compositional complexity and coiled coils, respectively, and are not particularly informative in the context of defining protein families.
An E value threshold of 10−3 was applied to the remaining data sources, except for SUPERFAMILY, HMMPanther, Gene3D and HMMPIR did not need a set threshold, as domains from these resources had an E value less than 10−3. The threshold of 10−3 was chosen based on the following observation. From the InterProScan domain assignment file for The Arabidopsis Information Resource, genome release version 9 (TAIR9) proteins, there were 3816 domain assignments from HMMPfam with an E value larger than 0.1, 1625 assignments with an E value between 0.1 and 0.01 and 1650 assignments with an E value between 0.01 and 0.001. We looked at the type of domains that had an E value between 0.1 and 0.01 and also domains with E value between 0.01 and 0.001. We observed that at least 80% of these domains were some type of repeat domains (Pentatricopeptide repeat (PPR), Kelch, Leucine Rich Repeat (LRR), Armadillo, Tetratricopeptide repeat (TPR), etc.) or short protein motifs (different types of zinc fingers, EF-hand, Helix Loop Helix (HLH), etc.). It is reasonable to believe that, at an E value greater than 0.001, the majority of the domains are likely to be spurious matches due to the sequence nature (low complex and short) of these domains. Hence, we decided to consider domains from HMMPfam that had an E value of 0.001 or smaller. Given this observation, we may miss a small number of real domains if we choose 0.001 as our E value threshold. However, we point out that this threshold is not hard wired into GFam, rather it is a parameter than can be adjusted for each assignment source to suit the user’s needs.
We allowed GFam to perform three passes on the list of domain assignments obtained using the steps earlier. The first and second passes did not consider HMMPanther and Gene3D assignments; among all resources, HMMPanther had the smallest number (7.5%) of signatures integrated into InterPro; Gene3D was similar to SUPERFAMILY in the nature of assignment they provide (HMMs based on proteins of known structure), although the sequence and residue coverage provided by SUPERFAMILY (56% and 42%, respectively) were much better than Gene3D (45% and 28%, respectively). Thus, GFam offers the flexibility to incorporate user-specific informed choices to chain domains. The third stage considers all the data sources.
The maximum overlap we allowed between two domains of the same source (excluding complete insertions which were always accepted) was 30 amino acids. This was based on the distribution of domain overlap lengths for the different resources.
The stages and the E value thresholds are configurable in the configuration file.
Step 3—finding unassigned sequence fragments
This step begins the exploration for novel, previously uncharacterized domains among the sequence fragments left uncovered by the preliminary assignment that we calculated in Step 2. We modified the method described by Haas et al., (45) to identify novel domains. This step iterates over each sequence and extracts the fragments that are not covered by any of the domains in the preliminary domain assignment. Sequences or fragments that are too short are discarded, and the remaining fragments are written in FASTA format into an intermediary file. The sequence and fragment length thresholds are configurable. For the analysis of Arabidopsis sequences, the minimum fragment length was set to 75 amino acids.
Step 4—all-against-all BLAST comparison and filtering
This step uses the external NCBI BLAST executables (namely formatdb and blastall) to determine pairwise similarity scores between the unassigned sequence fragments. First, a database is created from all sequence fragments using formatdb in a temporary folder, and then a BLAST query is run on the database with the same set of unassigned fragments using blastall -p blastp. Matches with a sequence percent identity or an alignment length less than a given threshold are thrown away, so are matches with an E value larger than a given threshold. The user may choose between using un-normalized alignment lengths or normalized alignment lengths with various normalization methods (normalizing with the length of the smaller, the larger, the query or the hit sequence).
For Arabidopsis protein sequences from TAIR9 and TAIR10 genome releases, the following settings were used:
Minimum sequence identity: 45%.
Minimum normalized alignment length: 0.7 (normalization was done by the length of the query sequence).
Maximum E value: 10−3.
Step 5—calculation of similarity
After the Step 4, we have essentially obtained a graph representation of similarity relations between unassigned sequence fragments. In this graph representation, each sequence fragment is a node, and two fragments are connected by an edge if they passed the BLAST filter in Step 4. GFam looks for tightly connected regions in the graph to identify sequence fragments that potentially contain the same novel domain. We assume that if two sequences contain the same novel domain, their neighbour sets in the similarity graph should be very similar. To quantify the similarity between two sequences in the similarity graph, we use the Jaccard similarity of the neighbour sets of the sequences. More precisely, let i and j denote two nodes in a graph and let Γi denote the set consisting of i itself and i’s neighbours in the graph. The similarity of i and j is then defined as follows:
where we note that the formula above is essentially the Jaccard similarity of Γi and Γj. We calculate the similarity of each connected pairs of nodes and keep those which have a similarity larger than 0.66. This corresponds to keeping pairs where approximately 2/3rd of their neighbours are shared.
Step 6—identification of novel domains
Having obtained the graph filtered by Jaccard similarity in Step 5, we detect the connected regions of this graph by performing a simple connected component analysis. In other words, sequence fragments corresponding to the same connected component of the filtered graph are assumed to belong to the same novel domain. Note that these novel domains should be treated with care, as some may belong to those that were already characterized in the original input domain assignment file but were filtered in Step 2.
Novel domains are given temporary IDs consisting of the string NOVEL and a five-digit numerical identifier; for instance, NOVEL00042 is the 42nd novel domain found during this process. Components containing less than four sequence fragments are not considered novel domains. Again, the parameters we used for this analysis are not hard wired and can be changed depending on users’ needs.
Step 7—consensus domain architecture
This step determines the final consensus domain architecture for each sequence by starting from the preliminary domain architecture obtained in Step 2 and extending it with the novel domains found for the given sequence. The consensus domain architectures are written into two files, one containing a simpler flat-file representation of the consensus architectures suitable for further processing, whereas the other file contains a detailed domain architecture description with InterPro IDs and human-readable descriptions for each domain in each sequence. This latter file also lists the primary data source for the sequence, the coverage of the sequence with and without novel domains and also the number of the stage in which each domain was selected into the consensus assignment.
PSEUDO-CODE FOR GFam ALGORITHM
Main algorithm
Input
A, the list of domain annotations for the sequence being analysed. Each domain annotation contains the following information:
-
-
Index of the first residue
-
-
Index of the last residue
-
-
InterPro data source (e.g. HMMPfam, HMMTIGR and SUPERFAMILY) or ‘NOVEL’ if the domain is a putative novel domain found in the clustering step
-
-
ID of the domain type according to the original data source
-
-
ID of the domain type in InterPro (optional)
-
-
E value (optional)
S, a list of stages, each stage containing a list of data sources allowed in that stage
E, the list of InterPro data sources excluded from further analysis.
F, a set of E value filters for InterPro data sources.
o, the maximum allowed length of overlaps between two domains in the consensus architecture
Output
C, a subset of A that contains the consensus architecture.
Steps
-
1.
Let C = [] (i.e. an empty list).
-
2.
Let A1 = the annotations in A whose data source is not in E
-
3.
Let A2 = the annotations in A1 that do not contain E values or whose E values are lower than the thresholds prescribed by F (where the threshold may be infinite)
-
4.
Create a hash table H which maps InterPro data sources to the corresponding domain assignments in A2
-
5.
Find the data source DS that is in S[0] and whose assignments in H cover the most residues in the sequence.
-
6.
For every annotation ann in H[DS], try adding ann to C according to sub-algorithm Domain-addition(ann, C, o).
-
7.
For each stage starting from S[1], do:
-
7/a.
Let A3 = the annotations in A2 whose data sources are allowed in the stage being examined.
-
7/b.
Sort the annotations in A3 by decreasing length.
-
7/c.
For each annotation ann in A3, try adding it to C according to sub-algorithm Domain-addition(ann, C, o). Note that Domain-addition may refuse to add ann to C.
-
8.
Return C as the consensus domain architecture.
Domain addition
Input
ann, a domain annotation containing the same information as outlined in the main algorithm.
C, a partial consensus domain architecture containing zero or more annotations.
o, the maximum allowed overlap length
Output
True if the annotation can be added to C, false otherwise. When true is returned, C is also modified in place.
Steps
-
1.
If ann has exactly the same starting and ending indices as some other annotation in C, then ann is a duplicate annotation; return false.
-
2.
If there exists ann2 such that ann.start_index <= ann2.start_index and ann.end_index >= ann2.end_index, then do the following:
-
2/a.
If ann2 is from the same InterPro data source as ann, then this is a valid domain insertion (ann2 was inserted into ann); add ann to C and return true.
-
2/b.
Otherwise, return false.
-
3.
If there exists ann2 such that ann2.start_index <= ann.start_index and ann2.end_index >= ann.end_index, then do the following:
-
3/a.
If ann2 is from the same InterPro data source as ann, then this is a valid domain insertion (ann was inserted into ann2); add ann to C and return true.
-
3/b.
Otherwise, return false.
-
4.
If ann2.start <= ann.start and ann2.end <= ann.end and ann2.end >= ann.start, then do the following:
-
4/a.
If ann2 is from the same InterPro data source as ann, then this is a partial overlap between two detected domains where ann2 is to the left of ann. If the length of the overlap is at most o, add ann to C and return true.
-
4/b.
Otherwise, return false.
-
5.
If ann2.start >= ann.start and ann2.end >= ann.end and ann2.end <= ann.start, then do the following:
-
5/a.
If ann2 is from the same InterPro data source as ann, then this is a partial overlap between two detected domains where ann2 is to the right of ann. If the length of the overlap is at most o, add ann to C and return true.
-
5/b.
Otherwise, return false.
-
6.
Otherwise, there is no (partial or complete) overlap between ann and any of the annotations already in C, so add ann to C and return true.
RESULTS
In this work, we achieved the following goals that are practically useful for genome databases. We have developed GFam, a tool that integrates InterPro domains from 12 protein signature annotation resources using a greedy domain chaining algorithm followed by sequence-based clustering of InterPro un-annotated sequence fragments to derive a consensus domain architecture and families using those domain architectures. We used GFam to generate protein family assignments for several model organism genomes. This includes the mammalian model of Mus musculus (mouse) and non-mammalian models of Arabidopsis thaliana (mouse-ear cress), Arabidopsis lyrata (Lyre-leaved rock-cress), Neurospora crassa (filamentus fungi), Dictyostelium discoideum (social amoebae), Caenorhabditis elegans (round worm), Daphnia pulex (water flea), Drosophila melanogaster (fruit fly), Danio rerio (Zebrafish) and Gallus gallus (chicken). GFam assignments for all the above species are provided in the Supplementary Data. We obtained consistent results across all model genomes in terms of increased sequence and residue coverage relative to the best single-constituent resource within InterPro. For the purpose of this article, we have taken The Arabidopsis Information Resource (TAIR) as a model organism database and the proteome of the model organism Arabidopsis thaliana that TAIR annotates to demonstrate the practical utility of GFam. In addition to generating protein family assignments for Arabidopsis, we used short descriptions for proteins from the TAIR9 genome release (46), Uniprot protein descriptions for each protein, domain descriptions from InterPro and its member databases and Gene Ontology (GO) molecular function terms provided by InterPro2GO (17) annotation to derive functional labels for families generated by GFam for proteins in the TAIR9 release of the Arabidopsis genome. We also established a semi-automated system to transfer the curated functional labels generated for TAIR9 families to GFam-derived families for Arabidopsis proteome in the TAIR10 release. The families and their corresponding short descriptions have been incorporated into the TAIR10 genome update. We discuss these results in detail in the following sections.
Sequence and residue coverage for GFam
One of the most important objectives behind developing GFam as a unified system was to provide consensus domain architecture that maximizes annotation coverage provided by InterPro member databases in a meaningful way for a given sequence. This can be very useful and informative as the member databases have individual strengths and offer specific advantages in sequence annotation. Although all the methods aim to annotate and classify protein sequences, diagnostically, these resources have different areas of optimum application owing to the different underlying assumptions, data sources and analysis methods (17). PANTHER, PIRSF, Pfam, SMART, TIGRFAMs, Gene3D and SUPERFAMILY as a group of methods use HMMs to identify and annotate remote homologous relationships. Pfam is a widely used, comprehensive database of almost 12 000 conserved protein families across all kingdoms of life (11). SMART is a database that provides annotation of domains from signalling and extracellular protein sequences and allows the identification and annotation of genetically mobile domains and the analysis of domain architectures in multi-cellular organisms (13). PANTHER (Protein ANalysis THrough Evolutionary Relationships) is a database of phylogenetic trees of protein-coding gene families where likely functional divergence events are identified and used to classify protein families (19). SUPERFAMILY (12) and Gene3D (14) are based on a collection of HMMs derived using protein domains at the superfamily/H level based on the hierarchical protein structure classification schemes SCOP (47,48) and CATH (49), respectively. The annotation resource PIRSF (20) uses HMMs over the full length of a protein rather than on the component domains. By integrating these individual resources, InterPro (17) capitalizes on their specific advantages, producing a powerful integrated database along with a search tool InterProScan (40). To assess how well GFam captures such integrated annotations, we calculated sequence and residue coverage for TAIR9 and TAIR10 proteins for all member resources in InterPro and from GFam. Tables 1 and 2 describe coverage statistics for both sequence and residue, for each of the 12 individual member resources, all resources pooled together and for consensus domain architecture derived using GFam for TAIR9 and TAIR10 data sets, respectively. To understand the contribution of novel domains and also to avoid any type of bias towards GFam, we calculate four types of sequence and residue coverage for annotation for GFam. (i) GFam_NoFilter: We calculated coverage provided by GFam considering domain annotation provided by member resources as is, without any filtering on E values or domain lengths. In addition, we also included coverage provided by novel domains. (ii) GFam_NoFilter_No-novel: This is similar to (i) but we excluded the coverage provided by novel domains. (iii) GFam_WithFilter: we calculated coverage using filters we described previously. Specifically, we excluded domains from HAMAP, PatternScan and FPrintScan; we applied an E value threshold of 10−3 to collect domains from the various data sources. (iv) GFam_WithFilter_No-novel: this is similar to (iii) but excluding novel domains. For calculating novel domains, we used a minimum sequence identity of 45%, minimum normalized alignment length of 0.7, maximum E value of 10−3 and a Jaccard coefficient of 0.66.
Table 1.
Coverage statistics for TAIR9 assignment
| InterPro data source | Total annotated sequences (27 379) | Sequence coverage | Residue coverage |
|---|---|---|---|
| BlastProDom | 425 | 0.0155 | 0.0031 |
| FPrintScan | 3686 | 0.1346 | 0.0285 |
| Gene3D | 12 293 | 0.4490 | 0.2799 |
| HAMAP | 133 | 0.0049 | 0.0035 |
| HMMPIR | 1228 | 0.0449 | 0.0469 |
| HMMPANTHER | 14 973 | 0.5469 | 0.4687 |
| HMMPfam | 20 859 | 0.7619 | 0.4120 |
| HMMSMART | 7809 | 0.2852 | 0.1120 |
| HMMTIGR | 3105 | 0.1134 | 0.0874 |
| PatternScan | 5221 | 0.1907 | 0.0116 |
| ProfileScan | 8798 | 0.3213 | 0.1466 |
| SUPERFAMILY | 15 399 | 0.5624 | 0.4174 |
| All | 22 591 | 0.8251 | 0.6932 |
| GFam_NoFilter | 22 826 | 0.8337 | 0.6147 |
| GFam_NoFilter_No-novel | 22 591 | 0.8251 | 0.5906 |
| GFam_WithFilter | 22 634 | 0.8267 | 0.6065 |
| GFam_WithFilter_No-novel | 22 382 | 0.8175 | 0.5809 |
Sequence coverage from GFam output for TAIR9 proteome was calculated from the number of sequences having at least one domain divided by the total number of sequences (the number in parenthesis in the table header). Residue coverage was calculated from the number of residues covered by at least one domain divided by the total number of residues in all the sequences. GFam_NoFilter describes coverage provided by GFam considering domain annotation provided by member resources as is. In addition, we also included coverage provided by novel domains. GFam_NoFilter_No-novel is similar to GFam_NoFilter after excluding coverage from novel domains. GFam_WithFilter describes coverage calculated after using filters (described in the text). GFam_WithFilter_No-novel is similar to GFam_WithFilter after excluding coverage from novel domains.
Table 2.
Coverage statistics for TAIR10 assignment
| InterPro data source | Total annotated sequences (27 416) | Sequence coverage | Residue coverage |
|---|---|---|---|
| BlastProDom | 425 | 0.0155 | 0.0031 |
| FPrintScan | 3687 | 0.1345 | 0.0283 |
| Gene3D | 12 308 | 0.4489 | 0.2796 |
| HAMAP | 145 | 0.0053 | 0.0039 |
| HMMPIR | 1238 | 0.0452 | 0.0472 |
| HMMPanther | 14 998 | 0.5471 | 0.4684 |
| HMMPfam | 20 889 | 0.7619 | 0.4113 |
| HMMSMART | 7828 | 0.2855 | 0.1123 |
| HMMTIGR | 3102 | 0.1131 | 0.0871 |
| PatternScan | 5216 | 0.1903 | 0.0115 |
| ProfileScan | 8821 | 0.3217 | 0.1464 |
| SUPERFAMILY | 15 420 | 0.5624 | 0.4170 |
| All | 22 622 | 0.8251 | 0.6924 |
| GFam_NoFilter | 22 866 | 0.8340 | 0.6142 |
| GFam_NoFilter_No-novel | 22 622 | 0.8251 | 0.5898 |
| GFam_WithFilter | 22 680 | 0.8273 | 0.6057 |
| GFam_WithFilter_No-novel | 22 419 | 0.8177 | 0.5798 |
Sequence coverage from GFam output for TAIR10 proteome was calculated from the number of sequences having at least one domain divided by the total number of sequences (the number in parenthesis in the table header). Residue coverage was calculated from the number of residues covered by at least one domain divided by the total number of residues in all the sequences. GFam_NoFilter describes coverage provided by GFam considering domain annotation provided by member resources as is. In addition, we also included coverage provided by novel domains. GFam_NoFilter_No-novel is similar to GFam_NoFilter after excluding coverage from novel domains. GFam_WithFilter describes coverage calculated after using filters (described in the text). GFam_WithFilter_No-novel is similar to GFam_WithFilter after excluding coverage from novel domains.
For TAIR9 and TAIR10 data sets, novel domains contribute to ∼5% of the total sequence coverage and 4% of the total residue coverage (Tables 3 and 4) provided by GFam. The filters used in our analysis do not affect the coverage significantly as the difference in sequence and residue coverage between (i) and (iii) and (ii) and (iv) contribute ∼1%. Although the choice of filters used in our work do not affect the final coverage much, they can contribute to providing a clean set of families by avoiding false positive domain annotation. To assess the coverage provided by GFam with respect to the member resources individually, we describe the coverage given by GFam_NoFilter. As Tables 1 and 2 reveal, the sequence coverage provided by GFam is 7.2% more than the next best coverage (provided by Pfam), whereas the residue coverage provided by GFam is 14.6% more than the next best coverage (provided by HMMPANTHER) within InterPro data sources. If we exclude the contribution of novel domains, the sequence coverage provided by GFam is 6.3% more than the best single-constituent domain annotation resource within InterPro (Pfam), whereas the residue coverage is 12.2% higher compared with the best single-constituent resource (HMMPANTHER).
Table 3.
Contribution of individual resources to GFam residue coverage for TAIR9 proteome
| InterPro data source | Total domains from Inter ProScan output | Total domains after GFam | Total residues from domains after GFam | Residue coverage |
|---|---|---|---|---|
| BlastProDom | 434 | 139 | 13 448 | 0.0019 |
| FPrintScan | 19 462 | 483 | 8702 | 0.0013 |
| Gene3D | 17 619 | 26 | 2414 | 0.0003 |
| HAMAP | 133 | 55 | 14 600 | 0.0021 |
| HMMPIR | 1228 | 1163 | 498 641 | 0.0718 |
| HMMPANTHER | 25 216 | 467 | 109 131 | 0.0157 |
| HMMPfam | 36 617 | 8939 | 1 466 666 | 0.2113 |
| HMMSMART | 15 630 | 1965 | 163 298 | 0.0235 |
| HMMTIGR | 7430 | 1625 | 459 902 | 0.0663 |
| PatternScan | 7323 | 56 | 870 | 0.0001 |
| ProfileScan | 19 072 | 4576 | 328 557 | 0.0473 |
| SUPERFAMILY | 22 405 | 16 382 | 3 607 647 | 0.5197 |
| Novel | NA | 1530 | 267 824 | 0.0386 |
| Total | 172 569 | 37 406 | 6 941 700 |
The number of domains from InterProScan output for each of the 12 resources, the number of domains that were incorporated into the final GFam assignment and their residue coverage.
Table 4.
Contribution of individual resources to GFam residue coverage for TAIR10 proteome
| InterPro data source | Total domains from InterPro Scan output | Total domains after GFam | Total residues from domains after GFam | Residue coverage |
|---|---|---|---|---|
| BlastProDom | 519 | 139 | 13 468 | 0.0019 |
| FPrintScan | 24 917 | 475 | 8585 | 0.0012 |
| Gene3D | 23 290 | 26 | 2414 | 0.0003 |
| HAMAP | 191 | 59 | 16 304 | 0.0023 |
| HMMPIR | 1700 | 1172 | 503 950 | 0.0726 |
| HMMPANTHER | 33 878 | 472 | 109 076 | 0.0157 |
| HMMPfam | 46 991 | 8933 | 1 467 716 | 0.2114 |
| HMMSMART | 20 682 | 1962 | 164 732 | 0.0237 |
| HMMTIGR | 8660 | 1610 | 459 488 | 0.0662 |
| PatternScan | 9662 | 56 | 867 | 0.0001 |
| ProfileScan | 24 147 | 4630 | 328 655 | 0.0473 |
| SUPERFAMILY | 29 568 | 16 394 | 3 610 190 | 0.5201 |
| Novel | NA | 1546 | 270 894 | 0.0390 |
| Total | 224 205 | 37 474 | 6 956 339 |
The number of domains from InterProScan output for each of the 12 resources, the number of domains that were incorporated into the final GFam assignment and their residue coverage.
Although gaining 7.2 and 6 percentage points (from GFam_NoFilter and GFam_NoFilter_No-novel, respectively) in sequence coverage can be useful for functional annotation, we believe that the true power of GFam lies in maximizing the annotation provided by the different resources when the resources cover different regions of a sequence. This is evident, as, even if we ignore the contribution of novel domains, the residue coverage that GFam provides is at least 12.2 percentage points higher than the best single-constituent database within InterPro. Tables 3 and 4 provide the individual contribution of each of the 12 member resources towards total residue coverage for TAIR9 and TAIR10 GFam assignments, respectively. SUPERFAMILY has by far the biggest contribution to total residue coverage (52%) followed by Pfam (21%); this observation remains consistent for TAIR9 and TAIR10 GFam assignment. Tables 3 and 4 also reveal that 21% (TAIR9) and 16% (TAIR10) of the total InterPro domains assigned to the proteome become part of the final domain architecture. Table 5 provides a summary of GFam sequence and residue coverage values for several model genomes. On average, GFam provides 8.4% increase in sequence coverage and 7.4% increase in residue coverage compared with the best single-constituent domain annotation resource within InterPro. Details of annotation including domain architectures, various coverages and contribution of individual resources can be obtained from Supplementary Data provided at the website mentioned under the section ‘Availability and Requirements’.
Table 5.
GFam sequence and residue coverage for model genomes
| Species | Sequence coverage |
Residue coverage |
||||
|---|---|---|---|---|---|---|
| A | B | C | A | B | C | |
| Mus musculus | 0.6790 | 0.6367 | 0.5915 (HMMPanther) | 0.6497 | 0.6233 | 0.5861 (HMMPanther) |
| Danio rerio | 0.8904 | 0.8767 | 0.8440 (HMMPanther) | 0.8229 | 0.8053 | 0.7672 (HMMPanther) |
| Neurospora crassa | 0.6979 | 0.6975 | 0.6310 (HMMPfam) | 0.5977 | 0.5974 | 0.5023 (HMMPanther) |
| Caenorhabditis elegans | 0.6831 | 0.6742 | 0.5884 (HMMPfam) | 0.6638 | 0.6574 | 0.6050 (HMMPanther) |
| Drosophila melanogaster | 0.7235 | 0.6955 | 0.6437 (HMMPanther) | 0.6739 | 0.6193 | 0.5851 (HMMPanther) |
| Dictyostelium discoideum | 0.7020 | 0.6698 | 0.5400 (HMMPfam) | 0.6673 | 0.6370 | 0.5674 (HMMPanther) |
| Gallus gallus | 0.7737 | 0.7596 | 0.7276 (HMMPanther) | 0.8089 | 0.8012 | 0.7687 (HMMPanther) |
| Daphnia pulex | 0.5109 | 0.5109 | 0.4230 (HMMPfam) | 0.5041 | 0.4873 | 0.4165 (HMMPanther) |
Sequence and residue coverage for several model genomes using GFam and the best single-constituent InterPro resource. A, GFam_NoFilter; B, GFam_NoFilter_No-novel and C, best single-constituent resource within InterPro.
Although GFam can (by design) capture 100% sequence coverage provided by all 12 InterPro data resources, obtaining 100% residue coverage in a non-redundant manner is still a challenge. The choice of parameters for chaining domains during the primary and expansion phases of the algorithm can have varied effects on the final residue coverage. Also, domain boundaries predicted by HMM-based models can have large overlaps (>30 amino acids), and although GFam allows setting overlap threshold, it is possible that we may miss coverage on complicated cases of domain fusions. We hope to document these cases systematically and look into incorporating these aspects in future.
The increased sequence and residue coverage that we obtain after final GFam assignment is purely a result of integrating InterPro annotation without compromising the accuracy of annotation. The sensitivity and accuracy for GFam annotation that involves any InterPro domain (other than a novel domain as defined by GFam) is determined by the InterPro data source that provided the domain annotation in the first place. GFam uses InterPro annotation ‘as is’ although the choice of data source to consider and E value threshold is configurable. As we explain in the section ‘Implementation of GFam: steps of the GFam pipeline’, using stringent filters on InterPro annotation did not affect sequence coverage when compared with using InterPro domain annotation ‘as is’ for final GFam assignment. There are several examples (full details can be obtained from the Supplementary Data Files) to demonstrate that the increase in sequence and residue coverage provided by GFam relative to the best single-constituent database within InterPro is after integration of the various data sources.
During Step 2, where the preliminary domain architecture for each sequence is determined, the first pass of GFam pipeline (InterPro data parsing) selects one single data source that gives the highest residue coverage for the sequence on its own. This step alone ensures that the overall residue coverage has to be better than the coverage relative to the best single-constituent InterPro data source.
For the TAIR9 proteome final GFam assignment, 21 086 sequences use one data source, 1255 sequences use two data sources and 41 sequences use three data sources. For the TAIR10 proteome final GFam assignment, 21 128 sequences use one data source, 1249 sequences use two data sources and 42 sequences use three data sources. We excluded the contribution of novel domains for these calculations. Thus, for the 1296 sequences in TAIR9 proteome and 1291 sequences in TAIR10 proteome, GFam integrates and captures annotation provided by two or three resources that result in an increased residue coverage that would otherwise be missed if only the best single-constituent database within InterPro was used. For instance, the data sources HMMPfam and SUPERFAMILY provide non-overlapping domain annotation for the sequences AT4G14230.1 and AT2G02650.1.
In addition, there are 3212 sequences in the TAIR9 proteome InterPro assignments and 3999 sequences in the TAIR10 proteome InterPro assignments that are annotated by only 1 of the 12 InterPro data sources. For the Arabidopsis protein sequence AT5G53760.1, HMMPfam is the only method that provides any domain annotation (total length, 573; annotated residues, 14–478; E value, 1.8e-165; label, Mlo-related protein). Similarly, for the sequence AT5G45030.1, SUPERFAMILY is the only method that provides any domain annotation (total length, 607; annotated residues, 206–424; E value, 3.2e-8; label, serine/cysteine peptidase).
The sensitivity and accuracy of annotation is intrinsic to the InterPro member resources and the curation efforts of the InterPro Consortium and not imposed by GFam. Thus, these coverages are additive in nature and GFam tries to maximize the coverage by considering non-overlapping InterPro annotation for a given sequence.
Curating functional labels for Arabidopsis protein families
TAIR assumed primary responsibility for annotating the genome of Arabidopsis following the complete re-annotation of the genome by The Institute for Genome Research (TIGR) in 2005. TAIR has, since then, produced major updates to the structural and functional content of the genome through its annual genome releases. Today, the number and quality of gene models have significantly improved providing plant biologists with a mature set of protein products for Arabidopsis. However, there remains a need for curated short descriptions for the majority of Arabidopsis gene products that lack direct experimental data or function. On the other hand, curation efforts by Uniprot and the growth and development of InterPro as a meta-resource for protein domain/family annotation along with its member databases offered us an opportunity to generate a curated set of functional labels for Arabidopsis protein families.
We obtained InterPro (release 25.0) domain assignments using InterProScan for 27 379 protein sequences (from representative gene models) in the TAIR9 genome release and subsequently ran GFam on the InterProScan output using the parameters described earlier. For TAIR9 proteome, there are 20 156 sequences that comprise 2558 GFam families with two or more members, 2478 singletons and 4745 unassigned sequences (i.e. sequences with neither InterPro nor novel domains assigned). We used the curated InterPro2GO terms along with domain descriptions from InterPro member databases (primarily Pfam and SUPERFAMILY), computational descriptions for proteins in TAIR9 release and descriptions from Uniprot to curate GFam-derived families with two or more members for proteins in the TAIR9 release. We followed a set of simple guidelines to assign functional labels to these 2558 TAIR9 protein families.
We created functional labels to read as general as possible and aimed at the superfamily level, the highest level of evolutionary relatedness. For example, there were 127 members of the family that contained InterPro domain IPR016177 as the only identified protein domain with protein descriptions that were as varied and diverse as ethylene-responsive element/factor, AP2-domain containing protein, methyl-CpG-binding domain-containing protein and Drought Responsive Element (DRE)-binding transcription factor. This family was named as integrase-type deoxyribonucleic acid (DNA)-binding superfamily. There were 122 members annotated to SUPERFAMILY domain identifier SSF53335 with ∼20 different keywords or short descriptions (embryo abundant protein related, NOL1/NOP2/sun family protein, methyltranferase related, S locus-linked protein, spermidine synthase 2, protein arginine N-methyltransferase, etc.); all these proteins were given the label S-adenosyl-l-methionine-dependent methyltransferases superfamily, same as the SUPERFAMILY model they mapped to.
When possible, labels were chosen to provide an idea of the molecular function of the component domains. For instance, there were 103 members with the InterPro domain IPR003441 that were previously named as the NAC family for No Apical Meristem (NAC). Although the short description is not suggestive of the actual function of the protein, InterPro annotation suggests that these proteins are plant-specific transcriptional regulators. NAC proteins are involved in developmental processes, including the formation of the shoot apical meristem, floral organs and lateral shoots, and in plant hormonal control and defence (50,51). Hence, we labelled these members as NAC (No Apical Meristem) domain transcriptional regulator superfamily. The 27 members that were previously named as belonging to Universal stress protein family were named as adenine nucleotide alpha hydrolases-like superfamily protein based on matches to SUPERFAMILY model of the same description.
Although an existing label conveyed the molecular function, we improved term description by adding specific terms. For instance, the 26 members that were identified as simply proteasome subunit protein were labelled as N-terminal nucleophile aminohydrolases (Ntn hydrolases) superfamily protein based on matches to SUPERFAMILY domain of the same description. Similarly, the 17 members of strictosidine synthase family were renamed as calcium-dependent phosphotriesterase superfamily.
Incorporation of keywords from multiple resources has an advantage in cases when databases such as Pfam cannot provide a meaningful short description. For example, there are 21 members that belong to the Pfam family DUF573, a family of uncharacterized proteins. However, looking at the existing descriptions from TAIR9, we noticed that 9 of the 21 members were named as DNA-binding storekeeper protein-related transcriptional regulators. This name was adopted for the family, thus providing a meaningful label for the 21 members.
Descriptions based on mutants were replaced with more meaningful names. For example, 18 members belonging to senescence-associated family proteins were renamed as Tetraspanin family proteins based on the InterPro domain description for IPR018499. Although the new label falls short of providing clues as to the molecular function of the family, we think that biologists can relate to the Pfam family description (as Pfam is a widely cited resource) based on which we adopted the label. Similarly, there were 27 members labelled previously as scarecrow transcription factor family. We labelled this family as GRAS (products of GAI, RGA, SCR) family transcription factor based on the short description from Pfam for the corresponding domain.
Portability of family annotation across genome releases
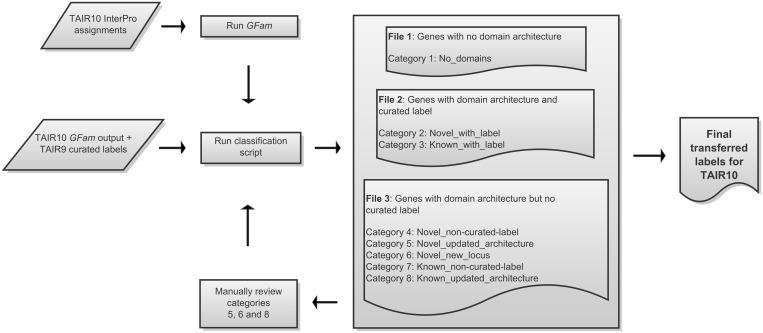
In this section, we describe the procedure we implemented to generate a new set of protein families for TAIR10 proteins and transfer functional labels generated for TAIR9 families to TAIR10 proteome and families. GFam generates domain-based family assignments and provides domain information from the respective InterPro data source (Pfam, SUPERFAMILY, HMMPANTHER, etc.). A database resource will frequently want to associate more descriptive labels to the families. The TAIR curated labels described earlier represent a good starting point for many database projects; however, we recognize that these labels will require refinement over time as new (or organism specific) information becomes available. Given that there are periodic updates to the InterPro database and also the gene models in genome databases, one of the important tasks in reconciling families across two releases is to identify gene models that have been updated and new domain architectures not associated with a curated label. GFam currently does not have an inbuilt mechanism to maintain stable identifiers for novel domains across two sets of family assignments. As such, we propose the following workflow to identify updates to curated labels and applied this process when generating the TAIR10 genome release (Figure 2).
Run GFam on the set of protein sequences to generate domain-based family assignments.
Run classification/labelling script. This step associates a curated label (from supplied file) to every gene and outputs three files with each gene classified into one of eight categories based on presence of known/novel domains, curated label and updated domain architecture.
Manual review of selected categories to verify the current label is appropriate for the updated architecture or to add a new label for novel genes. The curated labels file used in step 2 is updated.
Re-run classification/labelling script using updated labels file to generate final set of gene descriptions.
Figure 2.
Schematic of the work flow adopted to transfer curated labels from TAIR9 GFam families to TAIR10 GFam families.
In this section, we describe the implementation of this procedure to transfer functional labels generated for TAIR9 families to TAIR10 proteins.
Step 1
We used GFam to build families for 27 416 proteins in the TAIR10 genome release. We obtained 5098 distinct domain architectures (i.e. families), of which 3136 architectures have a curated functional label.
Step 2
We wrote a classification/label association script to assign a curated label to the TAIR10 families by transferring the functional labels we generated for TAIR9 based on shared domain architecture. The script takes two files, the GFam output of genes/domain architectures and a file of curated domain architectures with associated labels. In the context of a genome database, the latter would be the set of curated or existing functional labels from a previous release. In our case, this was the set of labels generated for TAIR9 proteins.
The classification/labelling script generates three files with each gene classified into one of eight categories:
-
1.
Genes with no known domains/architectures.
Category 1. NO_DOMAINS—genes for which no domains were identified by InterProScan or GFam’s novel domain assignment procedure.
-
2.
Genes with domain architecture and a curated functional label.
Category 2. NOVEL_WITH_LABEL—genes containing a novel domain with an associated label.
Category 3. KNOWN_WITH_LABEL—genes containing known domains only, which also have associated labels.
-
3.
Genes with domain architecture but no curated functional label.
Category 4. NOVEL_NON-CURATED-LABEL—genes containing a novel domain with no associated label.
Category 5. NOVEL_UPDATED_ARCHITECTURE—gene containing a novel domain with an updated architecture.
Category 6. NOVEL_NEW_LOCUS—new genes (novel identifiers) that contains a novel domain.
Category 7. KNOWN_NON-CURATED-LABEL—genes containing known domains only with no curated label.
Category 8. KNOWN_UPDATED_ARCHITECTURE—gene containing known domains only with an updated architecture.
The classification distinguishes those genes with novel domains (based on all-against-all BLAST followed by clustering) from known domains (based on chaining InterPro domains). To accomplish this task, we verified whether a gene had (i) updated domain architecture, (ii) novel domain(s) and (iii) a curated label. Additionally, when checking for updated domain architecture, we have to distinguish between genes containing only known domains and those that contain a mix of known and novel (or only novel) domains. For genes with known domain architectures, labels are associated to the new genes based solely on identical domain architecture. As GFam generates a new set of identifiers for novel domains in each run, in these cases we cannot map to curated labels based solely on domain architecture. We, therefore, mapped between the genes in the GFam TAIR10 family assignment and the curated labels from the TAIR9 family assignment based on gene identifier and then secondly confirm whether the domain architecture is unchanged. For example, the GFam output may give domain architecture, for instance, IPR022364-IPR017451-NOVEL4 for the gene AT1GXXXXX. Using the gene identifier, we extracted the domain architecture from the set of TAIR9 curated labels, and if this was also IPR022364-IPR017451-NOVEL9, we would deem the architecture to be unchanged (this assumes the novel domain is the same) and associate the label.
Step 3: review
We then reviewed Categories 5, 6 and 8 to either verify that the current label is appropriate for the updated architecture or to add a new label for the novel genes. After review, the curated labels file was updated, and the association script run again (the above three categories should, therefore, not be found in the second run). The NOVEL_NON-CURATED-LABEL (Category 4) and KNOWN_NON-CURATED-LABEL (Category 7) sets represent cases where domain information is present, but a label has not been curated.
Step 4: re-run classification/label association script
This step produces the final set of families with assigned labels. From the total of 27 416 protein-coding genes, 19 367 gene models/loci were assigned a label (Category 3, KNOWN_WITH_LABEL) and 1341 genes contain a novel domain identified by GFam (Category 2, NOVEL_WITH_LABEL). This process leaves 1936 genes with domain architectures that lack a curated label and 4767 genes that had no domains identified.
In the review step 3, we examined the following categories. Of the 825 genes in Category 8 (KNOWN_UPDATED_ARCHITECTURE), 690 had an existing curated label and 135 had no curated label; we also reviewed 136 genes in Category 5 (NOVEL_UPDATED_ARCHITECTURE) and 11 genes in Category 6 (NOVEL_NEW_LOCUS). It needs to be mentioned that the last category mentioned earlier is not the number of new genes in TAIR10 as many would have known domain structures.
A total of 972 genes were examined to see whether the family label required updating. This set, therefore, represents the number of genes with updated/novel domain architectures between TAIR9 and TAIR10.
Hardware requirements
In this section, we describe the nature of hardware we used for all our calculations and provide an idea of computational timescale for the various steps involved in using GFam. To obtain InterPro domain assignments, InterProScan was run on Sun V20z servers with 24 machines, each with two AMD64 processors. Each machine had its own instance of InterProScan installed on it with complete databases. The runs were parallelized over query, i.e. each of the 24 machines got its own subset of sequences to process (in reality, these subsets were created dynamically to ensure proper load balancing using a special dispatcher program that does this). Using this type of parallelization, in an ideal case, the calculation of timing scales linearly with the number of query sequences. For 27 379 sequences in TAIR9 genome release, we used 24 machines for 12 h; the average time per sequence was 37s. Please note that, for TAIR10 release, the number of protein sequences (35 381) used for the InterProScan run is more than what we describe in the article (27 416) as the former includes all protein-coding gene models (i.e. inclusive of splice variants at the time the calculations were done). In this work, we describe calculations for only the representative protein-coding gene models in both TAIR9 and TAIR10 genome releases. For each protein-coding gene loci, TAIR defines a representative gene model as the gene model with the longest coding sequence (with very few exceptions).
GFam was run on an AMD Phenom II X4 955 processor, 3.2 GHz, four Central Processing Unit (CPU) cores with 12 GB of memory although one core runs only at 800 MHz. The all-against-all BLAST part of the GFam pipeline has an option to include the number of CPU cores users want to deploy for processing although the other steps never use more than one core because (i) Python itself is single threaded due to the global interpreter lock and (ii) the time requirements of other steps is insignificant compared with the InterProScan run and all-against-all BLAST steps.
For the 27 379 sequences in TAIR9 proteome, filtering InterProScan input and computing preliminary domain architecture took 9 s, finding unassigned regions took 3 s, slicing unassigned regions into a separate FASTA file took 35 s, all-against-all BLAST took 49 min and 25 s, connected component analysis took 1 s and the final domain architecture calculation took 13 s. The processing time was almost identical for the 27 416 sequences in TAIR10 proteome.
The GFam part of the computation is mostly IO (Input/Output) bound, and the Random Access Memory (RAM) and CPU requirements are pretty low. The two CPU-intensive aspects are external to GFam, namely obtaining InterProScan output for the sequences of interest and running all-against-all BLAST for InterPro un-annotated sequence fragments. Thus, as for hardware requirements, any standard desktop computer with 1-GB RAM and a 2-GHz single-core processor is more than enough to run GFam and will be sufficient to obtain assignment for any one genome at a given time.
DISCUSSION
In summary, we have demonstrated through this work that genome initiatives, both nascent and mature, can adopt GFam to identify domain-based families, create functional labels for families and transfer such annotation seamlessly across genome updates. We have demonstrated that GFam has the potential to serve as a one-stop source for domain-based family assignment for new genome initiatives and model organism resources.
Our hybrid approach increases the sequence coverage by 7.2 percentage points and residue coverage by 14.6 percentage points higher than the coverage provided by the best single-constituent database within InterPro. A linear list of functional assignments through protein domains, regions, signatures and sites provided by 12 independent resources, perhaps, will be useful in the context of a sequence search to understand the putative molecular and biochemical function of a protein. However, it is not helpful if one is interested in the larger organization and arrangement of evolutionarily conserved domains. This will be particularly useful in the context of a collection of sequences derived through a genome sequencing project or a transcriptome project. The objective of providing a consensus architecture is to reduce such a diverse collection (although by and large integrated by InterPro) of functional assignments into a single assignment that captures as much as annotation and also provide a complete functional picture of the protein sequence. The added value of this exercise is a natural grouping of proteins that share a common domain arrangement that can then be used for comparative genomics studies. The addition of novel domains as through our clustering step adds further value, by improving sequence and residue coverage, to this rich functional data provided by InterPro. The novel domains as defined by GFam are putative functional sites that are shared by a set of proteins. In some cases, it is possible that these are extensions of an existing domain. In such cases, we think that these GFam novel domains will help refine domain boundaries for curated domains.
Although GFam as a method can be considered stable and mature, it also offers scope for adding practical functionalities for genome databases. It would be useful to incorporate an internal functionality as part of GFam to compare and contrast two slightly different annotations; for instance, family annotations between two genome releases. Such functionality would also be useful in the situation of comparing very highly similar genomes (strains, sub-species, etc.). Creating sequence profiles of novel domains and using such profiles through HMM methods such as HMMER3 to search for close and remote homologous members is another way to provide consistent annotation of such novel domains. Given the wide variety and sources of annotation of a protein’s molecular function, sub-cellular location and biological role, it is also desirable to integrate such annotations and provide a meaningful label automatically. This would substantially reduce the amount of time required for manual curation of functional labels. Such automatically generated labels would also be easily tractable across different releases or sets of families generated at different time points.
AVAILABILITY AND REQUIREMENTS
The source code for GFam, the association script, the protein sequences, raw InterPro output, families and functional labels for TAIR9 and TAIR10 proteins can be downloaded from http://www.paccanarolab.org/software/gfam/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Newton International Fellowship Scheme of the Royal Society [NF080750 to T.N.]. Funding for open access charge: Biotechnology and Biological Sciences Research Council [BB/F00964X/1 to A.P.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Christopher Wilks for his technical help in integrating the family annotation into the TAIR10 genome release and TAIR curators for providing valuable feedback on the project. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation. We wish to thank Robert Bukowski at Cornell University for his help with InterPro assignments. R.S. designed and oversaw the project, developed the domain chaining algorithm, tested the software, generated curated labels for TAIR9 families and together with T.N. wrote the manuscript and software documentation; T.N. implemented the software; D.S. established a system to transfer functional labels from one genome release to the next and wrote the section on portability and E.H. and A.P. gave valuable project ideas and contributed to writing the manuscript. All authors read and approved the manuscript.
REFERENCES
- 1.Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg I. Automated protein function prediction—the genomic challenge. Brief Bioinform. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 3.Hughes TR, Roth FP. A race through the maze of genomic evidence. Genome Biol. 2008;9(Suppl. 1):S1. doi: 10.1186/gb-2008-9-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970. [Google Scholar]
- 5.Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- 6.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 7.Teichmann SA, Park J, Chothia C. Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc. Natl Acad. Sci. USA. 1998;95:14658–14663. doi: 10.1073/pnas.95.25.14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS One. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, Buell CR. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol. Biol. 2010;10:41. doi: 10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, Ouyang S, Egan A, Nobuta K, Haas BJ, Zhu W, Gu X, Silva JC, Meyers BC, Buell CR. Characterization of paralogous protein families in rice. BMC Plant Biol. 2008;8:18. doi: 10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lees J, Yeats C, Redfern O, Clegg A, Orengo C. Gene3D: merging structure and function for a Thousand genomes. Nucleic Acids Res. 2010;38:D296–D300. doi: 10.1093/nar/gkp987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 17.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corpet F, Servant F, Gouzy J, Kahn D. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pipenbacher P, Schliep A, Schneckener S, Schonhuth A, Schomburg D, Schrader R. ProClust: improved clustering of protein sequences with an extended graph-based approach. Bioinformatics. 2002;18(Suppl. 2):S182–S191. doi: 10.1093/bioinformatics/18.suppl_2.s182. [DOI] [PubMed] [Google Scholar]
- 25.Krause A, Stoye J, Vingron M. Large scale hierarchical clustering of protein sequences. BMC Bioinformatics. 2005;6:15. doi: 10.1186/1471-2105-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nepusz T, Sasidharan R, Paccanaro A. SCPS: a fast implementation of a spectral method for detecting protein families on a genome-wide scale. BMC Bioinformatics. 2010;11:120. doi: 10.1186/1471-2105-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paccanaro A, Casbon JA, Saqi MA. Spectral clustering of protein sequences. Nucleic Acids Res. 2006;34:1571–1580. doi: 10.1093/nar/gkj515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittkop T, Baumbach J, Lobo FP, Rahmann S. Large scale clustering of protein sequences with FORCE—a layout based heuristic for weighted cluster editing. BMC Bioinformatics. 2007;8:396. doi: 10.1186/1471-2105-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petryszak R, Kretschmann E, Wieser D, Apweiler R. The predictive power of the CluSTr database. Bioinformatics. 2005;21:3604–3609. doi: 10.1093/bioinformatics/bti542. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell RB, Ponting CP. Protein fold irregularities that hinder sequence analysis. Curr. Opin. Struct. Biol. 1998;8:364–371. doi: 10.1016/s0959-440x(98)80071-7. [DOI] [PubMed] [Google Scholar]
- 34.Aroul-Selvam R, Hubbard T, Sasidharan R. Domain insertions in protein structures. J. Mol. Biol. 2004;338:633–641. doi: 10.1016/j.jmb.2004.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell RB. Domain insertion. Protein Eng. 1994;7:1407–1410. doi: 10.1093/protein/7.12.1407. [DOI] [PubMed] [Google Scholar]
- 36.Jung J, Lee B. Circularly permuted proteins in the protein structure database. Protein Sci. 2001;10:1881–1886. doi: 10.1110/ps.05801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heger A, Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Aravind L. Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene. 469:18–30. doi: 10.1016/j.gene.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anantharaman V, Aravind L. The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol. 2006;3:18–27. doi: 10.4161/rna.3.1.2548. [DOI] [PubMed] [Google Scholar]
- 40.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scordis P, Flower DR, Attwood TK. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15:799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- 42.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 43.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 44.Consortium TU. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith RK, Jr, Maiti R, Chan AP, Yu C, Farzad M, Wu D, et al. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 49.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duval M, Hsieh TF, Kim SY, Thomas TL. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002;50:237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- 51.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]