Abstract
Transcripts have been found to be site selectively edited from adenosine-to-inosine (A-to-I) in the mammalian brain, mostly in genes involved in neurotransmission. While A-to-I editing occurs at double-stranded structures, other structural requirements are largely unknown. We have investigated the requirements for editing at the I/M site in the Gabra-3 transcript of the GABAA receptor. We identify an evolutionarily conserved intronic duplex, 150 nt downstream of the exonic hairpin where the I/M site resides, which is required for its editing. This is the first time a distant RNA structure has been shown to be important for A-to-I editing. We demonstrate that the element also can induce editing in related but normally not edited RNA sequences. In human, thousands of genes are edited in duplexes formed by inverted repeats in non-coding regions. It is likely that numerous such duplexes can induce editing of coding regions throughout the transcriptome.
INTRODUCTION
Adenosine-to-inosine (A-to-I) RNA editing is catalyzed by a family of enzymes called ADARs, (adenosine deaminases that act on RNA) (1). Two enzymes, ADAR1 and ADAR2, have been proven to have catalytic activity on substrates in the mammalian brain [reviewed in (2)]. These enzymes convert A-to-I within structured RNA that is largely double stranded. A-to-I editing can recode an mRNA since I is interpreted as a guanosine (G) by the translation machinery. Any perfectly duplexed RNA containing adenosines can be a substrate for A-to-I editing in vitro, although some preferential selection exists (3). However, perfect stem loop structures are rare in vivo and the ADAR enzymes also recognize specific adenosines for deamination within double-stranded RNA structures that are interrupted by bulges and loops. In fact, we have previously shown that bulges and internal loops are important for editing specificity in a natural substrate but not for binding (4,5). The number of edited sites in an ADAR substrate usually increases with the length of the duplex [reviewed in (6)]. A-to-I editing can therefore be categorized into two types: (i) ‘hyper’-editing of multiple adenosines in longer almost completely duplexed structures, which has been found almost exclusively within untranslated regions and for which the functional consequences mostly is elusive (7–11); and (ii) ‘site selectively’ edited substrates where a few adenosines are targeted within an imperfect RNA foldback structure. The properties that make an RNA molecule/sequence prone to site selective editing are still not fully understood but the assumption is that internal mismatches and bulges within an RNA duplex are important for ADAR selectivity (4,12–14).
Most of the known site selectively edited pre-mRNAs encode proteins that are expressed in the central nervous system. ADAR-mediated editing alters the function of ligand- and voltage-gated ion channels as well as of G-protein-coupled receptors and give rise to diversified protein isoforms, essential for balanced neuronal kinetics (15–20). GABAA receptors are the main mediators of fast inhibitory neurotransmission in the mammalian nervous system [reviewed in (21)]. We have previously shown that the mammalian Gabra-3 transcript coding for the α3 subunit of the GABAA receptor is selectively A-to-I edited at one site (22). The edited A is situated in exon 9 at the third position of an isoleucine codon. Thus, upon editing, the sequence recodes for a methionine at this site. The editing event, referred to as the I/M site, is predicted to be situated within transmembrane region 3 of the α3 subunit. I/M editing is developmentally regulated and increases with age to a level of 92% edited transcripts in the adult brain (23). It has been postulated that this editing event modifies the kinetics of the receptor (24,25). Furthermore, editing has a negative effect on the cell surface presentation of α3 containing receptors (26). Unlike most other site selectively edited substrates, consisting of both exonic and intronic sequence, the putative stem loop structure required for Gabra-3 editing is formed by exonic sequence entirely. The importance of this stem loop structure for editing has been thoroughly described (25,27). The I/M site can be efficiently edited by both ADAR1 and ADAR2 (22). Both structure and sequence in the vicinity of the editing site are evolutionarily conserved and species from human to chicken have been shown to edit the I/M site (6,25).
In this work we have examined the influence of intronic sequence downstream of the I/M site on editing efficiency. A conserved intronic duplex of about 150 nt was found in intron 9, over a hundred bases downstream of the I/M site in the Gabra-3 transcript. We show that this intronic stem loop works as an editing inducer that is required for efficient site selective I/M editing. Furthermore, we demonstrate that in the presence of this intronic duplex, related transcripts not edited in vivo can be edited, suggesting that the duplex works as an editing inducer RNA.
MATERIALS AND METHODS
Plasmids and substrate mutagenesis
The ADAR1 expression vector pCS DRADA-FLIS6 (28) was a kind gift from Mary O’Connell. The ADAR2 expression vector pcDNA3 FLAG/rADAR2 and the Gabra-3 editing reporter construct pGARα3-I/M (Gabra-3 exon + intron) generated from mouse sequence has previously been described (22,29). The Gabra-3 cDNA expression vector (Gabra-3 exon) pRK5-α3 was a kind gift from Hartmut Lüddens (University of Mainz, Germany). The chicken Gabra-1 and pig Gabra-3 editing reporters were generated by polymerase chain reaction (PCR) amplification from the genomic gabra-1 and the gabra-3 gene, respectively, and were cloned into pcDNA3 FLAG. Primer sequences were as follows: chicken Gabra-1 Forward (FW), 5′-gagtgacaactgtcctaaca-3′ and Reverse, (RE), 5′-acccaatttcatatataggc-3′; pig Gabra-3 FW, 5′-tgctgaccatgacgaccctcag-3′ and RE, 5′-gcagaaggcacactgcgtggtg-3′. The pGARα3-I/M/splice reporter (Gabra-3 exon–intron–exon) where generated by two step PCR from the genomic mouse Gabra-3 and consist of exon 9, 5′ splice site, polypyrimidine tract, 3′ splice site and part of exon 10. To generate this 8 kb truncated Gabra-3 exon–intron–exon construct, two separate PCR reactions were done using following primers Gabra-3 (5′) FW, 5′-gtgtcaccactgttctcaccatg-3′ and RE, 5′-caggaaccacagcctccttcccggtgtggccttgttgaagtagg-3′; Gabra-3 (3′) FW, 5′-cctacttcaacaaggccacaccgggaaggaggctgtggttcctg-3′ and RE, 5′-cccggttcacatatgtagccc-3′. The PCR-generated 5′ and 3′ fragments where fused by the second PCR reaction using 5′ FW and 3′ RE primers. The fragments were cloned into pcDNA3 FLAG. The deletion mutants described were made using QuickChange site-directed mutagenesis (Stratagene). All mutants were verified by Sanger sequencing (Eurofins MWG operon).
Transfections
Reporter constructs (1.25 µg) were co-transfected with ADAR1 or ADAR2 (2.75 µg) into HEK293 cells. For endogenous editing, the reporter constructs (4 µg) were transfected into HeLa cells. For the transfections, 10 µl LIPOFECTAMINE™ 2000 (Invitrogen) was used. Control transfections using an empty expression vector with or without the substrates were done for each experiment. RNA was isolated 48 h (HEK293) and 72 h (HeLa) after transfection using GenElute™ mammalian total RNA isolation (Sigma), and treated with DNase (TURBO DNA-free; Ambion). The cDNA was generated using random hexamer deoxyoligonucleotides and SuperscriptIII RT (Invitrogen). The following PCR was made using Taq (Invitrogen). Primers used for the PCR reactions were: mouseGabra-3 (exon + intron), FW, 5′-ggtgtcaccactgttctcacc-3′ and RE, 5′-gctgtggatgtaataagactcc-3′; Gabra-3 exon, FW (as above Gabra-3 exon + intron) and RE, 5′-gagcacagggaagatgatgcggg-3′; Gabra-3 exon–intron–exon, FW (as above Gabra-3 exon–intron) and RE, cccggttcacatatgtagccc; ChickenGabra-1 FW, 5′-gagtgacaactgtcctaaca-3′ and RE, 5′-acccaatttcatatataggc-3′; pigGabra-3 FW, 5′-tgctgaccatgacgaccctcag-3′ and RE, 5′-gcagaaggcacactgcgtggtg-3′. The PCR products were gel-purified and editing was determined by Sanger sequencing (Eurofins MWG Operon).
Analysis of Gabra-3 intronic stem RNA editing in vivo
Total RNA from adult brain was isolated from human (cerebellum), rat (cerebellum) and mouse (total brain) using the TRIzol reagent (Invitrogen) and treated with DNase 1 (Sigma). First-strand cDNA synthesis was made as above. Primers specific for exon 9 and the downstream intron including the intronic stem of the pre-mRNA Gabra-3 in different species were amplified by PCR as above. Primers used for mouse and pig Gabra-3 were as above. Primers used for human Gabra-3 pre-mRNA were FW, 5′-ggtgtcaccactgtgcttacc-3′ and RE, 5′-ctgggttgaagatagagtcc-3′ and for ratGabra-3 pre-mRNA FW, 5′-ggtgtcaccactgttctcacc-3′ and RE, 5′-ggtcaagggatagaagattgtgc-3′.
In vitro RNA editing assay
The Gabra-3 sequence equivalent to the Gabra-3 mini-gene sequence was PCR-amplified and inserted into the pGEM-T vector (Promega). The leader sequence after the T7 promoter was deleted. For in vitro transcription, sequence was cleaved with EcoR1 and SpeI. The Gabra-3 Δ149 template for the in vitro transcription was made by PCR using the Δ149 expression vector as the template. The sequence of the T7 promoter was fused to the 5′ primer used for the PCR reaction. Transcripts for in vitro editing were made using MEGAshortscript T7 (Ambion). Whole cell extracts were made from HEK293 cells transfected with 4 µg of the ADAR2 expression vector. After 48 h the cells were lysed using Lysis-M containing a protease inhibitor cocktail (Roche). The buffer was exchanged to the ADAR modification buffer in a MicroSpin G-25 column (GE healthcare). In vitro editing was performed in the ADAR modification buffer (250 mM K+-Glutamate,10 mM Tris-Base, 1 mM DTT, 1 mM Phenylmethylsulfonyl fluoride (PMSF), Protease inhibitor cocktail, 0.1 mg/ml BSA, 1 U/ul RNaseOUT™ (Invitrogene), 0.125 ug/ul tRNA and 10% Glycerol) at pH 7 with 20 fmol RNA and 10 µl of ADAR2 cell extract. The reaction was incubated for 1 h and stopped by adding proteinase K followed by phenol extraction. Samples were precipitated and treated with DNase I (TURBO DNA-free; Ambion) followed by RT-PCR for editing determination.
454 sequencing of Gabra-3 in porcine brain tissues
Tissues dissected from embryonic Day 115 porcine brain were transferred to RNAlater-ICE (Ambion) and high molecular weight RNA (≥ 200 nt.) was purified using the MirVana kit (Ambion). RNA samples were DNase treated using the TURBO DNA-free Kit (Ambion). Primers used for RT-PCR of Gabra-3 were: Gabra3_FW, 5′-gagaaagcttatgacgaccctcagtatcagtgcc-3′ and Gabra-3_RE,
5′-aggactcgagtggtgaaatagttgactgtggcaaactc-3′. Both primers introduce a restriction site in the first 10 nt of the 5′-end (HindIII for FW and XhoI for RE primer). RNA samples were reverse transcribed with M-MLV Reverse Transcriptase (RT) (Invitrogen) using the Gabra-3_RE primer for gene specific RT. PCR was done using HotStart Taq Plus polymerase (Qiagen) and PCR products were purified using Ultra-Sep Gel Extraction Kit (E.Z.N.A.) as per the manufacturer’s directions with the exception that purification was done directly on the PCR product not on a gel slab. Purified RT-PCR products were 454 sequenced at Research Centre Foulum, Denmark. Reads from 454 sequencing were mapped to the Gabra-3 amplicon using BLAT (BLAST-Like Alignment Tool). Reads not containing insertions or deletions in the immediate vicinity of the I/M editing site were inspected for the presence of either A or G at the editing site. The percentages of A-to-G events were calculated.
Calculation of editing frequency
To evaluate the amount of I/M edited Gabra-3 transcripts, RNA from at least three independent experiments were sequenced. Editing was determined by measuring the ratio between the A and G peak height in individual chromatograms using FinchTV. Percent editing was calculated as the peak height of G/(A + G) × 100. In addition, the percent of editing evaluated by the chromatograms were compared with our previously published 454-high-throughput amplicon sequencing of individual Gabra-3 transcripts during brain development (23). To compare editing ratios, RNA derived from the same source as used for the 454-amplicon sequencing was Sanger sequenced at the I/M site. According to this data, Sanger sequencing show some inconsistencies in the height of the mixed A/G peaks compared with the 454 data where hundreds individual transcripts were sequenced (Supplementary Figure S1). To correct the variance, we grouped the level of edited transcripts according to the mean value of the G-peak derived from triplicates of the experiments into non (< 10%), low (10–25%), medium (25–50%), high (50–75%) and full (75–100%) editing.
Software
RNA secondary structure predictions of the Gabra-3 transcript were made through the Mfold (30) and Sfold web server available at http://sfold.wadsworth.org (31,32).
RESULTS
An intronic sequence downstream of the I/M site is required for efficient editing
Since most site selective editing events require a complementary intronic sequence to form the double-stranded structure needed for editing, it has been postulated that editing precedes splicing by targeting the pre-mRNA. However, one rare exception is the exonic duplex structure formed at the edited I/M site in the Gabra-3 transcript. It can therefore be hypothesized that ADAR editing at this site can occur as a post-splicing event. To investigate this, we used a construct containing the mouse Gabra-3 cDNA (lacking intronic sequences) as a template in transient co-transfections with either ADAR1 or ADAR2. To determine the editing efficiency, we used Sanger sequencing after RT-PCR on extracted total RNA. The ratio between the A and the G peak heights at the I/M site was used as a measurement of editing levels as I is interpreted as G by the RT. To get accurate values of editing levels, the A/G peak heights were compared with known editing levels at the I/M site of Gabra-3 from previous analysis using 454 high-throughput amplicon sequencing (Supplementary Figure S1). Editing levels were thereafter classified into 5 groups: non (<10%), low (10–25%), medium (25–50%), high (50–75%) and full (75–100%) (for calculations see ‘Materials and Methods’ section).
Interestingly, a substantial decrease in editing efficiency was detected in transcripts derived from the cDNA construct compared with a wild-type (WT) Gabra-3 mini gene that includes part of the downstream intron. In transient co-transfections of the mini gene (WT, Gabra-3 exon + intron), the efficiency of I/M editing by ADAR1 or ADAR2 was determined as fully edited (75–100%), whereas editing in absence of intronic sequence (Gabra-3 exon) was low (10–25%) (Figure 1A). To ensure that over-saturated levels of enzymes do not mask potential differences in editing efficiency, the reporter constructs were transfected into HeLa cells, expressing endogenously active ADAR enzymes. Here a medium (25–50%) number of edited transcripts were obtained from the Gabra-3 exon + intron reporter, whereas no editing (< 10%) could be observed when the intron sequence was omitted (Gabra-3 exon) (Figure 1A). These results indicate that intronic sequences contribute to the editing event in the exon, even if no potential duplex forming interactions between the sequences could be predicted.
Figure 1.
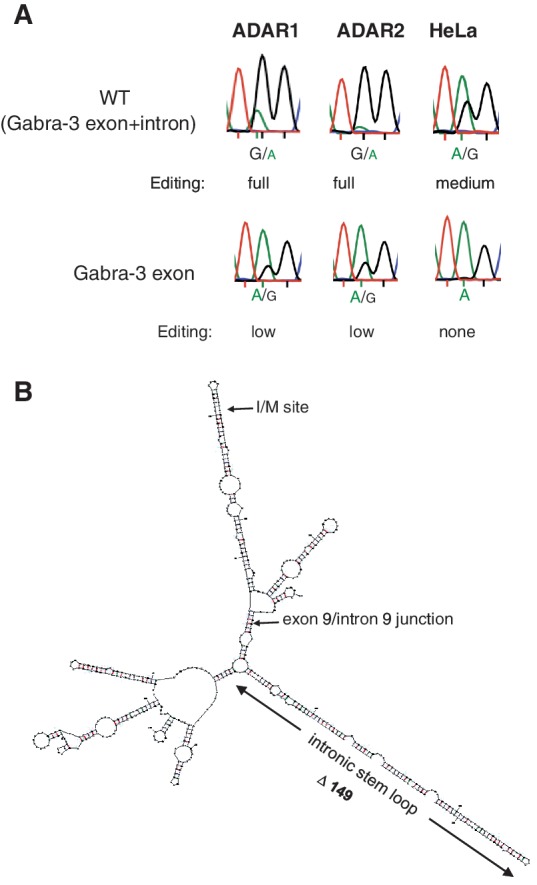
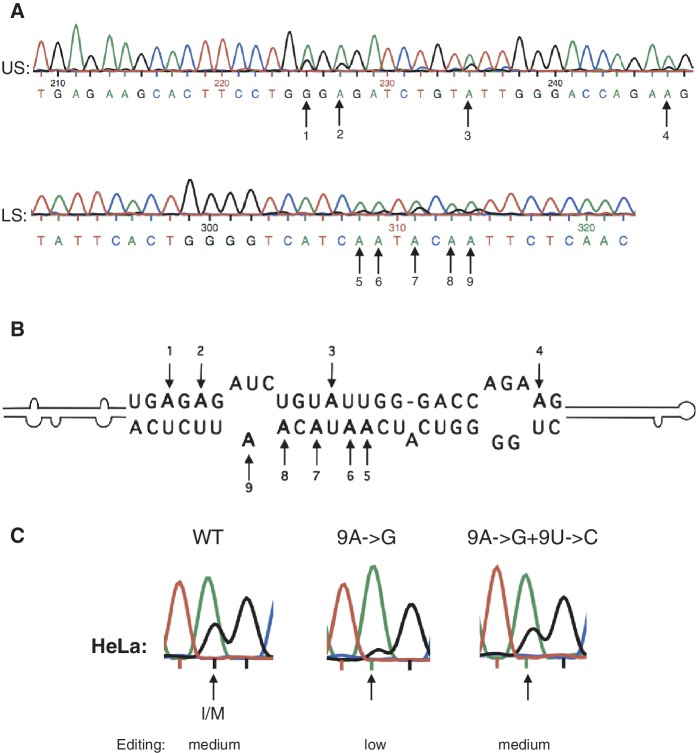
Efficiency of editing at the I/M site using different Gabra-3 constructs. (A) Sanger sequencing results after RT-PCR from co-transfections of ADAR1 or ADAR2 with Gabra-3 editing reporter constructs with or without intronic sequence. Editing is detected as a dual A and G peak at the I/M site and the amount of edited transcripts is determined by measuring the ratio between the A and G peak heights. Reproducible triplicates of these measurements were compared with known levels of editing at the I/M site using 454 HTP sequencing (see ‘Materials and Methods’ section; Supplementary Figure S1). For endogenous editing the reporters were transfected into HeLa cells. (B) Putative RNA secondary structure of the Gabra-3 transcript at exon 9 and part of the intron 9. The edited I/M site as well as the exon/intron junction are indicated. The conserved intronic duplex structure is located 150 nt downstream of the I/M site.
A putative intronic stem loop structure induces editing at the I/M site
When analyzing the RNA structure of intron 9 using RNA structure prediction programs (see ‘Materials and Methods’ section), we found a long stem loop structure located 150 nt downstream of the I/M site (Figure 1B). To investigate if this intronic putative stem loop influences editing efficiency at the I/M site, the Gabra-3 WT mini gene, including the intronic duplex, was used for deletion analysis in ADAR-transfected HEK293 cells and endogenous editing in HeLa cells. When the intronic duplex consisting of 149 nt was deleted from this construct (Δ149) (Figure 1B), the efficiency of editing vastly decreased (Figure 2A). The most prominent effects were seen for ADAR1 co-transfections and endogenous editing. In the ADAR1 co-transfection, the level of edited transcripts decreased from full (75–100%) to medium (25–50%) after removal of the intronic duplex. In HeLa cells no editing could be observed at the I/M site in absence of the intronic stem loop, whereas a medium (25–50%) level of editing was seen in the WT reporter. A small but consistent decrease in I/M editing in the Δ149 reporter (high, 50–75%) compared with WT (full, 75–100%) could also be observed in the ADAR2-transfected cells. This relatively small effect could be due to the non-physiological high concentration of ADAR2 enzyme produced in the transfected HEK293 cells. To ensure that the effect was caused specifically by the intronic stem loop deletion, 149 nt were deleted further downstream in the intron (Δ149 d.s. int.). This deletion had no effect on editing efficiency at the I/M site (Figure 2A). We next carried out partial deletions of the intron stem, which led to a gradual decrease in editing efficiency correlated with the length of the stem (Supplementary Figure S2). From these analyses we conclude that the intronic stem loop has a major impact as an editing inducer at the upstream I/M site.
Figure 2.
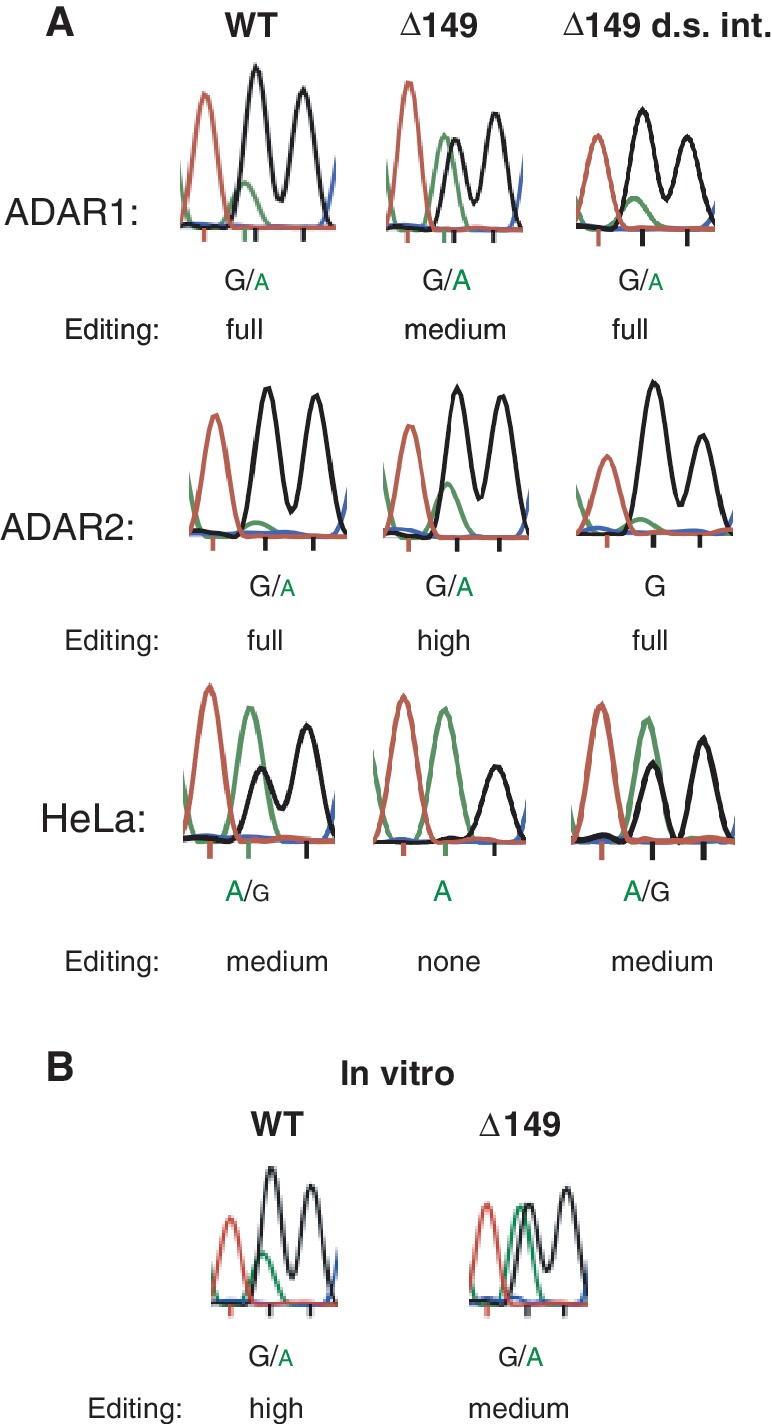
Editing efficiency at the I/M site is lower in Gabra-3 transcripts in the absence of the intronic hairpin. (A) The editing levels in transcripts derived from transfected Gabra-3 constructs and edited by co-expressed ADAR1 or ADAR2 in HEK293 as well as endogenous editing in HeLa cells. Editing was determined by Sanger sequencing measuring the ratio of the A and G peak after RT-PCR and compared with known levels of editing using 454 high throughtput (HTP) sequencing (see ‘Materials and Methods’ section; Supplementary Figure S1). Reproducible triplicates were made for each sample shown in the figure. The Δ149 lane indicates a reporter construct where 149 nt and thereby the entire intronic stem loop was deleted. As a control 149 nt downstream of the intronic stem was deleted (Δ149 d.s. int). (B) The intronic duplex influences editing of an in vitro transcribed Gabra-3 RNA. A 532 nt long transcript including Gabra-3 exon 9 and part of intron 9 (WT) was used in an ADAR2 modification assay in vitro. This was compared with in vitro editing of a reporter lacking the intronic hairpin of 149 nt (Δ149). Editing was detected by Sanger sequencing after RT-PCR.
The intronic stem can enhance editing also in vitro
To exclude cellular effects on I/M editing efficiency, we analyzed the influence of the intronic stem on editing in vitro. A mouse Gabra-3 transcript was made by in vitro transcription using the T7 polymerase. This 532 nt long transcript contained the entire stem loop structure at the I/M site as well as the 149 nt long intronic stem loop and was equivalent to the transcript produced in the transfected cells (see Figure 1B). ADAR2-transfected whole cell extract was used in the in vitro editing assay as described in the ‘Materials and Methods’ section. In brief, the in vitro transcribed Gabra-3 RNA was incubated with the whole cell extract for 1 h at 25°C. After RT-PCR, editing was determined by Sanger sequencing. This transcript was highly edited at the I/M site in vitro (Figure 2B). To determine if the intronic stem plays a role in editing efficiency in vitro, the entire stem loop structure of 149 nt was deleted (Δ149). When this transcript was used in the in vitro editing assay, the extent of edited transcripts decreased from high to medium (Figure 2B). This result indicates that the intronic stem loop has a positive effect on editing also in vitro.
The intronic stem is edited at several sites
To further investigate the role of the intronic stem in Gabra-3 editing, we analyzed if this structure could act as a substrate for ADAR1 and/or ADAR2. This duplex consists of a long double-stranded stem that is interrupted by mismatches and bulges, a hallmark for ADAR substrates (Figure 1B). Indeed, when analyzing the Gabra-3 pre-mRNA in the adult mouse brain, we found that at least 9 nt were edited in the intron (Figure 3A). Furthermore, the homologous RNA sequences from human and rat brain were also edited at the same sites (Supplementary Figure S3). Co-transfection experiments with either ADAR1 or ADAR2 revealed that the different enzymes have both overlapping and specific editing activity on sites in the intronic stem (Supplementary Figure S4).
Figure 3.
Editing at several sites in the putative intronic stem loop. (A) Chromatogram showing A-to-I editing in the intronic stem of the Gabra-3 transcripts derived from mouse brain. US indicates the upper strand, while LS equals the lower strand according to the cartoon in B. Arrows indicate the edited adenosines. (B) Cartoon showing the sequence of the edited part in the intronic hairpin. Arrows indicate the edited adenosines. (C) Sanger sequencing chromatograms showing I/M editing of the wild-type Gabra-3 editing reporter in HeLa cells compared with a reporter where 9 edited adenosines were mutated to guanosines (9A->G) and a reporter with compensatory mutations, restoring the base pairs in the intronic stem (9A->G+9U->C).
We asked if the editing events in the intron influence the editing efficiency at the I/M site? A construct that mimics a fully edited intron stem was made, where the 9 adenosines edited in mouse (Figure 3B) were mutated to guanosines (9A- > G), presumably leading to a less stable duplex structure with 9 A:U bp changed to more unstable G:U bp. These mutations led to a decrease in editing frequency at the I/M site from medium to low in HeLa cells (Figure 3C). At least two potential mechanisms could explain the intronic element effect on upstream exonic editing: (i) the intronic stem enhances I/M editing by attracting the ADAR enzyme to a nearby site or (ii) the intronic stem stabilizes the duplex structure at the I/M site and thereby facilitates editing. To analyze if the intronic double-stranded structure per se induced editing at the I/M site, we made compensatory mutations in the editing reporter (9A- > G) that restored the disrupted bp and created 9 new G:C bp in the stem (9A- > G + 9U- > C). Restoration of the intronic stem loop by changing 9 G:U to G:C bp led to an increase in editing efficiency at the I/M site to a level that is comparable with the WT (Figure 3C). Moreover, as mentioned above, when deleting only 20 nt of the stem (Δ20, Supplementary Figure S2), thereby removing the edited sites 5–9 and the editing complementary sequence (ECS) for site 2 and 3, the stem could still induce editing at the I/M site. When deleting as much as 70 nt of the stem, removing 8 out of 9 sites of editing, the stem length was reduced from 66 to 29 bp. Still the intron stem could induce I/M editing to some extent (Supplementary Figure S2). However, a deletion of all editing sites (Δ100) had an equal effect on I/M editing as removal of the entire stem loop (Δ149).
Taken together, pre-edited intronic events disrupting the duplex structure decrease the editing efficiency. Compensatory mutations reveal that the structure of the intronic stem rather than its sequence, induces editing at the upstream I/M site and that the induction is cis-acting. Furthermore, the gradual increase in editing induction is proportional to the stem length and the number of ADAR editing/binding sites on the duplex.
Splicing does not interfere with the editing inducer
In all editing reporter constructs described so far, splicing that removes intron 9 was impaired. To investigate if the intron 9 editing inducer is functional also in the presence of splicing, a reporter was made where a truncated intron 9, containing the editing inducer, and exon 10 was added (see ‘Materials and Methods’ section; Supplementary Figure S2A). As seen in Figure 4A, the intronic hairpin concurrent to splicing can induce editing at the I/M site. Moreover, RT-PCRs using primers specific for spliced and pre-mRNA transcripts, respectively, indicate that the editing inducer has a positive effect also on splicing (Figure 4B).
Figure 4.
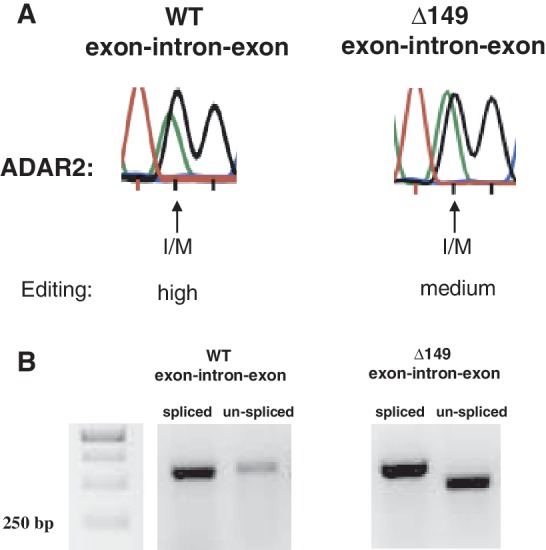
The intronic hairpin can induce editing at the I/M site prior to splicing. (A) Co-expression of the Gabra-3 exon–intron–exon constructs with the ADAR2 expression vector in HEK 293 cells. The chromatograms after sequencing show that the editing efficiency at the I/M site decrease from high to medium in the absence of the intronic editing inducer when spliced. (B) Detection of spliced and non-spliced Gabra-3 exon–intron–exon products after RT-PCR from the same samples as in (A). Deletion of the editing inducer (Δ149 exon–intron–exon) reduces the splicing efficiency compared to the WT exon–intron–exon reporter.
Gabra-3 is not edited at the I/M site in porcine brain
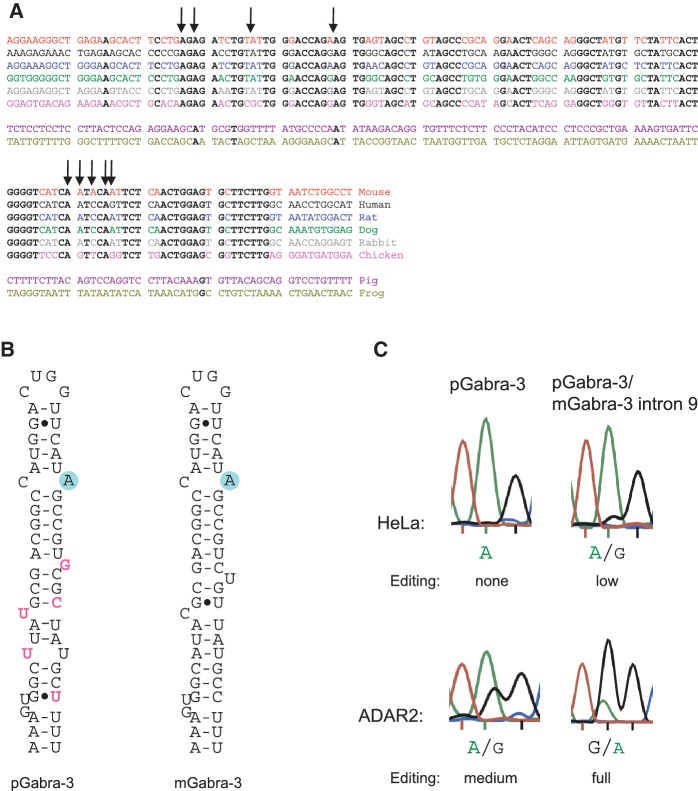
To further investigate the significance of the intronic stem loop, we aligned the mouse gabra-3 intron 9 with other species and found that the sequence is conserved in all animals with an edited I/M site in exon 9 (26) (Figure 5A). Mouse, human, rat, dog, rabbit and chicken in which the I/M site is edited, the intronic editing inducer duplex is highly conserved, whereas in frog (Xenopus tropicalis), that encodes an unusual genomic G at the I/M site, has a more divergent intron 9 sequence. In a more extended phylogenetic screen, we found that pig also has a divergent sequence in intron 9. The stem loop at the I/M site in exon 9 is conserved, differing only at a few nucleotides compared with the mouse sequence (Figure 5B). To determine whether the I/M site of Gabra-3 is edited in pig, despite the absence of the intronic stem, RNA from five different tissues (brain stem, cortex, cerebellum, basal ganglia and hippocampus) were analyzed by 454 sequencing of RT-PCR amplicons at the I/M site. As shown in Table 1, only a few transcripts were edited and only by sequencing thousands of reads we were able to estimate the editing level to 0.2–0.6% in newborn pigs as compared with 50% in newborn mice (23). This suggests that endogenous editing of the I/M site in Gabra-3 is directly correlated to the down stream intronic element.
Figure 5.
Induced editing of a non-ADAR substrate in presence of the conserved Gabra-3 intronic duplex. (A) Alignment of sequences from different species of gabra-3 intron 9 at the location of the intronic duplex. Letters in bold indicate conserved nucleotides. The structure as well as the nucleotide sequence is conserved between species that are edited at the I/M site whereas little conservation can be found in non-edited species like frog and pig. Arrows indicates the location of edited adenosines. (B) Comparing the putative RNA secondary structure of Gabra-3 from pig (pGabra-3) and mouse (mGabra-3) at the I/M site. The I/M site is indicated with a circle in blue. Divergent nucleotides are in pink. (C) Sanger sequencing showing the I/M site after transfection of a pGabra-3 reporter and a construct where the mouse Gabra-3 I/M stem loop was exchanged for the pGabra-3 sequence (pGabra-3/mGabra-3 intron 9). Editing after transfection into HeLa cells and co-transfection with an ADAR2 expression vector into HEK293 cells are shown.
Table 1.
Detection of edited Gabra-3 transcripts in pig brain
| Tissuea | No. of reads | No. of edited transcripts | % editing |
|---|---|---|---|
| Brain stem | 2449 | 7 | 0.29 |
| Cortex | 4291 | 9 | 0.21 |
| Cerebellum | 1539 | 6 | 0.39 |
| Basal ganglia | 3311 | 13 | 0.39 |
| Hippocampus | 3227 | 21 | 0.65 |
aEditing in pig brain at developmental day E115 measured by 454 amplicon sequencing.
The intron sequence can induce editing in transcripts that are normally not edited
To investigate if editing at the I/M site in pig Gabra-3 could be induced in the presence of the intronic stem, we transfected a reporter construct containing 54 nt of the pig I/M stem loop fused to the mouse intron 9 sequence, containing the intronic stem loop. As shown by Sanger sequencing after RT-PCR, the transcript of this reporter was edited by the endogenous ADARs in HeLa cells (see pGabra-3/mGabra-3 intron 9 in Figure 5C), whereas no editing at the pig I/M site was observed in a reporter with the WT pig intron 9 sequence. This was clearer when the reporters were co-transfected with ADAR2 in HEK293 cells. With ADAR2 over expression, the WT pig Gabra-3 transcript was moderately edited at the I/M site, whereas the site was fully edited in presence of the mouse intron hairpin (Figure 5C). These results show that the I/M site in pig can be recognized as a substrate for editing if induced by the intronic element and provide evidence that the distant stem loop is a requirement for efficient editing at the I/M site of Gabra-3.
After confirming that the sequence of intron 9 induces I/M editing, we wanted to know whether this induction was specific to the Gabra-3 transcript or could it induce editing also in other RNA structures. When analyzing the structure and sequence of the genes coding for other α subunits of the GABAA receptor, we discovered that the Gabra-1 transcript, coding for the chicken α1 subunit, can form a stem loop structure similar to the Gabra-3 structure (Supplementary Figure S5A). The Gabra-1 transcript also harbors an A–C mismatch at the position equivalent to the I/M site in Gabra-3. Nevertheless, no editing could be detected in Gabra-1 transcripts from chicken brain and the stem loop is not tailed by an intronic hairpin (Supplementary Figure S5B). This was also true for a Gabra-1 reporter, co-transfected with ADAR1 or ADAR2. The sequence analysis revealed that neither ADAR1 nor ADAR2 target this as a substrate (Supplementary Figure S5B). Similar results have been revealed in an oocyte editing assay (27). To analyze if the intronic Gabra-3 editing inducer could also stimulate editing of Gabra-1, 54 nt of the Gabra-1 sequence including the I/M stem loop was fused with the sequence of the Gabra-3 intronic hairpin (cGabra-1/3). Indeed, sequence analysis revealed that Gabra-1 is edited at the I/M site when it is in the context of the Gabra-3 intron background (Supplementary Figure S5B). This result indicates that the intronic stem loop structure in Gabra-3 is not specific for the I/M site of Gabra-3 as it can induce editing also in other related RNA structures. Hence the Gabra-3 intron element may work as a general enhancer for ADAR editing.
Sequence requirements for I/M editing in the presence of the intronic stem
The sequence and the putative stem loop structure in intron 9 of Gabra-3 is conserved in all species that edit the I/M site in exon 9. In the vicinity of the I/M site, a stem consisting of 38 bp interrupted by bulges and internal loops, is predicted by both the Mfold and Sfold RNA structure prediction programs (Figure 6A). A previous analysis indicates that the first 11 bp of the stem followed by a non-conserved region of 10 nt is important for editing (27). We aimed to further determine how many base pairs of the I/M stem are required for efficient editing and whether this is dependent on the intronic editing inducer. Deletions were performed on the Gabra-3 WT mini gene and the effects were analyzed by co-transfection into HEK 293 cells together with either the ADAR1 or ADAR2 expression vector. Deletion of the first 16 bp of the stem had no effect on the editing efficiency (Figure 6B). Further analyses indicated that the minimal stem loop structure required for efficient editing is 33 nt long. This hairpin consists of 13 bp, interrupted by two bulges and connected by a loop of 4 nt (Figure 6C). Further shortening of the stem gave rise to low or no editing at the I/M site (Figure 6D and E). To reveal the importance of the distant intronic hairpin on editing efficiency of this minimal 13 bp-long stem, we deleted the 149 nt long intron duplex (Figure 6C′). Upon this deletion, editing decreased from high to low, showing the impact of this distant hairpin on the efficiency of I/M editing. Taken together, our results indicate that even a very short duplex, not recognized as a substrate on its own, can be edited if it is in the vicinity of another longer stem loop structure.
Figure 6.
Deletion analyses to determine the shortest sequence for efficient I/M editing of Gabra-3. Editing reporter constructs (A–E) expressing exon 9 and part of intron 9 were co-transfected into HEK 293 cells together with an ADAR1 or ADAR2 expression vector. The chromatograms after sequencing of the RT-PCR products at the I/M site are shown on top. Below are the putative secondary structures that can be formed at the I/M site. The I/M site is indicated in grey. (C′) indicates the same construct as in (C) but where 149 nt of the intronic hairpin is deleted. The number of deleted nucleotides within each reporter construct is shown below the putative structures.
DISCUSSION
In the present study we have analyzed the importance of intronic sequences for the efficiency of editing within coding sequences. Most site selective editing, occurring within coding sequence, depend on the presence of an editing complementary sequence located in the downstream intron. It is therefore conceivable that editing occurs prior to splicing in these substrates. ADAR editing has also been suggested to occur as a co-transcriptional event, coordinated with the splicing machinery (33–35). Nevertheless, the I/M site of Gabra-3 and its predicted ECS are both located within exon 9 of the transcript. In principle it should therefore be possible for editing to occur as a post-splicing event catalyzed by the cytoplasmic ADAR1 p150 (36). However, in the present study we show that editing at the I/M site requires a long hairpin structure located within the downstream intron. In porcine Gabra-3, we show that the I/M site is poorly edited even though the stem loop at the I/M site is conserved. Furthermore, we demonstrate that the intronic editing inducer element is absent in pig Gabra-3. This observation indicates that the intronic hairpin is an absolute requirement for I/M editing in vivo. The absence of intronic editing inducer in pig is puzzling since it is present in all other species from human to chicken. A reasonable explanation is that pigs lost the genomic sequence containing the long intronic hairpin when domesticated. It has previously been shown that domestication of pigs have led to changes particularly in genes related to brain and neuron functions (37).
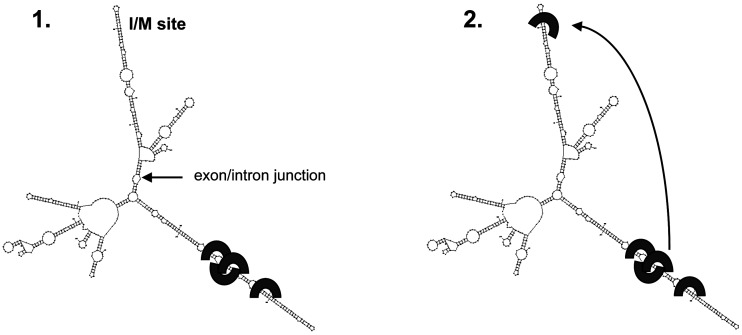
How does the intron hairpin then induce editing at the upstream I/M site? We hypothesize that the 66 bp-long intronic stem helps recruiting the ADAR enzymes to the transcript and thereby increase the local concentration of ADAR, thus facilitating efficient editing (Figure 7). This theory is supported by the observation that the intronic stem is edited at 9 sites, a proof of interaction with the ADAR enzyme. However, these 9 adenosines, in A:U bp, could be exchanged for G:C bp without an effect on editing efficiency at the I/M site, indicating that it is the structure of the hairpin rather than the intronic editing events that enhances I/M editing. Since the editing enzyme will interact with any double-stranded RNA, we therefore suggest that non-specific ADAR binding rather than editing is important for the recruitment. Furthermore, the intronic hairpin can induce I/M editing also in vitro (Figure 2B). This result implies that recruitment by the intronic hairpin is not dependent on other cellular factors.
Figure 7.
Model for induced editing at the I/M site of Gabra-3 by the downstream intronic editing inducer element. ADAR enzymes (black bow) are recruited to the transcript by the long intronic hairpin. Increased local concentration of ADAR leads to more efficient editing at the I/M site.
Another possible mechanism for editing induction by the intron duplex is by stabilizing the upstream stem loop containing the I/M site. However, the distance between the I/M site and the intronic hairpin is about 150 nt, and at least 28 nt near the I/M site could be deleted without a major effect on the induction of editing (Figure 6C). Furthermore, when the intronic stem was shortened by 20 bp, it still induced I/M editing but partial deletions of the intron stem caused a gradual decrease in I/M editing induction that was correlated with the number of edited/binding sites deleted (Supplementary Figure S2B). Moreover, since the intronic hairpin can induce editing at sites that are not recognized as substrates in vivo, a structural interaction is less likely (Figure 5; Supplementary Figure S5). These results further points to the recruitment theory.
The finding that a distant duplex can increase the efficiency of site selective A-to-I editing is novel and interesting in a more global perspective. In other transcripts, conserved intron sequences have been found downstream of several other exons to A-to-I editing. One example is the GluR-C (GluA3) transcript edited at the R/G site, with a largely conserved intron sequence downstream of the R/G editing site which is not part of the ECS (38). By screening sequences for stem loop structures in the vicinity of known recoding editing sites, we identified conserved downstream duplexes in all GluR transcripts that undergo editing (data not shown). Furthermore, it is well-known that a substantial number of long stem loop structures are located in non-coding sequences in primates. These hairpins are formed by inverted repeats of Alu transposable elements. Several groups have shown that thousands of these hairpins are subjected to hyper-editing (7–11,39). We predict that many of these long stem loop structures located close to exon sequences function as inducers of exonic editing. It is therefore possible that this is a general mechanism used to recognize editing substrates. Since long hairpin structures formed by inverted repeats are particularly common in human, it would be interesting to analyze if these give rise to elevated levels of editing elsewhere in the transcriptome. Therefore, in future studies of A-to-I editing predictions, stable stem loop structures located in untranslated regions should be taken into account as editing inducers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Swedish Research Council [K2011-X-20702-04-6 to M.Ö.]; Lennanders stiftelse, Uppsala (to C.D.). Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jakob Skou Pedersen and Lars Wieslander for fruitful discussions and Gilad Silberberg for critically reading the paper.
REFERENCES
- 1.Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 2.Jepson JE, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Källman AM, Sahlin M, Öhman M. ADAR2 A–>I editing: site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003;31:4874–4881. doi: 10.1093/nar/gkg681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Öhman M, Källman AM, Bass BL. In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA. 2000;6:687–697. doi: 10.1017/s1355838200000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahlstedt H, Öhman M. Site-selective versus promiscuous A-to-I editing. Wiley Interdiscip Rev RNA. 2011;2:761–771. doi: 10.1002/wrna.89. [DOI] [PubMed] [Google Scholar]
- 7.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 10.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmi S, Borukhov I, Levanon EY. Identification of widespread ultra-edited human RNAs. PLoS Genet. 2011;7:e1002317. doi: 10.1371/journal.pgen.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 13.Stephens OM, Haudenschild BL, Beal PA. The binding selectivity of ADAR2’s dsRBMs contributes to RNA-editing selectivity. Chem. Biol. 2004;11:1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 16.Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 17.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 19.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 20.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 21.Farrant M, Kaila K. The cellular, molecular and ionic basis of GABA(A) receptor signalling. Prog. Brain Res. 2007;160:59–87. doi: 10.1016/S0079-6123(06)60005-8. [DOI] [PubMed] [Google Scholar]
- 22.Ohlson J, Pedersen JS, Haussler D, Öhman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wahlstedt H, Daniel C, Ensterö M, Öhman M. Large-scale mRNA sequencingdetermines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nimmich ML, Heidelberg LS, Fisher JL. RNA editing of the GABAA receptor a3 subunit alters the functional properties of recombinant receptors Neurosci. Res. 2009;63:288–293. doi: 10.1016/j.neures.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J. Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel C, Wahlstedt H, Ohlson J, Björk P, Öhman M. Adenosine-to-inosine RNA editing affects trafficking of the gamma-aminobutyric acid type A (GABA(A)) receptor. J Biol Chem. 2011;286:2031–2040. doi: 10.1074/jbc.M110.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian N, Yang Y, Sachsenmaier N, Muggenhumer D, Bi J, Waldsich C, Jantsch MF, Jin Y. A structural determinant required for RNA editing. Nucleic Acids Res. 2011;39:5669–5681. doi: 10.1093/nar/gkr144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desterro JM, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 29.Bratt E, Öhman M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 34.Laurencikiene J, Källman AM, Fong N, Bentley DL, Öhman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryman K, Fong N, Bratt E, Bentley DL, Öhman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amaral AJ, Ferretti L, Megens HJ, Crooijmans RP, Nie H, Ramos-Onsins SE, Perez-Enciso M, Schook LB, Groenen MA. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One. 2011;6:e14782. doi: 10.1371/journal.pone.0014782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aruscavage PJ, Bass BL. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA. 2000;6:257–269. doi: 10.1017/s1355838200991921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl Acad. Sci. USA. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.