Abstract
Many tools are available to analyse genomes but are often challenging to use in a cell type–specific context. We have developed a method similar to the isolation of nuclei tagged in a specific cell type (INTACT) technique [Deal,R.B. and Henikoff,S. (2010) A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell, 18, 1030–1040; Steiner,F.A., Talbert,P.B., Kasinathan,S., Deal,R.B. and Henikoff,S. (2012) Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res., doi:10.1101/gr.131748.111], first developed in plants, for use in Drosophila neurons. We profile gene expression and histone modifications in Kenyon cells and octopaminergic neurons in the adult brain. In addition to recovering known gene expression differences, we also observe significant cell type–specific chromatin modifications. In particular, a small subset of differentially expressed genes exhibits a striking anti-correlation between repressive and activating histone modifications. These genes are enriched for transcription factors, recovering those known to regulate mushroom body identity and predicting analogous regulators of octopaminergic neurons. Our results suggest that applying INTACT to specific neuronal populations can illuminate the transcriptional regulatory networks that underlie neuronal cell identity.
INTRODUCTION
The nervous system provides a striking example of cellular diversity, with myriad neuronal, glial, and other cell types organized into neural circuits. The identity of these cell types, established during development and maintained throughout adulthood, requires the expression of unique combinations of genes (1,2). These combinations include genes that implement a particular biochemical or signaling function (e.g. ion channels, neurotransmitter receptors) and other regulatory genes (e.g. transcription factors) that control when, where and at what level each gene is expressed (2). Understanding how these transcriptional networks are established and then control the phenotype of a specific cell type is a fundamental problem in modern molecular biology. This challenge also has practical implications for molecular neuroscience, where characterizing the molecular components of individual neuronal cell types will improve our ability to dissect neural circuits.
In principle, genome-wide methods allow systematic characterization of these regulatory networks (3). However, applying these techniques to specific cell types requires a method for the isolation of a homogeneous population of cells in quantities sufficient to produce a robust signal. Solutions to this problem, particularly for transcript analysis, include cell purification techniques (e.g. fluorescent activated cell sorting, laser capture micro-dissection and manual sorting) and biochemical purification strategies that rely on cell type–specific labeling of core machinery, including ribosomes (translating ribosome affinity purification) and the Argonaute complex (microRNA tagging-affinity-purification) (4–8). It would, however, be advantageous to use a single isolation method to characterize cell type–specific gene expression, chromatin modifications, transcription factor binding and other types of genome-wide profiles.
One promising approach is the isolation of nuclei tagged in a specific cell type (INTACT) strategy, first described in Arabidopsis and extended to Caenorhabditis elegans (9,10). This method marks the nucleus of a specific cell type with a genetically encoded tag. After these labeled nuclei are purified, cell type–specific transcriptional profiles and chromatin maps can be constructed. Another approach involves the cell type–specific expression of a GFP-histone H2B fusion protein, which was used to isolate nuclei from Drosophila by fluorescent activated cell sorting (11). Both of these approaches have been used to characterize embryonic mesoderm in Drosophila.
We are interested in adult Drosophila neuronal cell types and would like to take advantage of an extensive collection of GAL4 lines that target sparse sub-populations of neurons (12). Toward this end, we have independently developed an INTACT procedure that permits the isolation of nuclei from the brains of adult flies. Unlike the original INTACT approach, our system does not rely on streptavidin-mediated capture of biotinylated nuclei (9). Instead, nuclei are immunoaffinity purified by magnetic beads adsorbed to an antibody that recognizes our tag. In addition, we describe a rapid isolation procedure that allows the purification of nuclei from adult flies at reasonable yields with high purity. Finally, because tag expression is driven by the GAL4/UAS system, it can be used in any class of neuron for which a suitable driver is available (13).
We present a proof of principle study profiling gene expression (RNA-seq) and histone modifications (ChIP-seq) in three Drosophila neuronal populations ranging from 100 000 to 130 cells per brain. We describe the observed differential expression profiles in the context of known marker genes. We further describe patterns of differential histone modifications that indicate active promoter (H3K4me3), open chromatin (H3K27ac) and polycomb group (PcG)-mediated transcriptional silencing (H3K27me3). In particular, we observed strong cell-specific repression of a small number of transcription factors in one population, a concomitant cell-specific activation in the other population and a consistent differential expression pattern. We close by discussing the utility of our approach for characterizing the regulatory networks that control neuronal cell identity.
MATERIALS AND METHODS
DNA constructs
A synthetic linker encoding the following amino acids: LAAASGGGGSGGGGSLAAASEFSAAALSGGGGSGGGGSAAAL was inserted into the unc84 (NP_001024707.1) reading frame between amino acid 1111 and the stop codon. Two copies of the super folder GFP variant were then cloned into the centrally located EcoRI site (amino acids EF in the linker) (14) to produce UNC-84-2XGFP. The UNC-84-tdTomFL construct used the same linker strategy except that the fluorescent protein cassette carried a restriction site at its 3′ end that allowed the addition of a C-terminal 3XFlag epitope tag.
Fly stocks
P(GawB)ey[OK107-GAL4] (#854) and P(Tdc2-GAL4.C)2 (#9313) were obtained from the Bloomington stock center. R57C10-GAL4 is a promoter fusion of the GAL4 coding region and an 824 bp upstream fragment of the n-synaptobrevin gene, defined by the primers atttcccaccccttggccatcggca and gttctagagggttgcgctctcagtg, and was constructed as described previously (12). Similarly, both the UAS_unc84-2XGFP and UAS_unc84-tdTomFl cassettes were inserted into the attP2 site using phi31-mediated recombination (15).
Cell transfection
ML-DmBG3-c2 cells were transfected with the same UAS constructs that were used to make transgenic flies by the Effectene method (Qiagen: 301425). Expression was driven by ubiquitin-GAL4.
Magnetic bead preparation
300 μl of Dynal Protein-G beads (Invitrogen: 100-03D) were adsorbed to either 5 μg of anti-GFP antibody (Invitrogen: G10362) or 10 ug of anti-Flag antibody (Sigma: F7425) in 600 μl PBS/0.1%Tween 20 for 30 min at 4°C. Beads were then washed once in PBS/0.1%Tween-20 and stored in 300 μl of 10 mM β-glycerophosphate pH 7, 2 mM MgCl2.
Immunopurification of nuclei
Adult flies were anesthetized by CO2 and flash frozen in liquid N2. Heads were separated from thoracicoabdominal segments, wings and legs by vigorous vortexing followed by separation over dry ice cooled sieves. In all, 600–10 000 frozen heads were added to 100 ml of 10 mM β-glycerophosphate pH 7, 2 mM MgCl2, 5 mM sodium butyrate, 1X complete protease inhibitor cocktail (Roche: 11873580001), and the suspension was passed over a Yamato continuous flow homogenizer, set at 100 rpm, five to seven times. The homogenate was filtered over Miracloth (EMD Biosciences: 475855) and brought to 0.7 mM β-mercaptoethanol and 0.5% NP-40. After six tractions in two 40 ml Dounce homogenizers (tight-pestle B), 600 μl of antibody-adsorbed beads were added to 100 ml of lysate. The binding reaction was performed at 4°C for 30 min with constant end-over-end agitation. Beads were then collected on a magnet (Invitrogen: 123-02D) and washed three to four times in 50 ml 10 mM β-glycerophosphate pH 7, 250 mM sucrose, 2 mM MgCl2, 25 mM KCl and 5 mM sodium butyrate. Bead-bound nuclei in 20 ml of wash buffer were then passed over a 20 um nylon mesh (Small Parts: B001D8ECDE), returned to the magnet stand and resuspended in 1 ml of 10 mM β-glycerophosphate pH 7, 250 mM sucrose, 2 mM MgCl2, 25 mM KCl and 5 mM sodium butyrate. Sodium butyrate and the protease inhibitor cocktail are omitted from all buffers, if nuclei were to be used for transcript profiling (RNA-seq).
Transcript analysis by RNA-seq
Bead-bound nuclei collected on a magnet stand (Invitrogen: 123-21D) or whole dissected brains were resuspended in 400 μl of 100 mM Tris pH 7, 4 M guanidinium thiocyanate. After 30 min of agitation at 4°C (in the case of bead-bound nuclei), the supernatant containing nuclear RNA was removed from the beads and extracted with an equal volume of phenol:CHCl3. After the addition of 0.1 volume 3 M sodium acetate pH 5, the sample was extracted with an equal volume of acid phenol:CHCl3 (Invitrogen: AM9722). The aqueous layer was recovered and brought to 400 μl by the addition of H2O. The Agencourt RNA-Advantage kit (Beckman Coulter: 47942) was then used to further purify the sample. Briefly, 100 μl of the lysis buffer supplied with the kit was added to the aqueous layer that resulted from the acid extraction step. After brief centrifugation to remove insoluble material, the samples were then processed exactly as directed by the kit’s instructions (including DNaseI treatment). Nuclear RNA (10–50 ng) was then converted to complementary DNA using a Nugen Ovation RNA-seq v2 kit (Nugen: 7102). Amplified complementary DNA (2 μg) was then sheared in a Covaris S2 instrument (duty cycle = 10%; intensity = 5; cycles/burst = 100; time = 5 minutes; volume = 120 μl). In all, 200 ng of sheared DNA was then end-repaired, linker-adapted and sequenced on an Illumina HiSeq 2000 to 50 bp read length. The library synthesis steps are exactly those recommended by Illumina in the Genomic DNA Sample Preparation Kit except that Qiagen column purification was substituted with Agencourt AMPure magnetic bead purification.
Chromatin profiling by ChIP-seq
Bead-bound nuclei were collected on a magnet stand (Invitrogen: 123-21D) and re-suspended in 1 ml of 15 mM Hepes pH 7, 1 mM KCl, 5 mM MgCl2, 2 mM CaCl2, 340 mM sucrose, 0.5 mM spermidine, 0.15 mM spermine, 5 mM sodium butyrate. The sample was then split into two 500 μl volumes, and nuclei were digested for 15 min at 37°C after the addition of micrococcal nuclease (Worthington: LS004798) to 0.025 units/μl. The reaction was terminated by the addition of EGTA at 2 mM. Nucleosomes were then extracted on ice for 30 min in 200–400 μl of 15 mM Hepes pH 7, 200 mM NaCl, 1 mM KCl, 5 mM MgCl2, 2 mM EGTA, 340 mM sucrose, 0.5 mM spermidine, 0.15 mM spermine and 5 mM sodium butyrate. The extraction was repeated with the same buffer adjusted to 400 mM NaCl. The supernatant from the second extraction was combined with the first and dialyzed for 2 hours at 4°C against 15 mM Hepes pH 7, 25 mM KCl, 1 mM β-mercaptoethanol, 1 mM PMSF, 5 mM sodium butyrate. Greater than 70% of the nucleosomes prepared in this manner are monosomes.
The following antibodies were used to detect modified histones: H3K4me3 (Abcam: 8580), H3K27Ac (Abcam: 4729) and H3K27me3 (Millipore: 07-449). In all cases, 10 μg of antibody was adsorbed to 3 mg Dynal Protein-G beads in 600 μl 1XPBS, 5 mg/ml BSA for 4–8 hours at 4°C. After washing the beads on a magnet stand 3X in 1XPBS, 5 mg/ml BSA, they were resuspended in 50 ul of the same buffer before ChIP.
Purified nucleosomes (1–5 μg) were brought to 500 μl in 15 mM Hepes pH 7, 25 mM KCl and 5 mM sodium butyrate. In all, 50 μl of this material was removed and stored as the non-enriched input sample, whereas the remaining 450 μl portion was adjusted to 600 μl by the addition of 150 μl of 34 mM Hepes pH 7, 9 mM EDTA, 4% Triton X-100, 0.4% deoxycholate, 4X complete protease inhibitor cocktail. Finally, 50 μl of antibody adsorbed Dynal Protein-G beads were added to the nucleosome preparation, and ChIP was carried out at 4°C for 12 hours under constant end-over-end agitation. Bead-bound nucleosomes were then washed on a magnet stand 8X in 50 mM Hepes pH 8, 1 mM EDTA, 1% IGEPAL, 0.7% deoxycholate, 0.5 M LiCl, 1X complete protease inhibitor cocktail. After a single wash in TE, beads were pelleted at 4000 rpm for 3 min in a microcentrifuge and then incubated in 170 μl of 1X TE/1% SDS for 30 min at 65°C. After brief centrifugation, 150 μl of 400 ug/ml glycogen, 933 ug/ml proteinase K was added to the supernatant fraction, and the sample was incubated at 37°C for 2 hours. Nucleic acid was recovered by extracting the sample once with phenol, followed by an additional extraction with phenol: CHCl3 and precipitation after the addition of NaCl to 0.2 M. Finally, the sample was incubated in 50 μl of TE containing RNAse A at 330 ug/ml for 30 min at 37°C for 1 hour, followed by purification on Agencourt AMPure magnetic beads (16) (Beckman Coulter: A63880). Enriched immunoprecipitated and non-enriched input DNA was end-repaired, linker-adapted and sequenced on an Illumina HiSeq 2000 to 50 bp read length (17). The library synthesis steps are exactly those recommended by Illumina in the Genomic DNA sample preparation kit except that Qiagen column purification was substituted with Agencourt AMPure magnetic bead purification.
RNA-seq data analysis
5′ ends of all reads were trimmed by five nucleotides to remove artifacts of the Nugen Ovation kit (FASTX; http://hannonlab.cshl.edu/fastx_toolkit). Reads were then aligned to the annotated transcriptome (FlyBase r5.41) (18) of the fly genome (UCSC dm3), using the TOPHAT splice-aware aligner (v1.4.0) (19). Pairs of libraries were analysed using CUFFDIFF v1.3.0 to estimate the abundance of each isoform and identify differentially expressed genes at a 1% false discovery rate (20). Fragment bias correction, multi-hit read correction and a mask of mitochondrial and non-coding transcripts were used to improve robustness of the expression levels, which were estimated in terms of reads per kilobase of exon model per million. Genome tracks of RNA-seq reads were created by counting read alignments per genomic position using BEDTools (v 2.15) (21) and scaling these counts to 10 million total read alignments using a custom Perl script. Gene ontology analysis was performed with the FlyMine web server (22). A list of candidate transcription factors (n = 749) in the Drosophila genome was obtained from FlyTF (23,24).
ChIP-seq data analysis
ChIP-seq and input library reads were aligned to the fly genome using BOWTIE (v0.12.7) (25), keeping only those that mapped uniquely to a single position in the genome. For visualization, the reads were extended to the mean length of the library fragments (200 bp), the number of extended reads covering each genomic position counted using BEDTools (21), and these counts scaled to a total number of 10 million read alignments using a custom Perl script.
We counted the number of reads in each ChIP and input library within a 10kb window scanned across the genome in 5kb increments using BEDTools. These counts were converted to a Z-score using chromosome-specific mean and standard deviations of window counts. To compare marks between cell types, differences in corresponding Z-scores were computed and then plotted on a Hilbert curve representing the euchromatic Drosophila genome (2L, 2R, 3L, 3R, 4, X) (26).
Each annotated FlyBase isoform was assigned a score representing the intensity of each mark by counting the number of reads mapping to the gene body or promoter (1-Kb window surrounding the TSS), and converting these counts to Z-scores using the mean and standard deviation of corresponding counts across all genes. These per-gene scores were corrected by subtracting the corresponding Z-score from an input library of the same cell type.
Data analysis was performed using a combination of the aforementioned utilities and custom Perl scripts. Plots were made using the R project (R Development Core Team, 2010) and genome landscapes visualized using the Broad Integrated Genomics Viewer (27). Hilbert curves were visualized using the HilbertVis R package (26).
Reagent and data distribution
The DNA constructs described in this article are available at Addgene (http://www.addgene.org). All data have been deposited in National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GSE37033).
RESULTS
A Drosophila INTACT tag
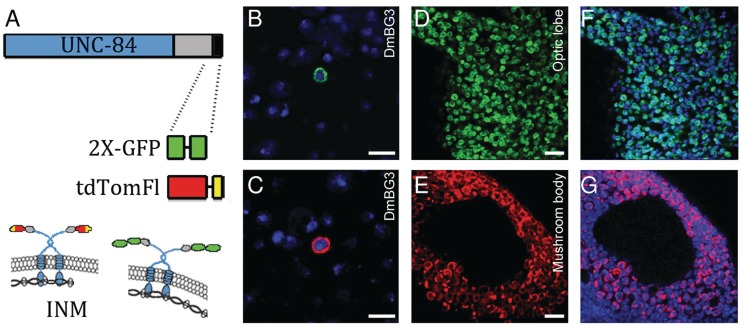
When nuclei are harvested in the presence of non-ionic detergents, the outer nuclear membrane is stripped away from the nucleus; thus, our strategy takes advantage of the SUN domain family of proteins, which are embedded in the inner nuclear membrane of all eukaryotes (28). We evaluated several candidate SUN domain proteins for their ability to both localize to the nuclear envelope and to have minimal effects on the viability of flies. In the end, we selected a construct based on the C. elegans protein UNC-84 because both the mouse and Drosophila SUN homologues failed to support efficient tag localization in transfected Drosophila cells (29). For a GFP-based tag (UNC84-2XGFP), two copies of the fluorescent protein were used to increase both the antigenicity and brightness of the tag (Figure 1A). A tdTomato-based tag was also constructed that contained a C-terminal 3XFlag epitope tag (UNC84-tdTomFlag) (Figure 1A). In each tag, the fluorescent protein/epitope tag is oriented into the lumenal space of the nuclear envelope, which requires the removal of the outer nuclear membrane for detection (Figure 1A). The expression of both the red and green tags was driven by the GAL4/UAS system, and proper localization at the periphery of the nucleus was observed in both transfected cultured Drosophila cells and in neurons of the adult fly (Figure 1B–G) (13).
Figure 1.
Construction and localization of an UNC-84 based nuclear tag. (A) Either two copies of GFP (2XGFP) or Flag-tagged tdTomato (tdTomFl) were placed at the C-terminus of UNC-84. The gray box denotes the conserved SUN domain and the black box the area in the protein where the tag is inserted. At the bottom of (A) the topological distribution of the fusion proteins, in the inner nuclear membrane, are indicated (30). ML-DmBG3-c2 cells expressing either a GFP (B) or tdTomato based tag (C). Localization of the two tags in adult Drosophila brains (D–G). Expression of either the green (D, F) or red (E, G) tag was driven by the pan-neuronal driver R57C10-Gal4 (12). In (D, F) the medial edge of the optic lobe is shown, whereas the Kenyon cell population of the mushroom body is shown in (E, G). Nuclei are labeled by Draq5 and indicated in blue (B, C, F and G). Scale bars: 10 μm.
Purity and yield
We developed a bead-based immunoaffinity purification scheme and tested its yield and purity in a reconstruction experiment. An equivalent number of nuclei from two populations of transfected cultured Drosophila cells, one expressing UNC84-2XGFP and the other UNC84-tdTomFlag, were mixed and subjected to bead-based immunoaffinity purification (Figure 2A, D). As expected, beads adsorbed to α-GFP antibody selectively capture GFP labeled nuclei (Figure 2B), and α-Flag beads specifically bind to nuclei tagged with tdTomatoFlag (Figure 2E). At subsaturating (ratio of nuclei to beads) conditions, the capture of UNC84-2XGFP tagged nuclei is more efficient than UNC84-tdTomFl tagged nuclei, as seen in the unbound fractions of nuclei (compare Figure 2C and F).
Figure 2.
The purification of tagged nuclei. (A–F) Separate populations of green and red tagged ML-DmBg3-c2 cells were prepared by transfection. The cells were mixed, nuclei harvested and the sample split into two identical inputs (A, D). α-GFP adsorbed beads (B) were used to capture nuclei in one sample, and α-Flag beads were used in the other (E). The unbound nuclei that fail to be captured in either the α-GFP or α-Flag binding reactions are shown in (C) and (F) respectively. As indicated in the wash samples the capture of GFP tagged nuclei is typically more efficient (compare C with F). For in vivo binding studies, tag expression was driven by R57C10-GAL4 (G), OK107-GAL4 (H) and Tdc2-GAL4 drivers (I) (12,31,32). Schematic of the INTACT procedure using in vivo tagged nuclei (J). In the diagram, tagged nuclei are indicated by the green and red patches inside of the heads of the two flies. The method involves two steps: first, nuclei are obtained from tagged flies; second, magnetic beads are used to purify tagged nuclei. The gray ellipse denotes the magnetic bead and either the green or red ‘T’ the particular antibody used for capture (J). Nuclei were stained with Draq5 and are indicated in blue (G-I). Scale bars: 40 μm.
An important requirement for our method is the ability to isolate nuclei from flies where a small number of nuclei are tagged per brain. For the experiments described in this report, we used three GAL4 driver lines that express in a range of cell numbers per brain. Pan-neuronal expression was driven with the R57C10-GAL4 driver, which uses the neuron-specific enhancer of the n-synaptobrevin gene; OK107-GAL4 was used to target the Kenyon cell population of the mushroom body, and octopaminergic neurons were targeted with a Tdc2-GAL4 line (Figure 2G-I) (12,31,32). To test the sensitivity of our INTACT procedure, we used a bead binding assay (Figure 2J) that allowed us to quantitate yields from flies, where either green or red tag expression was driven by either the pan-neuronal or octopaminergic drivers. We estimate that the R57C10 driver targets 105 nuclei and that the Tdc2 driver targets 100–150 nuclei per brain (33,34). When INTACT was performed on 600 pan-neuronally tagged heads, 1.1 × 107 (three trials: 1.5, 1.0, 0.9 × 107) and 1.6 × 107 (three trials: 2.2, 0.9, 1.8 × 107) green and red nuclei, respectively, were recovered at 15–20% yield. The same experiment using the octopaminergic driver resulted in the recovery of 4.1 × 104 (three trials: 4.0, 4.1, 4.1 × 104) and 1.1 × 104 (three trials: 0.9, 0.9, 1.5 × 104) green and red nuclei, respectively, at approximately 15–50% yield. The lower yields associated with the pan-neuronally tagged brains result from saturation of the binding reaction (ratio of nuclei to magnetic beads), whereas the recovery of nuclei from sparsely tagged brains is more efficient especially when the green tag is used.
The specificity of INTACT was measured in a mixing experiment where UNC84-2XGFP tagged nuclei were mixed with an excess of UNC84-tdTomFl tagged nuclei. The mixture was generated by mixing green nuclei obtained from heads with octopaminergic tag expression and red nuclei obtained from an equal number of heads with pan-neuronal tag expression. Thus, the input mixture contained a ratio of 130/105 green versus red nuclei. After capture of these nuclei with beads adsorbed to an α-GFP antibody, the exact number of correctly captured green and incorrectly captured red nuclei was determined. These experiments showed that our technique is capable of recovering the approximately 130 Tdc2 cells per brain in 99% purity at 50% yield (Table 1). Because we can scale the assay to tens of thousands animals, we can isolate hundreds of thousands of nuclei from flies where similar numbers of neurons have been tagged.
Table 1.
INTACT specificity
| Input | Recovered | Fold purification | Yield | |
|---|---|---|---|---|
| A | ||||
| Green | 8.2 × 104 | 5.4 × 104 | 990 | 65% |
| Red | 6.0 × 107 | 400 | ||
| Fraction (Green) | 0.001 | 0.99 | ||
| B | ||||
| Green | 8.2 × 104 | 4.6 × 104 | 76 | 56% |
| Red | 6.0 × 106 | 250 | ||
| Fraction (Green) | 0.013 | 0.99 |
(A) A mixture of GFP and tdTomato tagged nuclei was generated by mixing ∼300 red tagged (pan-neuronal driver) and 300 green tagged (octopaminergic driver) adult heads. (B) Same as in (A), except that ∼300 red tagged (pan-neuronal driver) thoracicoabdominal segments and 300 green tagged (octopaminergic driver) adult heads were mixed. The mixture was subjected to INTACT and the number of bead-bound green and red nuclei determined by manually counting a dilution of the purified material. We assumed that each head had 105 neurons, each thoracicoabdominal segment 104 neurons and that the Tdc2 tagged brain had 137 neurons (33,34).
Gene expression profiling
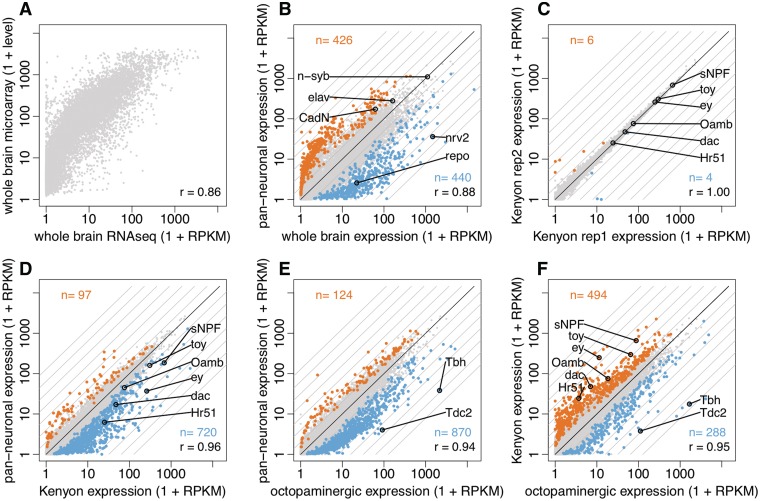
One of the main goals of our method is to characterize gene expression in individual neuronal cell types. Although it is already established that nuclear RNA is sufficient to transcriptionally profile a cell type (9,10), we performed a series of experiments to confirm that RNA-seq can be performed with nuclei isolated from Drosophila neurons (35). However, before doing so, we assessed the performance of our RNA-seq procedure in the absence of INTACT, by first profiling whole-cell RNA isolated from whole dissected brains and compared the resulting expression levels with microarray results in the FlyAtlas compendium (Figure 3A) (36). Of the 27 tissue profiles in FlyAtlas, our brain RNA-seq levels were most correlated to microarray levels measured from the adult brain (Pearson’s r = 0.86, Figure 3A), followed by the adult thoracicoabdominal ganglion (r = 0.84) and larval central nervous system (r = 0.74). These correlation values are in line with previous studies comparing RNA-seq and microarrays, suggesting our RNA-seq procedure is valid (37).
Figure 3.
Gene expression profiles of neuronal subpopulations. (A) Gene expression levels estimated from RNA-seq of dissected whole brain correlate well with microarray levels. (B) Pan-neuronal nuclear RNA is enriched for known markers of neurons (n-syb, elav, CadN) and significantly depleted for glial markers (repo, nrv2). Differentially expressed genes (q < 0.01) are shown in color. (C) Gene expression levels estimated from biological replicates of INTACT isolated Kenyon cell nuclei are highly correlated. (D) Kenyon cell nuclei and (E) octopaminergic nuclei are both enriched for known markers relative to pan-neuronal nuclei. (F) Comparing the expression profiles of Kenyon cell vs. octopaminergic nuclei correctly recovers known markers.
Next, we used the INTACT method with RNA-seq to characterize gene expression in nuclei isolated from all neurons, Kenyon cells and octopaminergic cells, using R57C10-GAL4, OK107-GAL4 and Tdc2-GAL4 drivers respectively (Figure 2G–I) (12,31,32). In the first experiment, RNA obtained from bulk neuronal nuclei (pan-neuronal INTACT) was compared with RNA harvested from whole brain (without INTACT), revealing 426 neuronally enriched genes and 440 depleted genes (CUFFDIFF q-value < 0.01) (Figure 3B). If INTACT works as anticipated, we expect pan-neuronal nuclear RNA to be enriched in transcripts that encode genes that are involved in neuronal function and depleted in transcripts that are known to be expressed in non-neuronal cell types like glia. Gene ontology (GO) analysis (38) revealed that neuronally enriched genes (pan-neuronal INTACT) were significantly over-represented for ion channel activity (Holm-Bonferonni P = 10−7; n = 24), whereas neuronally depleted genes (relative to whole dissected brains) were over-represented in active transmembrane transporter activity (P = 10−7, n = 42) and gliogenesis (P = 0.04; n = 11). Transcripts that were identified in a screen for genes enriched in glia were also significantly over-represented in the depleted pool (P = 10−7, n = 16) (39). In addition to this broad-scale functional analysis, we checked the levels of genes known to be specific to neurons or glia. The pan-neuronal sample is relatively enriched for transcripts that encode the neuron-specific genes elav (277 versus 161 Fragments Per Kilobase of transcript per Million mapped (FPKM) in neurons vs. whole brain) and cadN (172 versus 61 FPKM). Neither of these markers reaches the threshold for differential expression, which is not surprising, given that 90% of cells in the fly brain are thought to be neuronal (34). In contrast, the glial markers repo (1.6 vs. 21 FPKM in pan-neuronal nuclei vs. whole brain) and nrv2 (35 vs. 1474 FPKM) are significantly depleted in the neuronal sample (Figure 3B) (40–43). It is possible that some of the observed transcriptional differences result from the retention of specific mRNAs inside of the nucleus, which has been demonstrated for a small population of mRNAs (44).
Given that 90% of the brain is estimated to be neuronal, the maximum attainable enrichment would seem to be 1.1X; thus, we were surprised at the number of genes (n = 426) that were significantly enriched in the pan-neuronal sample relative to the whole dissected brain (34). We believe that the main reason for this apparent discrepancy is that the INTACT procedure was performed on whole heads (not dissected brains), which contain not only neurons in the brain but also R57C10-GAL4 expressing neurons found in peripheral sensory structures. Supporting this explanation, the pan-neuronal sample is significantly enriched in the mechanosensory channel nompC (7.3-fold) expressed in the antennae (45), and several chemosensory receptors including ionotropic receptors (Ir47a, 11.5x; Ir56a, 1.2x; Ir76b, 35.8x), gustatory receptors (Gr47a, 15.4x; Gr64b, 33.6x), odorant receptors (Or45a, 26.4x; Or98a, 31.1x) and the chemosensory protein CheB74a (9.5x). Biological replicates showed that RNA-seq on INTACT samples are reproducible (Figure 3C).
We next asked whether RNA-seq of INTACT isolated nuclei is as efficient as conventional RNA-seq of whole cells, given that we are sequencing a more complex nuclear RNA population that also contains introns. To address this issue, we analysed the genomic distribution of RNA-seq reads from each sample. Fewer of the RNA-seq read alignments from the INTACT nuclear RNA samples occurred over exons (pan-neuronal: 63%, 63%; Kenyon cells: 72%, 79%; Octopaminergic neurons: 77%, 85%) when compared with whole-cell RNA alignments (whole brain: 91%). The relatively small decrease in exon-mapped reads is consistent with the finding that splicing occurs co-transcriptionally (46,47). These observations suggest that roughly 25% more RNA-seq reads are necessary to achieve exon coverage of nuclear RNA comparable with whole-cell RNA.
Having established that RNA-seq of INTACT isolated nuclei is a reproducible and efficient means of transcriptional profiling, we next asked whether this approach could provide functional insight into neuronal subpopulations. To address this question, we analysed the transcriptional profile of two neuronal subpopulations: Kenyon cells and octopaminergic cells. We first checked whether their profiles were individually enriched in a functionally diverse set of genes previously shown to express in these two cell types. For example, Kenyon cells express a trio of transcription factors (ey, dac and toy), short neuropeptide F (sNPF) and the octopamine receptor of the mushroom body (OAMB), all of which we see significantly enriched in Kenyon cells versus pan-neuronal nuclei (Figure 3D) (48–51). Octopaminergic cells express two enzymes required for the biosynthesis of octopamine: tyrosine decarboxylase 2 (Tdc2) and Tyramine β-hydroxylase (Tbh), both of which we see significantly enriched in nuclear RNA harvested from octopaminergic neurons relative to pan-neuronal nuclei (Figure 3E) (52). The transcript levels of these markers were also appropriately enriched, when we directly compared the Kenyon cell and octopaminergic populations (Figure 3F). For a more systematic analysis, we also compared our expression data with FlyBase annotations of gene expression in each cell population (18). We observed at least moderate expression (FPKM > 10) in Kenyon cells, for 53 of 66 genes (80%) reported to express in the adult mushroom body (Fisher exact test P = 10–56), and 8 of 10 genes (80%) reported to express in adult Kenyon cells (P = 10–8). Tbh, which is strongly expressed in octopaminergic RNA-seq data, is the only gene reported in FlyBase to express in octopaminergic neurons.
We next turned to the question of what neurotransmitters operate in the two profiled cell types. As expected, the octopaminergic profiles were enriched for the biosynthetic enzymes Tdc2 (58-fold vs. pan-neuronal nuclei) and Tbh (30x) and for the vesicular transporter of octopamine, Vmat (‘HIDATA’–CUFFLINKS is unable to reliably estimate an expression level because of the high number of RNA-seq reads). To a lesser extent, the octopaminergic profile was also enriched for genes involved in glutamate synthesis (Got2, 2.7×) and transport (VGlut, 1.7x; Eaat2, 1.7×), in line with previous reports of octopamine and glutamate co-transmission (53,54). In contrast to the clear signal in the octopaminergic profile, no single group of neurotransmitter genes was strongly enriched in the Kenyon cell profile. The Kenyon cell profile was also enriched for portabella (CG10251, 5.5×), a recently identified vesicular transporter that expresses in the mushroom body, but whose substrate is unknown (55). Our Kenyon cell data should contribute a rich set of candidate genes to help identify the portabella ligand.
Chromatin profiling
As chromatin profiling has shown promise for systematically identifying transcriptional regulatory regions (e.g. enhancers), we tested its feasibility on INTACT samples (56,57). ChIP-seq was used to profile histone modifications associated with active promoters (trimethylation of histone H3 on lysine 4, H3K4me3) (58), open chromatin (acetylation of histone H3 on lysine 27, H3K27ac) (59) and Polycomb group (PcG)-mediated silencing (trimethylation of histone H3 on lysine 27, H3K27me3) (60). We quantified the level of H3K4me3 modification over promoters, as this signal correlates with gene expression (58). In contrast, H3K27me3 occurs in broad domains that often span the entire body of Polycomb target genes (61). H3K27ac is enriched over active promoters and can also mark whole gene bodies (61). For this reason, both H3K27me3 and H3K27ac levels were quantified over gene bodies. Although assigning a single value to each gene does not capture the subtleties of the histone modification pattern, this representation provides a convenient and compact way of interpreting the signal in a genome-wide manner. We first profiled pan-neuronal nuclei and found that all three histone modifications were reproducibly detected using our ChIP-seq protocol (Figure 4A–C).
Figure 4.
Cell type–specific chromatin profiles of neuronal subpopulations. ChIP-seq profiling of active promoter [(A) H3K4me3], active chromatin [(B) H3K27me3] and silenced chromatin [(C) H3K27me3] obtained from INTACT isolated nuclei is strongly reproducible. (D–F) Comparing the chromatin profiles of Kenyon cells vs. octopaminergic cells correctly recovers known markers (orange, Kenyon cell; blue, octopaminergic neurons). (G–I) Differential histone modification is weakly, but significantly, correlated with differential gene expression. Genes with enriched expression in Kenyon cells are shown in orange, and those enriched in octopaminergic cells are shown in blue. In all panels, a point is shown only for the most highly expressed isoform of each annotated gene.
When we profiled the histone modifications in octopaminergic and Kenyon cell neurons (Figure 4D–F), we found that nearly all the marker genes were differentially modified in the appropriate population, but with far less enrichment than observed in the RNA signal (Figure 3F). For example, the mushroom body marker ey is more actively marked at its promoter (H3K4me3) and gene body (H3K27ac) in Kenyon cells. Consistent with their proposed active and repressive roles, we observed a statistically significant, although weak, correlation between differential histone modification and differential gene expression in the octopaminergic and Kenyon cell populations (Figure 4G–I). Although most markers were differentially modified in a direction consistent with their expression, we were surprised to see that the biosynthetic enzymes Tbh and Tdc2 were not differentially marked by the PcG-mediated H3K27me3 modification (Figure 4F and I).
As the octopaminergic biosynthetic factors did not appear to be differentially PcG-repressed, we decided to take a closer look at the genes that are targeted by this silencing mechanism. In both the octopaminergic and Kenyon cell populations, repressed loci (H3K27me3 z ≥ 2) were significantly enriched for transcriptional regulators (Figure 5A; Octopaminergic cells, n = 168 of 561 genes, P = 10−92; Kenyon cells, n = 168 of 596 genes, P = 10−88), which is in line with previous studies that have shown PcG-mediated silencing to target developmentally regulated transcription factors (60). The silenced genes were also enriched for several GO terms that are associated with neuronal cell fate determination. For example, the genes silenced in octopaminergic neurons were enriched for CNS development (P = 10−20, n = 46), cell fate specification (P = 10−20, n = 31), cell fate commitment (P = 10−17, n = 63), generation of neurons (P = 10−16, n = 72), neuron differentiation (P = 10−10, n = 59) and neuron projection development (P = 10−5, n = 39). The genes silenced in Kenyon cells showed a similar enrichment profile. Based on this observation, we hypothesized that perhaps transcription factors that are required for the establishment or maintenance of neuronal identity undergo PcG-mediated repression in cell types where they have no function (i.e. cell types where ectopic expression would alter their identity). To address this hypothesis, we studied PcG silencing over transcription factors found to be differentially expressed by RNA-seq (Figure 5A, colored points). This analysis revealed only a handful of differentially expressed transcription factors that were significantly repressed in one cell type but not the other. Consistent with our hypothesis, transcription factors known to regulate mushroom body development (ey, toy and dac) are repressed in octopaminergic nuclei, but lack repression in Kenyon cell nuclei. Based on this observation, we predict that the less-studied factors dmrt99b, Fer2, CG4328 and fd59A are responsible for establishing or maintaining the identity of octopaminergic neurons (Figure 5A).
Figure 5.
Cell type–specific chromatin silencing of transcription factors. (A) The main scatterplot depicts the level of repressive H3K27me3 histone modification over all annotated gene isoforms (grey points) as measured in octopaminergic (x-axis) and Kenyon cells (y-axis). The grey curve drawn above the scatterplot depicts the distribution of H3K27me3 marking over all genes in octopaminergic. The black curve represents the distribution for the subset of genes that are transcription factors. Similar curves drawn to the right of the plot reflect the H3K27me3 levels observed in Kenyon cells. A subset of transcription factors is strongly silenced (H3K27me3 z ≥ 2), as seen by the bimodal distribution (black line). Differentially expressed transcription factors are colored according to the population in which they are enriched (blue = octopaminergic, orange = Kenyon cells, point size indicates the magnitude of the difference in expression). This unbiased analysis reveals a handful of transcription factors that are enriched in one population, whereas repressed in the other (gray quadrants). These genes include Hr51, toy, dac and ey, all expressed in Kenyon cells and dmrt99b, Fer2, CG4328 and fd59A, which are expressed in octopaminergic neurons. (B) In Kenyon cells, ey is highly expressed from an active promoter, within an open chromatin domain that is not silenced. In contrast, this locus is not expressed, is not active (low H3K4me3, H3K27ac), and is strongly silenced (high H3K27me3) in octopaminergic neurons. (C) The dmrt99b locus exhibits the opposite pattern.
The striking pattern of differential repression and activation is evident when we look at genome landscapes incorporating all of our ChIP-seq and RNA-seq data for the two most differentially modified loci: dmrt99b and ey (Figure 5B and C). Expression of ey in Kenyon cells is consistent with the promoter of the gene being actively marked, the gene body sitting in an open chromatin domain and the locus lacking PcG-mediated silencing (H3K4me3+, H3K27Ac+, H3K27me3−) (Figure 5B). Ey is not expressed in octopaminergic neurons, supported by the promoter and gene body lacking active histone modifications and the locus sitting under a broad island of PcG-mediated silencing (H3K4me3−, H3K27Ac−, H3K27me3+). The dmrt99b locus exhibits the complementary pattern of expression and repression, as the gene is expressed in octopaminergic neurons and repressed in the mushroom body (Figure 5C). A feature present at both the ey and dmrt99b loci is that in measurements from bulk neuronal nuclei, there is low-level expression and strong repression over the gene bodies, as one would expect from a mixed population of cells—detecting transcripts from expressing cells while detecting repression in other non-expressing cells. We reason that genes that show both expression and PcG repressive marks indicate (i) that the cell population is mixed and (ii) that the gene plays an important role in the specification of cell type.
If this hypothesis is true, then a combination of active and repressive histone modifications could systematically identify such developmentally important genes. We next asked if there are other genomic regions where a gene is actively marked (H3K27ac) in one population of neurons and repressed (H3K27me3) in the other. We first quantified the level of each modification observed in the two cell populations over a 10kb window scanned in 5kb increments across the whole genome (Figure 6A and B, top left). We chose a 10kb window to identify broad patterns, as H3K27me3 has been shown to mark the genome in broad domains of tens to hundred kilobases (61). H3K27ac can also mark the genome in broad domains, although it is also enriched at active promoters (61). The majority of genomic windows were similarly modified in the two cell populations (Figure 6A and B, top right). To provide a genome-wide view of the differential modification, we projected the data onto a Hilbert curve (Figure 6A and B, bottom). The Hilbert curve representation essentially folds the entire genome onto itself in a self-similar, or fractal, manner that fits into a two-dimensional image where neighboring pixels are typically also close in genomic sequence. Coloring this curve according to a genomic signal, such as differential modification, enables one to visualize its genome-wide spatial distribution in a compact manner. It is clear from these plots that differences in histone modifications between the two cell types occur in broad domains rather than individual windows (Figure 6A and B, bottom). As expected, the differential H3K27me3 modification occurs in broader domains than H3K27ac (57,61). We next asked how often a stronger H3K27ac signal in one cell type accompanies a stronger H3K27me3 signal in the other cell type. To address this issue, we calculated a correlation score between the differential H3K27ac and H3K27me3 modification levels measured in each genomic window (Figure 6C, top). Projecting this score onto a Hilbert curve indicates only a few discrete loci in the genome with strongly opposing differential H3K27me3 and H3K27ac signals in octopaminergic neurons versus Kenyon cells. These regions cover roughly 700 kb of the genome and contain 16 genes, including 10 that are significantly differentially expressed, such as the mushroom body regulators (dac, toy, ey) and the vesicular transporter for octopamine (Vmat) (Figure 6C, bottom). Performing this series of analyses at a 1kb window scale does not significantly change the results. As expected, the Hilbert images become more punctate and the colors more intense; however, the distributions of histone modification levels and the broad domains of differential modification remain similar.
Figure 6.
Genome-wide comparison of active versus silenced chromatin in octopaminergic and Kenyon cells. (A) Scatterplot: Each point represents the intensity of H3K27ac modification over a 10kb window (5kb increments) in the genome, as measured in octopaminergic (x-axis) and Kenyon cell (y-axis) populations. The points are colored to reflect the cell type with higher levels of modification (blue = octopaminergic cells; orange = Kenyon cells). The distribution of differential modification in each genomic window is shown in a histogram colored on the same scale, with each numeric label denoting the number of windows represented in each bar. The larger image represents the intensity of differential modification across all windows in the genome, colored on the same scale and organized into a Hilbert curve. A Hilbert curve is a self-similar, or fractal, curve that essentially folds a line representing the genome sequence onto itself and packs it into a two-dimensional image. Coloring this folded line according to a genomic signal, such as histone modification, offers a convenient way of visualizing genome-wide data (26). The curve begins at the top left corner of the image, with the first window of chromosome 2L, and winds counter-clockwise in an intricate pattern that ends in the top right corner with the last window of chromosome X. (B) A similar representation as (A) depicts the repressive H3K27me3 modification in the two cell populations. (C) A comparison of the levels of differential H3K27ac and H3K27me3 modification as measured in panels (A) and (B). The colors range from purple in windows with anti-correlated modifications (i.e. strong acetylation in one cell population and strong trimethylation in the other) to green in windows with strongly correlated modifications (more acetylation as well as trimethylation in the same population). The genes that fall under the most anti-correlated (dZ ≥ 1.5) loci are labeled below the Hilbert curve. The numbers of RNA-seq reads aligning to Vmat and the nested CG13331 gene in the octopaminergic population were so high that CUFFLINKS (20) was unable to reliably estimate expression levels and instead reported a ‘HIDATA’ signal. (D) Ranking the differentially expressed genes by the strength of this anti-correlation score enriches for transcription factors, including those known to regulate mushroom body development (ey, toy, dac, Hr51).
We then returned to the list of differentially expressed genes (Figure 3F) and ordered them by the anti-correlation of their differential H3K27me3 and H3K27ac signals (Figure 6D). We found, as we previously observed (Figure 5A), that many of the anti-correlated genes were transcription factors. In the case of the Kenyon cell population, four factors known to play a role in mushroom body development were highly ranked by this analysis (ey, toy, dac, Hr51) (48,62). Similarly, the two most anti-correlated loci in octopaminergic cells were CG4328, a homeobox transcription factor, and dmrt99b, a doublesex-related transcription factor.
DISCUSSION
Our version of the INTACT method enables both the isolation of specific neuronal cell types in Drosophila and their characterization by RNA-seq, ChIP-seq and other systematic genomic methods. Expression of our UAS-nuclear tag cassettes can be driven by any GAL4 line, such as those described in large systematic collections of drivers that have been screened for specific neuronal expression patterns (12). We showed that we can isolate tagged nuclei in high yields (∼50%) at high purity (∼99%) from sparse lines where a few 100 neurons (Tdc2) are tagged per brain. Because the purification protocol starts from frozen adult flies, we can amass many thousands of frozen animals, if necessary to obtain sufficient numbers of cells, either from a sparsely expressing line or for a genomic analysis that requires a large amount of input material (such as ChIP-seq). An additional advantage of starting with frozen flies is that in cases where the expression of a GAL4 driver is only characterized at the level of the brain (12), exogenous expression in the thoracicoabdominal region of the body can be ignored because the heads of frozen flies can be isolated by passing dissociated bodies over cooled sieves. We expect that the protocol will work on lines that are sparser than Tdc2, but the exact limit of sensitivity is unknown at this time.
The most immediate application we envision for this technology is the generation of cell type–specific gene expression profiles of specific Drosophila neuronal cell types by INTACT/RNA-seq. High resolution anatomical descriptions of specific cell types in neuronal circuits has been made possible by the systematic identification of cell type–specific GAL4 lines (12), which can be used to drive the expression of a nuclear tag, thus enabling the generation of cell type–specific profiles. This will allow the systematic characterization of the neurotransmitters, receptors, peptides and transcription factors expressed by the individual neurons that populate a neuronal circuit. Our data show that such gene expression profiles can be obtained by either RNA-seq or ChIP-seq, but RNA-seq gives better signal/noise and requires less input material (102–103 nuclei for RNA-seq versus 105–106 nuclei for ChIP-seq).
An advantage of isolating nuclei (either by INTACT or other sorting approaches) is that one can apply high-throughput genomic characterization protocols to isolated nuclei, beyond just transcriptional profiling. Our experiments demonstrate the reliability and feasibility of chromatin profiling by INTACT/ChIP-seq, and we also expect to be able to apply a variety of other methods, such as DNAse-seq, Gro-seq, Nascent-seq, Hi-C and ChIA-PET (43,63–66). We therefore expect to gain access not only to gene expression profiles but also to the transcriptional regulatory networks that are necessary for driving the expression profile. Such information has proven critical to the study of the mechanisms that control neuronal identity. For example, in the worm C. elegans, excellent progress has been made in the identification of terminal selector transcription factors, which maintain the identity of differentiated neurons (67–69). These factors were identified by first generating a list of genes specifically expressed in the neuron of interest (a gene battery), followed by a thorough experimental analysis to identify regulatory regions and binding sites around the loci of members of the gene battery. By enabling comprehensive application of the same basic idea, we expect that INTACT should facilitate such efforts in Drosophila neurons.
When we compared the chromatin profiles of Kenyon cells (OK107) with octopaminergic neurons (Tdc2), we fortuitously noticed a pattern that suggests a means of screening for key transcription factors that are involved in either the establishment or maintenance of neuronal identity. PcG-mediated trimethylation of histone H3 on lysine 27 has been implicated in the regulation of transcription factors that are known to play an important role in development (70,71), and we observed selective PcG-silencing of transcription factors in differentiated neurons. In fact, some of these loci show a strongly anti-correlated H3K27me3 and H3K27ac signal in octopaminergic neurons (Tdc2) and Kenyon cells (OK107). We imagine that key transcription factors, potentially capable of altering cell fate, must be silenced in cell types where they should be off, and thus they are targeted with an additional layer of repression (PcG-mediated). We hypothesized that we can enrich for these factors by identifying loci that show expression (measured by RNA-seq) and H3K27ac marking in one cell type along with an anti-correlated lack of expression and PcG-mediated silencing in the other cell type. When we do this for Kenyon cells, a small set of transcription factors are identified, including ey, dac, toy and Hr51, all of which are known to play a role in the development of the mushroom body (48,62). When we do the reverse comparison for octopaminergic neurons, where much less is known about their transcriptional program, we identify a different set of genes including the presumptive transcription factors dmrt99B, fd59A, Fer2 and CG4328. Consistent with the hypothesis that these factors play a role in the specification of octopaminergic neurons, all four are expressed on the embryonic midline (72,73), from which the octopaminergic cell population arises (74). It is not uncommon for the same transcriptional regulatory network to play a role both in the early development and adult maintenance of a neuronal cell type as has been described for Tv neuropeptidergic cells (75). A role for PcG-silencing in the specification of cell types, in particular specific subsets of neurons, has been suggested by others (76–80).
Our PcG-silencing data can also be used to characterize the heterogeneity of a population of neurons. In bulk neuronal nuclei (57C10), we see many genetic loci that show signatures of being both active and repressed (active: RNA-seq, H3K4me3, H3K27ac; repressed: H3K27me3). A simple explanation, which has been previously observed in other systems (81), is that the bulk population is a mixture composed of expressing and non-expressing/repressed cells. For example, in bulk neuronal nuclei (57C10) the ey locus appears to be active and repressed because the gene is known to be expressed in a specific group of cells in the adult brain (82). In the Kenyon cell population (OK107) where ey is broadly expressed, the locus is active and lacks repression, which is consistent with the OK107-GAL4 line being an enhancer trap near the ey locus (31).
A major limitation of INTACT involves its application to sparsely tagged lines (1–10 neurons) or to cell types found at earlier stages of development where freezing the animals is not possible (larval stages of development). For example, some of the downstream genomic protocols, such as ChIP-seq, typically require 105–106 cells. We expect this barrier to drop as more sophisticated methodologies for amplification are interfaced with the technique. For example, a method has been described that allows ChIP-seq to be performed on 103 cells (83). Another solution for the isolation of nuclei from sparsely tagged lines might involve the generation of a second generation of tags that have increased antigenicity or that enable two-step purification procedures similar to those used in proteomic assays that rely on tandem affinity purification (84).
FUNDING
Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Julide Bilen and Todd Laverty for having the patience to teach us how to work with Drosophila; Amanda Cavallaro, Dona Fetter, Jennifer Jeter, Kevin McGowan, Andrey Revyakin, Zhengjian Zhen, Tim Brown, the Eddy lab, Julie Simpson, Jim Truman, Aljoscha Nern, Barret Pfeiffer, Gerry Rubin and David Stern were also helpful at various points in the development of this technique. We also thank Goran Ceric for managing Janelia Farm’s high-performance computing resources.
REFERENCES
- 1.Davidson EH. The regulatory genome: gene regulatory networks in development and evolution. Amsterdam: Elsevier, Acad. Press; 2006. [Google Scholar]
- 2.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annual Ann. Review of Cell and Developmental. Biology Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 3.Roy S, Ernst J, Kharchenko PV, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 5.Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krasnow MA, Cumberledge S, Manning G, Herzenberg LA, Nolan GP. Whole animal cell sorting of Drosophila embryos. Science. 1991;251:81–85. doi: 10.1126/science.1898782. [DOI] [PubMed] [Google Scholar]
- 7.Sugino K, Hempel CM, Miller MN, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 8.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012 doi: 10.1101/gr.131748.111. 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonn S, Zinzen RP, Girardot C, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer BD, Jenett A, Hammonds AS, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 14.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 15.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrera LO, Li Z, Smith AD, et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Tweedie S, Ashburner M, Falls K, et al. FlyBase: enhancing drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Williams B, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyne R, Smith R, Rutherford K, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfreundt U, James DP, Tweedie S, et al. FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 2010;38:D443–447. doi: 10.1093/nar/gkp910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hens K, Feuz JD, Isakova A, et al. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat. Methods. 2011;8:1065–1070. doi: 10.1038/nmeth.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–1235. doi: 10.1093/bioinformatics/btp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crisp M, Liu Q, Roux K, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 30.Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell. Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- 31.Adachi Y, Hauck B, Clements J, et al. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech. Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Cole SH, Carney GE, McClung CA, et al. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 2005;280:14948–14955. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- 33.Busch S, Selcho M, Ito K, Tanimoto H. A map of octopaminergic neurons in the Drosophila brain. J. Comp. Neurol. 2009;513:643–667. doi: 10.1002/cne.21966. [DOI] [PubMed] [Google Scholar]
- 34.Ito K, Urban J, Technau GM. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Roux’s Arch. Dev. Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 35.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 36.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 37.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 40.Górska-Andrzejak J, Salvaterra PM, Meinertzhagen IA, Krzeptowski W, Görlich A, Pyza E. Cyclical expression of Na+/K+-ATPase in the visual system of Drosophila melanogaster. J. Insect Physiol. 2009;55:459–468. doi: 10.1016/j.jinsphys.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwai Y, Usui T, Hirano S, et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 42.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 43.Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 44.Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, Moon S, Cha Y, Chung YD. Drosophila TRPN(=NOMPC) channel localizes to the distal end of mechanosensory cilia. PLoS ONE. 2010;5:e11012. doi: 10.1371/journal.pone.0011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khodor YL, Rodriguez J, Abruzzi KC, et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurusu M, Nagao T, Walldorf U, et al. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc. Natl Acad. Sci. USA. 2000;97:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;127:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- 50.Johard HAD, Enell LE, Gustafsson E, et al. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. J. Comp. Neurol. 2008;507:1479–1496. doi: 10.1002/cne.21636. [DOI] [PubMed] [Google Scholar]
- 51.Han KA, Millar NS, Davis RL. A novel octopamine receptor with preferential expression in Drosophila mushroom bodies. J. Neurosci. 1998;18:3650–3658. doi: 10.1523/JNEUROSCI.18-10-03650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vömel M, Wegener C. Neuroarchitecture of aminergic systems in the larval ventral ganglion of Drosophila melanogaster. PLoS ONE. 2008;3:e1848. doi: 10.1371/journal.pone.0001848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greer CL, Grygoruk A, Patton DE, et al. A splice variant of the Drosophila vesicular monoamine transporter contains a conserved trafficking domain and functions in the storage of dopamine, serotonin, and octopamine. J. Neurobiol. 2005;64:239–258. doi: 10.1002/neu.20146. [DOI] [PubMed] [Google Scholar]
- 54.Rodríguez-Valentín R, López-González I, Jorquera R, et al. Oviduct contraction in Drosophila is modulated by a neural network that is both, octopaminergic and glutamatergic. J. Cell. Physiol. 2006;209:183–198. doi: 10.1002/jcp.20722. [DOI] [PubMed] [Google Scholar]
- 55.Brooks ES, Greer CL, Romero-Calderón R, et al. A putative vesicular transporter expressed in Drosophila mushroom bodies that mediates sexual behavior may define a neurotransmitter system. Neuron. 2011;72:316–329. doi: 10.1016/j.neuron.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 57.Kharchenko PV, Alekseyenko AA, Schwartz YB, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 59.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc. Natl Acad. Sci. USA. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nekrasov M, Klymenko T, Fraterman S, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz YB, Kahn TG, Stenberg P, et al. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin S, Huang Y, Lee T. Nuclear receptor unfulfilled regulates axonal guidance and cell identity of Drosophila mushroom body neurons. PLoS ONE. 2009;4:e8392. doi: 10.1371/journal.pone.0008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyle AP, Davis S, Shulha HP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leiberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 2012;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 71.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo dna methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 72.Tomancak P, Berman BP, Beaton A, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev. Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Busch S, Tanimoto H. Cellular configuration of single octopamine neurons in Drosophila. J. Comp. Neurol. 2010;518:2355–2364. doi: 10.1002/cne.22337. [DOI] [PubMed] [Google Scholar]
- 75.Eade KT, Fancher HA, Ridyard MS, Allan DW. Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS Genet. 2012;8:e1002501. doi: 10.1371/journal.pgen.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Juan AH, Derfoul A, Feng X, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smouse D, Goodman C, Mahowald A, Perrimon N. polyhomeotic: a gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes Dev. 1988;2:830–842. doi: 10.1101/gad.2.7.830. [DOI] [PubMed] [Google Scholar]
- 78.Smouse D, Perrimon N. Genetic dissection of a complex neurological mutant, polyhomeotic, in Drosophila. Dev. Biol. 1990;139:169–185. doi: 10.1016/0012-1606(90)90286-r. [DOI] [PubMed] [Google Scholar]
- 79.Parrish JZ, Emoto K, Jan LY, Jan YN. Polycomb genes interact with the tumor suppressor genes hippo and warts in the maintenance of Drosophila sensory neuron dendrites. Genes Dev. 2007;21:956–972. doi: 10.1101/gad.1514507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J, Lee CHJ, Lin S, Lee T. Steroid hormone-dependent transformation of polyhomeotic mutant neurons in the Drosophila brain. Development. 2006;133:1231–1240. doi: 10.1242/dev.02299. [DOI] [PubMed] [Google Scholar]
- 81.Akkers RC, van Heeringen SJ, Jacobi UG, et al. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Callaerts P, Leng S, Clements J, et al. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J. Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 83.Shankaranarayanan P, Mendoza-Parra MA, Walia M, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat. Methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 84.Rigaut G, Shevchenko A, Rutz B, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]