Abstract
Small RNAs have been implicated in numerous cellular processes, including effects on chromatin structure and the repression of transposons. We describe the generation of a small RNA response at DNA ends in Drosophila that is analogous to the recently reported double-strand break (DSB)-induced RNAs or Dicer- and Drosha-dependent small RNAs in Arabidopsis and vertebrates. Active transcription in the vicinity of the break amplifies this small RNA response, demonstrating that the normal messenger RNA contributes to the endogenous small interfering RNAs precursor. The double-stranded RNA precursor forms with an antisense transcript that initiates at the DNA break. Breaks are thus sites of transcription initiation, a novel aspect of the cellular DSB response. This response is specific to a double-strand break since nicked DNA structures do not trigger small RNA production. The small RNAs are generated independently of the exact end structure (blunt, 3′- or 5′-overhang), can repress homologous sequences in trans and may therefore—in addition to putative roles in repair—exert a quality control function by clearing potentially truncated messages from genes in the vicinity of the break.
INTRODUCTION
The integrity of our genome is constantly challenged by both extrinsic and intrinsic factors, which can change the nucleotide sequence or induce DNA breaks. Transposable elements and retroviruses can cause insertional mutagenesis. Furthermore, stalling and breakdown of the DNA replication fork can also result in broken DNA (1). Intricate surveillance systems detect and distinguish the various types of DNA damage. They induce either the appropriate repair processes or drive the cell into apoptosis (2). Following DNA damage, extensive modifications and changes of the local chromatin structure take place, most of which are triggered by action of the key signaling kinases ATM and ATR (3).
In the case of transposable elements, a different strategy is used. Rather than waiting for the damaging integration event, cells must prevent the accumulation of RNA transcripts, which upon translation will produce the enzymes and substrate for integration. Here, the small RNA silencing system provides an important line of cellular defense. Complexes composed of an Argonaute-family protein endowed with nuclease activity and a small RNA that programs this nuclease to target perfectly complementary sequences restrict the accumulation of transposon messenger RNA (mRNA) and induce repressive chromatin structures at genomic loci of homologous sequences (4–6). Somatic Drosophila cells repress transposon activity through corresponding endogenous small interfering RNAs (siRNAs) (7). This pathway is genetically separable from the micro RNA (miRNA) system that regulates gene expression by translational control and mRNA degradation. Their biogenesis requires a double-stranded (ds) RNA precursor, which is processed by Dicer-2 and then loaded into the cleavage-competent Ago2 effector (7–11). Recent publications describe that in Arabidopsis and vertebrate cells, a double-strand break elicits a small RNA response that is required for recognition of the γ-H2Ax foci by downstream effectors (12) and efficient repair by homologous recombination (13). In Arabidopsis, the damage recruits RNA pol IV enzymes and their transcripts are subsequently converted into dsRNA by RNA-dependent RNA polymerase (RdRP). In the case of vertebrates, which lack both RNA pol IV and RdRP, it is unknown how the precursor for damage-induced siRNAs is generated. We have observed an analogous small RNA response to DNA ends in cultured Drosophila cells. This response requires a break in both DNA strands, depends on endo-siRNA factors, is stimulated by active transcription in the vicinity of the break and has the capacity to silence transcripts with homologous sequence in trans.
MATERIALS AND METHODS
Cell culture, RNAi, transfection and reporter assays
Drosophila S2-cells were cultured in Schneider’s medium and transfected with Fugene-HD as previously described (14). For plasmid linearization, the vectors were cut with the indicated restriction endonucleases at 37°C overnight, then the linearized DNA was gel purified. For reporter assays, 100 ng of linearized green fluorescent protein (GFP) expression vector or 50 ng each of linear and circular luciferase expression vectors were transfected per well of a 96-well plate. For deep sequencing analysis, we transfected 300 ng each of circular plasmid, linear plasmid and polymerase chain reaction (PCR) product in 1 well of a 6-well dish.
Depletion of RNAi factors was performed by soaking of previously validated (15), in vitro-transcribed dsRNA triggers at a concentration of 20 µg/ml for 3 days. The cells were subsequently diluted to a density of 0.5 × 106 cells/ml, transfected as described above and reporter gene expression was determined 3 days after transfection. GFP fluorescence intensity was measured on a Becton Dickinson FACSCalibur flow cytometer, non-transfected cells were excluded and the mean fluorescence value of the transfected cells was calculated. Luciferase activity was measured with the Dual Luciferase assay system (Promega) in a plate-reading luminometer (Berthold).
RNA isolation and deep sequencing
RNA was isolated and libraries were prepared as previously published (16) with indexes for multiplexing appended during the final PCR step. Up to four libraries were pooled and sequenced on an Illumina GAIIx Genome Analyzer at the Gene Center core facility. The reads were mapped using Bowtie (17) with no mismatch allowed. Pre-computed index-files for the Drosophila genome and all mature miRNAs were used to determine the number of reads matching these target sequence collections. For the transfected construct, a custom index was created based on the sequences of the GFP expression vector pKF63, the yeast plasmid pRS425 and the Firefly luciferase open reading frame we amplified by PCR. Since reads mapping to the plasmid origin or AmpR gene cannot be unambiguously assigned, we excluded all reads that did not map uniquely among all three constructs. The sequence data have been deposited in the NCBI GEO database with the accession number GSE38967.
Reporter gene vectors
The GFP expression vector with ubiquitin-promotor and the SV40 3′-untranslated region (UTR) was described previously (15). To generate an analogous myc-tagged Renilla luciferase expression vector, we excised the myc-GFP coding sequence with BamHI/NotI and inserted an accordingly digested PCR product obtained with oligonucleotides 5′-CAGGATCCTAATCCAAAATGGAACAGAAACTGATTAGCGAAGAAGATCTGGCTTCCAAGGTGTACGACC-3′ (fwd) and 5′-ATGCGGCCGCTTACTGCTCGTTCTTCAGCAC-3′ (rev). For the untagged N-terminal truncation, we used oligonucleotide 5′-CAGGATCCATCAACTACTATGATTCCGAG-3′ as the forward primer instead. For the firefly luciferase expression vector, we used a previously described backbone containing the tubulin promotor and 3′-UTR (16), then used KpnI/NotI to insert a PCR product coding for Flag-tagged firefly luciferase obtained with oligonucleotides 5′-CAGGTACCTAATCCAAAATGGATTATAAAGATGATGATGATAAAGCCGATGCTAAGAACATTAAG-3′ (fwd) and 5′-ATGCGGCCGCTTACACGGCGATCTTGCCGC-3′ (rev).
RESULTS
RNAi-mediated repression close to a double-strand break, but not nicked DNA
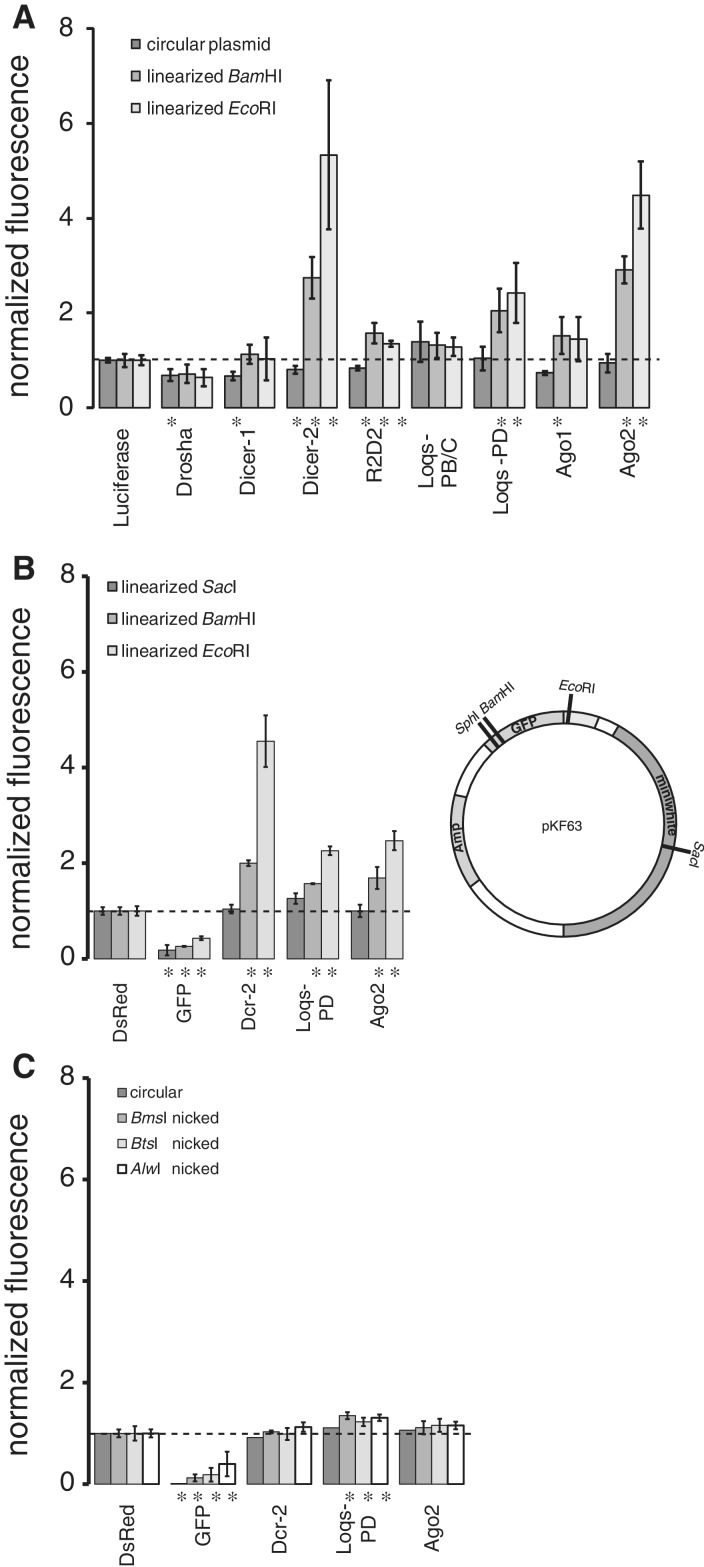
We had previously noticed a limited production of small RNAs from a transiently transfected, circular plasmid containing a GFP expression cassette in Drosophila S2-cells (14). An independent study (9) observed a much more pronounced siRNA generation in response to transiently transfected plasmid DNA. In our efforts to understand the differences, we were motivated by the discovery of a DNA-damage-induced small RNA response in Neurospora to examine whether a damage of the plasmid DNA may boost the endo-siRNA response. Analogous to elegant experiments performed with budding yeast (e.g. (18)), we transfected cells with a linearized plasmid coding for GFP. Flow cytometry measurements indicated strongly reduced, but clearly detectable expression (Supplementary Figures S1 and S2A). Unlike, e.g. treatment with ionizing radiation, this experimental system is precise yet very flexible as it allows varying the position and nature of the lesion in a controlled manner. When combined with prior depletion of small RNA biogenesis factors through RNAi, we observed an increase of GFP expression specifically for the linearized vector in the absence of Dcr-2, Loqs-D and Ago2 (Figure 1A) as well as a smaller amount of de-repression in the absence of Ago1 and Dcr-1. This profile is characteristic for the endo-siRNA pathway (8,9,11,14,19) and we therefore refer to the damage-induced small RNAs as endo-siRNAs in the remainder of this article. We observe measurable effects of Drosha and Dicer-1 depletion on reporter gene expression levels from the circular as well as linearized reporters (see also Figure 2B), but these treatments also induce differences in the cell-cycle distribution (data not shown). Since the cell-cycle distribution likely affects both DNA repair and expression kinetics from transfected constructs, we refrain from interpreting these effects for the time being.
Figure 1.
Linearized plasmids provoke an endo-siRNA response. (A) Drosophila S2-cells were treated with dsRNA targeting the indicated factors, transfected with either circular or linearized plasmids and the resulting GFP expression was measured by flow cytometry. Measurement values were normalized to control treatment (three independent experiments, mean ± SD; * below bar indicates P < 0.05, t-test unequal variance). (B) Linearization in the downstream ‘mini-white’ marker gene by a SacI-cut (1940 nt downstream of the polyadenlyation signal) does not induce an endo-siRNA response targeting GFP (three independent experiments, mean ± SD; * below bar indicates P < 0.05, t-test unequal variance). (C) Drosophila S2-cells were treated with dsRNA as in Figure 1A, then transfected with either circular or nicked GFP expression plasmids (three independent experiments, mean ± SD, circular plasmid: one experiment for each knock-down condition). The GFP fluorescence intensities were normalized to the control (three independent experiments, mean ± SD; * below bar indicates P < 0.05, t-test unequal variance).
Figure 2.
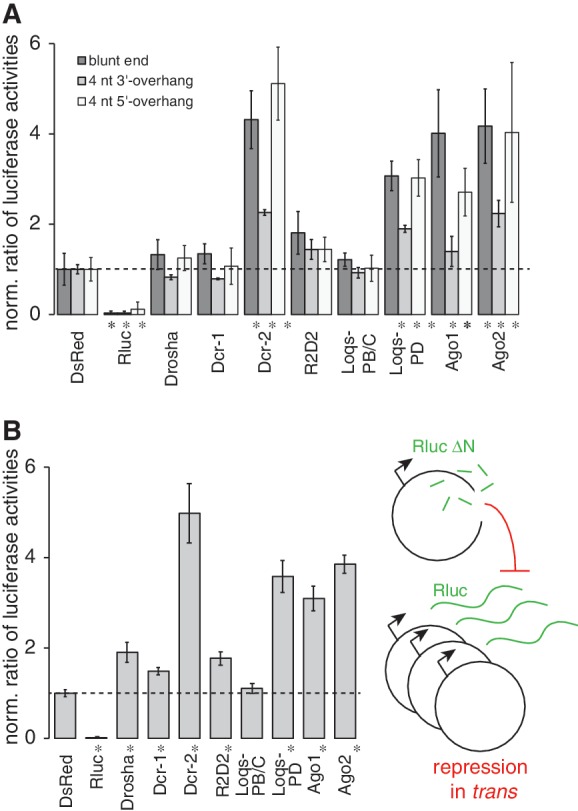
The small RNA response is independent of the end structure and can affect gene expression at other loci with homologous sequence. (A) The plasmid encoding Renilla luciferase was either linearized with EcoRI, or converted to a blunt end enzymatically, or cleaved with SphI (327 nt upstream of the BamHI site) to generate a 4-nt 3′-overhang. In all three cases, de-repression occurred upon prior depletion of endo-siRNA factors (three independent experiments, mean ± SD; * below bar indicates P < 0.05, t-test unequal variance). (B) A plasmid with a deletion of ∼300 nt at the N-terminus of Renilla luciferase, which renders the enzyme inactive, was used to generate break-induced endo-siRNAs by linearization with EcoRI. Their ability to repress reporter gene expression in trans was assessed with the help of a co-transfected, circular Renilla luciferase expression vector (three independent experiments, mean ± SD; * below bar indicates P < 0.05, t-test unequal variance).
In contrast to the effect of plasmid linearization within the GFP transcription unit, a cut in the ‘mini-white’ marker gene ∼2 kb downstream of the GFP insert revealed no dependence on RNAi factors (Figure 1B, SacI-cut). The break-induced repressive response thus has a limited range along the DNA. To determine whether a single-strand break could also trigger a repressive response, we treated our GFP-expression plasmid with a series of nicking endonucleases that cleave only one strand of the DNA. We could not detect a consistent de-repression of the signal upon depletion of endo-siRNA factors, only depletion of Loqs-PD led to a marginal de-repression (Figure 1C). The endo-siRNA response is thus specific to a double-strand break.
Blunt, 5′- and 3′-recessed DNA ends trigger an endo-siRNA response
To rule out that the effects we observed only affect the GFP reporter or the ubiquitin promotor, we replaced the GFP coding sequence with the one of Renilla luciferase. Upon co-transfection of a distinct plasmid coding for firefly luciferase, which carries the tubulin promotor and 3′-UTR and thus shares no common sequence with the relevant region of the Renilla vector, both vectors elicited an endo-siRNA response of comparable strength when linearized (Supplementary Figure S3). We used the luciferase system to test whether specific DNA end structures are required to trigger the endo-siRNA response. We modified our assay by either filling in the 5′-overhang generated by the EcoRI cut or by cleaving the vector with SphI (322-nt upstream of the BamHI site, see Figure 1B) to generate a 4-nt 3′-overhang. In both cases, the endo-siRNA repression remained detectable (Figure 2A), indicating that generation of a dsRNA precursor is independent of the precise structure of the DNA end. The magnitude of the response appears lower in the case of the SphI cut, but this may be due to the position of the cut (it is comparable in strength and position with the BamHI-cut; Figure 1A). In conclusion, the small RNA response requires a double-strand break but can initiate at any of the end structures we tested. The simplest explanation for this finding is that recognition and potentially initial processing of the double-strand break can occur before the generation of small RNAs. This is consistent with the observation of Wei et al. (13) that the formation of γ-H2Ax foci is still possible when generation of a small RNA response is blocked.
Break-derived endo-siRNA can repress gene expression in trans
To test whether regulation by damage-induced endo-siRNAs can occur in trans, we deleted ∼300 nt of the Renilla luciferase coding sequence, leading to an N-terminally truncated protein. This vector generated only very low luciferase activity when transfected alone, but after linearization induced an endo-siRNA response that repressed luciferase expression from a co-transfected, circular vector containing a full-length Renilla luciferase coding sequence (Figure 2B). The repressive response originating at a double-strand break can therefore act in trans, most likely through the well-characterized degradation pathway for perfectly matched target mRNAs. However, we do not want to exclude the possibility that the small RNA response also influences chromatin structure either in cis or in trans.
Transcriptional activity controls the endo-siRNA response
The work of Wei et al. (13) profiled the small RNAs generated upon induction of a double-strand break in vivo. In Arabidopsis, the small RNA response required RNA pol IV and RdRP, appeared strongest in the region upstream (with respect to transcription) of the cleavage site and did not extend beyond the mRNA 3′-end. An analogous process has been proposed for the fungus Neurospora crassa: here, a DNA-damage-induced small RNA response arises from the ribosomal DNA (20) due to an aberrant transcript (aRNA) generated by the DNA/RdRP Qde-1 (21). In Drosophila—as in humans—there is no RNA pol IV or RdRP homolog; a different mechanism must therefore produce the dsRNA precursor required for endo-siRNA biogenesis. For direct analysis, we deep sequenced small RNAs from cells transfected with linear and circular GFP expression plasmids (size: 10 800bp), then mapped the uniquely matching reads onto the plasmid sequence with no mismatch allowed. Compared with the circular plasmid, the BamHI linearized plasmid generated 13.5-fold more endo-siRNAs, the EcoRI-linearized vector 11.5-fold more and the NotI-linearized vector 6.5-fold more endo-siRNAs (Table 1). We observed that endo-siRNAs were predominantly produced from the region between the promotor (the three annotated transcription start sites are indicated in Figure 3) and a downstream cut within the GFP transcriptional unit, while the region corresponding to the mRNA downstream of the cut is not enriched for small RNAs (Figure 3). Furthermore, only a 2-fold increase of small RNAs was detected upon a SacI-cut in the mini-white gene (Figure 3 and Table 1), which is only weakly transcribed in S2-cells. The association of the small RNA response with an active transcriptional unit containing the double-strand break strongly suggests that the mRNA produced from the locus contributes one strand of the dsRNA precursor for endo-siRNA generation. Consistent with the reporter assays presented in Figure 1C, we observe no preferential generation of endo-siRNAs in cells transfected with AlwI-nicked plasmid (Table 1 and Supplementary Figure S4). The size distribution of plasmid-matching reads showed a peak at 21 nt characteristic for siRNAs (Supplementary Figure S5).
Table 1.
Deep sequencing count data
| Total reads | 21–23-nt Length | Genome matching (% of 21–23) | Mature miRNAs (% of gen. mat.) | pKF63 (‰ of gen. mat.) | pRS425 (‰ of gen. mat.) | Fluc | |
|---|---|---|---|---|---|---|---|
| pKF63 BamHI; pRS425 circ.; Fluc PCR | 9 036 646 | 3 465 173 | 2 581 985 (74.5%) | 1 301 398 (50.4%) | 6931 (2.7‰) | 423 (0.16‰) | 0 |
| pKF63 NotI; pRS425 circ.; Fluc PCR | 8 224 052 | 3 909 854 | 2 947 756 (75.4%) | 1 605 762 (54.5%) | 3739 (1.3‰) | 149 (0.05‰) | 0 |
| pKF63 EcoRI; pRS425 circ.; Fluc PCR | 8 391 521 | 2 699 100 | 1 971 598 (73.1%) | 965 493 (49.0%) | 4544 (2.3‰) | 123 (0.06‰) | 5 |
| pKF63 SacI; pRS425 circ.; Fluc PCR | 7 515 383 | 3 931 978 | 2 887 215 (73.4%) | 1 545 121 (53.5%) | 1091 (0.4‰) | 167 (0.06‰) | 3 |
| pKF63 AlwI; pRS425 circ.; Fluc PCR | 9 381 901 | 3 394 873 | 2 480 116 (73.1%) | 1 473 759 (59.4%) | 606 (0.2‰) | 218 (0.08‰) | 2 |
| pKF63 circ.; pRS425 BamHI; Fluc PCR | 20 644 453 | 7 794 275 | 5 781 192 (74.2%) | 2 775 175 (47.9%) | 1075 (0.6‰) | 3034 (0.52‰) | 0 |
| Untreated cells | 21 055 737 | 5 435 317 | 4 938 222 (90.1%) | 2 864 713 (58.0%) | 38 (0.01‰) | 110 (0.02‰) | 0 |
Figure 3.
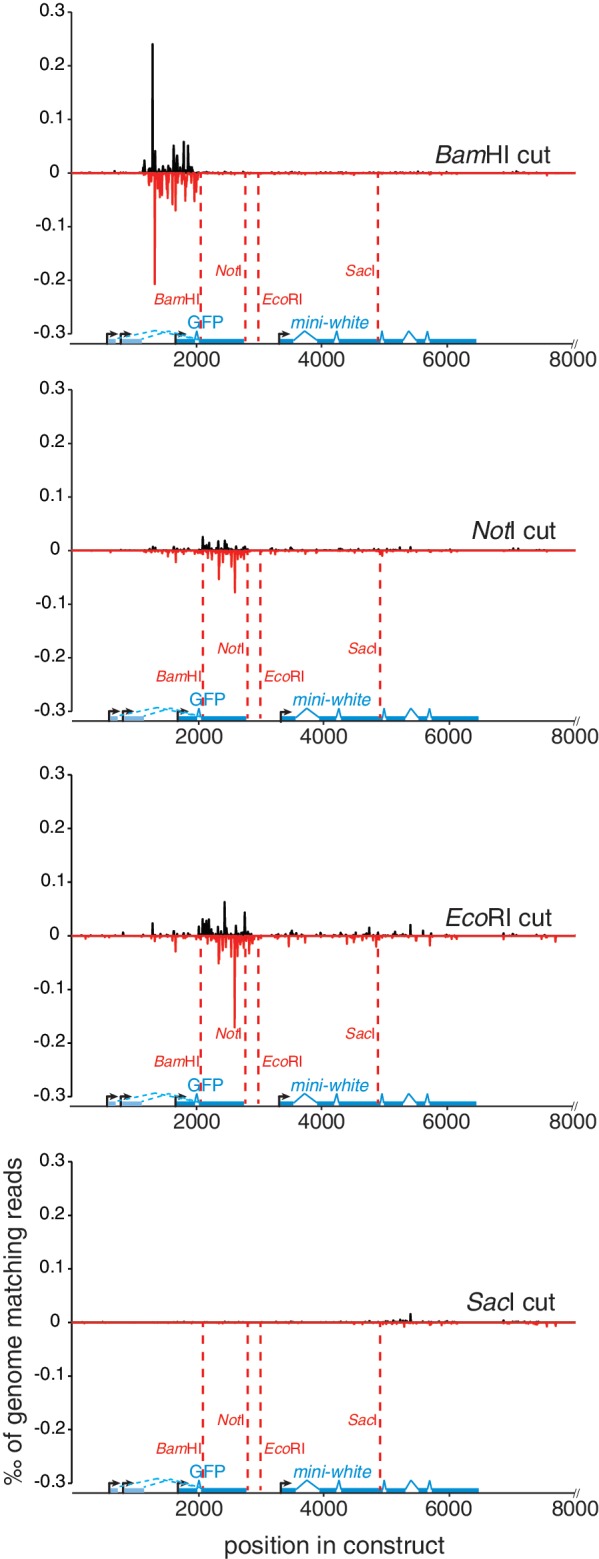
Profiling of break-induced endo-siRNAs by deep sequencing. We transfected our GFP expression vector after linearization at the indicated positions, deep sequenced small RNAs and determined the uniquely mapping reads with no mismatches allowed. Their distribution along the plasmid sequence was calculated by binning into non-overlapping 5-nt intervals and normalized to the number of genome-matching reads in each library. The graphs depict the sense (black) and antisense (red) matching reads as ‰ of genome matching 21–23-nt reads in the respective library. The three annotated transcription start sites for the ubiquitin-promotor are indicated.
For control purposes, we had co-transfected equal amounts of an unrelated yeast plasmid (pRS425, 6849 nt) and a PCR product comprising the firefly luciferase coding sequence (2387 nt). We observed a low but reproducible amount of endo-siRNA generation from the circular yeast plasmid; their levels increased upon linearization of this plasmid but showed no enrichment around the cleavage site (Table 1 and Supplementary Figure S4). We conclude that small RNA generation triggered by a double-strand break (DSB) can occur also in the absence of promotor activity, reminiscent of the small RNAs reported by Francia and colleagues (12) at a break in a non-transcribed sequence in human cells. Possibly non-specific, cryptic transcription or a low-level, non-physiologic recognition of the yeast promotors by the Drosophila transcriptional machinery is contributing the sense strand to dsRNA generation. Importantly, the firefly luciferase PCR product did not elicit a significant endo-siRNA response in any of the libraries examined. Break-induced endo-siRNA generation therefore requires sequence elements that mediate the assembly of at least a rudimentary chromatin structure (this might be, e.g. the yeast promotor sequences or the bacterial replication origin present on the plasmids) in cis to the break. Alternatively, a minimal DNA length or distance between DSBs that lies between 2387 and 6849 nt is needed.
DISCUSSION
Importance of small RNAs in DSB recognition and repair pathways
A double-strand break must first be recognized, then a signal is generated that arrests the cell cycle, induces the repair process and finally either vanishes when the damage is repaired or induces apoptosis if this cannot be reached. Is the small RNA response an important player in one of these processes?
Wei and colleagues have demonstrated that the radiation-induced formation of γ-H2Ax foci is independent of the small RNA response in Arabidopsis, arguing that recognition and signaling to the DSB repair machinery is still functioning without the small RNAs (13). However, DSB repair efficiency by homologous recombination was reduced in the absence of small RNA biogenesis factors in both Arabidopsis and human cells. In particular, lack of pol IV completely blocked DSB-induced RNA generation and reduced the DSB repair efficiency by a factor of 5. In human cells, repair efficiency was about 2-fold lower upon depletion of Dicer or Ago2. Due to the incomplete removal of protein inherent to the use of RNAi, it is unclear whether this reflects the full extent of siRNA implication. Since the frequency of homologous recombination can be modulated to a similar extent by shifts in cell-cycle distribution (22), indirect effects cannot be fully excluded. DNA damage induces qiRNAs in Neurospora and cells with mutations in qiRNA biogenesis factors are sensitive to DNA-damaging agents. However, mutations in the key DNA-damage signaling kinase atm display an even more pronounced sensitivity (20). The Drosophila endo-siRNA system is separate from the bulk of miRNA biogenesis, thus allowing a relatively straightforward interpretation of genetic data. Loss of the ATM and Mre11 proteins (tefu and mre11) leads to late pupal lethality in Drosophila primarily due to telomere fusions (23,24), and the DSB repair factor spnA, a Rad51 homolog, is essential for female fertility (25). The endo-siRNA factors dcr-2, ago2 and loqs (isoform D), however, can be inactivated without causing lethality, sterility or any morphological phenotype (reviewed in (7)). Thus, the phenotypes of RNAi mutants are clearly weaker. Although a genome-wide RNAi screen for function of the DSB-triggered G2/M checkpoint in Drosophila cells recovered many known and novel components of the checkpoint signaling systems, the endo-siRNA factors did not score positive (26).
In fact, one might even speculate that meiotic DSBs are protected to some degree from generating an endo-siRNA response: both injected dsRNA (27) and transgenic hairpin-generating constructs are inefficiently inducing RNAi in the germ line during oogenesis. For the transgenic approach, this can be alleviated either by overexpressing Dcr-2 (5,28,29) or by using artificial miRNAs, which may be processed through Dcr-1 (30–32), indicating that the messages are not per se inaccessible to the RNAi effector complexes. It is thus conceivable that the siRNA system is not active enough to generate a robust small RNA response at meiotic double-strand breaks. Beyond this particular situation, changes in chromatin structure, transcriptional activity and mitotic chromosome condensation have been reported for Drosophila dcr2 and ago2 mutants (33–36). A contribution of the endo-siRNA response to the efficiency of DSB repair through effects on local chromatin structure is therefore possible. Furthermore, cell-type-specific differences in the small RNA response pathways, e.g. a putative piRNA response to breaks in meiosis or early embryonic development (37) may also account for weaker phenotypes. Our observation that small RNA generation initiates independently of the precise end structure is consistent with a model where DSB recognition occurs before the induction of antisense transcription. We speculate that after initial end processing, the 3′-single-stranded overhangs generated by DSB resection serve as a platform to recruit one of the cellular DNA-dependent RNA polymerases and initiate antisense transcription close to the DNA break. This break-induced antisense transcript may then form dsRNA with the mRNA ‘stump’ produced by normal RNA Pol-II transcription (Figure 4).
Figure 4.
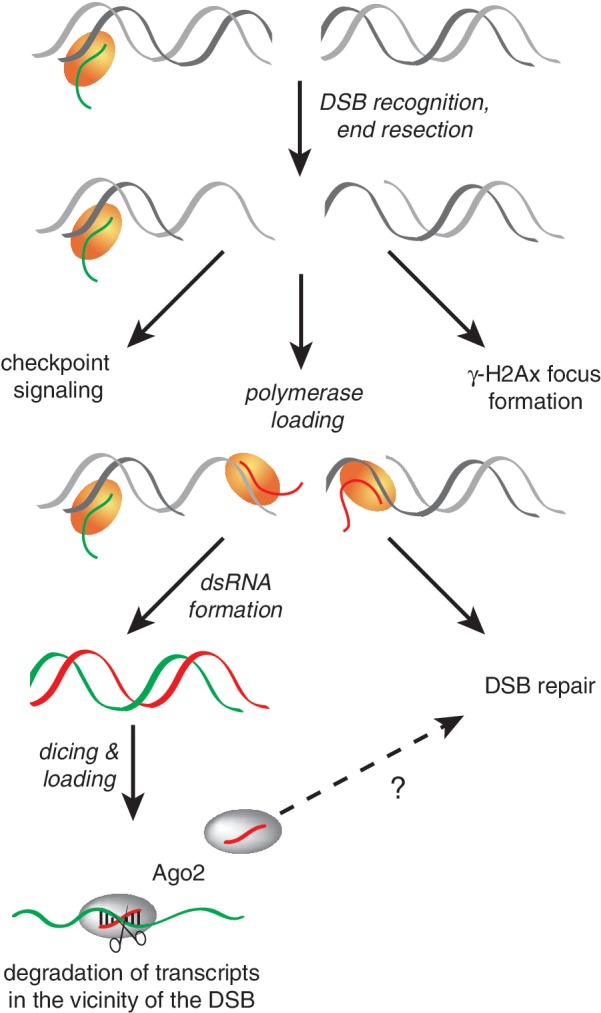
Model for the generation of break-induced endo-siRNAs. This figure depicts our hypothesis for break-induced endo-siRNA generation in Drosophila. Upon the creation of a double-strand break, recognition and initial end processing likely occur independently of and prior to the generation of small. The DNA ends recruit one of the cellular DNA-dependent RNA polymerases (yellow), which then generates a break-induced transcript that may be conceptually analogous to the aberrant RNA described in Neurospora or the damage-induced RNA pol IV transcript in Arabidopsis. If normal mRNA transcription occurs toward the break, the two transcripts produced from the locus form dsRNA, which is then processed and loaded into Ago2. Since a PCR product comprising only the firefly luciferase coding sequence did not generate any corresponding small RNAs, the process may only operate in the context of chromatin. We have not addressed the question whether the efficiency of DNA repair is affected by the presence of small RNAs. However, the small RNAs are capable of repressing gene expression at other sites with homologous sequences. It is therefore possible that they participate in a quality-control process that clears potentially truncated messages.
A novel player in the DSB response: transcription initiation at the break
We report that in cultured somatic cells of Drosophila, a double-strand break within a transcribed gene leads to the initiation of a cellular RNA polymerase, which then continues to transcribe in the direction away from the break. To our knowledge, this event has not been described so far and constitutes a new aspect of signaling by double-strand breaks: By forming dsRNA with the normal sense transcript of the locus, this antisense transcript activates the RNAi system and thereby has a signaling and/or effector function, akin to the activation of a protein kinase. The concept of a transcript with signaling function is well established in plants and the fungus Neurospora crassa, where specialized RNA polymerases (RNA pol IV and V) are dedicated to the synthesis of transcripts that activate and maintain the non-coding RNA response (38). Accordingly, the requirement for RNA pol IV in the Arabidopsis break-induced small RNA response implied that this polymerase is recruited to the lesion and produces a transcript that is recognized as aberrant (13). It is unclear, however, where exactly RNA pol IV initiates and in what direction this first transcript is generated—toward or away from the break. Further characterization of genetic requirements for the small RNA response may elucidate whether the molecular events leading from the DNA break to antisense transcription in Drosophila and RNA pol IV recruitment in Arabidopsis are conserved. Signaling of DNA damage by RNA polymerases is well established in the case of nucleotide excision repair: here, the stalling of RNA polymerase II as it encounters a lesion either triggers transcription-coupled nucleotide excision repair through the cockayne syndrome protein A/cockayne syndrome protein B complex, or leads to ubiquitylation and degradation of the polymerase (reviewed in (39)). Whether small RNAs are produced in this context has not been addressed.
The fact that Drosophila break-induced small RNAs can silence in trans indicates that they may have a function beyond the repair process by repressing transcripts that might be affected by the DNA break. Such transcripts could have adverse effects on the cell, e.g. if transcripts truncated within an intron had an increased propensity to engage in non-physiologic trans-splicing and thereby acquire export- and translation competence. Since only actively transcribed genes have the potential to generate significant amounts of dsRNA upon damage, this quality-control system would be automatically restricted to its appropriate targets.
In summary, we propose that double-strand break recognition and potentially initial processing occur prior to generation of small RNAs. Recruitment of a cellular RNA polymerase then initiates transcription of antisense RNA that can form dsRNA if a corresponding sense transcript is made in the vicinity of the break. Dicing and loading are performed by the Drosophila endo-siRNA pathway and—besides putative roles in DNA repair—the resulting small RNAs may serve a quality-control purpose to protect the cell from potentially truncated mRNA (Figure 4).
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Deutsche Forschungsgemeinschaft [SFB646, FO360/2-1 to K.F.]; DFG Cluster of Excellence ‘Munich Center for integrated Protein Science’ CiPSM and an LMU Excellent (to K.F.). Human Frontier Science Program Career Development award (to K.F.). Funding for open access charge: LMU München.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Stefan Krebs and Dr Helmut Blum/LAFUGA, Gene Center, for deep sequencing services.
REFERENCES
- 1.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 5.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl Acad. Sci. USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 9.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carnici P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartig JV, Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahmed K, Seth A, Nitiss KC, Nitiss JL. End-processing during non-homologous end-joining: a role for exonuclease 1. Nucleic Acids Res. 2011;39:970–978. doi: 10.1093/nar/gkq886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HC, Aalto AP, Yang Q, Chang SS, Huang G, Fisher D, Cha J, Poranen MM, Bamford DH, Liu Y. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8:e1000496. doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieckmann T, Kriegs M, Nitsch L, Hoffer K, Rohaly G, Kocher S, Petersen C, Dikomey E, Dornreiter I, Dahm-Daphi J. p53 modulates homologous recombination at I-SceI-induced double-strand breaks through cell-cycle regulation. Oncogene. 2012 doi: 10.1038/onc.2012.123. April 9 (doi:10.1038/onc.2012.123; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 23.Bi X, Wei SC, Rong YS. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 2004;14:1348–1353. doi: 10.1016/j.cub.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 24.Song YH, Mirey G, Betson M, Haber DA, Settleman J. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr. Biol. 2004;14:1354–1359. doi: 10.1016/j.cub.2004.06.064. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Reyes A, Elliott H, St Johnston D. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development. 1997;124:4927–4937. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- 26.Kondo S, Perrimon N. A genome-wide RNAi screen identifies core components of the G-M DNA damage checkpoint. Sci. Signal. 2011;4:rs1. doi: 10.1126/scisignal.2001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 29.Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haley B, Foys B, Levine M. Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila. Proc. Natl Acad. Sci. USA. 2010;107:11435–11440. doi: 10.1073/pnas.1006689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haley B, Hendrix D, Trang V, Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshpande G, Calhoun G, Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 39.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.