Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that function in literally all cellular processes. miRNAs interact with Argonaute (Ago) proteins and guide them to specific target sites located in the 3′-untranslated region (3′-UTR) of target mRNAs leading to translational repression and deadenylation-induced mRNA degradation. Most miRNAs are processed from hairpin-structured precursors by the consecutive action of the RNase III enzymes Drosha and Dicer. However, processing of miR-451 is Dicer independent and cleavage is mediated by the endonuclease Ago2. Here we have characterized miR-451 sequence and structure requirements for processing as well as sorting of miRNAs into different Ago proteins. Pre-miR-451 appears to be optimized for Ago2 cleavage and changes result in reduced processing. In addition, we show that the mature miR-451 only associates with Ago2 suggesting that mature miRNAs are not exchanged between different members of the Ago protein family. Based on cloning and deep sequencing of endogenous miRNAs associated with Ago1–3, we do not find evidence for miRNA sorting in human cells. However, Ago identity appears to influence the length of some miRNAs, while others remain unaffected.
INTRODUCTION
Small noncoding RNAs including microRNAs (miRNAs) and siRNAs regulate gene expression on posttranscriptional levels (1,2). miRNAs are endogenously transcribed from specific genes generating capped and polyadenylated primary miRNA transcripts (pri-miRNA), which are characterized by hairpins containing the miRNAs flanked by single-stranded RNA segments (2,3). In the nucleus, the microprocessor, composed of several proteins including the RNase III Drosha, cleaves the pri-miRNA and generates a hairpin-structured miRNA precursor (pre-miRNA) that is subsequently exported from the nucleus. In the cytoplasm, the RNase III Dicer cleaves off the loop of the pre-miRNA forming a double-stranded short RNA that contains the mature miRNA as well as the opposing strand termed miRNA*. In subsequent steps, the two strands are separated and the mature miRNA is incorporated into a miRNA–protein complex [miRNP also referred to as microRNA-induced silencing complex (miRISC)], while the miRNA* is degraded by cellular nucleases (2–4).
Within the miRNP, the mature miRNA interacts with a member of the Argonaute (Ago) protein family (5–7). In mammals, Ago1–4 constitute the Ago subfamily of the Ago proteins. Ago proteins are characterized by PAZ (Piwi–Argonaute–Zwille), MID (located between the PAZ and the PIWI domain) and PIWI (P element-induced wimpy testes) domains. The PAZ domain is required for binding of the 3′ end of the small RNA, while the MID domain specifically anchors the 5′ end. The PIWI domain is structurally similar to RNase H and it has been experimentally demonstrated that some, but not all PIWI domains are endonucleolytically active (8). Among the mammalian Ago proteins, only Ago2 possesses cleavage activity and is therefore termed Slicer (9,10). Catalytically active Ago proteins also participate in RISC assembly. Ago proteins cleave the passenger strand in perfectly complementary siRNA duplexes leading to efficient RISC loading (11–13).
In addition to the canonical miRNA biosynthesis pathway, several noncanonical pathways exist. First, miRNAs can be generated from very short introns. After splicing, these introns, referred to as mirtrons, form pre-miRNA hairpins without Drosha processing (14–16). Second, some miRNAs are generated from small nucleolar RNAs (snoRNAs) without Drosha requirement (17–20). Finally, miR-451 is expressed independently of Dicer. Instead, Ago2 cleaves the pre-miR-451 hairpin to form the mature miR-451 (21,22). The precise mechanisms, however, are not fully understood.
miRNAs guide Ago proteins and associated factors to partially complementary target sites predominantly located in 3′ untranslated regions (UTRs) of mRNAs (23). As a consequence, the mRNA is degraded or its translation is inhibited. For mRNA degradation, a member of the GW (Glycin-trytophan) protein family (termed TNRC6A-C in mammals) interacts with the Ago protein and recruits the deadenylation machinery to the mRNA leading to poly(A) tail shortening. The mRNA is subsequently decapped and degraded by exonucleases (24,25).
In contrast to miRNAs, siRNAs are fully complementary to specific target RNAs and direct their sequence-specific cleavage (26). In mammals, endogenous siRNAs have only been reported in the germ line and siRNAs are predominantly used as research tool for sequence-specific gene knockdown (3). SiRNA strands are chemically synthesized and annealed to double-stranded molecules and after transfection, one strand (guide strand) is incorporated into the RISC and directly interacts with an Ago protein. For siRNA knockdown, short-hairpin RNAs mimicking pre-miRNAs have been developed and are widely used as well (27).
In Drosophila, miRNAs and siRNAs are sorted into specific Ago proteins (28,29), while sorting of miRNAs or siRNAs into distinct members of the mammalian Ago proteins remains elusive. In fact, a typical siRNA transfection experiment results in loading of all four mammalian Ago proteins (30) although only Ago2 is catalytically active and needed for gene-specific knockdown. SiRNAs may interact with partially complementary targets as well (30,31). The inactive Ago proteins (Ago1, Ago3, and Ago4) typically cause unwanted off-target effects by regulating unrelated targets in a miRNA-like manner (32). Therefore, developing siRNAs that are exclusively loaded into Ago2 would be a major step forward. In order to generate such a tool, we explored the miR-451 system. We find that miR-451 is indeed only bound by Ago2 and not by other Ago proteins. However, altering the miR-451 hairpin resulted in a reduced processing efficiency. Furthermore, we isolated endogenous Ago1–3 complexes and identified associated miRNAs. We do not find evidence for miRNA sorting in human cells. In addition, Ago identity appears to impact the length of some specific mature miRNAs, while others are not affected.
MATERIALS AND METHODS
Isolation of single cells and RNA from mouse organs
The mouse strain C57BL/6 J was used for all experiments with mouse tissues. For isolation of RNA, liver, brain, kidneys and spleen were shock frozen in liquid nitrogen and pulverized in a mortar with a pestle. The crushed tissue was lyzed immediately in peqGOLD TriFast (peqlab), rigorously mixed and incubated 5–10 min at room temperature. The RNA was isolated according to the manufacturer’s manual. After precipitation over night at −20°C, the RNA was pelleted for 30 min, 17 000g and 4°C. After washing with 1 ml ice-cold ethanol (80%), the RNA was centrifuged again for 30 min, 17 000g and 4°C. Pellets were dried for a short time at room temperature, suspended in ddH2O and solved at 70°C for 5 min while shaking (1000 rpm).
For preparation of single cell suspensions, livers and spleens of two mice were isolated. The livers were pressed through a cell strainer into a petri dish containing phosphate buffered saline (PBS). Remaining tissue was discarded. The spleens were crushed between the roughened surfaces of two microscope slides. The cells were washed off the slides with posphate buffered saline (PBS) and subsequently pipetted through a cell strainer (BD Falcon, REF 352350, 70 µm nylon). Cells in both suspensions were collected by centrifugation at 200g, 4°C for 5 min. To remove contaminating erythrocytes, cells were resuspended in 10–20 ml of ACK lysis buffer (150 mM NH4Cl, 1 mM KHCO3, 100 mM Na2EDTA) and incubated for 5 min at room temperature. Remaining liver and spleen cells, respectively, were collected again by centrifugation (200g, 5 min, 4°C). Lyzed (red) erythrocytes remained in the supernatant and were discarded. White pellets were washed once with cold PBS, before cells were lyzed in 25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5% (v/v) NP-40, 2 mM EDTA, 1 mM NaF for 20 min on ice. Cell debris was removed by centrifugation at 17 000g, 4°C for 45 min. The supernatant was shock frozen with liquid nitrogen and stored at −80°C.
Immunoprecipitations and cell lysis
Ago immunoprecipitations
Monoclonal antibodies for human Ago1 (4B8) (33), Ago2 (11A9) (34), Ago3 (5A3), Ago4 (6C10) (30,35) or mouse Ago2 (6F4) (36) were coupled to Protein G Sepharose beads over night at 4°C. Antibody-decorated beads were washed twice with lysis buffer [25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5% (v/v) NP-40, 2 mM EDTA, 1 mM NaF]. For immunoprecipitations (IPs) from mouse tissue, lysates from liver and spleen of two mice were applied. For IPs from transfected HEK 293, cells were grown on 15 cm plates to full density, washed with cold PBS and lyzed in 1 ml of lysis buffer. The lysates were cleared by centrifugation at 17 000g for 30 min, before addition to the beads. As a control for the IPs, beads without coupled antibody were used. After 4 h of rotation at 4°C, the beads were washed four times with wash buffer [300 mM NaCl, 50 mM Tris–HCl pH 7.4, 1 mM MgCl2 and 0.1% (v/v) NP-40] and once with PBS.
For RNA extraction, the immunoprecipitates were treated with Proteinase K (40 µg per sample, in 200 µl 300 mM NaCl, 25 mM EDTA, 2% SDS, 200 mM Tris pH 7.5) at 65°C for 15 min. The RNA was extracted with 200 µl of acidic phenol and precipitated with 2.5 volumes pure ethanol at −20°C overnight. For input samples, 100 µl of the respective lysate was denatured by addition of 1 ml of TriFast reagent. The RNA was subsequently extracted according the manufacturer’s instructions. After pelleting the RNA, it was washed once with cold 80% ethanol, dried and solved in H2O.
Small RNA cloning
Isolated RNA was ligated to a barcoded, adenylated 3′ adapter [5′-Phospho-T-4nt-Barcode-CGTATGCCGTCTTCTGCTTG-(C7amino)-3′] by a truncated T4 RNA Ligase 2, the 5′ RNA adapter (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) was added in a second ligation step by T4 RNA Ligase 1. The product was reverse-transcribed using the SuperScriptIII First Strand Synthesis Super Mix (Invitrogen) using a specific primer (5′-CAAGCAGAAGACGGCATACGA), followed by a PCR amplification (5′ Primer: 5′ AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGACGATC; 3′ Primer: 5′ CAAGCAGAAGACGGCATACGA). The samples were run on a 6% Urea–PAGE, the bands corresponding to small RNA containing ligation products were cut out and eluted over night in 300 mM NaCl, 2 mM EDTA. The libraries were precipitated with ethanol over night at −20°C, then collected by centrifugation and solved in water.
The libraries were measured on a Genome Analyzer GAIIx (Illumina) by Fasteris SA (Geneva) in 1 × 38 bp single-end runs.
Plasmids
Expression of shRNAs was achieved by using a modified pSuper plasmid (oligoengine). All sequences were cloned as oligonucleotides according to the manufacturer’s protocol using the restriction enzymes BglII and HindIII. All clones were verified by sequencing. The Imp8 shRNA has been reported before (35).
Primers used:
MiR-451 original sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTAAACTCAGTAATGGTAACGGTTTGGA; miR-451 classical sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTTTCAAGAGAAACTCAGTAATGGTAACGGTTTTTTTTA, antisense 5′ AGCTTAAAAAAAACCGTTACCATTACTGAGTTTCTCTTGAAAACTCAGTAATGGTAACGGTTTGGA; siGRK4 classical sense 5′ GATCTCCAACTGCCTGAAGAGACGTCTTTTCAAGAGAAAGACGTCTCTTCAGGCAGTTTTTTTA, antisense 5′ AGCTTAAAAAAACTGCCTGAAGAGACGTCTTTCTCTTGAAAAGACGTCTCTTCAGGCAGTTGGA; siGRK4 mimic sense 5′ GATCTCCAACTGCCTGAAGAGACGTCTTCACGTCTCTTCAGGCAGTCTTTTTTA, antisense 5′ AGCTTAAAAAAGACTGCCTGAAGAGACGTGAAGACGTCTCTTCAGGCAGTTGGA; siImp8 classical sense 5′ ACAGAGATCTCCTTAGTGAGAGTCCAATTAATTCAAGAGATTAATTGGACTCTCACTAATTTTTAAGCTTACAG, antisense 5′ CTGTAAGCTTAAAAATTAGTGAGAGTCCAATTAATCTCTTGAATTAATTGGACTCTCACTAAGGAGATCTCTGT; siImp8 mimic sense 5′ GATCTCCTTAGTGAGAGTCCAATTAACCATAATTGGACTCTCACTACTTTTTTA, antisense 5′ AGCTTAAAAAAGTAGTGAGAGTCCAATTATGGTTAATTGGACTCTCACTAAGGA; mutant #1 sense 5′ GATCTCCTAACCGTTACCATTACTGAGTTTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTAAACTCAGTAATGGTAACGGTTAGGA; mutant #2 sense 5′ GATCTCCAAACCGTAACCATTACTGAGTTTAGTAATGGTTATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATAACCATTACTAAACTCAGTAATGGTTACGGTTTGGA; mutant #3 sense 5′ GATCTCCAAACCGTTACCAATACTGAGTTTAGTATTGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCAATACTAAACTCAGTATTGGTAACGGTTTGGA; mutant #4 sense 5′ GATCTCCAAACCGTTAGCATTACTGAGTTTAGTAATGCTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTAGCATTACTAAACTCAGTAATGCTAACGGTTTGGA; mutant #5 sense 5′ GATCTCCAAACCGTTACGATTACTGAGTTTAGTAATCGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACGATTACTAAACTCAGTAATCGTAACGGTTTGGA; miR-451 loop +1 nt sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTAGAACTCAGTAATGGTAACGGTTTGGA; miR-451 loop +2 nt sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCGTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTACGAACTCAGTAATGGTAACGGTTTGGA; miR-451 loop +3 nt sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCGGTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTACCGAACTCAGTAATGGTAACGGTTTGGA; miR-451 loop +4 nt sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCGGGTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTACCCGAACTCAGTAATGGTAACGGTTTGGA; miR-451 loop +5 nt sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCGGGGTAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTACCCCGAACTCAGTAATGGTAACGGTTTGGA; 1st wobble sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTTAGTAATGGTAACGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCGTTACCATTACTAAACTCAGTAATGGTAACGGTTTGGA; 2nd wobble sense 5′ GATCTCCAAACCGTTACCATTACTGAGTTCAGTAATGGTAATGGTTCTTTTTTA, antisense 5′ AGCTTAAAAAAGAACCATTACCATTACTGAACTCAGTAATGGTAACGGTTTGGA; let-7c genomic sense 5′ GATCTCCTGAGGTAGTAGGTTGTATGGTTTAGAGTTACACCCTGGGAGTTAACTGTACAACCTTCTAGCTTTTTTTTA, antisense 5′ AGCTTAAAAAAAAGCTAGAAGGTTGTACAGTTAACTCCCAGGGTGTAACTCTAAACCATACAACCTACTACCTCAGGA; let-7c classical sense 5′ GATCTCCTGAGGTAGTAGGTTGTATGGTTTTCAAGAGAAACCATACAACCTACTACCTCATTTTTA, antisense 5′ AGCTTAAAAATGAGGTAGTAGGTTGTATGGTTTCTCTTGAAAACCATACAACCTACTACCTCAGGA; let-7c mimic sense 5′ GATCTCCTGAGGTAGTAGGTTGTATGGTTGTACAACCTACTACCTCATTTTTA, antisense 5′ AGCTTAAAAATGAGGTAGTAGGTTGTACAACCATACAACCTACTACCTCAGGA.
Cell culture, transfections
HEK 293 and H1299 cells were cultured under standard conditions (37°C, 5% CO2) using Dulbecco’s modified eagle medium (DMEM, PAA) supplemented with 10% FBS (Sigma) and Penicillin/Streptomycin (PAA).
For transfection of HEK 293, cells were plated and transfected 5–6 h later with 5 or 15 µg DNA for 10 and 15 cm plates, respectively, using calcium phosphate. Cells were incubated for 40 h and subsequently harvested.
H1299 cells were transfected using 2 µg DNA and 5 µl Lipofectamine 2000 (Invitrogen) per 6 well according to the manufacturer’s manual. For longer incubation periods, cells were expanded when necessary.
cDNA synthesis, quantitative real-time PCR (qPCR)
cDNA synthesis was carried out using 1 µg of total RNA with the First Strand cDNA Synthesis Kit (Fermentas), according to the manufacturer’s instructions.
For measurement of siRNA knockdown efficiencies by quantitative real-time PCR, the transcribed cDNA was diluted 1 : 10 and 5 µl of this dilution was mixed with 2 pmol of forward and reverse primer and 10 µl of 2× MESA Green qPCR MasterMix Plus (Eurogentec) in a total volume of 20 µl.
Primers used: GRK4 fwd 5′ AGGAGGAGAACCCTTCCAAA, rev 5′ TTTCCAGCCATTTCCATTGT; GAPDH fwd 5′ AATGGAAATCCCATCACCATCT, rev 5′ CGCCCCACTTGATTTTGG.
Northern blotting
Northern blotting was performed as described before (37). In short, either 10–20 µg of total RNA or all of immunoprecipitated RNA was separated on 12 or 18% urea gels (UreaGel System, National diagnostics). As a size marker, ribooligonucleotides with a length of 19, 21 and 24 nt were labeled with 32P prior to loading. After running the gel for 1 h (12%) or 3–4 h (18%) at 350–450 V, the RNA was semi-dry blotted onto an Amersham Hybond-N membrane (GE Healthcare) at 20 V for 30 min. The membrane was subsequently cross-linked using EDC-solution (0.16M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in 0.13M 1-methylimidazole pH 8.0) and incubated for 1 h at 50°C. The membrane was rinsed in water and subjected to prehybridization with hybridization solution. For labeling of a probe antisense to the respective miRNA or siRNA, 20 pmol of a DNA oligo were incubated with 20 µCi of 32P in a T4 PNK reaction (Fermentas). The labeled oligo was purified with a G-25 column (GE Healthcare), added to the membrane and together was incubated overnight at 50°C. The membrane was washed twice with 5× SSC, 1% SDS, once with 1× SSC, 1% SDS and wrapped in saran. Signals were detected by exposure to a screen and scanning with the PMI (Biorad).
RESULTS
Ectopically expressed miR-451 interacts with Ago2 only
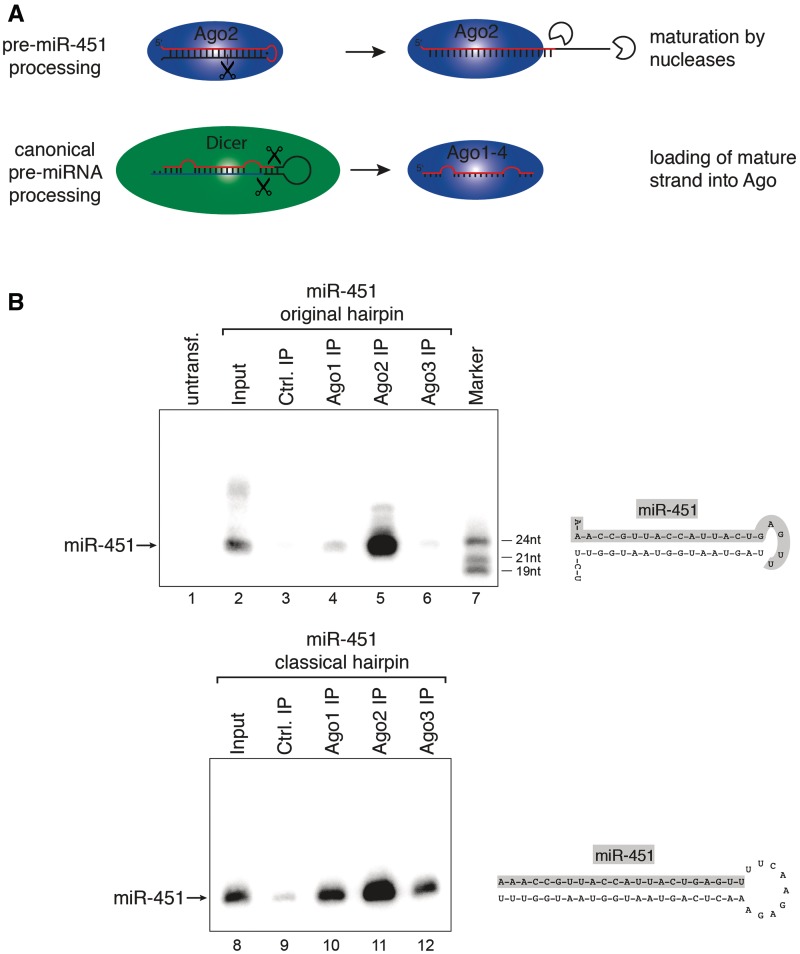
Ago2 processes miR-451 independently of Dicer (21,22). Since other human Ago proteins are catalytically inactive in target RNA cleavage, only Ago2 might be involved in pre-miR-451 processing and therefore loaded with miR-451 (Figure 1A). Short hairpins based on pre-miR-451 would be an attractive tool to generate siRNAs that are exclusively loaded into Ago2. To validate the applicability of the miR-451 system for short hairpin-based gene knockdown, we designed a vector that expresses the miR-451 hairpin. The plasmid was transfected into HEK 293 cells where miR-451 is not expressed endogenously and miR-451 expression was monitored by northern blotting (Figure 1B, upper panel). MiR-451 was efficiently transcribed and processed in this experiment (Lane 2). We next analyzed binding of the plasmid-derived miR-451 to Ago proteins. We have recently shown that only Ago1, Ago2 and Ago3 are significantly expressed on the protein level in HEK 293 cells (30). Therefore, endogenous Ago1–3 was immunoprecipitated using specific antibodies from cells transfected with the miR-451 construct. The co-immunoprecipitated miR-451 was analyzed by northern blotting (Lanes 4–6). Indeed, only the anti-Ago2 antibody co-immunoprecipitated mature miR-451 whereas only background signals were observed in the anti-Ago1 and anti-Ago3 IPs.
Figure 1.
miR-451 associates with Ago2 only. (A) Schematic representation of Ago2-mediated miR-451 processing (upper part) and canonical pre-miRNA processing by Dicer (lower part). (B) Constructs expressing wildtype (wt) pre-miR-451 (upper panel) or miR-451 embedded into a classical Dicer-dependent hairpin (lower panel) were transfected into HEK 293 cells. Ago1 (Lanes 4 and 10), Ago2 (Lanes 5 and 11) and Ago3 (Lanes 6 and 12) were immunoprecipitated using specific monoclonal antibodies. Co-immunoprecipitated miRNAs were analyzed by northern blotting against miR-451. Lanes 3 and 9 show control IPs, Lanes 2 and 8 show input samples and Lane 7 a size marker. (C) RNA was extracted from the indicated mouse tissues and miR-451 was detected by northern blotting. (D) Endogenous mAgo1 (Lanes 4 and 8) or mAgo2 (Lanes 5 and 9) were immunoprecipitated from spleen or liver and co-immunoprecipitated miR-451 (upper panel) or miR-19b (lower panel) was analyzed by northern blotting. Lanes 3 and 7 show control IPs, Lanes 2 and 6 input samples and Lanes 1 and 10 size markers.
Next, we incorporated the miR-451 sequence into a classical hairpin construct (Figure 1B, lower panel). The construct was transfected and miR-451 expression was analyzed by northern blotting. miR-451 expression from the miR-451 and the classical short hairpin construct were comparable (compare Lanes 2 and 8). However, mature miR-451 was loaded into Ago1, Ago2 and Ago3, when the classical hairpin construct was used (lower panel, lanes 10–12).
Mature miRNAs are not exchanged between Ago proteins
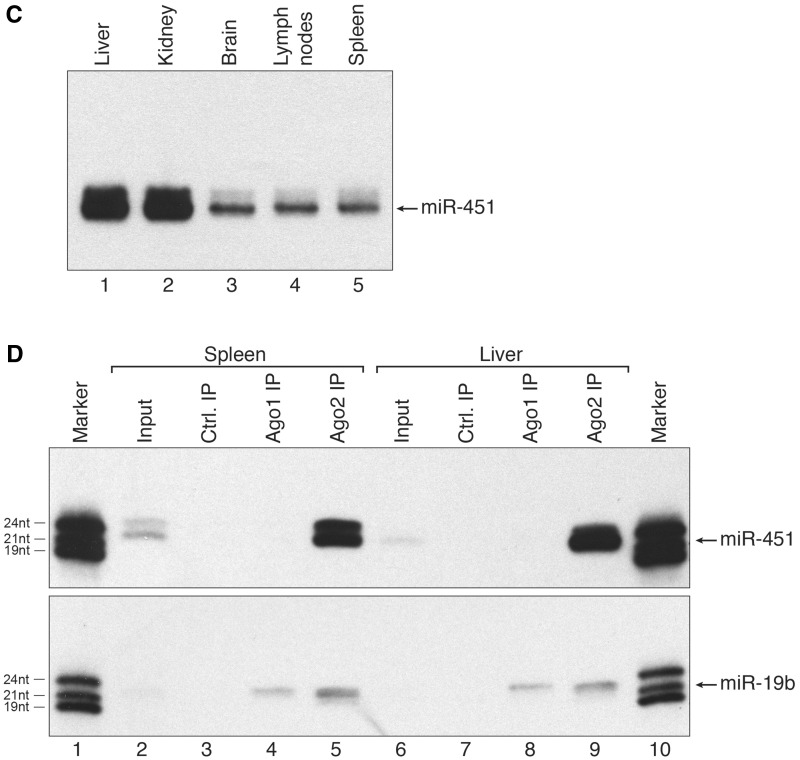
To test whether miR-451 associates exclusively with Ago2 in vivo as well, different mouse tissues were analyzed for miR-451 expression by northern blotting (Figure 1C). While miR-451 signals were observed in all tissues analyzed, the highest expression levels are found in kidney and liver. Extracts from liver and spleen were generated and endogenous Ago1 and Ago2 were immunoprecipitated using specific antibodies. Co-immunoprecipitated miR-451 was visualized by northern blotting (Figure 1D, upper panel). A strong miR-451 signal is observed in the anti-Ago2 IP, while miR-451 is not co-immunoprecipitated using anti-Ago1 antibodies indicating that mature miR-451 exclusively associates with Ago2 in vivo. As control, all samples were analyzed for the presence of miR-19b, which is a classical, Dicer-dependent miRNA. miR-19b was observed not only in the Ago2 but also in the Ago1 IPs (Figure 1D, lower panel).
In summary, our data provide evidence that exchange of mature miRNAs between different Ago proteins does not occur, a question that has not been addressed in the small RNA field so far.
The miR-451 hairpin system can be used for siRNA or ectopic miRNA expression
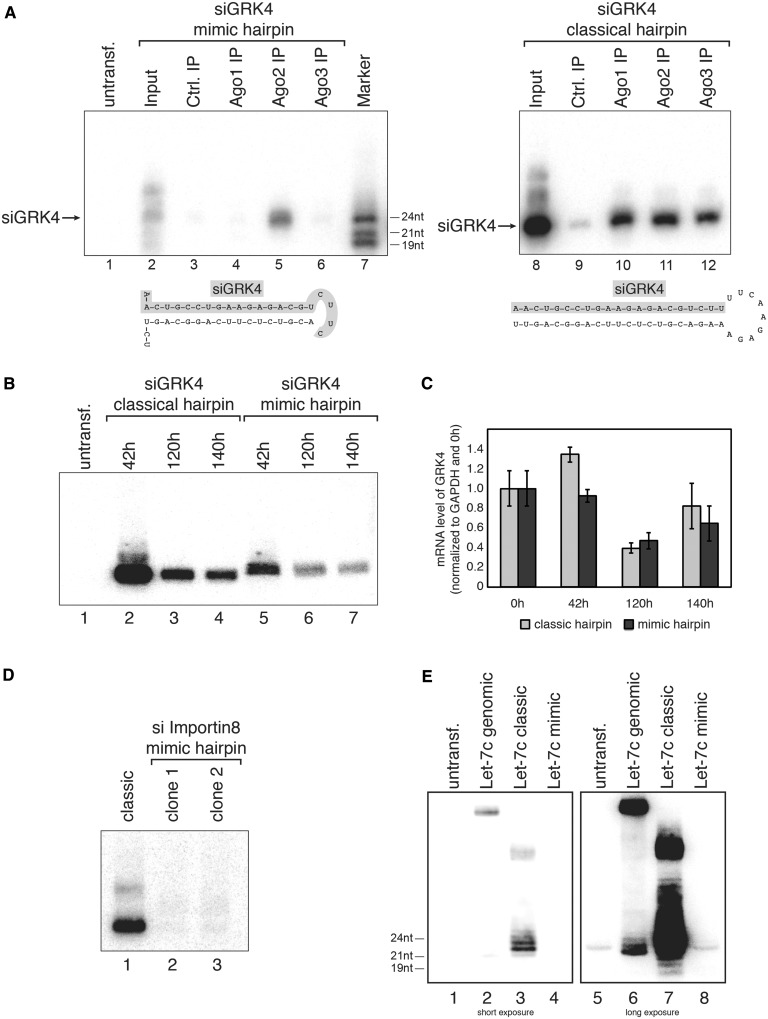
Since miR-451 is not only processed by Ago2 but also exclusively loaded into Ago2, we hypothesized that short-hairpin RNAs expressed from a miR-451-like hairpin would avoid catalytically inactive Ago proteins, which are not useful for RNAi experiments. We generated short-hairpin RNAs against GRK4 [G protein-coupled kinase 4 (38)] either as miR-451 mimic or as classical Dicer-processed shRNA (Figure 2A). Ago1–3 were immunoprecipitated and associated siRNAs were analyzed by northern blotting. Consistently with miR-451 loading, the siRNA derived from the miR-451 mimic construct accumulated only in Ago2 complexes (left panel), whereas the classical shRNA was loaded into all three Ago proteins (right panel). Notably, the mimic construct generated less siRNAs than the classical shRNA.
Figure 2.
Pre-miR-451-based shRNA expression. (A) An siRNA against GRK4 was either embedded into a pre-miR-451-like hairpin (left part) or a classical Dicer-dependent hairpin (right part). Both constructs were transfected into HEK 293 cells and Ago1 (Lanes 4 and 10), Ago2 (Lanes 5 and 11) or Ago3 (Lanes 6 and 12) was immunoprecipitated from the lysates. GRK4 siRNA incorporation was analyzed by northern blotting. Lanes 3 and 9 show control IPs, Lanes 2 and 8 input samples and Lane 7 a size marker. (B) Both GRK4 shRNAs described in (A) were transfected into HEK 293 cells. Total RNA was extracted at the indicated time points and GRK4 siRNA production was analyzed by northern blotting using a probe complementary to the guide strand of the GRK4 siRNA. (C) Samples described in (B) were analyzed by qPCR for knockdown of GRK4 at the indicated time points. (D) siRNAs against Importin 8 were embedded into a pre-miR-451-like hairpin (two different clones were tested, Lanes 2 and 3) or a classical Dicer-like shRNA (Lane 1). The constructs were transfected into HEK 293 cells and siRNA production was analyzed by northern blotting. (E) Relative expression levels of let-7c were tested by transfecting the genomically encoded (‘genomic’) (Lanes 2 and 6), the classical Dicer-like shRNA (‘classic’) (Lanes 3 and 7) or a pre-451-like hairpin (‘mimic’) (Lanes 4 and 8). As a background control, RNA from untransfected HEK 293 was used (Lanes 1 and 5).
We next analyzed knockdown efficiency in vivo. Both constructs were transfected into H1299 cells and siRNA production was assessed 42, 120 and 140 h after transfection by northern blotting. Again, the classic shRNA generated more siRNA compared with the miR-451 mimic construct (Figure 2B). For knockdown analysis, RNA was extracted and GRK4 levels were examined by qPCR (Figure 2C). Indeed, both constructs showed a comparable knockdown of the GRK4 gene suggesting that miR-451 mimic shRNAs could generally be used for knockdown experiments.
We realized that the GRK4 mimic construct was less efficiently processed than the classical hairpin. Therefore, we analyzed another mimic hairpin containing a siRNA directed against Importin 8 (Figure 2D). Two clones were transfected into HEK 293 cells and the processed siRNAs were analyzed by northern blotting. Indeed, processing of the mimic hairpin was much less efficient compared with the classical hairpin.
Finally, we analyzed whether the miR-451 hairpin system could be used to express other miRNAs, which has been demonstrated in mammalian cells (39) (Figure 2E). A genomic let-7c sequence, a let-7c containing a fully paired stem (let-7c classic) as well as a let-7c construct mimicking miR-451 were transfected into HEK 293 cells and processing was monitored by northern blotting. While processing from the classical, fully paired hairpin system as well as the genomic hairpin was rather efficient, only little let-7c expression was observed using the miR-451 mimic hairpin.
Taken together, our data indicate that both siRNAs and miRNAs can be placed into a miR-451 mimic hairpin for knockdown or miRNA expression. However, expression of such constructs is much weaker compared with classical, Dicer-dependent hairpins. In fact, it appears that miR-451 is evolutionarily optimized for efficient Ago2 cleavage. Changing the sequence seems to impair processing efficiency.
Structural requirements for Ago2-mediated miR-451 processing
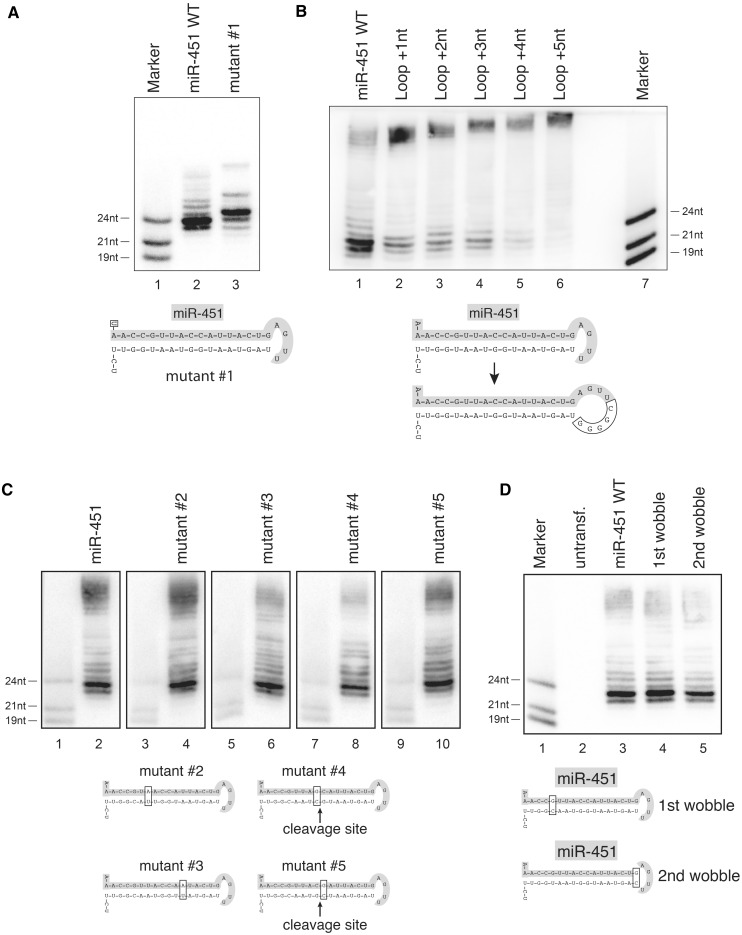
The surprising finding that processing of let-7c from a miR-451 mimic hairpin is much less efficient compared with miR-451 processing prompted us to investigate sequence requirements for pre-miR-451 cleavage (Figure 3). We first analyzed the 5′ end of miR-451 (Figure 3A). Constructs expressing wild-type miR-451 or an A to U mutation of the 5′-terminal nucleotide were transfected into HEK 293 cells and miRNA expression was analyzed by northern blotting. Structural studies have demonstrated that preferably uridines but sometimes also adenines are efficiently bound by Ago proteins (40). We find that both ends are accepted and the constructs are equally well processed. Of note, the construct with the U at the 5′ end generates a predominant product that is 1 nt longer than the wildtype (wt) miR-451 suggesting different anchoring on Ago2.
Figure 3.
Structure and sequence requirements of miR-451 for Ago2-mediated cleavage. (A) A wt miR-451 construct (Lane 2) or a mutant with a changed 5′ end (Lane 3, A to U mutation, mutant #1) were transfected into HEK 293 cells and processing was analyzed by northern blotting. Lane 1 shows a size marker. (B) Nucleotides were inserted into the loop of pre-miR-451 (Lanes 2 to 6) and the constructs were analyzed as described in (A). (C) Mutants of the wt pre-miR-451 hairpin were constructed to test the sequence specificity of Ago2-mediated processing. Constructs were transfected into HEK 293 cells and analyzed on individual northern blots, since each mutation alters the resulting mature sequence. Equal amounts of size marker were loaded to provide comparability. (D) G/U wobble base pairs at the 5′ end (Lane 4) and near the loop (Lane 5) were mutated to G/C Watson–Crick base pairs and the constructs analyzed as in (A).
Pre-miR-451 possesses a very short loop structure. We systematically extended the loop and analyzed pre-miR-451 processing by northern blotting (Figure 3B). Addition of 1–3 nt to the loop strongly reduces processing activity. When 4 or 5 extra nucleotides are added to the loop, processing is almost completely abolished. Our data indicate that the very short single-stranded loop of wt pre-miR-451 is essential for Ago2-mediated processing.
Finally, we analyzed sequence requirements within the double-stranded stem of the pre-miR-451 hairpin (Figure 3C). For this, we changed a number of nucleotides within the stem of the miR-451 hairpin but kept the double-stranded nature of the stem intact. We found that all these mutants are tolerated suggesting that the sequence within the miR-451 stem can be varied.
The wt miR-451 precursor contains two G/U wobbles: one at position 6 counting from the 5′ end of miR-451 and one at the distal end of the stem. Both G/U wobbles were mutated to G/C pairs (Figure 3D). While the G/U wobble at position 6 appears to be dispensable for Ago2 cleavage, changing the G/U wobble at the end of the stem significantly reduced cleavage efficiency. Our data suggest that the requirement of a G/U wobble at the distal end of the stem restricts the number of potential siRNA target sequences within one gene and might therefore be limited for a broad usage for gene knockdown.
Revisiting miRNA sorting into different Ago proteins in human cells
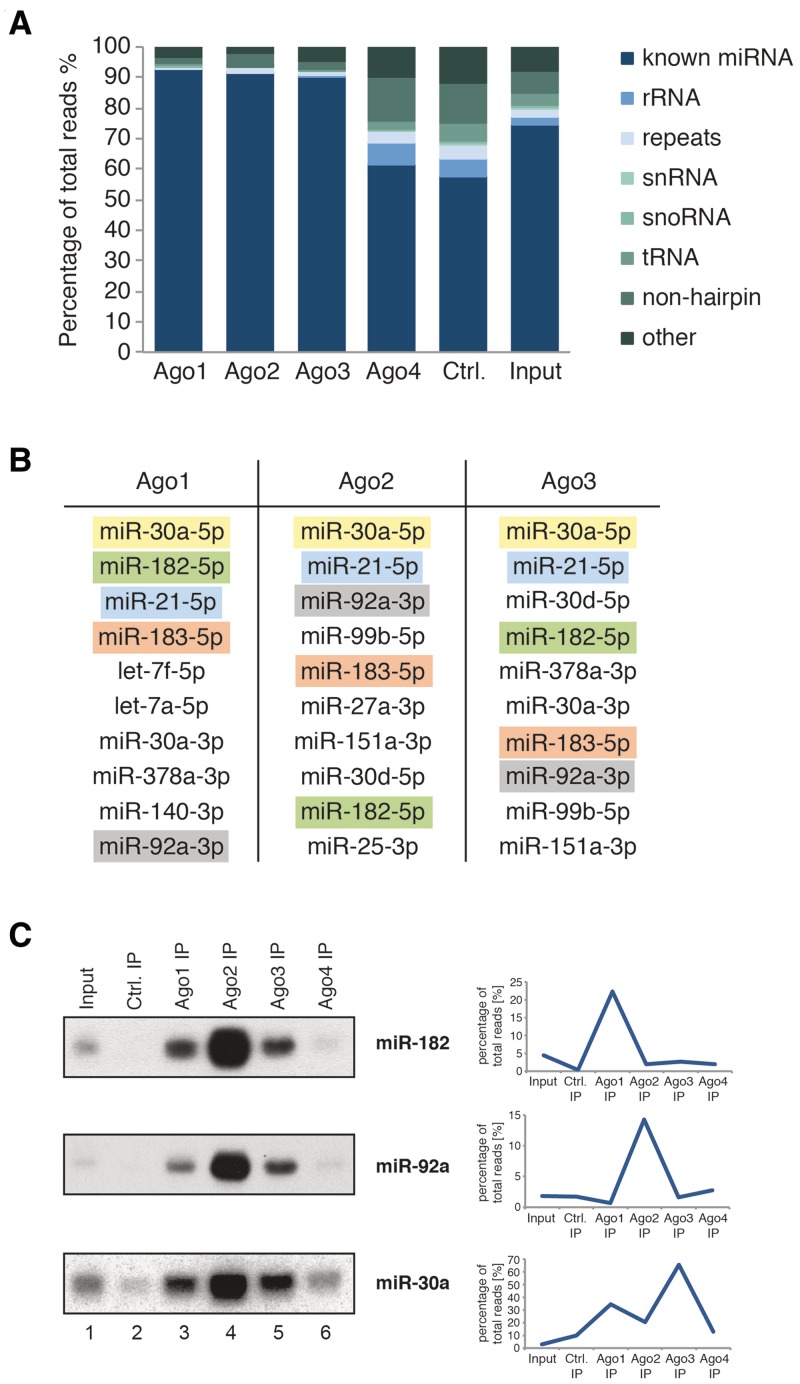
Since mature miRNAs are not transferred between the different human Ago proteins (Figure 1), we asked whether the Dicer-processing machinery selectively loads miRNAs into Ago proteins. Using overexpression of tagged Ago proteins followed by deep sequencing of associated miRNAs, it has been reported that miRNAs indeed preferentially associate with members of the Ago protein family (41). Using highly specific monoclonal anti-Ago antibodies (30,34,35), we immunoprecipitated endogenous Ago1–4 from HeLaS3 cell lysates, extracted the co-purified small RNAs, cloned and sequenced them (Figure 4A). We have shown previously that Ago4 protein is not found in HeLa lysates (30). Consistently, the small RNA profile obtained by anti-Ago4 IP is comparable with the control (an unrelated antibody against the complement system protein RmC from rat). Therefore, Ago4 was not included in our further analysis. Most of the small RNAs found in Ago1–3 were indeed known miRNAs (dark blue). We closely investigated the 10 most abundant miRNAs in Ago1, Ago2 and Ago3 and found that several highly abundant miRNA species are found associated with all three Ago proteins (Figure 4B, highlighted in color). However, many miRNAs appeared to be selectively associated with one Ago protein suggesting a sequence-specific distribution between the different Ago proteins. We next attempted to validate the sequencing data by northern blotting (Figure 4C). We analyzed miR-182, which was highly abundant in the Ago1 library (upper panel), miR-92a, which was found in high levels in the Ago2 library (middle panel) and miR-30a that dominated the Ago3 library (lower panel). All miRNAs that we analyzed in northern blots showed a distribution according to the Ago protein expression levels, i.e. Ago2 contributes 65%, Ago1 20% and Ago3 15% to the total Ago pool in HeLaS3 cells (30). Our northern blot validation therefore indicates that human miRNAs are not sorted into different Ago proteins.
Figure 4.
Deep-sequencing analysis of small RNAs associated with endogenous Ago proteins. (A) Composition of the small RNA libraries from the input sample and immunoprecipitated Ago1, Ago2, Ago3, Ago4 and control (an unrelated antibody against the complement system protein RmC from rat). (B) Table of the 10 most abundant miRNAs associated with Ago1, Ago2 or Ago3. The colors indicate the same miRNA. (C) Comparison of the sorting profile from the deep-sequencing data (right side) for miR-182 (top panel), miR-92a (middle panel, right) and miR-30a (lower panel) with profiles resulting from northern blotting (left side).
miRNA length variations in different Ago protein species
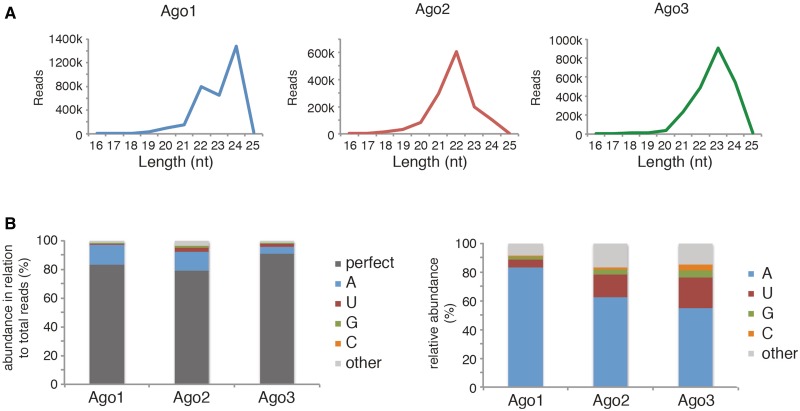
We further analyzed our small RNA libraries for Ago-specific small RNA features. Most miRNAs vary at their 3′ ends by 1–3 nt. The 3′ extended miRNAs are commonly referred to as isomirs (42). We analyzed the global miRNA length distribution among the different Ago proteins (Figure 5A). We find that all three Ago proteins bind miRNAs ranging from 20 to 24 nt. However, while Ago2-associated miRNAs peak at 22 nt (middle panel, red), Ago1 (left panel, blue) and Ago3-associated miRNAs (right panel, green) also peak at a length of 23 or 24 nt. We further analyzed the nature of the miRNA extentions at the 3′ end (Figure 5B). In all three Ago proteins, a large portion of the miRNAs perfectly matches the genome, i.e. they are not extended at the 3′ end (dark gray). However, a significant number of miRNAs contain nontemplated additions to their 3′ ends. Closer examination of the 3′ extensions revealed that an A is preferentially added as the first extra nucleotide to the 3′ end (Figure 5B, right panel, blue). For Ago1, for example, A addition is almost exclusively observed. While U addition is also frequently observed, G or C addition is rather rare. miRNAs with more than one extra nucleotide at the 3′ end constitute only a minor part of the isomirs (other, light gray). In summary, our data suggest that Ago proteins differentially tolerate miRNA length variations. Length variations in Ago proteins are dominated by a single A addition.
Figure 5.
Ago proteins show different global small RNA binding patterns and nontemplated 3′ extension profiles. (A) Length distribution patterns of Ago1 (left), Ago2 (middle) and Ago3 (right). (B) Left side: Ago1, Ago2 and Ago3 associated miRNAs were analyzed for nontemplated 3′ extensions, values were calculated in relation to all miRNA reads of the respective library. Perfect: no nontemplated 3′ extensions; A, U, G, C: amount of single nucleotide 3′ extensions; other: sum of all other 3′ extensions (≥2 nt). Right side: each single nontemplated 3′ extension was set in relation to the total number of nontemplated 3′ extensions in the respective library (Ago1, Ago2 or Ago3).
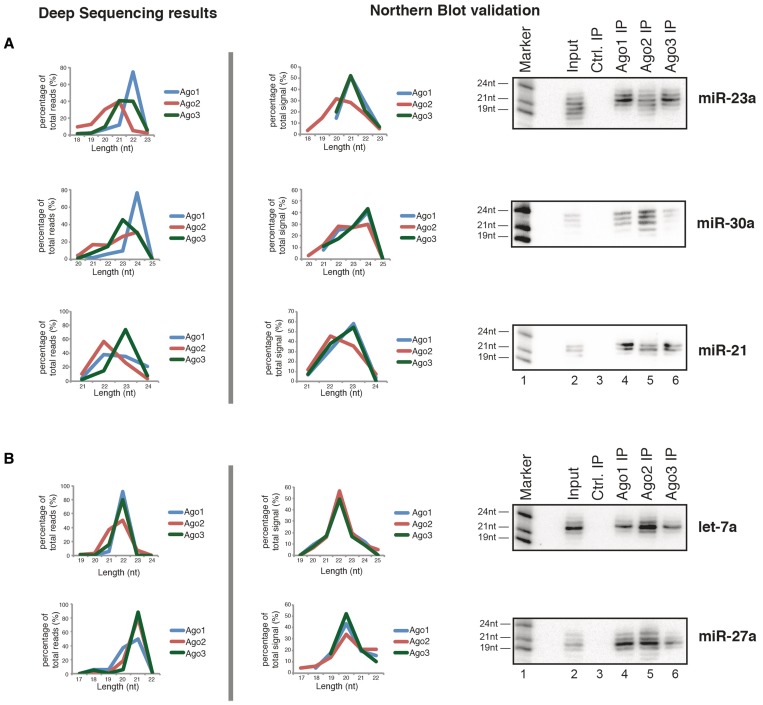
We next validated our data experimentally by analyzing the length distribution of individual miRNAs. Ago proteins were immunoprecipitated and miRNAs analyzed by northern blotting (Figure 6). The deep sequencing results for miR-23a showed a broader length variation in Ago2 compared with Ago1 or Ago3 (Figure 6A, upper panel, left graph). Northern blotting confirmed that Ago2 interacts with a broader range of miR-23a isomirs, while Ago1- and Ago3-associated miR-23a is characterized by a distinct length peak (upper panel, right part). Similar results were obtained for miR-30a and miR-21 (middle and lower panels).
Figure 6.
Ago proteins show distinct length profiles for individual miRNAs. The length pattern of individual miRNAs was extracted from the deep-sequencing data (left side) and compared against the length profiles seen by northern blotting (right side). For this, northern blotting signals for each miRNA band were quantified; the total signal from each lane/each Ago was set to 100%. The length of a miRNA in northern blotting was assessed by comparison with the size marker. miRNA length profiles were separated into: (A) miRNA length profiles showing divergent Ago association and (B) miRNA length profiles showing matching Ago1–3 association.
We next investigated miRNA examples that show no difference in their length distributions in our sequencing data sets (Figure 6B). Let-7a shows a distinct length for Ago1, Ago2 and Ago3 in the sequencing data (upper panel, left part). Indeed, using northern blotting we find that let-7a shows a very distinct length in all three Ago proteins (upper panel, right part). Similar results were obtained for miR-27a (lower panel).
Taken together, our results suggest that human miRNAs are characterized by individual isomir patterns with differential binding to Ago proteins.
DISCUSSION
An unsolved problem in RNAi experiments is that transfected synthetic or shRNA-derived siRNAs are loaded into all four Ago proteins (30,43). Only Ago2 is catalytically active and needed for efficient gene knockdown. Ago1, Ago3 and Ago4, however, may cause unwanted off-target effects by binding to partially complementary target sites in unrelated 3′ UTRs leading to miRNA-like gene silencing effects (30). It has been demonstrated that miR-451 processing is Dicer independent and pre-miR-451 is cleaved by Ago2 (21,22). Here we show that miR-451 is not only processed by Ago2 but also loaded exclusively into Ago2-containing RISCs. This experimental observation immediately points to an attractive method for specific knockdown experiments. ShRNAs mimicking the pre-miR-451 hairpin in size and structure but containing any given siRNA sequence might only interact with Ago2. We have tested this hypothesis and found that indeed siRNAs can be generated from pre-miR451-like shRNAs. However, processing as well as silencing activity was not as efficient as shRNAs that are processed by Dicer. Although pre-miR-451-like shRNAs are indeed loaded into Ago2-containing RISCs, we conclude from our study that such constructs are less efficient than classical Dicer substrates.
In Drosophila, small RNAs are sorted into different Ago proteins (28,29). However, miRNA sorting is less clear in mammals so far. Furthermore, it is not known whether miRNAs are able to dissociate from Ago proteins after processing and RISC loading allowing for an exchange of mature miRNAs between Ago proteins. It has been demonstrated that a protein termed hnRNPE2 can interact with a specific miRNA and sequester it from Ago proteins (44). We find that mature miR-451 associates exclusively with Ago2 indicating that dissociation and re-association with Ago proteins does not occur in case of miR-451. It is tempting to speculate that mature miRNAs are tightly anchored into Ago proteins and miRNA exchange between human Ago proteins is rather unlikely.
Using endogenous Ago protein isolation followed by cloning and deep sequencing, we found that miRNAs are not sorted into different human Ago proteins. It has been reported before that several miRNAs show distinct Ago association patterns (41). These studies used Ago overexpression and relied on deep-sequencing data. Validation using direct northern blotting has not been performed. It has been demonstrated that small RNA cloning might introduce both ligation and PCR biases into the library composition (45). This might explain our different findings. However, it is also conceivable that miRNAs might associate with Ago protein sequence specifically in other cell types or cell stages.
Our deep-sequencing approach of Ago-associated miRNAs revealed that Ago identity impacts the length of the bound miRNA. A very recent study showed that miRNAs are generally shortened during mammalian brain development (46). The shortening is due to a change in Ago protein expression suggesting that Ago identity affects the length of miRNAs. These findings are consistent with our results. However, in our study some miRNAs are characterized by a clear and distinct length (e.g. let-7a, Figure 6) independently of the Ago protein, which they bind to. These findings indicate that Ago identity is not the sole determinant of the miRNA length. So far unidentified factors might associate with Ago proteins under specific conditions and trim or extend the 3′ end of the miRNA.
A closer investigation of endogenous Ago-associated miRNAs revealed that a significant portion of miRNAs contain nontemplated A additions at the 3′ ends. It has been demonstrated that uridityl transferases such as TUT4 add poly-U stretches to precursors of members of the let-7 family leading to miRNA degradation (47–49). However, this mechanism seems to be let-7 specific and has not been found for other miRNA species. In contrast, mono- or di-nucleotides are often added to the 3′ end of other miRNAs (42,50–52). It has been shown recently, that miRNAs carrying a single A addition are preferentially bound by overexpressed myc-tagged Ago1 (53). Although we find a stronger enrichment of A addition in Ago1-associated miRNAs, extensions of Ago2- and Ago3-associated miRNAs is also dominated by A (Figure 5B). Therefore, we conclude that A and U are the predominant nucleotides that are added to the 3′ end of the miRNA. However, Ago protein identity has only a minor effect on nucleotide addition to miRNAs.
ACKNOWLEDGEMENTS
We thank Sigrun Ammon and Corinna Friederich for technical assistance and Sebastian Petri and Daniel Schraivogel for helpful discussions.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) (SFB 960 and FOR855); European Research Council (ERC grant ‘sRNAs’); European Union (FP7 project ‘ONCOMIRs’); German Bundesministerium für Bildung und Forschung (BMBF, NGFN+, FKZ PIM-01GS0804-5); Bavarian Genome Research Network (BayGene to G.M.); The Netherlands Organization for Scientific Research (NWO, VIDI grant to E.B.). Funding for open access charge: DFG via the open access publishing program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 5.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Ender C, Meister G. Argonaute proteins at a glance. J. Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 11.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. Rna. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 24.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 25.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 26.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 27.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 28.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petri S, Dueck A, Lehmann G, Putz N, Rudel S, Kremmer E, Meister G. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. Rna. 2011;17:737–749. doi: 10.1261/rna.2348111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr. Opin. Mol. Ther. 2007;9:248–257. [PubMed] [Google Scholar]
- 33.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 34.Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. Rna. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 36.Zhu JY, Strehle M, Frohn A, Kremmer E, Hofig KP, Meister G, Adler H. Identification and analysis of expression of novel microRNAs of murine gammaherpesvirus 68. J. Virol. 2010;84:10266–10275. doi: 10.1128/JVI.01119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 41.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 44.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. Rna. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juvvuna PK, Khandelia P, Lee LM, Makeyev EV. Argonaute identity defines the length of mature mammalian microRNAs. Nucleic Acids Res. 2012;40:6808–6820. doi: 10.1093/nar/gks293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38:e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]