Abstract
Family X DNA polymerases (PolXs) are involved in DNA repair. Their binding to gapped DNAs relies on two conserved helix-hairpin-helix motifs, one located at the 8-kDa domain and the other at the fingers subdomain. Bacterial/archaeal PolXs have a specifically conserved third helix-hairpin-helix motif (GFGxK) at the fingers subdomain whose putative role in DNA binding had not been established. Here, mutagenesis at the corresponding residues of Bacillus subtilis PolX (PolXBs), Gly130, Gly132 and Lys134 produced enzymes with altered DNA binding properties affecting the three enzymatic activities of the protein: polymerization, located at the PolX core, 3′-5′ exonucleolysis and apurinic/apyrimidinic (AP)-endonucleolysis, placed at the so-called polymerase and histidinol phosphatase domain. Furthermore, we have changed Lys192 of PolXBs, a residue moderately conserved in the palm subdomain of bacterial PolXs and immediately preceding two catalytic aspartates of the polymerization reaction. The results point to a function of residue Lys192 in guaranteeing the right orientation of the DNA substrates at the polymerization and histidinol phosphatase active sites. The results presented here and the recently solved structures of other bacterial PolX ternary complexes lead us to propose a structural model to account for the appropriate coordination of the different catalytic activities of bacterial PolXs.
INTRODUCTION
Maintenance of genome stability is essential for the survival of the organisms (1). The presence of DNA damage triggers the activation of specific DNA repair pathways including multiple enzymatic activities specialized in dealing with these lesions that otherwise could have deleterious effects in the replication and transcription processes. Among the varied enzymes involved in DNA repair, family X DNA polymerases (PolX), highly conserved in all the kingdoms of life, from human to viruses, stand out (2). These enzymes play a role in DNA synthesis during the base excision repair (3–6) and non-homologous end joining (7–11) pathways, acting on the short gapped intermediates that arise during these events. Their ability to fill a gapped DNA molecule relies on a common structural organization shared by most PolXs: the C-terminal polymerization domain formed by the fingers, palm and thumb subdomains, present in nearly all DNA-dependent DNA polymerases and responsible for the binding and further elongation of the 3′ terminus of the upstream primer strand and the N-terminal 8-kDa domain that contacts the downstream strand (6,12,13). In some cases, such as in mammalian Polβ and Polλ and yeast Pol4 and Trf4, the 8-kDa domain also contains an intrinsic 5′-deoxyribose 5′-phosphate lyase activity responsible for the release of the 5′-deoxyribose 5′-phosphate moiety during short patch base excision repair (4,6,14–16). Besides this common PolX core, several members of this family are endowed with accessory domains provided either with additional enzymatic activities, such as the 3′-5′ exonuclease and AP-endonuclease present in the C-terminal polymerase and histidinol phosphatase (PHP) domain of bacterial/archaeal PolXs (17–19), or with residues responsible for protein–protein/protein–DNA interactions, such as the N-terminal BRCT domain of Polλ, Polµ and TdT through which these polymerases interact with other non-homologous end joining factors, such as Ku, XRCC4 and ligase IV, placed at the ends of double strand breaks (20–22).
Binding of PolXs to DNA largely depends on the presence of two helix-hairpin-helix (HhH) motifs, one located at the 8-kDa domain and the other at the fingers subdomain of the polymerization domain. These motifs are stretches of ∼20 amino acids that bind to the DNA in a sequence independent manner; they are abundant in the prokaryotic and eukaryotic enzymes involved in DNA transactions, such as replication and repair processes (23). They are formed by highly conserved hydrophobic residues organized in two helices of defined length linked by a type-II β-hairpin. Hydrophobic residues form a contact surface between the two helices stabilizing the structure of the motif. The β-hairpin contains two glycine residues separated by a hydrophobic residue. The first glycine might be important in the hairpin formation, whereas the second one could provide a contact surface for DNA binding (23). Structural studies performed with Polβ bound to a nicked DNA showed that the 8-kDa and fingers HhH motifs interact with the DNA sugar–phosphate backbone of the downstream and upstream strands, respectively (12,24,25). Such interactions make the DNA to adopt a sharply bent conformation to expose the nascent base pair, allowing the right positioning of the 5′-phosphate and 3′-OH free ends at their respective active sites, 5′-deoxyribose 5′-phosphate lyase and polymerase. Besides the two universal HhH motifs described previously, bacterial and archaeal PolXs have a specifically conserved third HhH motif in the fingers subdomain (26) located three residues C-terminally from the common one (see Figure 1), and whose potential role in DNA binding has not been demonstrated so far.
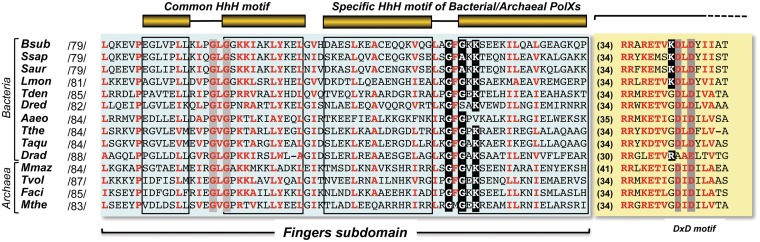
Figure 1.
Multiple amino acid sequences alignment of the fingers subdomain and the DxD motif of the palm subdomain of bacterial/archaeal PolXs. The common HhH motif shared by most of PolX members and the one specifically present in the bacterial/archaeal members are drawn. Numbers between slashes indicate the amino acid position relative to the N-terminus of each DNA polymerase. Numbers in parentheses indicate the length of the intervening amino acid sequence. Because of the large number of sequences, only selected representatives from the Eubacteria and Archea genus are aligned. Names of organisms are abbreviated as follows: Bsub, Bacillus subtilis (GenBank accession number NP_390737); Lmon, Lysteria monocytogenes (GenBank accession number YP_013839); Ssap, Staphylococcus saprolyticus (GenBank accession number YP_301742); Saur, Staphilococcus aureus (GenBank accession number YP_001246578); Dred, Desulfotomaculum reducens (GenBank accession number YP_001112987); Aaeo, Aquifex aeolicus (GenBank accession number NP_213981); Tthe, Thermus thermophilus (GenBank accession number YP_144416); Tden, Thiobacillus denitrificans (GenBank accession number AAZ97399); Mmaz, Methanosarcina mazei (GenBank accession number NP_633918); Faci, Ferroplasma acidarmanus (GenBank accession number ZP_01709777); Mthe, Methanothermobacter thermautotrophicus (GenBank accession number NP_275693); Dred, Deinococcus radiodurans (GenBank accession number NP_294190); Tvol, Thermoplasma volcanium (GenBank accession number NP_111375) and Taqu, Thermus aquaticus (GenBank accession number BAA13425). Conserved residues (≥80% of the aligned polymerases) are indicated with red letters. Conserved glysine and lysine residues of the specific HhH motif and the lysine residue preceding the first catalytic aspartic acid residue are indicated in white letters over a black background. Alignment was made by using the Multalin tool (http://multalin.toulouse.inra.fr/multalin/multalin.html) and was further adjusted by hand.
The palm subdomain of PolXs possesses the universal three catalytic aspartates that coordinate the divalent metal cations responsible for the nucleotidyl transfer reaction by the two metal ion mechanism. The first catalytic aspartate is preceded by a small residue (glycine) in eukaryotic Polβ and Polλ (27) that contacts the γ-phosphate of the incoming nucleotide (12,13,28,29). However, Polµ and TdT, endowed with a terminal transferase activity, contain a histidine residue instead (30). Crystallographic studies have shown that this histidine residue is positioned in such a way that it could form a hydrogen bond with the phosphate of the primer-terminal residue and the γ-phosphate of the incoming deoxyribonucleoside triphosphate (dNTP), enabling the nucleotidyl transfer reaction in the absence of a template nucleotide (31). Some bacterial PolXs have a moderately conserved lysine residue in that position (26) (see Figure 1).
Bacillus subtilis PolX (PolXBs) is a prototype of the bacterial/archaeal subgroup of PolXs that is potentially involved in base excision replair, as it is a strictly DNA-directed DNA polymerase, it is adapted to fill small gaps, allowing final sealing of the resulting nick by a DNA ligase (26). Additionally, like in most bacterial/archaeal PolXs, it has a C-terminal PHP domain (32) including highly conserved residues responsible for an Mn2+-dependent 3′-5′ exonuclease activity that is able to process unannealed 3′ termini (17) and an AP-endonuclease that confers the in vitro ability to recognize, incise and repair AP sites on the polymerase (18).
The aim of this work was to ascertain the role of the specifically conserved third HhH motif placed at the fingers subdomain of bacterial/archaeal PolXs, and the function of the lysine residue that precedes the first catalytic aspartate, using PolXBs as a model. The biochemical analyses of PolXBs mutants at residues Gly130, Gly132 and Lys134 (HhH motif) lead us to define a general DNA binding role for this motif during polymerization, 3′-5′ exonuclease and AP-endonuclease activities. Additionally, in vitro studies of mutants at residue Lys192 (preceding the first catalytic aspartate) allow us to predict a function for this lysine in the orientation of the DNA substrates at the polymerization and PHP active sites. Based on the results presented here and in light of the recently solved structure of the bacterial Thermus thermophilus PolX ternary complex (ttPolX) (33), we propose a structural model to account for the coordination of the different catalytic activities of bacterial PolXs.
MATERIALS AND METHODS
Nucleotides and proteins
Unlabelled nucleotides were purchased from GE Healthcare. [γ-32P]ATP (3000 Ci/mmol) was obtained from PerkinElmer. T4 polynucleotide kinase was purchased from New England Biolabs. Wild-type PolXBs was expressed and purified as described by Baños et al. (26).
Oligonucleotides
Oligonucleotides sp1 (5′-GATCACAGTGAGTAC) and sp1+3 (5′-GATCACAGTGAGTACCGG) were used as primer strands. Oligonucleotides sp1c+18 (5′-ACTGGCCGTCGTTCTATTGTACTCACTGTGATC) and sp1c+15 (5′-ACTGGCCGTCGTTTTGTACTCACTGTGATC) that have a 5′-terminal extension of 18 and 15 nt, respectively, in addition to the sequence complementary to sp1 and to the 5′ 15 nt of sp1+3, were used as the template strand. The 5′ phosphorylated oligonucleotide dws(P) (5′-AACGACGGCCAGT), complementary to the last 13 5′-nucleotides of sp1c+18 and sp1c+15, was used as a downstream oligonucleotide to construct gapped structures of five and two nucleotides, respectively. Oligonucleotides sp1 and sp1+3 were 5′-labelled using [γ-32P]ATP (3000 Ci/mmol) and T4 polynucleotide kinase. To analyse the DNA-dependent polymerization activity of the protein on different DNA-gapped structures and the 3′-5′ exonuclease activity, the labelled primers and the downstream dws(P) oligonucleotide were hybridized either to the template sp1c+18 (5-nt gap) or to sp1c+15 (2-nt gap). The substrate for the analysis of the AP-endonuclease activity was obtained by hybridizing 32P-labelled 5′ oligonucleotide THF (5′-GTACCCGGGGATCCGTACHGCGCATCAGCTGCAG), labelled as described previously where H stands for tetrahydrofuran (THF), with the complementary oligonucleotide THFc (5′-CTGCAGCTGATGCGCAGTACGGATCCCCGGGTAC). The oligonucleotides were purchased from Invitrogen. All the hybridizations were performed in the presence of 0.2 -M NaCl and 60-mM Tris-HCl, pH 7.5.
Site-directed mutagenesis of PolXBs
PolXBs mutants G130V, G132V, G130V/G132V, K134R, K134A, K192R and K192A were obtained by using the QuickChange site-directed mutagenesis kit provided by Amersham Pharmacia. Plasmid pET28-PolXBs containing the PolXBs gene was used as a template for the mutagenesis reaction. Expression and purification of the mutant proteins were performed as described for the wild-type PolXBs (26).
DNA binding assay
The assay was carried out using 5 nM of a 5′-labelled 2-nt gapped DNA as substrate. The substrate was incubated with 1 µM of the wild-type or mutant PolXBs for 10 minutes at 4°C in the presence of 50 mM Tris-HCl, pH 8, 25 mM NaCl, 1 mM dithiothreitol (DTT) and 1 mM MnCl2 and was filtered through nitrocellulose filters as described by Wong and Lohman (34). Radiolabelled DNA retained in each filter was quantified using the Cerenkov radiation and was represented as bars after subtracting the amount of DNA that remained bound to the filter in the absence of enzyme (background).
DNA polymerization assays on gapped DNA molecules
The 12.5 µl of incubation mixture contained 50 mM Tris-HCl, pH 7.5, 8 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 1.5 nM of the hybrid DNA indicated in each case and the specified concentration of polymerase and dNTPs. After incubation for the indicated period at 30°C, reactions were stopped by adding 10 mM of EDTA and analysed using 8 -M urea-20% PAGE and autoradiography.
Measurement of the Km value for the incoming nucleotide
The incubation mixture contained, in a final volume of 12.5 µl, 50 mM Tris-HCl, pH 7.5, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 8 mM MgCl2 and 1.5 nM of the 5′-labelled 2-nt gapped DNA. Reaction times and enzyme concentration were adjusted for each polymerase (3.75 nM, 6.25 nM and 10 nM of the wild-type, K192R and K192A polymerase, respectively) to optimize product detection while ensuring that all reactions were conducted in the steady-state. Only those reactions that fell within the linear range of substrate utilization (<30% primer extension) were used for analysis. Samples were incubated for 1 minute at 30°C in the presence of increasing concentration of the incoming nucleotide and were quenched by adding 10 mM EDTA. Reactions were analysed by electrophoresis in 8 -M urea-20% PAGE and were quantified using a Molecular Dynamics PhosphorImager. Formation of the extended product was plotted against dNTP concentration. Apparent value of Michaelis–Menten constant (Km) was obtained by least-squares non-linear regression analysis to a rectangular hyperbola using KaleidaGraph 3.6.4 software. Data are shown as mean ± SD corresponding to four independent measurements.
Exonuclease activity assays on 5′-labelled DNA substrates
The 12.5 µl of incubation mixture contained 50 mM Tris-HCl, pH 7.5, 1 mM MnCl2, 1 mM DTT, 4% glycerol, 0.1 of mg/ml BSA, 1.5 nM of the DNA molecule specified in each case and the indicated amount of the wild-type or mutant PolXBs. After incubation for the indicated period at 30°C, reactions were stopped by adding 10 mM EDTA and were analysed by 8 -M urea-20% PAGE and autoradiography.
AP-endonuclease activity assays
The 12.5 μl of incubation mixture contained 50 mM Tris-HCl, pH 7.5, 1 mM MnCl2, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 1.5 nM of 5′-labelled hybrid THF/THFc and 64 nM of the wild-type or mutant PolXBs. Samples were incubated at 30°C for the indicated period and stopped by adding 10 mM of EDTA. Reactions were analysed by 8 -M urea-20% PAGE and autoradiography.
RESULTS
Bacterial and archaeal PolXs possess a specific HhH motif
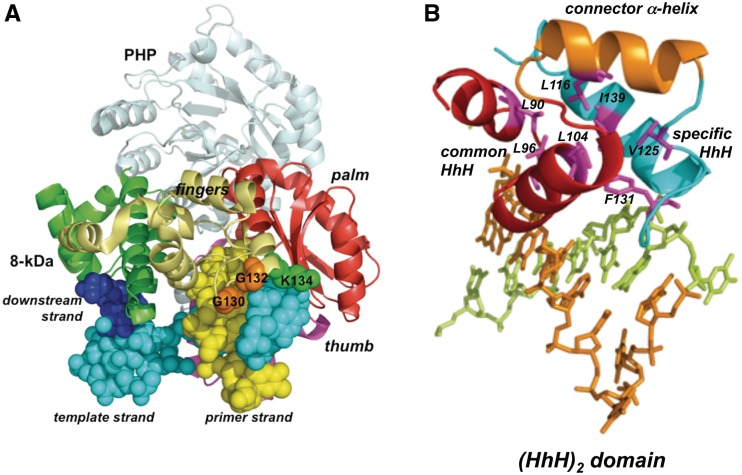
Figure 1 shows a sequence alignment of the well-delimited fingers subdomain of several representatives of bacterial/archaeal PolXs. This subdomain specifically contains, along with the common HhH motif that, by analogy with Polβ, would interact with the upstream primer strand of a gapped DNA (12), another HhH motif placed three amino acids C-terminally apart (26). Besides the two conserved glycine residues predicted to be critical for DNA–protein recognition, this motif contains a conserved lysine residue two nucleotides apart from the second glycine, as described for other HhH motifs (23). Crystallographic resolution of the structure of Deinococcus radiodurans PolX apoenzyme (PolXDr) showed the corresponding HhH motif formed by helices H and I of the fingers subdomain, the latter providing the link with the catalytic palm subdomain (35). The relative orientation of both subdomains compels PolXDr to adopt a non-standard extended conformation that makes a potential DNA binding role unpredictable for this HhH motif.
To ascertain the role of this motif of bacterial/archaeal PolXs, we introduced the aliphatic chain valine in lieu of the corresponding PolXBs residues Gly130 and Gly132 to obtain the mutants G130V, G132V and G130V/G132V, as such change has been previously reported in mutational studies carried out at the glycine residues of other HhH motifs (36,37), and because a valine residue is not found in the HhH motifs aligned in Doherty et al. (23). On the other hand, Lys134 was substituted by either a conservative (K134R) or non-conservative (K134A) residue. All these mutant polymerases were overexpressed and purified as described in ‘Materials and Methods’ section, and their catalytic activities were analysed by in vitro biochemical assays.
The HhH motif is essential for DNA binding
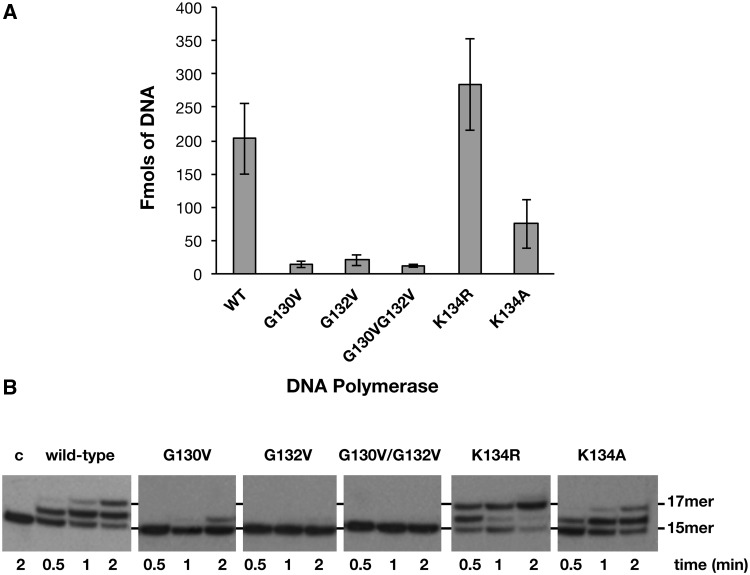
As most reported members of the PolX family, PolXBs acts preferentially on gapped DNA molecules (17,26). Because of the inefficiency shown by this polymerase to give rise to a stable retardation band in electrophoretic mobility shift assays (not shown), analysis of the efficiency of the wild-type and mutant derivatives to bind a 2-nt gapped DNA molecule was performed by nitrocellulose filter-binding assays (see ‘Materials and Methods’ section). As shown in Figure 2A, substitution of glycine residues in mutants G130V, G132V and G130V/G132V greatly impaired the binding capability of the polymerase, their efficiency being ∼10-fold lower than that displayed by the wild-type enzyme. Additionally, the presence of a positive charge in position 134 seems to be also important to maintain the stability of the PolXBs–DNA complex, as its removal in mutant K134A diminished DNA binding efficiency 2.7-fold with respect to the wild-type polymerase. Contrarily, mutant K134R showed an improved binding ability. Altogether, these results indicate a DNA-binding role for the specifically conserved HhH motif at the fingers subdomain of bacterial/archaeal PolXs.
Figure 2.
(A) Binding of the HhH motif mutants to a gapped DNA. The experiment was carried out as described in ‘Materials and Methods’ section, incubating 5 nM of 2-nt gapped DNA with 1 µM of either the wild-type or the indicated mutant PolXBs for 10 minutes at 4°C. The bars chart is the result of three independent experiments. (B) DNA polymerization activity on gapped DNA of HhH motif mutants. The assay was carried out as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 2-nt gapped DNA with 125 nM of either the wild-type or the indicated mutant PolXBs in the presence of 8 mM of MgCl2 and 50 µM of dNTPs at 30°C. The positions corresponding to the non-extended primer and to the filling-in products are indicated.
Mutant polymerases at the HhH motif have an altered polymerization activity on gapped DNA
PolXBs has been proposed to be involved in the repair of DNA damages because of its intrinsic ability to accommodate itself in gaps of one to few nucleotides, intermediates that arise during DNA repair processes (26). Figure 2B shows the effect of the mutations in filling a 2-nt gapped DNA substrate (see ‘Materials and Methods’ section). Whereas the wild-type PolXBs elongates about 50% of the initial substrate at the shortest elongation time assayed, filling the gap 1–2 minutes after starting the reaction, mutants G132V and G130V/G132V did not show any noticeable polymerization activity in the conditions assayed, mutant G130V giving rise only to a +1 faint elongation band that is 12% of the wild-type activity, in good correlation with their impaired DNA binding (aforementioned). Accordingly, the increased binding capability of mutant K134R (aforementioned) could be responsible for its higher gap-filling proficiency that enables the enzyme to synthesize the +2 product at the shortest elongation time assayed (see Figure 2B). Conversely, the diminished DNA binding capability of mutant K134A would account for its 2-fold lower polymerization activity than that of the wild-type polymerase.
The proportional increase in the polymerization reaction relative to the amount of DNA observed with mutants G130V and G132V is consistent with a defect in DNA binding as being responsible for the reduced polymerization activity (see Supplementary Figure S1).
Mutations introduced at the HhH motif affect the PHP-dependent activities of PolXBs
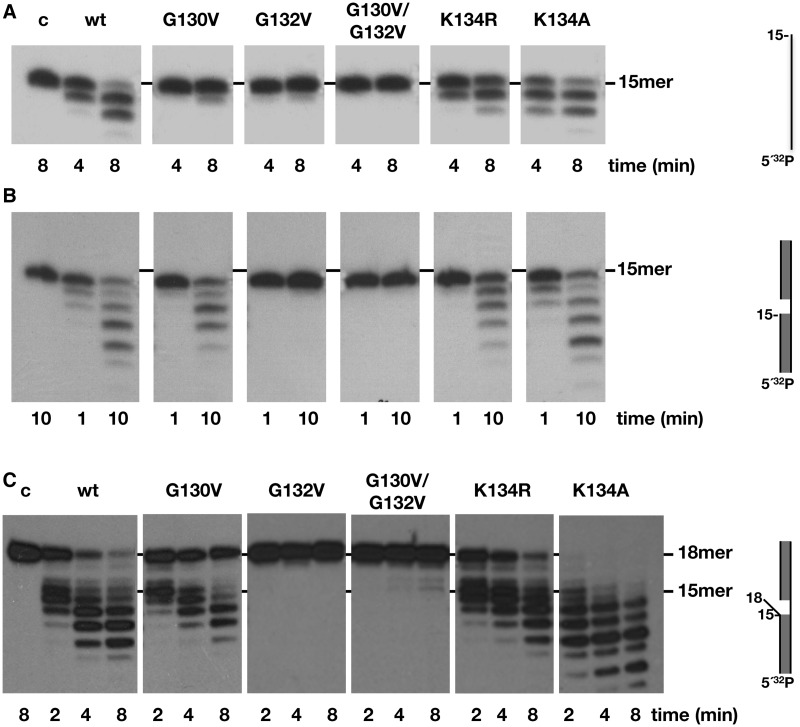
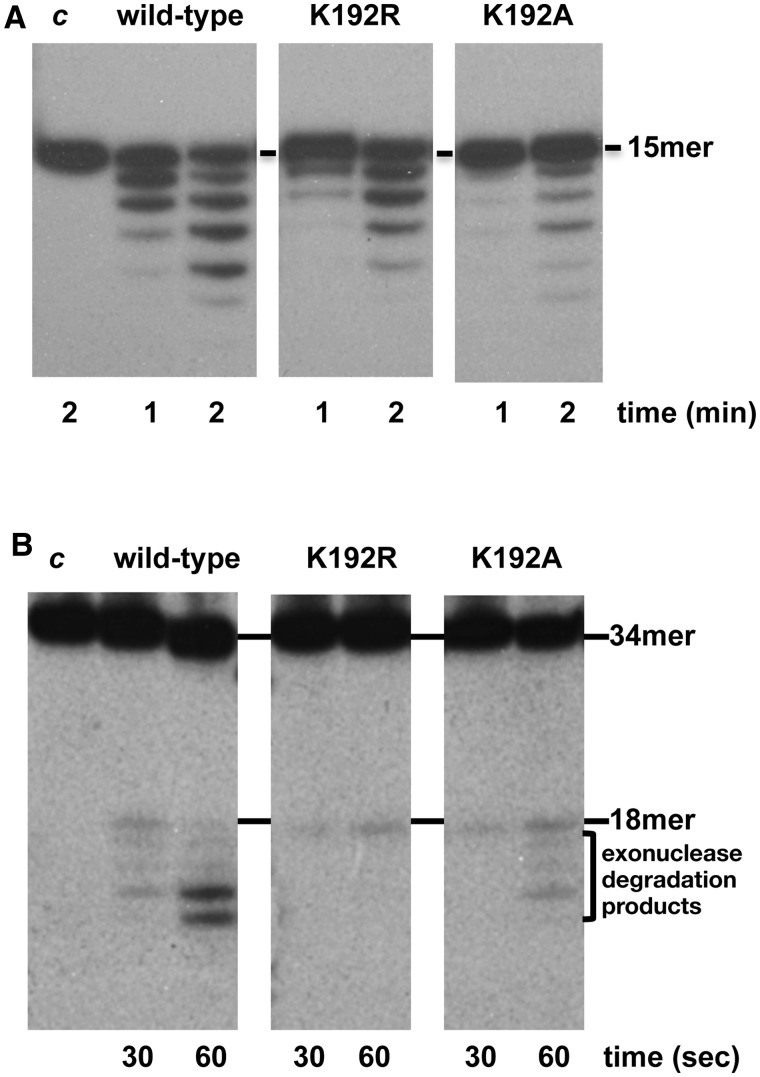
PolXBs has an Mn2+-dependent 3′-5′ exonuclease activity at its C-terminal PHP domain that capacitates the enzyme to resect mismatched 3′-termini in gapped DNA substrates (17). To find out whether the HhH residues under study could assist this nucleolytic activity, mutant polymerases underwent 3′-5′ exonuclease assays (see ‘Materials and Methods’ section). Unexpectedly, as shown in Figure 3, mutations introduced at residues Gly130 and Gly132 impaired the exonuclease activity of PolXBs when acting on ssDNA (Figure 3A) and a gapped DNA (Figure 3B) substrate. Thus, the results would suggest that these residues contact/stabilize the DNA substrate also when the 3′ terminus has to be allocated in the catalytic site of the structurally independent PHP domain. Interestingly, mutants at residue Lys134 displayed an activity opposed to their DNA-binding and polymerization capacities. Thus, whereas mutant K134R exhibited a slightly reduced exonuclease activity in comparison with the wild-type PolXBs, mutant K134A showed an exonucleolytic efficiency either higher than or similar to that of the wild-type enzyme when acting on ssDNA (Figure 3A) and gapped substrates (Figure 3B), respectively.
Figure 3.
3′-5′ exonuclease activity of the HhH motif mutants. (A) Exonuclease activity on ssDNA. The assay was performed as described in ‘Materials and Methods’ section, incubating 1.5 nM of the 15mer oligonucleotide with 125 nM of either the wild-type or the indicated mutant PolXBs in the presence of 1 mM of MnCl2 for the indicated period at 30°C. The unit length of the DNA molecule is indicated. (B) Exonuclease activity on 2-nt gapped DNA. The assay was performed as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 2-nt gapped DNA with 32 nM of either the wild-type or the indicated mutant PolXBs in the presence of 1 mM of MnCl2 for the indicated period at 30°C. (C) Exonuclease activity on 3′-mismatched gapped substrates. The assay was carried out as described in (B).
As aforementioned, previous studies showed that the 3′-5′ exonuclease activity preferentially degrades 3′-flap substrates (17). As it can be seen in Figure 3C, the wild-type enzyme efficiently removes the mispaired nucleotides, and the exonucleolytic rate slows down once it reaches the paired dsDNA region, as previously reported (17). On this substrate, mutants G132V and G130V/G132V did not render degradation products. The exonucleolytic activity shown by mutants G130V and K134R was also reduced on this DNA molecule, especially when the double-stranded region was reached. In contrast, mutant K134A presented an extremely high efficiency in degrading this kind of substrate (Figure 3C).
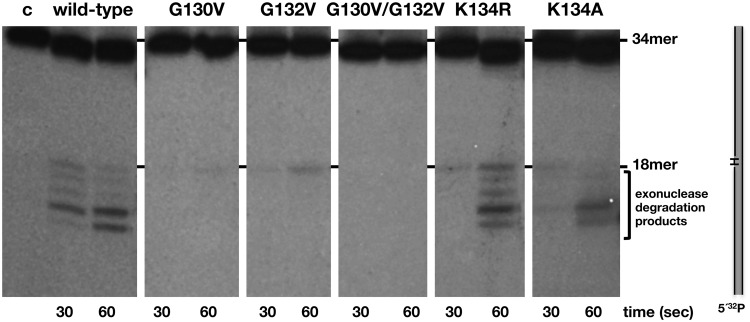
Recently, PolXBs has been demonstrated to be also endowed with an intrinsic AP-endonuclease activity at its PHP domain, sharing the same catalytic residues responsible for the 3′-5′ exonuclease activity (18). This activity qualifies PolXBs to perform in vitro recognition and incision at an AP site and further restoration of the original nucleotide in a stand-alone AP endonuclease independent way. To study the involvement of the HhH motif in aiding the AP endonuclease reaction, mutant polymerases were incubated in the presence of a DNA containing an internal THF (a stable analogue that mimics an abasic site) at the 19 position (see ‘Materials and Methods’ section). As observed in Figure 4, the wild-type enzyme promoted cleavage at the 5′ side of the THF position, rendering the expected 18mer product. The shorter bands observed are caused by the action of the 3′-5′ exonuclease activity on the 3′-end resulting after incision at the AP site, as previously described (18). As it is shown, mutations introduced in residues of the HhH motif impaired either severely (G130V, G132V and G130V/G1232V) or moderately (K134R and K134A) the AP-endonuclease activity of PolXBs, leading to conclude the importance of this HhH motif also in the stabilization of AP-bearing DNA substrates.
Figure 4.
AP-endonuclease activity of the HhH-motif mutants. The assay was carried out as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 34mer double stranded oligonucleotide containing in the 5′-labelled strand an internal THF (H) group, with 64 nM of either the wild-type or the specified mutant PolXBs in the presence of 1 mM of MnCl2 for the indicated period at 30°C.
We have previously shown that PolXBs can also introduce an internal nick on ssDNA substrates containing a THF much more efficiently than on dsDNA (18). On this substrate, mutant polymerases exhibited an AP-endonuclease activity similar to that of the wild-type enzyme, with the only exception of the double mutant G130V/G132V (see Supplementary Figure S2), strongly supporting that the defective phenotypes observed with mutant polymerases G130V and G132V is not because of a general misfolding of the protein. The poor efficiency observed with mutant G130V/G132V in all the assayed activities does not allow us to rule out a distortion of the overall structure in this case.
Several bacterial PolXs have a lysine residue preceding the catalytic aspartates
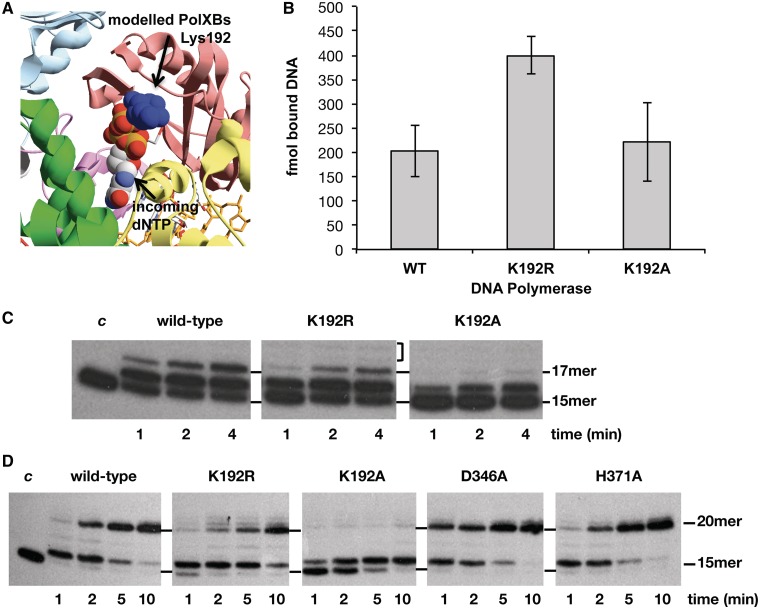
As described in the ‘Introduction’, eukaryotic Polβ and Polλ have a glycine residue preceding the first catalytic aspartate that interacts with the γ-phosphate of an incoming nucleotide, this residue being a histidine responsible for the terminal transferase activity of Polµ and TdT (31,38). Bacterial PolXs have a moderately conserved lysine/arginine residue in that position (Lys192 in PolXBs; see alignment in Figure 1). As there was not any structurally solved ternary complex of a bacterial/archaeal PolX when this work started, the palm subdomain of PolXBs was initially modelled on the Polβ ternary complex structure (39) (Figure 5A) showing a potential interaction between the incoming nucleotide and Lys192 of PolXBs, which precedes the catalytic Asp193 (26). To obtain further details about the role of this amino acid in the activity of PolXBs, the corresponding Lys192 was substituted either by a conservative (K192R) or non-conservative (K192A) residue. These mutants were overproduced and purified as described in ‘Materials and Methods’ section.
Figure 5.
(A) Potential interaction between PolXBs residue Lys192 and the incoming nucleotide. PolXBs was modelled using as template, the solved structure of PolXDr apoenzyme (PDB 2W9M) (35). Palm subdomains of modelled PolXBs and Polβ ternary complex (PDB 1BPY) (39) were further structurally aligned. (B) Binding of mutant polymerases to a gapped DNA. The experiment was carried out as described in ‘Materials and Methods’ section. Five nanomolar of the 2-nt gapped DNA was incubated with 1 µM of either the wild-type or the indicated mutant PolXBs for 10 minutes at 4°C. The bars chart is the result of three independent experiments. (C) Gap-filling reaction of mutants at residue Lys192 on a 2-nt gapped DNA. The assay was carried out as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 2-nt gapped DNA with 62 nM of either the wild-type or the indicated mutant PolXBs in the presence of 8 mM of MgCl2 and 1 µM of dNTPs for the indicated period at 30°C. The unit length of the primer molecule is indicated. (D) Gap-filling reaction of mutants at residue Lys192 on a 5-nt gapped DNA. The assay was performed essentially as described in (C), using as substrate 1.5 nM of a 5-nt gapped DNA (‘Materials and Methods’).
Lys192 is not involved in DNA binding
The ability of PolXBs mutants to bind a 2-nt gapped substrate was evaluated as described previously (‘Materials and Methods’ section). Figure 5B shows that removal of the positive charge in mutant K192A has no effect in PolXBs-DNA complex stability, ruling out a role for this residue in DNA binding. In contrast, substitution of lysine by the positively charged residue arginine provided mutant K192R with a DNA binding efficiency two-fold higher than that of the wild-type PolXBs.
Mutant K192R shows a strand displacement phenotype
Figure 5C shows the ability to fill the 2-nt gapped substrate by mutants at residue Lys192. As it can be observed, the lack of the positively charged side chain in mutant K192A greatly impaired the gap-filling reaction, despite its wild-type DNA binding ability (aforementioned). Interestingly, although mutant K192R filled the gap nearly as efficiently as the wild-type enzyme, DNA synthesis was not restricted to the completion of the gap, as faint bands corresponding to longer products could be observed (18–19mer bands in Figure 5C), suggesting the acquisition of a partial strand displacement capacity. Similar assays were also conducted on a 5-nt gapped DNA. As shown in Figure 5D, wild-type PolXBs filled the gap to completion, synthesis being restricted to the five nucleotides of the gap (20mer band). The specific blockage at the +1 position was previously described to be independent of the template sequence and length of the gap (26). It has been described in other DNA polymerases that their 3′-5′ exonuclease activity prevents strand displacement synthesis, such as Polδ during Okazaki fragment maturation (40). To analyse whether the absence of strand displacement synthesis by PolXBs is determined by its intrinsic 3′-5′ exonuclease activity, the gap filling activity of the exonuclease deficient PolXBs derivatives D346A and H371A (18) was also analysed. As shown in Figure 5D, mutant enzymes displayed a gap filling efficiency similar (H371A) or two-fold higher (D346A) than that exhibited by the wild-type enzyme, the longest elongation product being mainly the 20mer oligonucleotide. This result means that, at least under these assay conditions, exonuclease activity is not responsible for preventing strand displacement synthesis by PolXBs. Again, mutant K192R carried out proficient gap-filling reaction coupled to a partial downstream strand displacement, as deduced from the yield of 21–22mer products (Figure 5D). In contrast, mutant K192A had an impaired polymerization activity. Measurement of the Km value (in µM) for the incoming nucleotide showed no substantial differences among the wild-type and the two mutant polymerases K192R and K192A (see Table 1). This result and the nearly wild-type DNA binding capability of mutant K192A would suggest that its poor polymerization capacity could be because of a non-functional orientation of the primer-terminus at the polymerisation active site.
Table 1.
Incoming nucleotide affinity displayed by PolXBs and mutant derivatives K192R and K192A
| PolXBs | Wild-type | K192R | K192A |
|---|---|---|---|
| Km (µM) | 10.3 ± 1.75 | 13.9 ± 1.9 | 15 ± 3.9 |
Kinetic parameter was calculated from primer extension experiments, as described under ‘Materials and Methods’ section.
Km stands for the Michaelis–Menten constant.
Errors are calculated as mean values.
Changes introduced at Lys192 residue reduced the 3′-5′ exonuclease activity on gapped molecules (Figure 6A) and the AP-endonuclease one (Figure 6B). These results seem to indicate that this residue might be involved in orienting/stabilizing the DNA substrate also at the PHP active site.
Figure 6.
PHP-dependent activities of mutants at PolXBs residue Lys192. (A) 3′-5′ exonuclease activity. The assay was performed as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 2-nt gapped structure with 32 nM of either the wild-type or the indicated mutant PolXBs in the presence of 1 mM of MnCl2 for the indicated period at 30°C. (B) AP-endonuclease activity. The assay was carried out as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 34mer double stranded oligonucleotide containing an internal THF group with 64 nM of either the wild-type or the indicated mutant PolXBs in the presence of 1 mM of MnCl2 for the indicated period at 30°C.
DISCUSSION
PolXBs exhibits the general enzymatic features of the PolX members: dependence on divalent metal ions, requirement of a template strand to direct DNA synthesis, a distributive polymerization pattern, preferential use of gapped DNA structures bearing a downstream 5′-phosphate group and favourable insertion of the complementary deoxyribonucleoside monophosphate (26). This polymerase shares with most of bacterial/archaeal PolXs a C-terminal PHP domain. In PolXBs, this domain is endowed with an inherent 3′-5′ exonuclease activity, dependent on highly conserved residues, and that enables the enzyme to process unannealed 3′-termini and that made us to suggest the presence of this activity in the rest of PHP-containing bacterial/archaeal PolXs (17). In addition, PolXBs possesses also an intrinsic AP-endonuclease activity genetically linked to the exonucleolytic one and governed by the same metal ligands of the PHP domain (18). Altogether, these three different catalytic functions, polymerization, 3′-5′ exonucleolysis and AP-endonucleolysis capacitate PolXBs to perform, at least in vitro, recognition and incision at an AP site and further repair the original nucleotide in a stand-alone AP-endonuclease independent way (18).
Preferential binding of PolXs to gapped substrates largely relies on two universally present HhH motifs, one at the N-terminal 8-kDa domain and the other at the fingers subdomain (12,25,41,42). Crystallographic resolution of Polβ complexes has shown the backbone of the downstream DNA of the gap interacting with the protein through the HhH motif of the 8-kDa domain, whereas that of the fingers contacts with the sugar–phosphate backbone of the upstream primer strand stabilizing it at the polymerization active site (6,12,13,24,25,41). In addition, bacterial/archaeal PolX members also contain a third putative HhH motif whose function remained to be determined. When this work started, PolXDr apoenzyme was the only bacterial PolX member whose structure had been solved (35). In those crystals the PolX core of the polymerase exhibited a stretched out conformation instead of the commonly found closed right hand, the fingers subdomain being in a totally rotated position relative to the palm subdomain compared with other family X DNA polymerases (35). This fact made it difficult to envision a DNA-binding role for the specifically conserved HhH motif of the fingers subdomain of bacterial/archaeal PolXs.
The results presented here underline the importance of the glycine residues of the latter HhH motif to stabilize the binding of PolXBs to a gapped substrate and to an AP-containing DNA, allowing correct location of the primer-terminus at the polymerization and PHP active sites. In addition, the results obtained with mutants at residue Lys134 are also in accordance with a DNA binding role for this motif. Thus, mutant K134R showed an improved DNA binding capability that resulted in enhanced polymerization efficiency, although the exonuclease and AP- endonuclease activities were reduced in comparison to those of the wild-type enzyme, suggesting that the presence of an arginine residue in that position stabilizes the primer-terminus in a conformation prone to elongation. In this sense, the decreased DNA binding capability of mutant K134A caused a diminished polymerization activity. Here, the poorer stabilization of the primer strand at the polymerization site could be responsible for the enhanced 3′-5′ exonuclease observed, mainly on mispaired 3′ termini. Propensity of a lysine residue at this position, previously described for other HhH motifs, suggested that it could interact with DNA–phosphate groups in a similar manner to the same residues in a P-loop structure (23), as demonstrated for Lys68 of the Polβ 8-kDa HhH motif that was shown to contact the phosphate backbone of the downstream strand of a gapped molecule (24). Altogether, these results point to a role for the specifically conserved HhH motif of the fingers subdomain of bacterial/archaeal PolX in general stabilization of DNA substrates and in the coordination among the different activities of these enzymes, as well as to suggest a common DNA binding site for the three activities of these polymerases. Considering PolXBs apoenzyme folded as PolXDr, because of the homology shared between both enzymes, only a pronounced structural reorientation of the fingers subdomain on binding the substrates would account for the results presented here.
Recently, the crystallographic structure of ttPolX has been solved, showing that the complex with an incoming nucleotide (binary complex) adopts the same extended structure observed in the PolXDr apoenzyme (33). Comparison of the crystal structures of the ttPolX binary and ternary (with a 1-nt gapped DNA) complexes showed a fine superposition of the palm, thumb and PHP domains, in contrast to the 8-kDa domain and fingers subdomain that are shifted to a great extent to bind the DNA (33). Figure 7A shows a model of the PolXBs structure obtained with the SWISS-MODEL software (43,44), using ttPolX ternary complex as template. PolX core adopts the classical cupped shape observed in Polβ on binding DNA wrapping this substrate around. In this conformation, the HhH motif of bacterial/archaeal PolX fingers is now placed in such a way that the two glycines and the lysine residues would contact the upstream region of the gapped DNA, in good agreement with the DNA binding role for this HhH motif that is proposed here. In addition, the structural analysis of the common and specifically conserved HhH motifs of the fingers subdomain of ttPolX leads us to conclude that they are integrated as a part of a five-helical domain termed (HhH)2 (45). This domain classically consists of two consecutive HhH motifs linked by a connector helix, showing a conserved hydrophobic core comprising seven residues (see Figure 7B), one residue from each α-helix and each hairpin (45). The symmetric structure of this (HhH)2 domain would mirror the symmetry of the DNA double helix enabling strong non-specific binding.
Figure 7.
(A) Ribbon representation of the structural model of PolXBs bound to DNA. Model for PolXBs was provided by the homology-modelling server SWISS-MODEL, using the crystallographic structure of the ternary complex of ttPolX as template (PDB code 3AUO) (33). The PHP domain is shown in light blue, the 8-kDa domain in green, fingers in gold yellow, palm in red and thumb in magenta. Bound DNA is represented as spheres. The upstream, template and downstream strands are coloured in yellow, cyan and dark blue, respectively. Glycines and lysine of the specifically conserved HhH motif are represented as orange and green spheres, respectively. (B) Fingers subdomain of bacterial PolXs contains a (HhH)2 domain. PolXBs structure was modelled as described in (A). Common and specifically conserved HhH motifs are represented as red and cyan ribbons, respectively, whereas connector α-helix is coloured in orange. PolXBs residues forming the conserved hydrophobic core are represented as magenta sticks. Figure was made using PyMOL software (http://www.pymol.org).
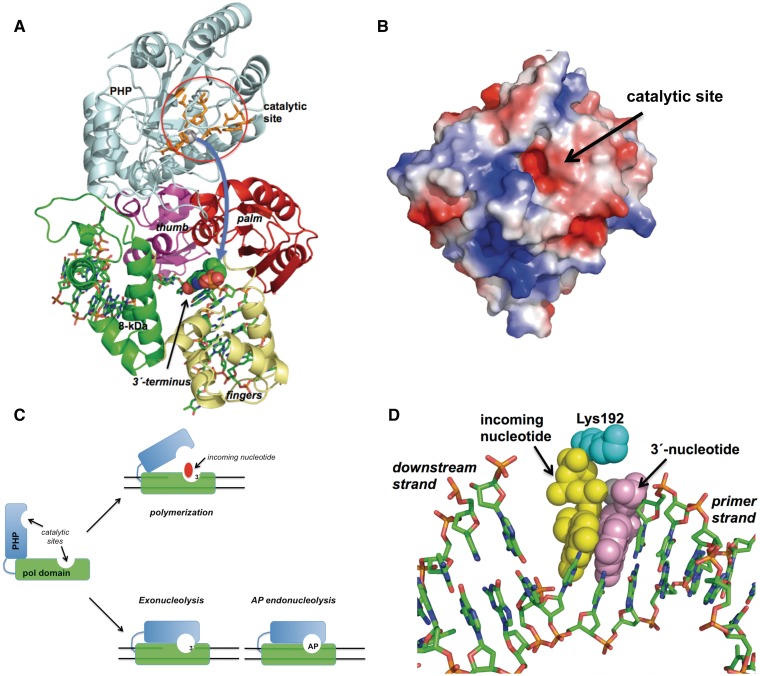
As shown in Figure 8A, the modelled PolXBs core is folded in a catalytically competent manner for polymerization in the ternary complex. The PHP domain is attached to the thumb subdomain by a 30 amino acids long linker, its β-barrel conformation forming an almost right angle with the longitudinal axis of the DNA bound to the PolX core. There is not any evident DNA binding cleft in the PHP domain, the catalytic residues being solvent exposed and placed close to the molecular surface of the domain but far away from the DNA bound to the only binding cleft of the polymerase (see Figure 8A). Hence, the relative orientation of the PolX core and the PHP domain could not account either for the appropriate coordination among the catalytic activities of the enzyme shown to occur during the resection of a misaligned 3′ terminus (17) and the repair of an abasic site (18) or for the effect that the HhH mutations studied here had on the different activities of PolXBs. Indeed, and as mentioned previously, our results strongly favour a common DNA binding site for the three enzymatic functions of PolXBs. If there is only a DNA binding cleft, and considering the disposal of the domains in the current structures of the homologous ttPolX, the question that arises is how the bound DNA can access to the polymerization and PHP active sites to coordinate polymerization, 3′-5′ exonuclease and AP-endonuclease activities.
Figure 8.
(A) Model of the PHP motion. The PHP and polymerization subdomains are coloured as in Figure 7A. Catalytic residues of the PHP domain are represented as orange sticks. The 3′ terminal nucleotide of the upstream primer strand is represented as spheres. Curved arrow indicates the proposed movement of the PHP domain. See main text for details. (B) Electrostatic surface of the modelled PHP domain of PolXBs. Superficial placement of the catalytic active site is indicated. (C) Scheme depicting the proposed PHP motion during the 3′-5′ exonucleolytic removal of mispaired 3′ termini and the repair of AP sites by PolXBs. PolX core and PHP domains are represented as green and cyan boxes, respectively. See main text for details. (D) Modelling of the PolXBs residue Lys192 at the polymerization active site. Incoming nucleotide, 3′ primer-terminus and Lys192 are represented as yellow, violet and cyan spheres. Figure was made using PyMOL software (http://www.pymol.org).
If the DNA does not go to the PHP domain, the PHP domain must go to the DNA
An appealing hypothesis would be that the incapacity of the DNA polymerase to elongate a primer, caused by the presence of a mismatched 3′ terminus at the polymerization active site, the absence of an incoming nucleotide or the presence of an AP site could promote a rotation of the PHP domain toward the upper surface (solvent accessible) of the PolX core. Such a rotation could be possible by virtue of the long linker that connects both polymerase portions, allowing the superficial PHP active site to reach and either remove a non-extendable 3′ terminus or catalyze hydrolysis at an AP site placed at the polymerization active site (see Figure 8A and C). Once the PHP domain goes back to the initial position, the entrance of an incoming nucleotide would promote further elongation of the resulting sanitized 3′ ends. The model proposed here could account for the role of the HhH motif in general DNA binding and would explain how the three enzymatic activities of PolXBs can be coordinated during the repair process using a single DNA binding cleft. In addition, it would give an explanation also for the following: (i) our previous results showing a higher efficiency of PolXBs to recognize and bind specifically and stably to an AP-containing dsDNA molecule, in comparison to a non-damaged dsDNA (18), (ii) the significant reduction of the 3′-5′ exonuclease activity of PolXDr because of mutations introduced at the common HhH motif of its fingers subdomain (36) and (iii) the tight interaction observed between the independently expressed polymerization and PHP domains of ttPolX when they were mixed, as the model described here would account for the large interface contact area proposed to exist (19). Although the PHP movement would be the major structural change required, the model demands for other rearrangements in the PolX core to allow the PHP domain to lie over the polymerization active site, mainly in the presence of an AP-containing DNA. In this case, the 8-kDa domain could not be oriented exactly as it is in the reported ternary complexes because a downstream 5′-phosphate end would result only after AP endonucleolysis. However, this kind of reorientations could be feasible, as a substantial movement of the 8-kDa and fingers subdomains has been shown to occur on binding DNA in ttPolX owing to their high flexibility (33).
Role of PolXBs residue Lys192
As mentioned previously, PolXBs possesses a lysine residue, moderately conserved among bacterial PolXs, preceding the two first catalytic aspartates. The results presented here rule out a role of this residue in general stabilization of the PolX–DNA complex, as the K192A mutant did not show a reduction in its DNA binding capability. However, in this case, the lack of the lysine side chain made the polymerase deficient in elongating the primer strand. Its nearly wild-type Km value for the incoming nucleotide suggests a non-suitable orientation and/or stabilization of the primer-terminus at the polymerization site as the most likely reason for its deficient polymerization activity. This hypothesis was initially substantiated by the PolXDr structure in which the corresponding Arg196 was postulated to stabilize the primer-terminus at the polymerization site through contacting with the 3′-phosphate, taking over the role of residues Arg254, Arg488 and Lys616 of Polλ, Polβ and PolIII, respectively (35). Additionally, further modelling of PolXBs residue Lys192 on the recently solved ttPolX ternary complex (which has a glycine residue instead) shows the side chain positioned just in between the primer-terminal residue and the incoming nucleotide (see Figure 8D), in a similar fashion to His229 in Polµ (31). It is noteworthy that in the absence of incoming nucleotide, mutant K192A also displayed a reduced 3′-5′ exonuclease and AP endonuclease activities, maybe because of the aforementioned distortion that would make the right allocation difficult either for the upstream 3′ terminus or for the AP site at the PHP catalytic centre, suggesting a participation of this residue in the coordination of the different activities of PolXBs. Introduction of an arginine in position 192 enhanced the DNA binding efficiency of the polymerase, mutant K192R showing a wild-type polymerization activity on gapped substrates. Interestingly, this mutant is endowed with the ability to displace partially the downstream strand. Contrarily to other DNA polymerases, such as Polδ, whose 3′-5′ exonuclease activity prevents strand displacement synthesis during Okazaki fragment maturation (40), the reduced exonuclease activity exhibited by mutant K192R does not seem to be responsible for its strand displacement capacity. Thus, these results could be pointing to a stronger stabilization of the primer-terminus in an orientation prone to polymerization that would impede dissociation of mutant PolXBs once the nucleotide has been inserted, compelling the polymerase to advance. In this sense, such an over-stabilization could hamper the access of the substrates to the PHP active site, explaining the reduced 3′-5′ exonuclease and AP endonuclease activities displayed by mutant K192R. Although this residue does not seem to be instrumental in general PolXBs–DNA complex stabilization, a fact that otherwise would undermine dissociation after gap-filling completion, compromising the final sealing step of the resulting adjacent 3′-OH and 5′-phosphate groups (17,26), the results support a role for Lys192 in the proper orientation of the DNA at both active sites to be functionally coordinated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Spanish Ministry of Economy and Competitiveness [BFU2011-23720 to M.V.]; the Spanish Research Council [200920I012 to M.V.]; the Spanish Ministry of Science and Innovation [BFU2008-00215 and Consolider-Ingenio CSD2007-00015 to M.S.]; Institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa.’ Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BFU2011-23720].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 3.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 4.García-Díaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J. Biol. Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 6.Moon AF, García-Díaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Lecointe F, Shevelev IV, Bailone A, Sommer S, Hübscher U. Involvement of an X family DNA polymerase in double-stranded break repair in the radioresistant organism Deinococcus radiodurans. Mol. Microbiol. 2004;53:1721–1730. doi: 10.1111/j.1365-2958.2004.04233.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee JW, Blanco L, Zhou T, García-Díaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 10.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nick McElhinny SA, Havener JM, García-Díaz M, Juárez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase beta. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 13.Yamtich J, Sweasy JB. DNA polymerase family X: function, structure, and cellular roles. Biochim. Biophys. Acta. 2010;1804:1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bebenek K, García-Díaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 15.García-Díaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat. Struct. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 16.Saxowsky TT, Matsumoto Y, Englund PT. The mitochondrial DNA polymerase beta from Crithidia fasciculata has 5′-deoxyribose phosphate (dRP) lyase activity but is deficient in the release of dRP. J. Biol. Chem. 2002;277:37201–37206. doi: 10.1074/jbc.M206654200. [DOI] [PubMed] [Google Scholar]
- 17.Baños B, Lázaro JM, Villar L, Salas M, de Vega M. Editing of misaligned 3′-termini by an intrinsic 3′-5′ exonuclease activity residing in the PHP domain of a family X DNA polymerase. Nucleic Acids Res. 2008;36:5736–5749. doi: 10.1093/nar/gkn526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baños B, Villar L, Salas M, de Vega M. Intrinsic apurinic/apyrimidinic (AP) endonuclease activity enables Bacillus subtilis DNA polymerase X to recognize, incise, and further repair abasic sites. Proc. Natl. Acad. Sci. USA. 2010;107:19219–19224. doi: 10.1073/pnas.1013603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakane S, Nakagawa N, Kuramitsu S, Masui R. Characterization of DNA polymerase X from Thermus thermophilus HB8 reveals the POLXc and PHP domains are both required for 3′-5′ exonuclease activity. Nucleic Acids Res. 2009;37:2037–2052. doi: 10.1093/nar/gkp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Ramsden DA. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- 22.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 23.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 25.Mullen GP, Wilson SH. DNA polymerase ß in abasic site repair: a structurally conserved helix-hairpin-helix motif in lesion detection by base excision repair enzymes. Biochemistry. 1997;36:4713–4717. doi: 10.1021/bi962363a. [DOI] [PubMed] [Google Scholar]
- 26.Baños B, Lázaro JM, Villar L, Salas M, de Vega M. Characterization of a Bacillus subtilis 64-kDa DNA polymerase X potentially involved in DNA repair. J. Mol. Biol. 2008;384:1019–1028. doi: 10.1016/j.jmb.2008.09.081. [DOI] [PubMed] [Google Scholar]
- 27.García-Díaz M, Domínguez O, López-Fernández LA, de Lera LT, Saniger ML, Ruiz JF, Párraga M, García-Ortiz MJ, Kirchhoff T, del Mazo J, et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 28.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat. Res. 2000;460:231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Arora K, Beard WA, Wilson SH, Schlick T. Critical role of magnesium ions in DNA polymerase beta’s closing and active site assembly. J. Am. Chem. Soc. 2004;126:8441–8453. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 30.Domínguez O, Ruiz JF, Lain de Lera T, García-Díaz M, González MA, Kirchhoff T, Martínez AC, Bernad A, Blanco L. DNA polymerase mu (Pol µ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon AF, García-Díaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase µ. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 32.Aravind L, Koonin EV. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakane S, Ishikawa H, Nakagawa N, Kuramitsu S, Masui R. The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA. J. Mol. Biol. 2012;417:179–196. doi: 10.1016/j.jmb.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leulliot N, Cladiere L, Lecointe F, Durand D, Hübscher U, van Tilbeurgh H. The family X DNA polymerase from Deinococcus radiodurans adopts a non-standard extended conformation. J. Biol. Chem. 2009;284:11992–11999. doi: 10.1074/jbc.M809342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blasius M, Shevelev I, Jolivet E, Sommer S, Hübscher U. DNA polymerase X from Deinococcus radiodurans possesses a structure-modulated 3′–>5′ exonuclease activity involved in radioresistance. Mol. Microbiol. 2006;60:165–176. doi: 10.1111/j.1365-2958.2006.05077.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang LK, Nair PA, Shuman S. Structure-guided mutational analysis of the OB, HhH, and BRCT domains of Escherichia coli DNA ligase. J. Biol. Chem. 2008;283:23343–23352. doi: 10.1074/jbc.M802945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrade P, Martín MJ, Juárez R, López de Saro F, Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl. Acad. Sci. USA. 2009;106:16203–16208. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 40.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase ß deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Maga G, Blanca G, Shevelev I, Frouin I, Ramadan K, Spadari S, Villani G, Hübscher U. The human DNA polymerase lambda interacts with PCNA through a domain important for DNA primer binding and the interaction is inhibited by p21/WAF1/CIP1. FASEB J. 2004;18:1743–1745. doi: 10.1096/fj.04-2268fje. [DOI] [PubMed] [Google Scholar]
- 43.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 44.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao X, Grishin NV. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 2000;28:2643–50. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]