Abstract
T-cell factor (Tcf)/lymphoid-enhancer factor (Lef) proteins are a structurally diverse family of deoxyribonucleic acid-binding proteins that have essential nuclear functions in Wnt/β-catenin signalling. Expression of Wnt/β-catenin target genes is highly dependent on context, but the precise role of Tcf/Lef family members in the generation and maintenance of cell-type-specific Wnt/β-catenin responses is unknown. Herein, we show that induction of a subset of Wnt/β-catenin targets in embryonic stem cells depends on Tcf1 and Tcf4, whereas other co-expressed Tcf/Lef family members cannot induce these targets. The Tcf1/Tcf4-dependent gene responses to Wnt are primarily if not exclusively mediated by C-clamp-containing Tcf1E and Tcf4E splice variants. A combined knockdown of Tcf1/Tcf4 abrogates Wnt-inducible transcription but does not affect the active chromatin conformation of their targets. Thus, the transcriptionally poised state of Wnt/β-catenin targets is maintained independent of Tcf/Lef proteins. Conversely, ectopically overexpressed Tcf1E cannot invade silent chromatin and fails to initiate expression of inactive Wnt/β-catenin targets even if repressive chromatin modifications are abolished. The observed non-redundant functions of Tcf1/Tcf4 isoforms in acute transcriptional activation demonstrated that the cell-type-specific complement of Tcf/Lef proteins is a critical determinant of context-dependent Wnt/β-catenin responses. Moreover, the apparent inability to cope with chromatin uncovers an intrinsic property of Tcf/Lef proteins that prevents false ectopic induction and ensures spatiotemporal stability of Wnt/β-catenin target gene expression.
INTRODUCTION
Embryogenesis and post-natal tissue homeostasis rely on dynamic and precisely coordinated alterations in stage- and tissue-specific gene expression. The Wnt/β-catenin signal transduction cascade belongs to a small group of signalling pathways with key roles in orchestrating dynamic gene expression changes throughout development and in adulthood (1,2). Wnt growth factor binding to Frizzled and low-density lipoprotein receptor-related protein (LRP) cell surface receptors triggers a series of events that ultimately lead to β-catenin accumulation in the nucleus where it serves as a cofactor for the T-cell factor (Tcf)/lymphoid-enhancer factor (Lef) family of sequence-specific deoxyribonucleic acid (DNA)-binding proteins (2,3). It is thought that Tcf/Lef proteins are positioned at Wnt/β-catenin target gene promoters in unstimulated cells, where they form complexes with co-repressor proteins to suppress basal transcription. Wnt-induced β-catenin binding to Tcf/Lef family members displaces the co-repressors and leads to promoter activation (4).
Cellular responses to Wnt/β-catenin signalling are highly dependent on context, and depending on the cellular background, the Wnt/β-catenin signal transduction pathway acts only on a subset of its potential targets. The cell-type-specific target gene selection and processes that preserve the silent state of Wnt/β-catenin target genes outside of their cognate expression domains, also in the presence of an active Wnt pathway, are guided by largely unknown mechanisms. One way in which promoter accessibility for Tcf/Lef proteins and Wnt target gene activation could be restricted depends on chromatin structural features (5,6). In support of this idea, we have previously shown that cell-type-specific loss of Wnt induction correlates with a high level of promoter DNA methylation and association with repressive trimethylation of lysine 27 in histone H3 (H3K27me3). Active histone modifications and Tcf/Lef proteins are absent from non-responsive Wnt/β-catenin target genes (5–7). However, causal relationships between repressive chromatin features and Tcf/Lef factor exclusion from their target genes have not been clarified.
In addition to chromatin structure, various observations suggest that Tcf/Lef family members play critical roles in shaping the tissue- and stage-specific transcriptional output from Wnt/β-catenin signalling. In mammals, the Tcf/Lef family consists of Tcf1, Tcf3, Tcf4 and Lef1, which are encoded by four genes: Tcf7, Tcf7l1, Tcf7l2 and Lef1, respectively. Tcf/Lef genes have tissue-specific expression patterns (8–10), and loss-of-function studies have demonstrated a unique requirement for individual Tcf/Lef genes in certain developmental processes (11–14). Moreover, changes in Tcf/Lef expression correlate with phenotypic changes in tumour cells (15–17). Where functional cooperation within the Tcf/Lef family has been observed, this seems to be highly dependent on context, and partnerships among Tcf/Lef genes change depending on cellular background (8,18–20). Overall, these observations suggest that Tcf/Lef family members are not fully interchangeable and are directed at different subgroups of Wnt/β-catenin target genes. In support of these observations, functional differences among Tcf/Lef family members in reporter gene assays (21–23) and differential promoter occupancy by Tcf/Lef family members have been observed (7), but firm evidence for the hypothesis that different Tcf/Lef family members control distinct groups of Wnt/β-catenin target genes is lacking.
Promoter-specific, non-redundant Tcf/Lef protein functions may arise from differences in their domain composition. Structural features common to all members of the Tcf/Lef protein family include an N-terminal β-catenin-binding domain, interaction sites for Groucho-related-gene (Grg)/transducin-like enhancer of split (TLE) transcriptional co-repressors, a high-mobility-group-box (HMG-box) DNA-binding domain and an adjacent nuclear localization signal (3,24). Outside of these domains, however, sequence similarity decreases considerably. Structural divergence among Tcf/Lef family members is further enhanced by alternative splicing. In particular, transcripts from the Tcf7 and Tcf7l2 genes undergo extensive and tissue-specific alternative splicing (23,25–27). The resulting differences in protein architecture can endow Tcf/Lef isoforms with unique properties in protein–protein interactions (28–30), DNA-binding (31) and posttranslational modifications (32). For example, the Tcf1E and Tcf4E splice variants are equipped with a second DNA-binding domain in addition to the HMG-box, the C-clamp (31), which facilitates composite DNA sequence motif recognition by Tcf1E and Tcf4E splice variants in vitro (23,31). Accordingly, extended DNA sequence recognition could underlie the Tcf1E and Tcf4E promoter specificity observed in reporter gene assays (7,23,31). However, this is a controversial issue (21–23), and selective promoter occupancy by Tcf/Lef isoforms with or without the C-clamp in living cells has not been demonstrated.
Molecular mechanisms that generate and maintain cell-type-specific Wnt/β-catenin transcriptional responses are poorly understood. Herein, we have explored the role of Tcf/Lef family members in these processes. We demonstrate an essential, non-redundant function specifically for Tcf1 and Tcf4 splice variants in acute transcriptional activation of T/Brachyury (T/Bra), Cdx2 and Sp5 in mouse embryonic stem cells (ESCs). Interestingly, an active chromatin conformation is maintained even without Tcf1 and Tcf4. Similarly, Wnt/β-catenin signalling cannot initiate expression of certain target genes de novo in neural progenitors and myoblasts even on repressive chromatin structure destabilization and ectopic overexpression of the most potent transactivator, Tcf1E. Overall, this identifies Tcf/Lef splice variants as critical determinants of cell-type-specific Wnt/β-catenin responses. In addition, the apparent inability of Tcf proteins to overcome restrictions imposed by chromatin structure uncovers intrinsic properties of Tcf/Lef family members that aid in maintaining stable, distinct expression territories for tissue-specific Wnt/β-catenin target genes.
MATERIALS AND METHODS
Cell culture
E14 ESCs, C17.2 neural progenitors (ECACC #07062902) and C2C12 myoblasts (ATCC #CRL-1772) were cultured as described previously (7,33,34). Cell lines stably expressing the hemagglutinin (HA)-tagged hTcf1E splice variant were generated previously (7). E14 ESCs expressing HA-tagged hTcf1B were produced in an analogous manner. To activate the Wnt/β-catenin signalling pathway, cells received 200 ng/ml recombinant Wnt3a (R&D Systems) or an equivalent quantity of Wnt3a partially purified from conditioned media 6 h before harvest.
siRNA transfections and rescue
For expression analyses after knockdown of Tcf/Lef or β-catenin, 0.5 × 106 E14 ESCs seeded in six-well plates were transfected with Lipofectamine 2000 (Invitrogen) 4 h after plating. Tcf/Lef-specific sets of four siRNAs (Dharmacon) or two different siRNAs against β-catenin (Qiagen) were transfected at 5 nM (Tcf1, Tcf3 and β-catenin) or 20 nM (Lef1, Tcf4), respectively. For rescue experiments, cells were co-transfected with subsets of Tcf1/4 siRNAs exclusively targeting mouse Tcf1 and Tcf4 transcripts with expression plasmids for human Tcf1E, Tcf1B, Tcf4E2 and Tcf4M1, respectively (23). To generate similar expression levels of the splice variants, the appropriate plasmid quantities were predetermined by western blot. For formaldehyde-assisted isolation of regulatory elements (FAIRE) and chromatin immunoprecipitation (ChIP), siRNA-mediated knockdown was performed in 10 cm diameter and 15 cm diameter plates, respectively, using SiLentFect (BioRad). For knockdown of polycomb repressive complex 2 (PRC2), 1 × 105 C17.2-Tcf1E and C2C12-Tcf1E cells in six-well plates were transfected with 20 nM per siRNA (siEzh2 set of 4, Dharmacon or two different siRNAs against Suz12, Qiagen) using Lipofectamine 2000. Cells were washed once with 1x PBS 16 h after transfection, fresh medium was added and cells were cultivated for additional 32 h until harvest.
Luciferase reporter assays
Using the FuGENE HD reagent (Roche Applied Science), 2 × 104 C17.2 and C2C12 cells and 1 × 105 E14 ESCs in 24-well plates were transfected. C17.2 and C2C12 cells received a mixture of 100 ng firefly luciferase reporter plasmid with Wnt target gene promoters as indicated, 10 ng of the Renilla luciferase expression vector pRL-CMV (Promega), 100 ng of the plasmid DNA to express a constitutively active form of β-catenin (22) and expression vectors for the Tcf/Lef family members and splice variants (23). Appropriate quantities of the empty expression vector pCS2+ were added to maintain a constant total quantity of transfected DNA. E14 ESCs were transfected with 3-fold more of the above plasmids. Reporter gene activities were determined as described (22), and Renilla luciferase activity was used for normalization.
Ribonucleic acid isolation, complementary DNA synthesis and reverse transcriptase-polymerase chain reaction
Where indicated, cells were treated with 10 µM 5-aza-2-deoxycytidine (Aza) for 72 h, 1 µM trichostatin A (TSA) for 24 h or a combination of both reagents. The total ribonucleic acid (RNA) was isolated using the NucleoSpin RNA II kit (Macherey and Nagel) or the peqGOLD total RNA kit (PeqLab). Subsequently, complementary DNA (cDNA) synthesis and reverse transcriptase(RT)-polymerase chain reaction (PCR) were performed as described (7). Quantitative PCR (qPCR) was performed in an IQTM5 multicolour real-time PCR detection system (BioRad) using SYBR green reaction mix (PeqLab) and an amount of cDNA equivalent to 20 ng RNA. Experiments were performed in duplicate, and the data were normalized to Gapdh expression. Primer sequences and PCR conditions for each primer set are listed in Supplementary Table S1.
Western blotting and immunodetection
To control knockdown efficiency before ChIP and FAIRE, an aliquot of cells was collected after formaldehyde fixation and analysed using sodium dodecyl sulphate (SDS) -polyacrylamide gel electrophoresis and western blotting. Whole cell lysate and nuclear extract preparation and immunodetection were performed as described (23). hTcf1/4 splice variants were transcribed and translated in vitro as described (23). Where indicated, equal quantities of recombinant hTcf1/4 proteins were analysed in parallel to nuclear extracts to serve as reference proteins. For immunodetection, the following antibodies were used: rabbit anti-Tcf1 (C63D9; Cell Signalling, 1:2000), rabbit anti-Lef1 (C12A5; Cell Signalling, 1:1000), goat anti-Tcf3 (M20; Santa Cruz, 1:1000), rabbit anti-Tcf4 (C9B9; Cell Signalling, 1:1000), mouse anti-β-catenin (no. 610 154; BD Transduction Laboratories, 1:1000), rat anti-HA (3F10; Roche, 1:2000), mouse anti-GSK3β (no. 610 201; BD Transduction Laboratories, 1:1000) and mouse anti-αTubulin (T9026; Sigma-Aldrich, 1:10 000).
Formaldehyde-assisted isolation of regulatory elements
FAIRE was performed as described (35). The samples were sonicated using a Branson W-450D, which yielded DNA fragments with an average size of 250–500 bp. qPCR was conducted as described earlier using 40 ng of DNA recovered from crosslinked cells and non-crosslinked reference cells. All samples were analysed in duplicate, and the data were calculated as percent input. Primer sequences and PCR conditions for each primer set are listed in Supplementary Table S2.
Chromatin immunoprecipitation
Chromatin from E14 ESCs, C2C12 myoblasts, C17.2 neural progenitors and their stably transduced descendents was prepared as described (7). After Tcf1/4 and β-catenin knockdown, respectively, the chromatin was prepared using the ChIP-IT kit with an enzymatic shearing module (Active Motif). For immunoprecipitation of histone modifications on knockdown of Tcf1/4 and β-catenin, digestion buffer was added to 50 µg chromatin aliquots for equal volumes and processed as described (7). All other ChIP analyses were performed with 50 µg chromatin in 1 ml sonication buffer except for the Tcf4 and Tcf4E analyses, which used 200 µg chromatin. Immunoprecipitates were washed twice for 10 min at 4°C with 1 ml of sonication buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) [pH 7.9], 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)), followed by wash buffer A (50 mM HEPES [pH 7.9], 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate and 0.1% SDS), wash buffer B (20 mM Tris, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate and 0.5 mM PMSF) and TE buffer (10 mM Tris/HCl pH 8.0 and 1 mM EDTA). The antibodies used were rat anti-HA (3F10; Roche), goat anti-Tcf4 (sc-8631; Santa Cruz), goat anti-Tcf3 (sc-8635; Santa Cruz), rabbit anti-H3K4me3 (#39159; Active Motif), rabbit anti-H3K9/14ac (06-599; Millipore) and rabbit anti-H3K27me3 (07-449; Millipore). qPCR was performed as described earlier with 1 µl of immunoprecipitated DNA and 2% input material as the templates. All samples were analysed in duplicate, and the data were normalized relative to an unspecific control region. Primer sequences and PCR conditions for each primer set are listed in Supplementary Table S2.
RESULTS
Cell-type-specific induction of Wnt/β-catenin target genes
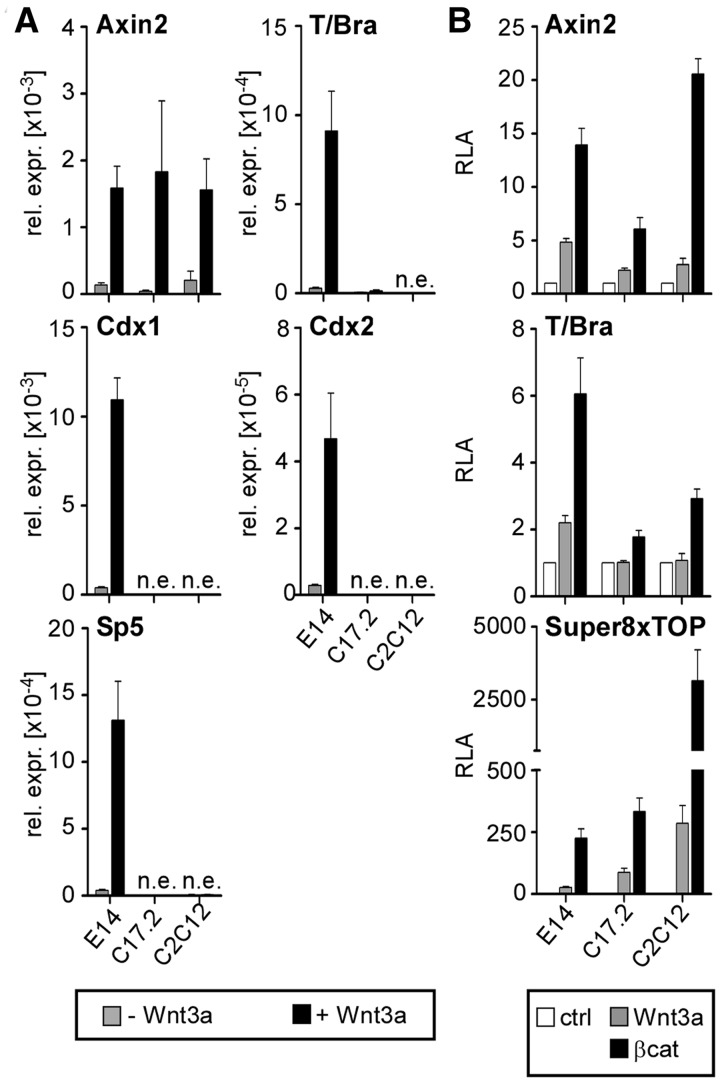
A cell culture model comprising the E14 mouse ESC line, a neural progenitor cell line (C17.2) derived from postnatal mouse cerebellum (33) and murine myoblasts (C2C12) was used to examine expression and regulation of the Wnt/β-catenin target genes Axin2, T/Bra, Cdx1, Cdx2 and Sp5. On Wnt pathway activation by application of recombinant Wnt3a, Axin2 was up-regulated in all three cell lines, whereas T/Bra, Cdx1, Cdx2 and Sp5 were only induced in E14 ESCs but not in C17.2 and C2C12 cells (Figure 1A). Cell-type-specific differential Wnt induction was also observed using luciferase reporter genes driven by the Axin2 and T/Bra promoters (Figure 1B). Transient transfection and treatment of transfected cells with Wnt3a induced a T/Bra reporter in E14 ESCs. However, Wnt3a failed to activate the reporter in C17.2 and C2C12 cells. Even overexpression of constitutively active β-catenin elicited only a reduced reporter response in C17.2 and C2C12 cells. Similar behaviour for a Cdx1 reporter construct was previously reported (7,23). In contrast, both an Axin2 reporter and the artificial pSuper8xTOPflash reporter (36) were robustly stimulated by Wnt3a addition or expression of β-catenin in E14 ESCs, C17.2 and C2C12 cells (Figure 1B), which indicates an intact signalling cascade in all three cell types. The parallels in the behaviour of the endogenous genes and corresponding episomal promoter regions suggest that trans-acting factors are involved in cell-type-specific induction of certain Wnt target genes.
Figure 1.
Cell-type-specific induction of Wnt/β-catenin target genes. (A) quantitative RT-PCR (qRT-PCR) analysis of target gene expression after Wnt3a stimulation in E14 ESCs, C17.2 and C2C12 cells. n.e., not expressed. The values are the expression levels for each gene normalized to Gapdh. (B) Assessment of Wnt induction in E14 ESCs, C17.2 and C2C12 cells using luciferase assay. Activation was analysed for Wnt-dependent reporter genes on treatment with Wnt3a or co-transfection of constitutively active β-catenin (βcat). RLA, relative luciferase activity. Unless indicated otherwise, quantitative data in this and all subsequent figures are derived from at least three independent experiments and are the mean values and corresponding standard errors (SEM).
Activation of T/Bra, Cdx2 and Sp5 by the Tcf1E and Tcf4E splice variants
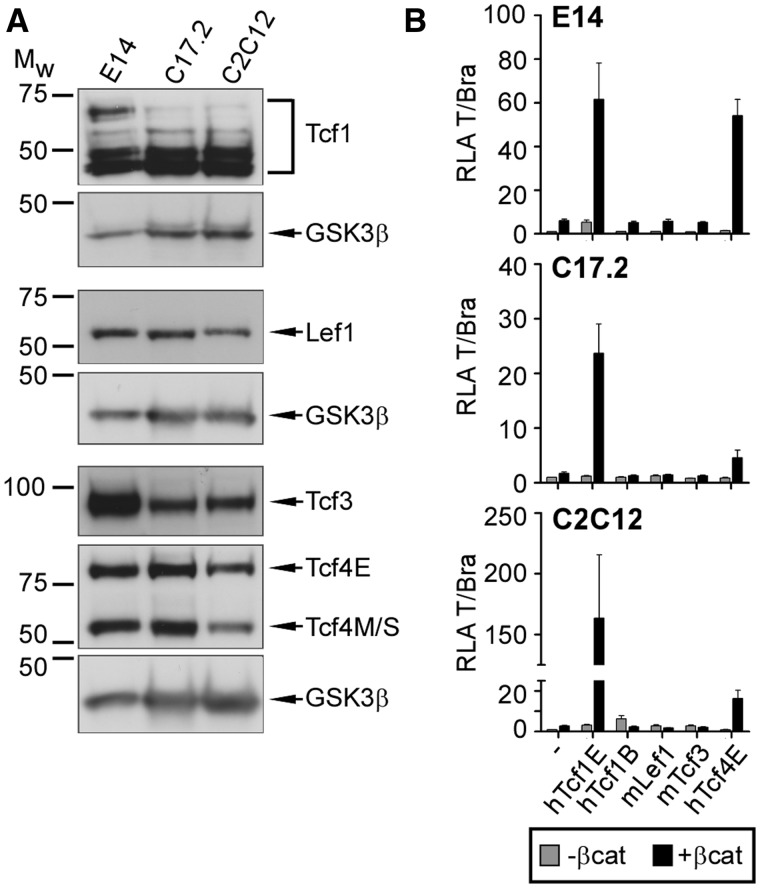
The Tcf/Lef family members are expressed in a cell-type-specific manner (8–10), and differences in their cooperation with β-catenin in activation of luciferase reporters driven by natural Wnt target gene promoters have been observed (7,21–23). Therefore, we sought to determine whether differences in Tcf/Lef expression could account for cell-type-specific induction of Wnt target genes. Therefore, we performed comparative expression analyses through western blotting using nuclear extracts from E14 ESCs, C17.2 and C2C12 cells (Figure 2A). These analyses demonstrated that all four family members are expressed in each cell line tested. No differences in Lef1 and Tcf4 expression were observed. However, consistent with previous studies, Tcf3 is more abundant in E14 ESCs compared with C17.2 and C2C12 cells (9,37). Similarly, among a cohort of Tcf1 splice variants detected, the largest variant, which is an isoform with an N-terminal β-catenin-binding domain and C-clamp-containing C-terminus, was markedly reduced in C17.2 and C2C12 cells compared with E14 ESCs (Figure 2A). Thus, changes in Tcf3 and Tcf1E expression correlate with loss of T/Bra, Cdx1, Cdx2 and Sp5 Wnt induction.
Figure 2.
Preferential activation of the T/Bra promoter by the hTcf1E and hTcf4E splice variants. (A) Western blot analysis of endogenous Tcf/Lef protein levels in nuclear extracts from E14 ESCs, C17.2 and C2C12 cells. GSK3β serves as a loading control. Mw, molecular weight standard in kDa. (B) Luciferase assay to test the capabilities of different Tcf/Lef family members and splice variants to activate the T/Bra promoter in different cell backgrounds without or with constitutively active β-catenin (βcat). RLA, relative luciferase activity.
On the basis of these results, we examined the importance of Tcf/Lef isoforms in cell-type-specific induction of Wnt target genes. Initially, we transiently transfected E14 ESCs, C17.2 and C2C12 cells with a T/Bra promoter-driven luciferase reporter and expression plasmids for different Tcf/Lef family members. Specifically, we analysed Lef1 and Tcf3, as well as the Tcf1B, Tcf1E and Tcf4E splice variants (see Figure 3C for structural representation). Co-expression of constitutively active β-catenin mimicked Wnt pathway activation. Under these conditions, Tcf/Lef family members exhibited strikingly different capacities to synergize with β-catenin for T/Bra reporter activation (Figure 2B). In all three cell lines, Tcf1E had the highest transactivation potential. Both Tcf4E and Tcf1E have a C-clamp at the C-terminus, and Tcf4E also enhanced T/Bra promoter activity in a β-catenin-dependent manner, but its potency varied depending on cellular context. Significantly, Lef1, the Tcf1B splice variant which did not show cell-type-specific expression differences, and Tcf3 failed to stimulate the T/Bra reporter.
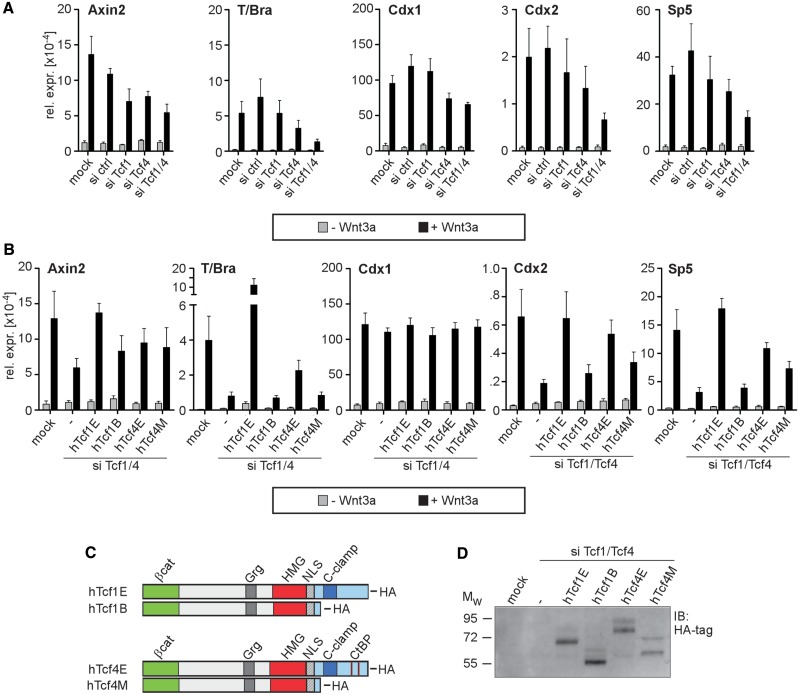
Figure 3.
The dependence of endogenous Wnt/β-catenin target gene expression on Tcf1/4 splice variants. (A) qRT-PCR showing the effect of a Tcf1/4 single (siTcf1 and siTcf4) or double knockdown (siTcf1/4) on Wnt/β-catenin target gene expression in E14 ESCs. (B) qRT-PCR showing the differential capabilities of human Tcf1 (hTcf1) and Tcf4 (hTcf4) splice variants to rescue the Tcf1/4 double knockdown phenotype in E14 ESCs. The values are the expression levels for each gene normalized to Gapdh. (C) Schematic representation of the HA-tagged hTcf1 and hTcf4 protein isoforms used for rescue experiments. The locations of the binding domains for β-catenin (βcat), Grg and CtBP co-repressors, the HMG-box, the nuclear localization signal (NLS) and the C-clamp are indicated. (D) E14 ESCs were simultaneously transfected with siRNAs and rescue plasmids coding for hTcf1 and hTcf4 protein isoforms as indicated. Subsequent western blot analysis with HA-specific antibodies confirmed similar expression levels for the hTcf1 and hTcf4 splice variants derived from the rescue plasmids. Mw, molecular-weight standard in kDa.
To test whether regulation of endogenous Wnt/β-catenin target genes also depends on specific Tcf/Lef family members, we performed loss-of-function analyses in E14 ESCs using RNA interference (RNAi). Transfection with specific siRNAs efficiently reduced levels of the corresponding target transcript, and no obvious cross-regulation was observed (Supplementary Figure S1). For Wnt/β-catenin target gene expression, we noticed that basal expression levels of Axin2 and Cdx1 in uninduced cells increased on knockdown of Tcf3 (Supplementary Figure S2A and C), which is consistent with the notion that Tcf3 functions primarily as a transcriptional repressor (3,32,37). However, individual knockdown of Tcf/Lef family members did not affect Wnt induction of T/Bra and Cdx1, and Wnt induction of Axin2, Cdx2 and Sp5 was mildly reduced at best (Supplementary Figure S2). In contrast, downregulation of β-catenin strongly impaired Wnt induction of all genes analysed (Supplementary Figure S2). Taken together, these results confirm Wnt/β-catenin-dependent expression of Axin2, T/Bra, Cdx1, Cdx2 and Sp5 in E14 ESCs and suggest that activation of these genes does not depend on cooperation between β-catenin and a single Tcf/Lef family member. Therefore, we performed double knockdown experiments for different combinations of Tcf/Lef factors (Figure 3A and Supplementary Figure S3). Based on the different levels of impaired Wnt induction by individual or combinatorial knockdown experiments, these results indicate that at least Tcf1, Tcf3 and Tcf4 act redundantly on Axin2, whereas Tcf1, Tcf3, Tcf4 and Lef1 control Cdx1 expression. Interestingly, Wnt induction of T/Bra, Cdx2 and Sp5 relies exclusively on Tcf1 and Tcf4 activity.
The siRNAs used in these knockdown experiments targeted all the Tcf1 and Tcf4 transcript variants. Thus, it is unclear whether specific isoforms are involved in Wnt induction of T/Bra, Cdx2 and Sp5. To address this issue, we performed Tcf1/4 double knockdown experiments with a reduced set of siRNAs, which specifically targeted the mouse messenger RNA transcripts, and we combined this experiment with plasmid-mediated rescue by knockdown-insensitive cDNAs for human Tcf1B, Tcf1E, Tcf4M and Tcf4E splice variants (Figure 3B–D) (23,27). Despite comparable expression levels (Figure 3D), Tcf1E exceeded the other isoforms tested and fully restored Wnt induction of Axin2, Cdx2 and Sp5 on Tcf1/4 double knockdown (Figure 3B). Tcf1E even hyperactivated the T/Bra gene. The Tcf1B, Tcf4E and Tcf4M isoforms had similar rescue potentials for the Axin2 gene and at least partially re-established its response to Wnt (Figure 3B). In contrast, attenuated Wnt induction of T/Bra, Cdx2 and Sp5 on Tcf1/4 double knockdown was restored more efficiently by Tcf4E compared with Tcf1B and Tcf4M, which were completely ineffective (Figure 3B, T/Bra) or much less potent (Figure 3B, Cdx2, Sp5). Cdx1 expression, which was the least sensitive to Tcf1/4 double knockdown in the previous series of experiments (Figure 3A), was unaffected in the combined double knockdown and plasmid-mediated overexpression of Tcf1/4 isoforms. This result underscores the specificity and significance of the effects observed for the other genes. In summary, the results from the RNAi and rescue experiments confirm the requirement for Tcf1 and Tcf4 in regulating T/Bra, Cdx2 and Sp5 expression through the Wnt/β-catenin pathway and demonstrate the transcriptional potency of the E-type splice variants, which is consistent with the Tcf1E and Tcf4E activities in reporter gene assays.
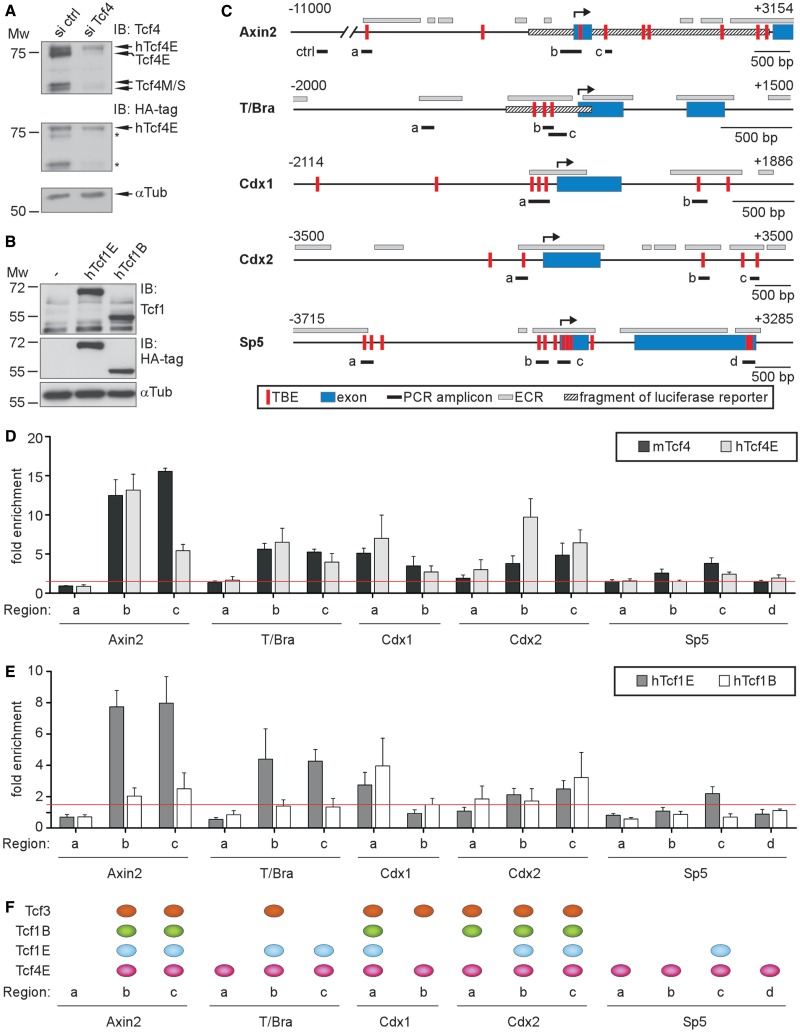
To obtain further insight into Wnt target gene regulation by Tcf1 and Tcf4, we determined their genomic distribution at the Axin2, T/Bra, Cdx1, Cdx2 and Sp5 loci using ChIP. Regions examined included known or predicted Tcf-binding elements (TBEs) situated in evolutionary conserved regions, which comprise DNA segments that likely have a regulatory function (38) (Figure 4C). The Tcf4-binding profiles were generated with antibodies that recognize all Tcf4 isoforms. To determine which sites were specifically occupied by Tcf4E, we used E14 ESCs that ectopically express HA-tagged human Tcf4E (7). Endogenous Tcf4 isoforms were knocked down in these cells to selectively retain the human variant (Figure 4A), and ChIP experiments were performed with anti-HA antibodies. ChIP analysis of endogenous Tcf1 was precluded because the antibodies performed poorly and Tcf1 abundance relative to Tcf4 was low (Supplementary Figure S4). Therefore, we generated E14 ESCs that stably overexpressed HA-tagged human Tcf1B or Tcf1E (Figure 4B), which facilitated a direct test of isoform-specific binding. Furthermore, ChIP analyses were extended to Tcf3 because it is also a Tcf/Lef family member but with different functional properties and without a C-clamp.
Figure 4.
Target gene promoter occupancy by Tcf1 and Tcf4. (A) Western blots confirming maintenance of ectopic hTcf4E expression on knockdown of endogenous Tcf4 in E14 ESCs stably expressing the HA-tagged Tcf4E splice variant. Asterisks indicate the reappearance of signals from the initial round of Tcf4 detection shown in the upper portion of the figure. α-Tubulin (αTub) serves as a loading control. (B) Western blots confirming ectopic expression of hTcf1 splice variants in stably transduced E14 ESCs. Mw, molecular-weight standard in kDa. (C) Schematic representation of the regulatory regions for Axin2, T/Bra, Cdx1, Cdx2 and Sp5. The transcription start sites (↱) and regions analysed in ChIP and FAIRE analyses using PCR (PCR amplicons) are labelled. The exons are shown as blue boxes, and the hatched bars represent DNA fragments used to drive expression of a luciferase reporter gene. TBE, Tcf-binding element (vertical red bar); ECR, evolutionary conserved region (light grey box); ctrl, control. (D, E) Quantitative chromatin IP (qChIP) in parental E14 ESCs or E14 ESCs with ectopic expression of HA-tagged hTcf4E, hTcf1E or hTcf1B variants. Tcf4- or HA-specific antibodies were used to detect Tcf4 (D) or Tcf1 (E) variant binding, respectively, to Axin2, T/Bra, Cdx1, Cdx2 and Sp5. The locations of the regions analysed are shown in (C). The data are shown as the fold enrichment over Gapdh. The red line indicates a 1.5-fold threshold level of enrichment. (F) A compilation of the genomic binding profiles for Tcf3, Tcf1B, Tcf1E and Tcf4E at the Axin2, T/Bra, Cdx1, Cdx2 and Sp5 loci. All positions are shown where the average value of enrichment for a given factor was greater than 1.5-fold.
In the ChIP experiments with isoform-non-selective Tcf4 antibodies (Figure 4D), we observed high occupancy at Axin2 regions b/c; intermediate occupancy at T/Bra regions b/c, Cdx1 regions a/b, Cdx2 regions b/c and Sp5 region c and low occupancy at T/Bra region a, Cdx2 region a and Sp5 region a/b/d. No binding was observed at Axin2 region a. The Tcf4E-specific binding pattern was similar except for statistically significant differences for two sites at Axin2 and Cdx2 (Figure 4D), which suggests that most of the genomic Tcf4-binding sites detected were preferentially or exclusively occupied by Tcf4E. Decreased association with Axin2 region c on knockdown of endogenous Tcf4 splice variants suggests differential, non-competitive occupancy by various Tcf4 isoforms. In contrast, the increase in Tcf4E occupancy at Cdx2 region b following knockdown of endogenous Tcf4 indicates competitive interactions and suppression of Tcf4E occupancy by Tcf4M/S isoforms at this site. Tcf1E-specific ChIP revealed a genomic binding pattern that differed from Tcf4E occupancy in several aspects (Figure 4E). Tcf1E had high and low levels of enrichment, respectively, at Axin2 region c and Cdx2 region b. Furthermore, in contrast to Tcf4E, we scored Tcf1E as absent from T/Bra region a, Cdx1 region b, Cdx2 region a and Sp5 regions a/b/d. Tcf1B had an even more restricted distribution and was entirely absent from T/Bra and Sp5. In addition, enrichment of Tcf1B was much lower at Axin2 and T/Bra regions b/c compared with Tcf1E and Tcf4E; although Tcf1B had occupancy levels at least equal to Tcf1E at Cdx1 and Cdx2. Lastly, we determined a binding profile for Tcf3 (Supplementary Figure S5B). Although enrichment at the target genes was lower compared with Tcf4 and Tcf1E, Tcf3 was observed at Axin2 regions b/c, T/Bra region b and Cdx2 regions a/b/c. The highest occupancy by Tcf3 was observed at Cdx1 regions a/b. A summary of the ChIP experiments and a qualitative illustration of the Tcf-binding profiles are shown in Figure 4F. Overall, Wnt/β-catenin target gene loci were occupied in complex patterns by Tcf/Lef family members and isoforms. Although we found that Sp5 exclusively associated with transactivation-competent Tcf1E and Tcf4E, the other genes examined had a more promiscuous occupancy by Tcf/Lef family members. Remarkably, non-activating Tcf3 and Tcf1B, which both lack a C-clamp, occupied T/Bra and Cdx2. Furthermore, even though both Tcf1E and Tcf4E are required for maximal Wnt induction of T/Bra, Cdx2 and Sp5, their genomic distribution at these loci differed, which supports the notion that the two factors perform different functions in target gene activation.
An active chromatin conformation at target gene promoters is preserved on Tcf1/4 double knockdown
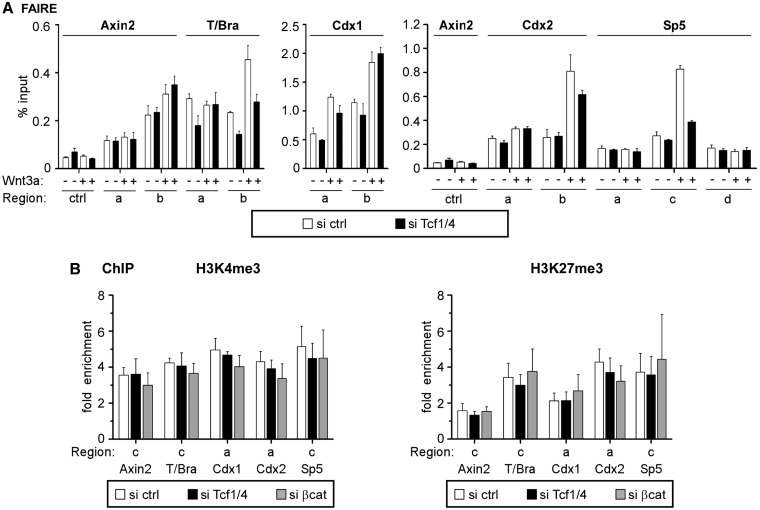
Having established the importance of Tcf1 and Tcf4 in transcriptional activation, we examined their role in maintaining a permissive chromatin status at Wnt-responsive genes using FAIRE, which yields structural information about regulatory regions similar to DNAseI hypersensitivity (35). Compared with a non-regulatory control, Tcf1/4-bound promoter-proximal and promoter-distal regions of Axin2, T/Bra, Cdx1, Cdx2 and Sp5 were highly enriched in FAIRE-DNA samples from resting E14 ESCs (Figure 5A), which indicated an open, active chromatin conformation. Wnt stimulation further opened the chromatin structure at T/Bra, Cdx1, Cdx2 and Sp5 but had less impact at Axin2 (Figure 5A). In resting cells, Tcf1/4 double knockdown (Supplementary Figure S6A) did not alter the active conformation of the Tcf1/4-bound regions and discernibly reduced FAIRE signal intensity only at T/Bra (Figure 5A). In contrast, Wnt-induced opening of the T/Bra, Cdx2 and Sp5 promoters depended on Tcf1/4, whereas changes in the Cdx1 promoter structure were not affected by the Tcf1/4 double knockdown.
Figure 5.
Tcf1 and Tcf4 are dispensable for maintenance of the active chromatin conformation. (A) FAIRE analyses to assess the chromatin conformation at Axin2, T/Bra, Cdx1, Cdx2 and Sp5 in E14 ESCs on Tcf1/4 double knockdown with or without concomitant stimulation by Wnt3a. The relative quantities of DNA recovered by FAIRE were determined using qPCR. Analysis of a control region (ctrl) was included to underscore specificity of the results. The data are shown as percent input. The locations of the regions analysed are shown in Figure 4C. (B) qChIP with antibodies specific for H3K4me3 and H3K27me3 to determine association with the promoter regions in Axin2, T/Bra, Cdx1, Cdx2 and Sp5 in E14 ESCs on Tcf1/4 double knockdown or knockdown of β-catenin. The data are shown as the fold enrichment over Gapdh.
To further investigate the impact of Tcf/Lef factors on the active chromatin state, we used ChIP to analyse trimethylation of histone H3 lysines 4 and 27 (H3K4me3 and H3K27me3) on Tcf1/4 double knockdown and β-catenin single knockdown in resting E14 ESCs (Supplementary Figure S6B). H3K4me3 is a histone mark that is characteristic of active promoter regions and may be reduced under conditions with impaired transcriptional activation, whereas the repressive mark H3K27me3 might concomitantly increase in these regions (39,40). However, levels of both H3K4me3 and H3K27me3 were unchanged at Axin2, T/Bra, Cdx1, Cdx2 and Sp5 without Tcf1/4 and β-catenin (Figure 5B). Thus, the results from our FAIRE and ChIP experiments support chromatin opening during Wnt-induced transcriptional activation that requires transactivation-competent Tcf/Lef family members, whereas active chromatin features at uninduced genes are maintained independent of β-catenin and these factors.
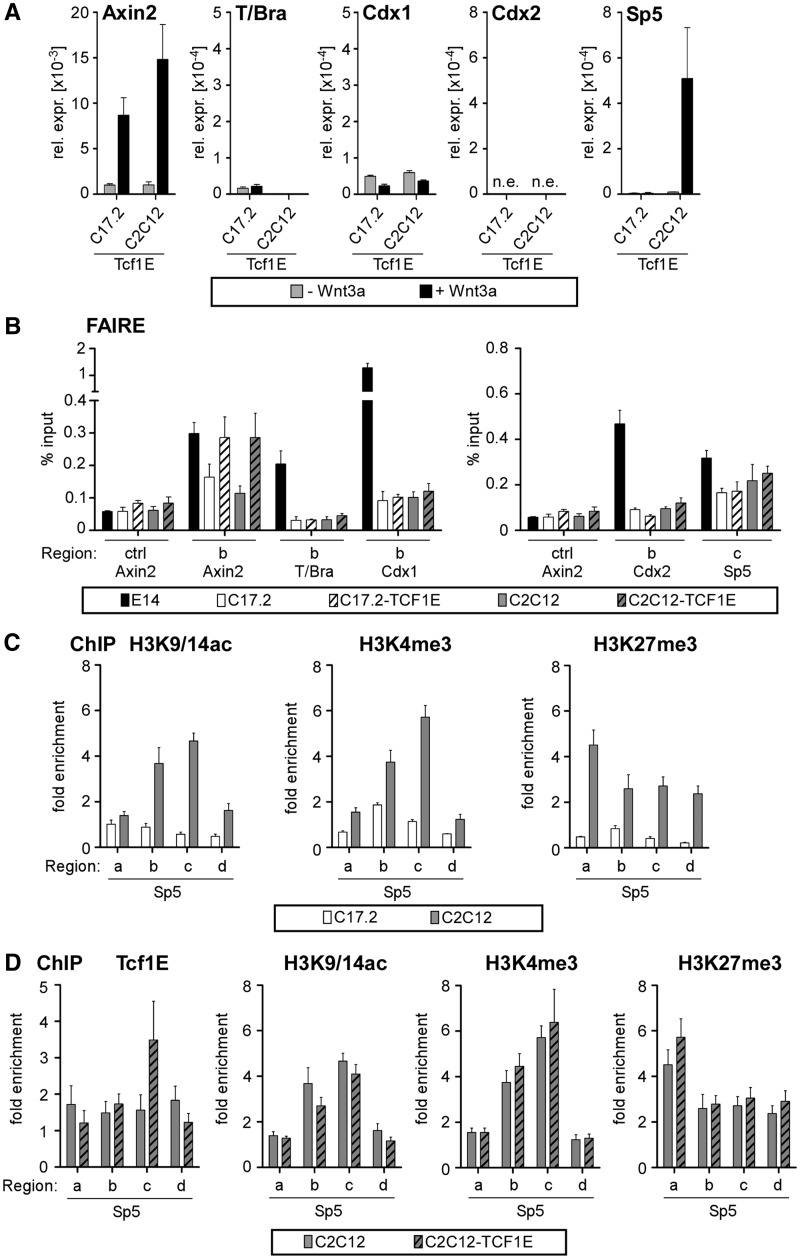
Inability of ectopic Tcf1E to open silent chromatin
Given its functional importance for Wnt induction of T/Bra, Cdx2 and Sp5 in E14 ESCs and its decreased expression in C17.2 and C2C12 cells, we further analysed the role of Tcf1E in cell-type-specific Wnt/β-catenin target gene regulation and investigated whether Tcf1E could establish Wnt induction de novo when ectopically overexpressed in C17.2 and C2C12 cells (Supplementary Figure S7). Wnt pathway functionality was maintained under these conditions as indicated by Wnt induction of Axin2 (Figure 6A), yet ectopic Tcf1E could not confer Wnt responsiveness to T/Bra, Cdx1 and Cdx2. Only Sp5 gained Wnt induction in C2C12-Tcf1E cells (Figure 6A). To determine whether Tcf1E could at least induce chromatin structural changes, we again used FAIRE. This experiment demonstrated that T/Bra, Cdx1 and Cdx2 are in a closed state in C17.2 and C2C12 cells, which is maintained even with excess Tcf1E (Figure 6B). Sp5 differs and has partially open chromatin in C17.2 and C2C12 cells, which may explain why Tcf1E can act on Sp5 in the C2C12 background. Similarly, the Axin2 promoter has an open conformation in C17.2 and C2C12 cells, and ectopic Tcf1E could further increase Axin2-derived FAIRE signals (Figure 6B), which suggests that Tcf1E can readily access regulatory elements if they are in the open conformation.
Figure 6.
A pre-existing open chromatin conformation is required for Tcf1E to access its target genes. (A) qRT-PCR analysis of target gene expression after Wnt stimulation in C17.2-Tcf1E and C2C12-Tcf1E cells. The values from all qRT-PCR analyses are the expression levels of each gene normalized to Gapdh. Note the much lower levels of T/Bra and Cdx1 expression compared with E14 ESCs (Figure 1). (B) qPCR after FAIRE analyses to monitor the chromatin conformation states at Axin2, T/Bra, Cdx1, Cdx2 and Sp5 in E14 ESCs, parental C17.2 and C2C12 cells, as well as C17.2-Tcf1E and C2C12-Tcf1E cells. The data are shown as percent input. (C) qChIP with antibodies specific for H3K9/14ac, H3K4me3 and H3K27me3 to determine association with the indicated genomic regions of Sp5 in C17.2 and C2C12 cells. The data are shown as the fold enrichment over Gapdh. (D) qChIP with antibodies specific for HA-tagged Tcf1E, H3K9/14ac, H3K4me3 and H3K27me3 to determine association with the indicated genomic regions for Sp5 in C2C12 and C2C12-Tcf1E cells. The data are shown as the fold enrichment over Gapdh.
A combined knockdown of Tcf1 and Tcf4 in ESCs had no impact on active and repressive histone modifications at Wnt/β-catenin target genes (Figure 5). Thus, the observation that ectopic Tcf1E conferred Wnt induction to Sp5 in C2C12 cells allowed us to complement our analysis in ESCs through gain-of-function studies and to investigate how Tcf1E in C2C12 cells affected histone modifications at a gene with newly gained Wnt induction. Comparative ChIP experiments demonstrated that the active histone modifications H3K9/14ac and H3K4me3 as well as the repressive mark H3K27me3 are present at the Sp5 locus in parental C2C12 cells, which indicates that this gene is bivalent (Figure 6C). In contrast, these histone modifications are much reduced or entirely absent from Sp5 in C17.2 cells. Furthermore, Tcf1E occupied the same region c in the first exon of Sp5 in C2C12-Tcf1E cells similar to ESCs (Figure 6D). However, ectopic expression of Tcf1E did not affect levels of H3K9/14ac, H3K4me3 and H3K27me3 at various regions in the Sp5 locus (Figure 6D). These observations corroborate our ESC observations and suggest that Sp5 is bivalent in C2C12 cells but not in C17.2, which likely is a prerequisite for promoter occupancy by Tcf1E. Tcf1E seemingly makes no additional contribution to the Sp5 chromatin conformation.
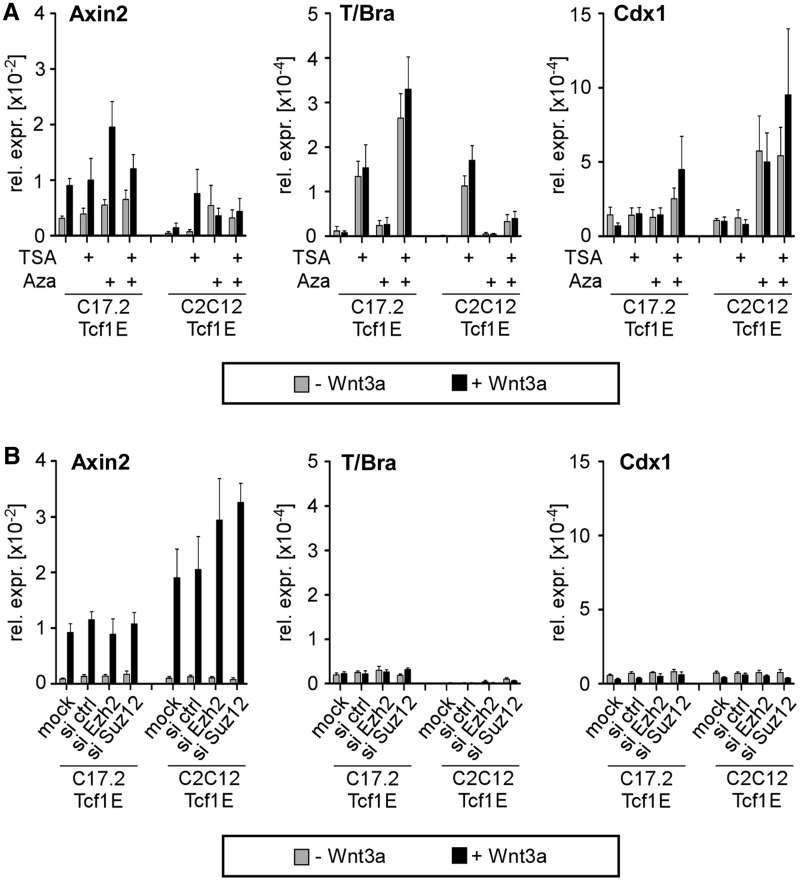
Cell-type-specific loss of T/Bra and Cdx1 Wnt induction correlates with reduced histone acetylation, hypermethylation of promoter DNA and increased trimethylation of H3K27 (7), which may block Tcf1E access to its target sites. Therefore, we investigated whether destabilizing repressive chromatin facilitates Tcf1E-promoted Wnt induction. C17.2-Tcf1E and C2C12-Tcf1E cells were treated with TSA and Aza to inhibit histone deacetylases and DNA methyltransferases, respectively. Knockdown of Ezh2 or Suz12, which are in the PRC2, was used to abrogate H3K27me3. However, despite efficacy of these strategies to weaken repressive chromatin features (Supplementary Figure S7) and unimpaired Wnt pathway activity, Wnt induction of T/Bra and Cdx1 could not be re-established (Figure 7). Only Wnt-independent basal transcription of T/Bra and Cdx1 was increased by TSA and Aza, respectively (Figure 7A), which is consistent with our previous observations in the parental C17.2 cells (7) and suggests that epigenetic mechanisms are involved in T/Bra and Cdx1 repression. The observation that Tcf1E, the key activator in ESCs, did not re-establish the Wnt response even after chromatin-based repression mechanisms are abolished suggests that additional mechanisms restrict promoter accessibility. Seemingly, Tcf/Lef family members inherently cannot initiate Wnt-dependent gene expression de novo.
Figure 7.
Interference with repressive chromatin structure does not enable ectopically expressed Tcf1E to restore Wnt induction of T/Bra and Cdx1 in C17.2 and C2C12 cells. (A) qRT-PCR analysis of target gene expression in C17.2-Tcf1E and C2C12-Tcf1E cells with or without Wnt3a and after treatment with TSA and Aza as indicated. (B) qRT-PCR analysis of target gene expression in C17.2-Tcf1E and C2C12-Tcf1E cells with or without Wnt3a and on Ezh2 or Suz12 knockdown as indicated. The values from all qRT-PCR analyses are the expression levels for each gene normalized to Gapdh. Note the much lower levels of T/Bra and Cdx1 expression compared with E14 ESCs (Figure 1).
DISCUSSION
Cell-type-specific expression of functionally distinct Tcf/Lef isoforms as determinant of contextual Wnt/β-catenin responses
The molecular mechanisms that generate and maintain the cell-type-specific response of Wnt/β-catenin target genes and prevent inappropriate expression outside of cognate expression domains even with an active Wnt/β-catenin pathway are largely unknown. Members of the Tcf/Lef transcription factor family are prime candidates for pathway-intrinsic regulators with potential roles in context-dependent control of Wnt/β-catenin target genes, and alterations in Tcf/Lef expression may contribute to the cell-type specificity of transcriptional responses. However, a prerequisite for this idea is that different Tcf/Lef family members facilitate expression of distinct Wnt/β-catenin target subgroups. The results from our study define for the first time the precise requirements for Tcf/Lef isoforms in Wnt/β-catenin target gene activation and provide important insights into the molecular basis of cell-type-specific responses to Wnt. We establish that Wnt induction of T/Bra, Cdx2 and Sp5 in E14 ESCs is specifically mediated by Tcf1 and Tcf4, and it primarily if not exclusively relies on the Tcf1E and Tcf4E splice variants. Despite co-expression, Lef1, Tcf3 and other Tcf1/4 splice variants are dispensable for the T/Bra, Cdx2 and Sp5 Wnt response. In contrast, Axin2 and Cdx1 have less stringent requirements for the Tcf/Lef isoforms. Thus, Tcf/Lef proteins affect different, albeit overlapping spectra of Wnt/β-catenin targets in a given cell background, and the cell-type-specific complement of Tcf/Lef isoforms may be a critical determinant of contextual Wnt/β-catenin responses.
Selective transcriptional activation of Wnt/β-catenin target genes by different Tcf/Lef isoforms has important implications. First, this hypothesis provides an alternative explanation for how different Tcf/Lef family members contribute to the same developmental processes. Aside from redundant, fully exchangeable regulation of the same genes, Tcf/Lef family members may synergize through parallel control of complementary genetic programs. Accordingly, cellular responses to Wnt/β-catenin pathway stimulation are likely to be modular and shaped by combined changes in expression of different gene sets, which are each activated by different Tcf/Lef isoforms. Second, isoform-specific transcriptional activation and the largely similar capacities of Tcf/Lef isoforms to occupy target genes facilitates finely tuned Wnt/β-catenin target gene expression through competitive binding by non-activating Tcf/Lef factors in the same cellular background (16,20,41,42). Consequently, variations in the expression of individual Tcf/Lef family members or single splice variants may shift the spectrum of actively transcribed Wnt/β-catenin target genes and, hence, the biological result of Wnt stimulation. This may be particularly significant given the changing activities of Wnt/β-catenin signalling for example in the intestine, where it controls not only stem cell maintenance but also progenitor proliferation and differentiation (43).
Role of the C-clamp-containing E-tails in target gene occupancy and transactivation by Tcf/Lef family members
Reporter gene assays with the T/Bra promoter and the combined double knockdown and rescue experiments revealed a superior transactivation capacity for the Tcf1E and Tcf4E splice variants. The activating function of Tcf4E that we observed at several endogenous Wnt/β-catenin target genes in mouse ESCs contrasts with the results of a previous study that reported a repressive influence of Tcf4 at an artificial Wnt reporter gene in a panel of human cancer cell lines and mouse fibroblasts (44). Apparently, some aspects of Tcf4 function seem to be promoter- and cell-type-specific (44). Several other studies also uncovered differences in transactivation potential among Tcf/Lef family members and demonstrated that Tcf1E and Tcf4E are uniquely capable of activating reporter gene constructs with the human LEF1 and mouse Cdx1 promoters (7,21–23,31). Mutation analyses linked the transactivation potential of Tcf1E and Tcf4E to the C-Clamp (21,22,31), which is an evolutionary conserved stretch of amino acids found in all invertebrate Tcf/Lef family members and a subset of vertebrate Tcf1 and Tcf4 splice variants but not in Lef1 and Tcf3 (23,31,45,46). In vitro, the C-clamp facilitates Tcf1E and Tcf4E splice variant recognition of composite DNA sequence motifs composed of canonical TBEs, which are contacted by the HMG-box and an adjacent 5′-RCCG-3′ element bound by the C-clamp (23,31). Similarly, dTcf/Pangolin makes dual DNA contacts with TBEs and TCF helper sites, which are related to the 5′-RCCG-3′ motif and are important for optimal induction of Wingless target genes in Drosophila (46).
The two DNA-binding domains in the Tcf1E and Tcf4E isoforms may be a simple explanation for their superior transactivation capacity. It is possible that only C-clamp-containing Tcf/Lef isoforms can bind to target genes that are dependent on Tcf1E/Tcf4E. Tcf/Lef isoforms without the C-clamp would not activate these genes because they cannot make the necessary DNA contacts. One corollary of this hypothesis is that Tcf1E/Tcf4E-specific target genes should consistently display composite Wnt response elements composed of classical TBEs and adjacent 5′-RCCG-3′ motifs. However, this is not the case. Examination of the T/Bra, Cdx2 and Sp5 DNA sequences using a published consensus sequence (31) revealed that 5′-RCCG-3′ motifs flank confirmed TBEs only at one of the three T/Bra promoter TBEs and at Sp5 region c. Furthermore, Tcf1E and Tcf4E also have superior transactivation capacity at target gene promoters that seemingly lack composite Wnt response elements (23, this study). Moreover, DNA-binding studies in vitro also do not support the concept that the C-clamp mediates differential target gene occupancy and thereby causes differences in transcriptional activation by Tcf/Lef family members. For example, Lef1 and Tcf1 have indistinguishable DNaseI footprint patterns at the human LEF1 promoter (21). Further, without a C-clamp, Lef1, Tcf3, Tcf4M and Tcf4S splice variants readily bind the Cdx1 and T/Bra promoters in vitro (22,23; Wallmen et al., unpublished). Clearly, both activating and non-activating Tcf/Lef isoforms associate with TBEs from differentially regulated target genes in vitro.
As an important step forward, we used ChIP to examine selective promoter occupancy by activating and non-activating Tcf/Lef isoforms at endogenous Wnt/β-catenin target genes in vivo. Significantly, the results from Tcf1B and Tcf3 at T/Bra and Cdx2, respectively, clearly show that both factors can associate with these genes even though they are much less potent in transactivation compared with Tcf1E or Tcf4E. Strikingly, Tcf1B and Tcf3 bind the Cdx1 promoter to an even greater extent than Tcf1E and Tcf4E, which is opposite to their transactivation potential observed with the Cdx1 luciferase reporter (23). Sp5 promoter region c is the only region with a strict parallel between transactivation and promoter occupancy. This notwithstanding and similar to the DNA-binding studies in vitro, the ChIP analyses also failed to demonstrate a clear correlation between promoter occupancy and the transactivation capacity of Tcf/Lef isoforms with or without a C-clamp.
On the basis of these observations and consistent with our previous work (23), we conclude that the C-clamp and 5′-RCCG-3′ motifs do not confer superior transactivation potential to Tcf1E and Tcf4E by promoting selective promoter recognition. Rather, we favour the concept that differences in the transactivation capacity of Tcf/Lef family members involve isoform-specific protein–protein interactions with transcriptional co-activators or DNA-binding proteins (22,47–49). For example, we have previously shown that the E-tail of Tcf4E interacts with the transcriptional co-activator p300/CBP and thereby promotes formation of a Tcf4E/β-catenin/p300 complex (22). Moreover, MUC1-C was identified as another protein interaction partner and co-activator for the Tcf4E C-clamp (50). Therefore, differences in the ability to participate in multimeric transcription factor complex formation may underlie differences in gene-specific regulatory potential of Tcf/Lef isoforms (22,47–49). This model could also explain certain quantitative differences in promoter occupancy by Tcf/Lef family members. Although all Tcf/Lef isoforms may be equally capable of initial DNA recognition, differences in post-DNA-binding events, such as protein–protein interactions, could generate isoform-specific stabilization of promoter association and, hence, stratify promoter occupancy patterns in vivo. Potential differences in protein–protein interactions between Tcf1E and Tcf4E may also explain the superior ability of Tcf1E to rescue Tcf1/4 double knockdown effects, further suggesting that Tcf1 and Tcf4 are functionally not entirely interchangeable and perform different tasks in Wnt/β-catenin target gene activation.
Activation of Cdx1-based reporter constructs showed requirements for Tcf1E and Tcf4E similar to the T/Bra promoter (23). Therefore, we were surprised to discover that the Tcf1/4 double knockdown had no effect on Wnt induction of Cdx1. Unlike the T/Bra promoter, all Tcf/Lef family members occupy the Cdx1 promoter in E14 ESCs (7), and Cdx1 can be recruited to the promoter for its gene by Lef1 to establish an autoregulatory loop (49). Accordingly, Cdx1 expression might alternatively be facilitated by the Tcf1/4 E-isoforms or Lef1/Cdx1 complexes, which would explain the disparate behaviour between the endogenous Cdx1 gene and the luciferase reporter only driven by the Cdx1 promoter.
Developmental priming of Wnt/β-catenin target genes and the importance of promoter accessibility by Tcf/Lef family members
It is increasingly recognized that lineage-specific expression of developmentally regulated genes can be presaged at time points much earlier than onset of transcription (51). Several mechanisms prime for later expression, including pioneer transcription factor binding to promoter-distal regulatory elements (52,53) or formation of an accessible promoter configuration as indicated by a paused RNA polymerase II, hypomethylation of promoter DNA and bivalent chromatin status (39,40,51,54–58). Differentiation processes are thought to resolve poised states into lineage-specific transcriptional activation or permanent silencing. Transition to an active transcription state is triggered by stage- and tissue-specific transcriptional activator binding to regulatory elements and is accompanied by acquisition of characteristic active histone modification patterns (53). In contrast, permanently silenced genes lack active chromatin marks, exhibit distinctive patterns of repressive histone modifications and may show DNA hypermethylation of promoter-associated CpG islands (5,6), which are thought to lock in the repressed state and prevent transcription factor access. How do Tcf/Lef family members and the target genes that we examined fit into these schemes? Based on structural hallmarks of chromatin, T/Bra, Cdx1, Cdx2 and Sp5 likely belong to the class of genes that feature promoter earmarking (7,40) and can exhibit repressive histone modifications and promoter DNA hypermethylation in their silent states (7). Given developmental control of Wnt/β-catenin target gene expression, our results confirm that Tcf/Lef proteins have essential functions in acute transcriptional activation. However, we could not detect any influence of the Tcf1/4 double knockdown on histone modifications and found only a limited impact on chromatin structure. Similarly, when ectopically expressed in C2C12 cells, Tcf1E readily occupied the Sp5 gene and conferred Wnt induction but did not noticeably alter the pre-existing active chromatin status. Other studies have also shown that histone modifications at Wnt/β-catenin target genes are primarily static and unaffected by Wnt/β-catenin pathway actuation (7,59). Overall, these results demonstrate that Tcf/Lef proteins are largely dispensable for maintenance of an active chromatin state and further suggest that generation of permissive chromatin structures is independent of Tcf/Lef family members.
Consistent with dispensability in maintaining active chromatin states, Tcf1E and Tcf4E could not invade silent chromatin even if the repressive DNA and histone modifications were abolished (this study; 7). An open chromatin conformation and DNA accessibility are likely prerequisites for binding site occupancy by Tcf/Lef family members. This property is seemingly shared with a large number of developmental regulators in Drosophila melanogaster (60). For Tcf/Lef proteins, the apparent inability to occupy chromatin-embedded binding sites was also observed in vitro (61) and may be due to the opposing DNA curvatures in complex with histones and the HMG-box, respectively (62). Thus, the failure of Tcf/Lef proteins to cope with chromatin per se suggests that repressive H3K27me3 and hypermethylation of promoter DNA at silent Wnt/β-catenin target genes (7) do not restrict accessibility for Tcf/Lef proteins. Instead, the repressive chromatin features could be a secondary event following lineage-specific inactivation of the mechanisms involved in Wnt/β-catenin target gene transcription poising (51,58).
Genetic studies in Drosophila melanogaster and mice show that Wnt/β-catenin pathway activity is required for maintaining gene expression but not for initiation (63–67). This hints at regulatory hierarchies and is consistent with the observation that master regulators of lineage-specific differentiation likely direct Tcf4 to its target genomic sites in intestinal epithelial cells and hematopoietic lineages (68,69). These observations are consistent with our finding that Tcf/Lef family members cannot generate active chromatin states alone de novo and likely rely on Wnt/β-catenin pathway-independent mechanisms to facilitate DNA accessibility. Thus, even though they play key roles in acute transcriptional activation, Tcf/Lef family members seem to lack pioneer factor properties. Given the wide-spread impact of Wnt/β-catenin signalling on a diversity of developmental processes, Tcf/Lef family members may have been selected for this feature to prevent erroneous ectopic induction and to ensure stable spatiotemporal control of Wnt target gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–7.
FUNDING
Deutsche Forschungsgemeinschaft [DFG He2004/8-1 and DFG CRC-850/B5 to A.H.]. Funding for open access charge: Institute of Molecular Medicine and Cell Research, Albert-Ludwigs-University Freiburg.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Rolf Kemler and Randy Moon for the gift of the T/Brachyury and pSuper8xTOPflash reporter constructs.
REFERENCES
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell. Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J. Cell. Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 4.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell. Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 5.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 6.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 7.Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol. Cell. Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a-/–like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 11.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 12.Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 13.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 14.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorieff A, Grosschedl R, Clevers H. Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(-/-)/Tcf1(-/-) embryos. EMBO J. 2004;23:1825–1833. doi: 10.1038/sj.emboj.7600191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell. Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 2003;278:16169–16175. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- 22.Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2003;278:3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 23.Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wohrle S, Hecht A. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964–1981. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 25.Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–3879. [PubMed] [Google Scholar]
- 26.Howng SL, Huang FH, Hwang SL, Lieu AS, Sy WD, Wang C, Hong YR. Differential expression and splicing isoform analysis of human Tcf-4 transcription factor in brain tumors. Int. J. Oncol. 2004;25:1685–1692. [PubMed] [Google Scholar]
- 27.Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 29.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 30.Valenta T, Lukas J, Korinek V. HMG box transcription factor TCF-4′s interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31:2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol. Cell. Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hikasa H, Sokol SY. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J. Biol. Chem. 2011;286:12093–12100. doi: 10.1074/jbc.M110.185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- 34.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 35.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 37.Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ovcharenko I, Stubbs L, Loots GG. Interpreting mammalian evolution using Fugu genome comparisons. Genomics. 2004;84:890–895. doi: 10.1016/j.ygeno.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vacik T, Stubbs JL, Lemke G. A novel mechanism for the transcriptional regulation of Wnt signaling in development. Genes Dev. 2011;25:1783–1795. doi: 10.1101/gad.17227011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 43.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 44.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc. Natl Acad. Sci. USA. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Archbold HC, Yang YX, Chen L, Cadigan KM. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta. Physiol. (Oxf) 2012;204:74–109. doi: 10.1111/j.1748-1716.2011.02293.x. [DOI] [PubMed] [Google Scholar]
- 46.Chang MV, Chang JL, Gangopadhyay A, Shearer A, Cadigan KM. Activation of wingless targets requires bipartite recognition of DNA by TCF. Curr. Biol. 2008;18:1877–1881. doi: 10.1016/j.cub.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl Acad. Sci. USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KW. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 49.Beland M, Pilon N, Houle M, Oh K, Sylvestre JR, Prinos P, Lohnes D. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol. Cell. Biol. 2004;24:5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi MD, Kufe D. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J. Biol. Chem. 2012;287:10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr. Opin. Genet. Dev. 2010;20:519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 55.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 56.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl Acad. Sci. USA. 108:3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaplan T, Li XY, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tutter AV, Fryer CJ, Jones KA. Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro. Genes Dev. 2001;15:3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 63.Danielian PS, McMahon AP. Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature. 1996;383:332–334. doi: 10.1038/383332a0. [DOI] [PubMed] [Google Scholar]
- 64.Dougan S, DiNardo S. Drosophila wingless generates cell type diversity among engrailed expressing cells. Nature. 1992;360:347–350. doi: 10.1038/360347a0. [DOI] [PubMed] [Google Scholar]
- 65.Galceran J, Hsu SC, Grosschedl R. Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proc. Natl Acad. Sci. USA. 2001;98:8668–8673. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siegfried E, Wilder EL, Perrimon N. Components of wingless signalling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- 67.Vincent JP, Lawrence PA. Drosophila wingless sustains engrailed expression only in adjoining cells: evidence from mosaic embryos. Cell. 1994;77:909–915. doi: 10.1016/0092-8674(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 68.Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, Dibiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl Acad. Sci. USA. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]