Abstract
Faithful chromosome segregation in meiosis is crucial to form viable, healthy offspring and in most species, it requires programmed recombination between homologous chromosomes. In fission yeast, meiotic recombination is initiated by Rec12 (Spo11 homolog) and generates single Holliday junction (HJ) intermediates, which are resolved by the Mus81–Eme1 endonuclease to generate crossovers and thereby allow proper chromosome segregation. Although Mus81 contains the active site for HJ resolution, the regulation of Mus81–Eme1 is unclear. In cells lacking Nse5–Nse6 of the Smc5–Smc6 genome stability complex, we observe persistent meiotic recombination intermediates (DNA joint molecules) resembling HJs that accumulate in mus81Δ cells. Elimination of Rec12 nearly completely rescues the meiotic defects of nse6Δ and mus81Δ single mutants and partially rescues nse6Δ mus81Δ double mutants, indicating that these factors act after DNA double-strand break formation. Likewise, expression of the bacterial HJ resolvase RusA partially rescues the defects of nse6Δ, mus81Δ and nse6Δ mus81Δ mitotic cells, as well as the meiotic defects of nse6Δ and mus81Δ cells. Partial rescue likely reflects the accumulation of structures other than HJs, such as hemicatenanes, and an additional role for Nse5–Nse6 most prominent during mitotic growth. Our results indicate a regulatory role for the Smc5–Smc6 complex in HJ resolution via Mus81–Eme1.
INTRODUCTION
The repair of DNA damage by homologous recombination (HR) is central to the faithful propagation of chromosomes during both the mitotic and meiotic cycles. During mitotic growth, exogenous or endogenous genotoxic agents can induce DNA double-strand breaks (DSBs), which may also arise from replication fork mishaps [reviewed in (1)]. The HR-based repair of these insidious forms of DNA damage is executed by a relatively well-defined and largely overlapping set of proteins. Broken DNA ends are first resected to generate a single-stranded (ss) 3′-overhang, which acts as a platform for the subsequent homology search and invasion steps of HR [reviewed in (2)]. The ss DNA-binding protein complex replication protein A initially coats the 3′-overhang, which is subsequently displaced by loading of the RecA homolog Rad51, also called Rhp51 in fission yeast, as well as Dmc1 during meiosis (3–5). The Rad51 nucleoprotein filament invades a homologous duplex, forming a displacement loop (D-loop) that is used to prime repair synthesis and, at a broken fork, to restart replication. Meiotically induced Rec12 (Spo11 homolog) and several partner proteins form DSBs during fission yeast meiosis, to form crossovers important for the proper segregation of homologs at the first meiotic division (6,7). Genomic regions that are hotspots for meiotic DSB formation by Rec12 require Rad51 for DSB repair and recombination; in regions with lower DSB levels, both Rad51 and Dmc1, a meiosis-specific paralog, are required (3–5).
Following D-loop formation, there are various postulated pathways for the completion of DSB repair. A major pathway, thought to be essential for crossover formation, requires the processing of four-way DNA junctions called Holliday junctions (HJs (8–11)). The nature of the initiating lesion and whether the cell is in mitosis or meiosis influence the mode of HJ processing. Closely spaced double HJs may be dissolved by the concerted action of a RecQ-related helicase and a topoisomerase partner (e.g. Sgs1–Top3 in budding yeast (12)). Notably, in fission yeast, the restart of broken replication forks critically depends on Mus81–Eme1 but not on the dissolution activity of Rqh1 (Sgs1 homolog)–Top3 (13). This likely reflects the formation of single, not double, HJs (or an HJ precursor) during restart of replication (12), because single HJs cannot be dissolved by a helicase and topoisomerase. Endonucleolytic HJ resolution facilitates crossing over between homologous chromosomes (homologs) and dominates during meiosis (8,14,15), when single HJs accumulate in mus81 mutants (8,15). Dissolution of double HJs without associated crossover seems to be the major pathway in mitotic cells of some species (12) and seems to play a role during meiosis in budding yeast (16).
HJs arising through HR-mediated DSB repair can be resolved by endonucleolytic activities such as Mus81–Eme1 ((8,15–19)), Slx1–Slx4 (20,21)) and Yen1 (19,22). Recently, it was shown in the budding yeast Saccharomyces cerevisiae that Mus81–Mms4 and Yen1 endonucleases collaborate during meiosis to resolve HJs and that their activity is carefully regulated during the cell cycle (19). Mms4 (Eme1 homolog) is phosphorylated and then hyperactivated during meiosis I, whereas Yen1 activity is inhibited until meiosis II.
Mus81–Eme1 is critical in the fission yeast Schizosaccharomyces pombe where it is the only known complex involved in meiotic HJ resolution (8,15,23). A Yen1 ortholog has not been identified in the S. pombe genome, and Slx1–Slx4 has no detectable defect in meiotic recombination (our unpublished data). The Mus81–Eme1 heterodimer is required for HR repair at stalled or broken replication forks and is also critical during mitotic (13,24,25) and meiotic HR (15,26). In vitro, Mus81–Eme1 can cleave various substrates that mimic stalled replication forks and nicked and intact HJs (15,27,28). Mus81–Eme1 not only has a binding and cleavage preference for nicked HJs but also has a robust cleavage activity on intact HJs (29). S. pombe meiotic cells deleted for mus81 accumulate single HJs, as shown by 2D gel analysis and electron microscopy (8). The accumulation of HJs in mus81Δ cells results in severe meiotic defects (15) and sensitivity to the topoisomerase I poison camptothecin (CPT (30)); these phenotypes can be partially suppressed by expression of the Escherichia coli HJ resolvase RusA. Therefore, RusA has been used extensively, and is widely accepted, to identify the structures accumulated in various DNA repair mutants (15,26,30–32).
As described above, the last decade has seen major advances in the identification of activities most proximally involved in the processing of joint molecules (JMs) formed during HR. However, currently, little is known about how JM processing enzymes are recruited to their substrates, and whether their activities are facilitated by additional chromatin-associated factors. Interestingly, in this regard, others and we have identified and characterized the Smc5–Smc6 holocomplex, which seems to play multiple roles in HR (33). The octameric Smc5–Smc6 complex is structurally related to the cohesin and condensin complexes but, uniquely, contains two subunits, Nse1 and Nse2, which exhibit E3 ligase activity for ubiquitin and SUMO (Small Ubiquitin-like Modifier), respectively (33,34). In addition to Smc5, Smc6, Nse1 and Nse2, the complex contains a melanoma antigen-domain protein Nse3, a kleisin-like protein Nse4, and two armadillo/Huntington, Elongation Factor 3, PR65A, TOR repeat proteins, Nse5 and Nse6. Notably, unlike in budding yeast (33), nse5Δ and nse6Δ mutants are viable and display indistinguishable sensitivities to all tested DNA-damaging agents (35). In addition, Nse5 and Nse6 form a stoichiometric heterodimer when purified from insect cells (35) or bacterial cells (our unpublished data), and furthermore, nse5Δ nse6Δ double mutants are no more sensitive to UV irradiation than either single mutant (35). Thus, Nse5 and Nse6 function as an obligate heterodimer.
Reflecting key HR roles of the complex, Smc5–Smc6 in budding yeast and human cells is loaded near an enzymatically induced DNA DSB and is important for HR-mediated repair of the break (36–38). In addition, smc5–smc6 mutation causes hypersensitivity to ionizing radiation (IR)-induced DSBs, which is not additive when combined with a rad51 deletion; smc5–smc6 mutants, like rad51Δ mutants, fail to restore chromosome integrity following IR (35,39–43). Intriguingly, the Smc5–Smc6 complex has been implicated in the processing of HR intermediates or suppression of their formation or both (reviewed in (33,44)). For example, smc5–smc6 hypomorphs, including nse2 E3 SUMO ligase-deficient cells, accumulate Rad51-dependent HR intermediates following replication stress, indicating a concerted action of the entire complex in the productive completion of HR (35,45–54).
What is the physical nature of the recombination structures that form when Smc5–Smc6 function is compromised? In budding yeast, a heterogeneous group of hemicatenane and converged replication fork structures arise during replication stress in Smc5–Smc6-deficient mitotic cells (45,47,48,55). These DNA structures appear similar to those that accumulate in sgs1Δ mutant cells, and thus, it has been proposed that Sgs1 acts in concert with Smc5–Smc6 and sumoylation in the removal of such linkages during vegetative growth (45).
Here, we identify a critical role for fission yeast Nse5–Nse6 in chromosome disjunction during meiosis. We use both genetic and physical assays to determine the underlying cause of meiotic failure in nse6Δ cells, the stage at which Nse5–Nse6 function is required, and the DNA structure(s) that prevent(s) chromosome segregation at the first meiotic division (MI) in nse6Δ cells. Our analyses demonstrate that Nse5–Nse6 acts after programmed meiotic DSB formation to drive the timely resolution of JMs whose persistence in nse6Δ cells causes the profound MI defects. The JMs observed in nse6Δ cells resemble the HJs that accumulate in mus81Δ cells, providing the first direct evidence for a role of the Smc5–Smc6 complex in HJ resolution. Although Nse5–Nse6 and Mus81 have some distinct roles, both proteins are essential to avoid an accumulation of JMs that impede meiosis. Here, we define Nse5–Nse6 as a novel regulator of nuclear division, which may act directly in the HJ resolution mechanism or by attracting the Smc5–Smc6 complex to its needed sites of action—mitotic or meiotic HJs.
MATERIALS AND METHODS
Standard Schizosaccharomyces pombe techniques
Standard fission yeast methods and media were used in these studies (56). CPT was obtained from Sigma-Aldrich (St. Louis, MO). Table 1 lists the S. pombe strains, and Table 2 lists the primers used.
Table 1.
Schizosaccharomyces pombe strains and primers
| Strain No. | Genotype |
|---|---|
| NBY128 | h+ mus81::kanMX6 |
| NBY185 | h− pat1-114 |
| NBY282 | h+ rec12-152::LEU2 |
| NBY355A | h− mus81::kanMX6 (pREP1:LEU2) |
| NBY355C | h+ mus81::kanMX6 (pREP1:LEU2) |
| NBY356A | h− mus81::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY356C | h+ mus81::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY364 | h− mus81::kanMX6 (pREP1-NLS-RusA-D70N:LEU2) |
| NBY365 | h+ mus81::kanMX6 (pREP1-NLS-RusA-D70N:LEU2) |
| NBY384 | h− ade7-152 ura4+ (pREP1:LEU2) |
| NBY420 | h+ ura1-61 |
| NBY780 | h+ |
| NBY781 | h− |
| NBY835 | h+ nse6::kanMX6 |
| NBY855 | h− rad51::ura4+ nse6::kanMX6 |
| NBY871 | h− nse6::kanMX6 |
| NBY883C | h+ rec12-152::LEU2 nse6::kanMX6 |
| NBY896 | h+ nse5::ura4+ |
| NBY897 | h− nse5::ura4+ |
| NBY917 | h+ nse6::kanMX6 (pREP1-NLS-RusA-D70N:LEU2) |
| NBY936 | h− nse6::kanMX6 (pREP1-NLS-RusA-D70N:LEU2) |
| NBY952 | h− rad51::ura4+ |
| NBY963 | h− nse6::kanMX6 ade7-152 leu1+ ura4+ |
| NBY991 | h− Ch16-MHH ade6-210 (pREP81X-HO:LEU2) |
| NBY1000 | h− nse6::kanMX6 Ch16-MHH ade6-210 (pREP81X-HO:LEU2) |
| NBY1484 | h+ nse6::kanMX6 (pE119:LEU2) |
| NBY1486 | h− nse6::kanMX6 (pE119:LEU2) |
| NBY1753 | h− rec12-152:LEU2 |
| NBY2027 | h+ rad51::ura4+ |
| NBY2051 | h+ rad51::ura4+ nse6::kanMX6 lacO::lys1 lacI-GFP::arg3+ |
| NBY2482 | h+ (pREP1:LEU2) |
| NBY2551 | h− dmc1::hphMX6 |
| NBY2589 | h− dmc1::hphMX6 nse6::kanMX6 |
| NBY2590 | h+ dmc1::hphMX6 nse6::kanMX6 |
| NBY2610 | h+ dmc1::hphMX6 |
| NBY2620 | h+ rad51::ura4+ dmc1::hphMX6 nse6::kanMX6 |
| NBY2621 | h− rad51::ura4+ dmc1::hphMX6 nse6::kanMX6 |
| NBY2622 | h+ rad51::ura4+ dmc1::hphMX6 |
| NBY2623 | h− rad51::ura4+ dmc1::hphMX6 |
| NBY2654 | h+ (pREP1-NLS-RusA:LEU2) |
| NBY2660 | h+ nse6::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY2750 | h− nse6::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY2893 | h+ ura1-61 nse6::kanMX6 |
| NBY2938 | h+ pat1-114 nse6::natMX6 |
| NBY2953 | h− ura4+ LEU2 his4-239 lys4-95 |
| NBY2962 | h− ura4+ LEU2 his4-239 lys4-95 nse6::kanMX6 |
| NBY3112 | h− ura4+ prh1:hphMX6 |
| NBY3114 | h− ura4+ nse6::kanMX6 prh1:hphMX6 |
| NBY3304 | h− nse6::natMX6 mus81::kanMX6 |
| NBY3311 | h+ nse6::natMX6 mus81::kanMX6 |
| NBY4176 | h+ nse6::natMX6 mus81::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY4178 | h+ nse6::natMX6 mus81::kanMX6 (pREP1:LEU2) |
| NBY4192 | h+ nse5::ura4+ (pREP1-NLS-RusA:LEU2) |
| NBY4194 | h+ nse5::ura4+ (pREP1:LEU2) |
| NBY4195 | h− nse5::ura4+ (pREP1-NLS-RusA:LEU2) |
| NBY4197 | h− nse5::ura4+ (pREP1:LEU2) |
| NBY4198 | h− nse6::natMX6 mus81::kanMX6 (pREP1-NLS-RusA:LEU2) |
| NBY4200 | h− nse6::natMX6 mus81::kanMX6 (pREP1:LEU2) |
| NBY4298 | h− rad51::hphMX6 Ch16-MGH ade6-210 (pREP81X-HO:LEU2) |
| GP1456 | h− rec12-152::LEU2 ura4-294 ade6-52 |
| GP5082 | h−/h− ade6-216/ade6-210 pat1-114/pat1-114 +/ura1-61 mbs1-24/mbs1-25 his4-239/+ +/lys4-95 mus81::kanMX6/mus81::kanMX6 |
| GP5086 | h−/h− ade6-216/ade6-210 pat-114/pat-114 +/ura1-61 mbs1-24/mbs1-25 |
| GP6234 | h−/h− ade6-216/ade6-210 pat1-114/pat1-114 lys3-37/+ +/ura1-61 mbs1-24/mbs1-25 nse6::kanMX6/nse6::kanMX6 |
| GP6656 | h−/h− ade6-3049/ade6-3049 bub1-243(L)/+ +/vtc4-1104(R) pat1-114/pat1-114 lys3-37/+ +/ura1-61 mbs1-24/mbs1-25 his4-239/+ +/lys4-95 |
| GP6657 | h−/h− ade6-3049/ade6-3049 bub1-243(L)/+ +/vtc4-1104(R) pat1-114/pat1-114 lys3-37/+ +/ura1-61 mbs1-24/mbs1-25 his4-239/+ +/lys4-95 mus81::kanMX6/ mus81::kanMX6 |
| GP7765 | h+/h+ ade6-3049/ade6-3049 bub1-243(L)/+ +/vtc4-1104(R) pat1-114/pat1-114 lys3-37/+ +/ura1-61 mbs1-24/mbs1-25 his4-239/+ +/lys4-95 mus81::kanMX6/ mus81::kanMX6 nse6::kanMX6/nse6::kanMX6 |
| GP7773 | h− ade6-3049 pat1-114 nse6::kanMX6 rec12-171::ura4+ |
Table 2.
Primers used to integrate the hphMX6 marker between prh1 and SPAC2G11.10c (primers 1–4) and to replace the dmc1 gene with the hphMX6 marker (primers 5–8)
| Primer 1 | 5′-AATTGAGCTCTATTTCTGAG-3′ |
| Primer 2 | 5′-TTAATTAACCCGGGGATCCGCTTTCATTTCAGTACTTCAATCC-3′ |
| Primer 3 | 5′-GTTTAAACGAGCTCGAATTCCATGGAGGTAATTATTGGTTG-3′ |
| Primer 4 | 5′-CTTTCTGGGCTTTCCTCACA-3′ |
| Primer 5 | 5′-GCGACGCGTTCATTGTTAC-3′ |
| Primer 6 | 5′-TTAATTAACCCGGGATCCGTGCACTTTATTTTTATATTGAAC-3′ |
| Primer 7 | 5′-GTTTAAACGAGCTCGAATTCTTGATTTTCTACCTATTCCA-3′ |
| Primer 8 | 5′-AGTTGCTTTTGGGGGTTTG-3′ |
Unless otherwise indicated, all NBY strains are ura4-D18 and leu1-32. pREP1-RusA, pREP1-RusA-D70N and pREP1 were described in (15). pE119, containing LEU2 and GST, was used as a control vector (57). The minichromosome assay using Ch16-MHH, Ch16-MGH and pREP81X-HO:LEU2 has been described in (58). Mutations other than mating type and commonly used auxotrophies are described in previous studies: mus81::kanMX6 (15), nse6::kanMX6 (35), prh1::hphMX6 (this study), rad51::ura4+ (59), dmc1::hphMX6 (this study), rec12-152::LEU2 (60), mbs1-24 and mbs1-25 (61), bub-1243, vtc4-1104 (9), lacO::lys1 lacI-GFP::arg3+ (62) and nse6::natMX6 (the kanMX6 marker in strain NBY835 (35) was switched to natMX6 as described in (64)). In Table 1 ‘:’ indicates marked by the following gene, and ‘::’ indicates that the preceding gene is replaced by the following marker.
Polymerase chain reaction (PCR) was used to create the hphMX6 cassette flanked by genomic DNA, to enable integration into the middle of the 1.2 kb intergenic region between the prh1 and SPAC2G11.10c loci. The hphMX6 cassette from pCR2.1 hphMX6 (63) was amplified in two steps, using a combination of the primers shown in Table 2, as described in (63,64). A stable transformant was obtained with the hphMX6 cassette integrated between prh1 and SPAC2G11.10c loci, approximately 70 kb to the right of the mbs1 meiotic DSB hotspot on chromosome I (61,65).
PCR was used to create the hphMX6 cassette flanked by genomic DNA, to replace the dmc1 gene. The hphMX6 cassette from pCR2.1 hphMX6 (63,64) was amplified in two steps, using a combination of the primers shown in Table 2, as described in (63,64).
HO-induced DSB repair assays
Cells were cultured in repressive (+ thiamine; B1) medium (EMM2), and a sample was plated onto non-selective medium (EMM2) for the 0 h time point. Cells were then cultured in repressive or de-repressive (−thiamine) media for 48 h and plated onto non-selective media. Colonies were counted after 3 days growth at 30°C. Replica plating onto media that contained hygromycin (or kanamycin for the rad51Δ mutant), or lacked either adenine or histidine, allowed calculation of the frequency of marker loss. Data are means and standard error of the mean from three independent assays, as described in (58).
Meiotic crosses and recombination assays
Cells from equal volumes (10 µl) of each parental haploid culture were mixed, washed, resuspended in water and plated onto supplemented sporulation agar plates (66). After 2 days at 25°C, the cell–ascus mixture was observed by microscopy or processed further for viability assays. Meiotic 4′,6-diamidino-2-phenylindole (DAPI) staining was performed essentially as described in (15). Zygotic asci were fixed in methanol (−80°C) for 10 min and then washed three times with phosphate-buffered saline (1× PBS). The fixed asci were treated with Zymolyase 100T (0.1 mg/ml) for 10 min at 37°C. Asci were collected by centrifugation and resuspended in 0.1% Triton X-100 for 2 min at room temperature. Asci were then washed three times (in 1× PBS) and resuspended in a drop of 1× PBS containing 0.5 μg/ml DAPI. Asci were photographed using a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix CCD camera. Images were acquired with IPlab Spectrum software (Signal Analytics Corporation).
For viability assays, the cell–ascus mixture was suspended in 1 ml of H2O and treated with 2% glusulase overnight at room temperature. Addition of ethanol to 30% for 1 h killed remaining vegetative cells, essentially as described in (67). To assess spore viability, spores were counted using a hemocytometer, diluted and plated onto the appropriate media. Spore viability was scored following incubation for 5–6 days at 30°C. For meiotic recombination assays, an appropriate dilution of spores was plated onto non-selective (YES) media and replica plated to appropriate test media: EMM2 (66) ± adenine for ade7-152; ± leucine for leu1-32; ± lysine for lys4-95; ± histidine for his4-239; ± uracil for ura1-61 and YES ± hygromycin (100 μg/ml) for hphMX6.
Analysis of recombination intermediates
pat1-114 strains were thermally induced for meiosis, and DNA was extracted from cells embedded in agarose plugs (68). The DNA was digested with appropriate restriction enzymes and analyzed by gel electrophoresis and Southern blot hybridization (8,69) and quantification (9).
The sensitivity of DNA to S1 nuclease (Roche) and to RuvC (gift of Ken Marians, Memorial Sloan-Kettering Cancer Center) was assayed by digesting the DNA, in plugs, with PvuII restriction enzyme, followed by washing twice with TE (Tris-EDTA; 10 mM Tris–HCl, pH 7.0, 1 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0) and twice with nuclease S1 buffer (33 mM sodium acetate, 100 mM NaCl, 0.033 mM ZnSO4, pH 4.5 at room temperature) or RuvC buffer (50 mM Tris–acetate pH 7.5, 10 mM Mg(OAc)2, 1 mM DTT, 50 μg of BSA per ml); duration for each wash was 20 min at room temperature. The plugs were incubated in 100 μl of buffer with the indicated amount of S1 for 2 h at 4°C or RuvC for 4 h at 4°C, then for 1 h at 37°C (S1) or for 4 h at 55°C (RuvC) to facilitate branch migration of HJs embedded in agarose; the digestions were stopped by adding 2.5 μl of 500 mM EDTA pH 8.0 and placing the incubation tubes on ice for 10 min. 2D electrophoresis was performed, and recombination intermediates were detected at mbs1 as described in (8).
RESULTS
Roles of Nse5–Nse6 in mitotic DNA double-strand break repair
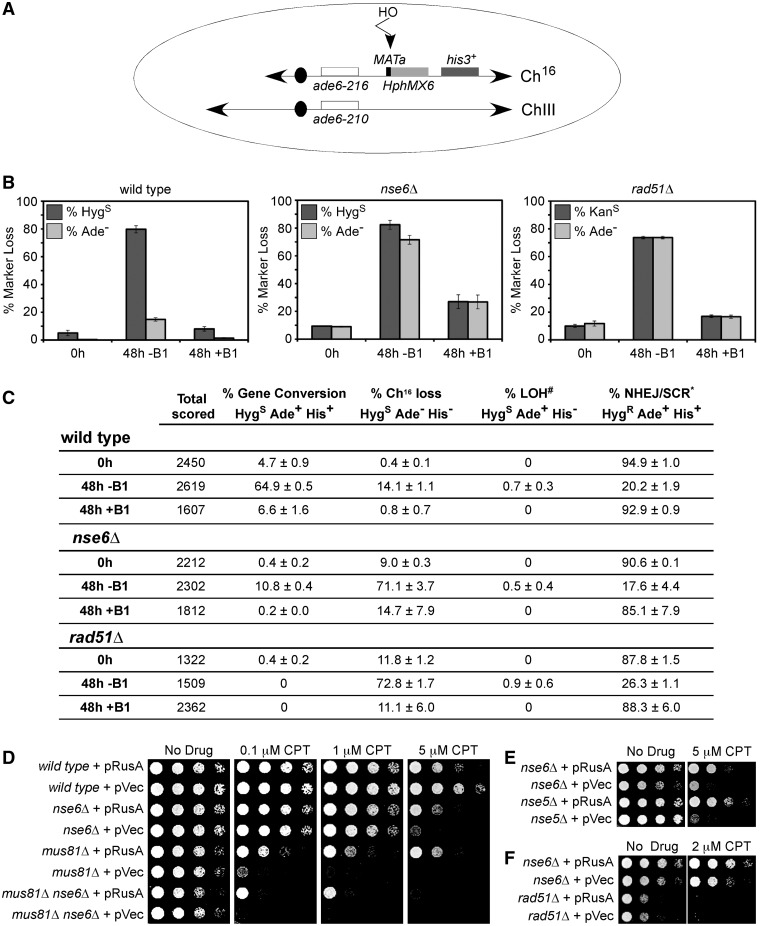
The budding yeast and human Smc5–Smc6 complexes have been implicated in the efficient repair of an enzymatically induced DNA DSB (36–38). To determine whether the fission yeast Nse5–Nse6 functionally interdependent heteromeric complex (35) plays an analogous role, we tested the DSB repair capacity of nse6Δ cells using a derivative of the non-essential minichromosome Ch16, which contains the MATa target site for the S. cerevisiae HOmothallic switching (HO) endonuclease (see schematic in Figure 1A, described in (58)). Given the relatively mild IR sensitivity of fission yeast nse6Δ cells versus the extreme sensitivity of rad51Δ cells (35), it was surprising that both nse6Δ and rad51Δ cells were highly defective in the HR-based repair of the HO-induced break. Unlike wild-type cells, which repaired the majority of the HO-induced DSBs by gene conversion, the nse6Δ and rad51Δ mutants instead lost nearly all of the HO cut minichromosomes (Figure 1B and C (58,70)). This result indicates that in fission yeast, Smc5–Smc6, is required for the repair of enzymatically induced DSBs.
Figure 1.
Mitotic roles for the Smc5–Smc6 complex. (A) Schematic of the HO-induced DSB minichromosome system. The minichromosome Ch16 and full-length Ch III with their centromeric regions (black ovals), histidine marker his3+ (dark grey box) and complementing ade6 heteroalleles (ade6-216 and ade6-210 (86); white boxes) are shown along with a MATa site for HO DSB formation (black rectangle) and the adjacent hphMX6 (hygromycin)-resistance marker (light gray box) for wild-type and nse6Δ cells. As rad51 was deleted using the hygromycin-resistance marker, a minichromosome bearing a kan (kanamycin)-resistance marker was used in this background. MATa is ∼25 kb from the ade6 marker. Removal of thiamine (B1) derepresses the HO gene, whose product generates a DSB at the MATa target site (vertical arrowhead). HO-induced DSBs can result in (i) loss of Ch16 with diagnostic Ade− and HygS phenotype, (ii) repair of the DSB by interchromosomal gene conversion using Ch III as a template, resulting in the loss of MATa and hphMX6 cassette and an Ade+ and HygS phenotype or (iii) maintenance of the initial Ade+ HygR phenotype, if the DSB is repaired by either non-homologous end-joining (NHEJ) or sister chromatid repair. The HO system can also assay levels of spontaneous minichromosome Ch16 loss, by scoring marker loss in the absence of HO induction (0 h, and 48 h culturing in thiamine). (B) and (C) Genetic analysis of site-specific DSB repair in wild-type (left panel), nse6Δ (middle panel) and rad51Δ (right panel) backgrounds. Percentage marker loss is given for cells grown in repressive (0 h and 48 h, +B1) or derepressive media (48 h, –B1). Data are means and standard errors from three independent experiments. #Loss of heterozygosity (LOH) can occur through various mechanisms, as described in (58). *Sister chromatid repair (SCR) also results in a HygR Ade+ phenotype. (D) RusA expression partially rescues the CPT sensitivity of nse6Δ, mus81Δ and nse6Δ mus81Δ mutants. (E) and (F) RusA expression partially rescues the CPT sensitivity of nse5Δ and nse6Δ, but does not rescue that of rad51Δ cells. (D–F) Five-fold serial dilutions of the indicated strains were plated onto medium that lacked thiamine to derepress plasmid-borne gene expression. Cells were either untreated or treated with the indicated concentrations of CPT and grown at 32°C for 3 days. pVec denotes an empty vector (87).
To test for an additional role of Nse6 in mitotic DNA break repair, we determined the sensitivity of nse6Δ mutants to CPT, which stabilizes topoisomerase I–DNA covalent complexes and can induce fork breakage during replication (30). The nse6Δ mutant was sensitive to CPT, although not as sensitive as mus81Δ or the mus81Δ nse6Δ double mutant [(71) and Figure 1D]. Although the expression of E. coli RusA HJ resolvase is mildly toxic to wild-type cells, as observed previously (72), the CPT sensitivities of both nse6Δ and mus81Δ cells were suppressed by RusA expression (Figure 1D). Notably, the nse5Δ mutant has a CPT sensitivity similar to that of nse6Δ, which is also rescued by RusA expression (Figure 1E). This result indicates a role for Nse5–Nse6, like Mus81, in HJ resolution. The mus81Δ nse6Δ double mutant was also suppressed by expression of RusA, although not as well as the single mutants, indicating that Nse5–Nse6 and Mus81 have distinct and overlapping roles. Although nse6Δ behaves like rad51Δ in the HO-induced DSB assay (Figure 1B and C (58)), the hypersensitivity of rad51Δ cells to CPT was not suppressed by RusA expression (Figure 1F).
Nse5–Nse6 executes an essential function during meiosis
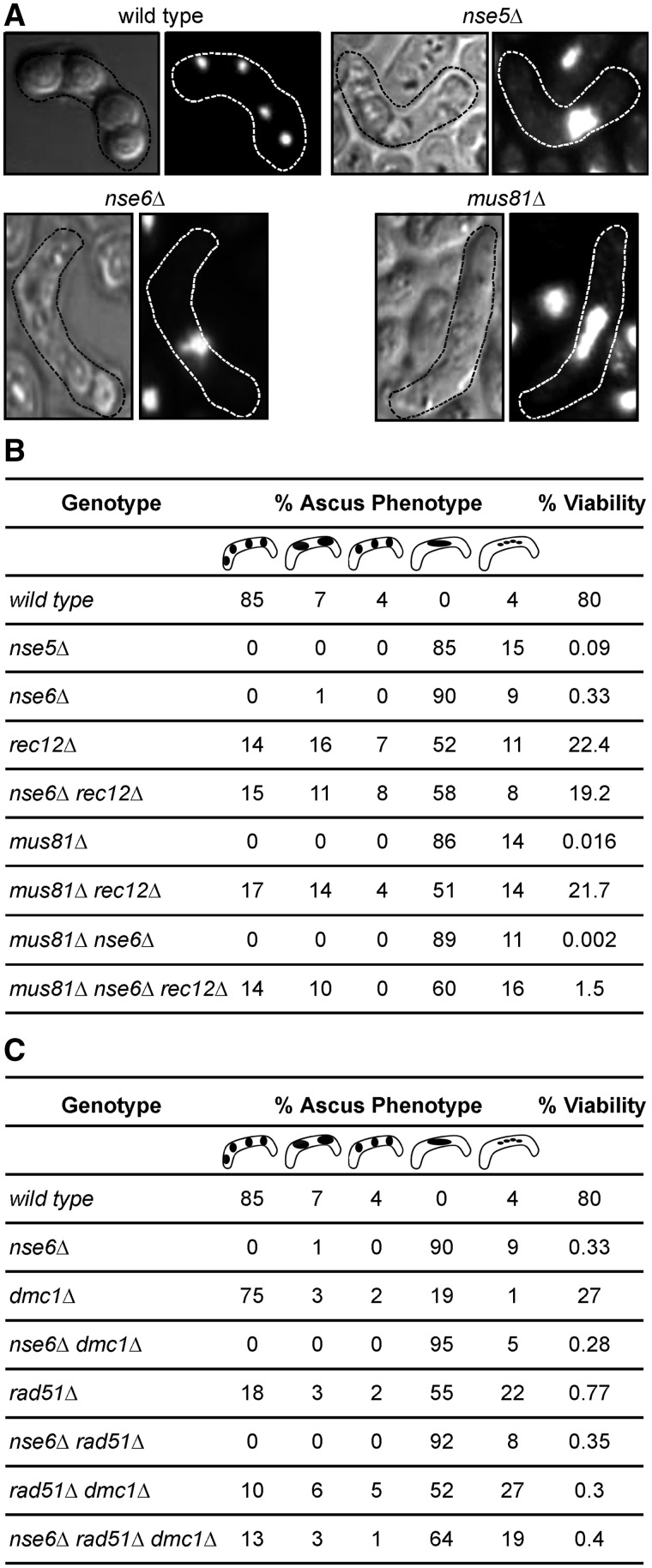
Due to the similarity of the mitotic phenotypes of nse5Δ, nse6Δ and mus81Δ cells, we tested the role of Nse5–Nse6 in meiosis, an HJ-generating process that is heavily dependent on Mus81–Eme1 resolution activity (8,15,23). Wild-type meiosis produces asci with four haploid spores, with DNA present in each spore (Figure 2A). Asci from nse5Δ and nse6Δ mutant crosses were largely devoid of well-defined spores but infrequently contained one single large spore (Figure 2A), much like mus81Δ asci (15). This result shows that Nse5–Nse6 plays an important role in meiosis.
Figure 2.
The Smc5–Smc6 complex acts after DSB formation and strand invasion during meiotic recombination. (A) Cells lacking Nse6 or Nse5 form aberrant asci following meiosis. Mature asci (phase microscopy on the left or DAPI stained on the right) outlined with a dashed line, from genetic crosses of wild-type, nse5Δ, nse6Δ and mus81Δ cells. Two days after mating, wild-type asci show four spores of equal size, each containing DNA. nse5Δ, nse6Δ and mus81Δ mutants do not form well-defined spores, with progeny often having only one large spore. (B) The low spore viability of nse6Δ, mus81Δ and the nse6Δ mus81Δ double mutant is rescued by deleting rec12. The percentage of asci with either a wild-type appearance (four well-defined spores of equal size) or aberrant phenotypes including triads, dyads (typical for rec12Δ), or a single large spore, or four very small spores (as observed in nse6Δ cells) was determined. Spore viabilities were scored for crosses of the indicated genotypes. Data shown are the averages from three independent crosses, and around 100 asci were counted for each cross. (C) Nse6 acts after the Rad51 and Dmc1 strand-exchange proteins. The percentage of asci produced from a genetic cross of rec12Δ, rad51Δ, dmc1Δ and mus81Δ strains (either as single or double mutants) in the presence or absence of Nse6 is shown. Black areas indicate spores of various sizes and shapes.
If Nse5–Nse6, like Mus81–Eme1, promotes JM resolution, then preventing the early stages of meiotic recombination should render Nse5–Nse6 unnecessary for meiotic nuclear division. For these tests, we first eliminated Rec12, which is essential for meiotic DSB formation and recombination (68). Asci from rec12Δ crosses were very heterogeneous: Some asci had four equal-sized spores like wild-type, some lacked spores or had only one like nse6Δ and many had two spores (dyads) like rec12Δ (Figure 2B; (7,15)). As anticipated, spore viability from homozygous nse5Δ or nse6Δ crosses was extremely low: Only ∼0.5% of spores germinated and produced colonies, compared to ∼80% for wild-type (Figure 2B (15)). Notably, both rec12Δ and rec12Δ nse6Δ crosses yielded similar spore viabilities of ∼20%, and, furthermore, their ascus morphologies were indistinguishable (Figure 2B). The same was true for rec12Δ and rec12Δ mus81Δ crosses. The rec12Δ nse6Δ and rec12Δ mus81Δ double mutants had indistinguishable defects from those observed in a rec12Δ deletion, indicating that rec12Δ is epistatic to both nse6Δ and mus81Δ in meiosis.
The morphology of asci produced by crosses of nse6Δ, mus81Δ and mus81Δ nse6Δ double mutants were indistinguishable (Figure 2B). However, the viability of spores from mus81Δ nse6Δ crosses was ∼8-fold lower than that of spores from crosses of the single mutants (Figure 2B). Deletion of rec12 rescued the mus81Δ nse6Δ meiotic ascus phenotype, but only partially rescued the low spore viability phenotype, between 1 and 2% spore viability. Nevertheless, the spore viability in the mus81Δ nse6Δ rec12Δ triple mutant was about 1000-fold higher than that in the mus81Δ nse6Δ double mutant (P < 0.0001). These results indicate that Nse5–Nse6, like Mus81, acts in meiotic recombination after DSB formation by Rec12 (i.e. in the repair of DSBs).
Next, we exploited the aberrant ascus morphology of nse6Δ cells to determine whether Nse5–Nse6 acts before or after strand invasion promoted by the RecA homologs Rad51 and Dmc1 (4,5). Cells lacking Rad51 fail to repair many meiotic DSBs (73) but can produce asci containing four discrete, although mostly inviable, spores (15), presumably because there are no physical linkages, such as HJs, between the chromosomes to prevent their segregation (Figure 2C). Although not essential for meiotic progression and abundant formation of viable spores, Dmc1 is required for normal levels of meiotic recombination in most genetic intervals (4,9). In contrast to rad51Δ or dmc1Δ single mutants, four-spore asci were not observed in the nse6Δ single or rad51Δ nse6Δ or dmc1Δ nse6Δ double mutants (Figure 2B and C). However, the dmc1Δ rad51Δ double and nse6Δ dmc1Δ rad51Δ triple mutants produced similar asci that sometimes contained four spores, which are not observed for the nse6Δ single mutant (Figure 2B and C). Thus, suppression of the nse6Δ phenotype (to a level equivalent to that of the dmc1Δ rad51Δ double mutant) requires elimination of both strand-exchange proteins. These results indicate that Nse6 acts after strand exchange by Rad51 and Dmc1, perhaps during JM resolution.
Genetic and physical analyses of meiotic crossing over in nse6Δ cells
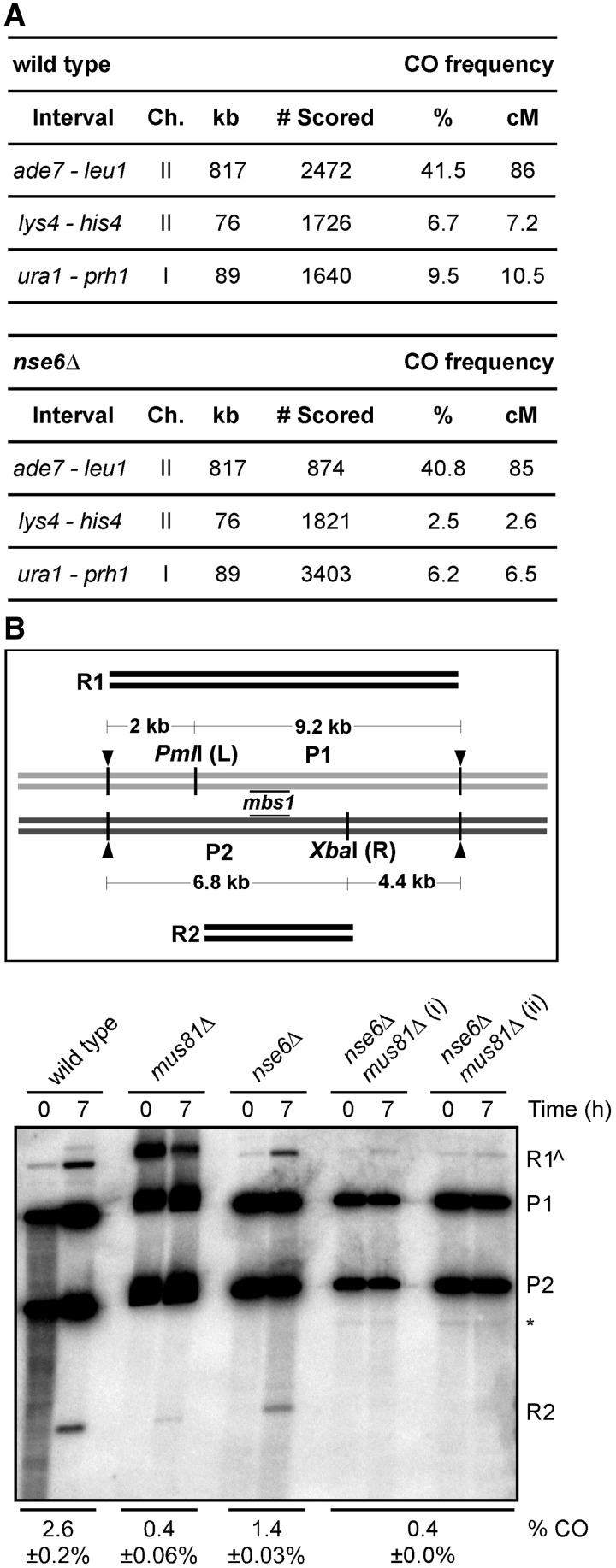
Next, we tested the impact of absence of Nse6 on meiotic recombination in three genomic intervals. In the large ade7–leu1 interval, frequency of crossover among viable spores from a wild-type cross was 41.5%, not significantly different from the 40.8% in nse6Δ mutants (Figure 3A). In two shorter intervals, lys4–his4 and ura1–prh1 (which contains the mbs1 DSB hotspot used in DNA analyses below (61)), frequencies of crossover were reduced by a factor of about 2 in nse6Δ mutants, relative to wild-type (Figure 3A), a statistically significant reduction (P < 0.0001). However, in contrast to the reductions by factors of 20–100 in mus81Δ cells (8,26), the meiotic crossover defects of nse6Δ cells are mild.
Figure 3.
Effect of nse6Δ mutation on the frequency of meiotic crossovers. (A) Crossover (CO) frequencies in three intervals in the wild-type and nse6Δ backgrounds. The percent of recombinant spore colonies was converted into genetic distance in centimorgans with Haldane’s equation. The number of colonies analyzed is the total from at least three independent experiments. (B) Amount of crossover DNA generated at the mbs1 hotspot (8). Diploid pat1-114 wild-type (GP6656), mus81Δ (GP6657), nse6Δ (GP6234) and nse6Δ mus81Δ (GP7765) strains with heterozygous restriction sites PmlI (L) and XbaI (R) flanking mbs1 were assayed. Digestion with these enzymes and PvuII (black arrowheads), which cuts outside L and R, gives two parental DNA fragments [9.2 kb (P1) or 6.8 kb (P2)] and two recombinant DNA fragments [11.2 kb (R1) or 4.8 kb (R2)] detected by the Southern blot hybridized probe (black lines at mbs1). The crossover DNA was measured at 7 h, after JM resolution in wild-type strains (Figure 4A, top); a pre-meiotic 0 h is shown as a control. Because R1 can also arise from incomplete digestion (R1^) at either L or R, the frequency of crossover DNA (% CO) was calculated as two times the amount of R2 DNA divided by the amount of total DNA. DNA from two independent inductions of strain GP7765 (nse6Δ mus81Δ) are shown. The means of two to three experiments (some from additional unpublished experiments) with the range or standard error of the mean are given. The asterisk indicates a non-specific cross-hybridization band.
Through genetic methods, we can measure crossovers only in the ∼0.5% of spores that are viable after nse6Δ meiosis; thus, our observed frequencies may overrepresent the frequencies in most of the cells. To circumvent this limitation, we physically measured the crossovers between two markers closely flanking the DSB hotspot mbs1 in the entire meiotic population, as previously analyzed in mus81Δ mutants (8). The crossover-specific product R2 (Figure 3B) was quantified in wild-type, mus81Δ, nse6Δ and nse6Δ mus81Δ strains at 7 h after induction of synchronous meiosis. Consistent with previous data (8), there was a severe defect, a reduction by a factor of 7, in crossover formation in mus81Δ and nse6Δ mus81Δ cells (Figure 3B). Importantly, in nse6Δ cells, crossover formation was reduced by a factor of about 2, as in the genetic assay for ura1–mbs1–prh1 recombinants (Figure 3A), suggesting that Nse5–Nse6 promotes but is not absolutely essential for meiotic crossover formation (Figure 3B).
Recombination intermediates (DNA joint molecules) accumulate in nse6Δ mutants
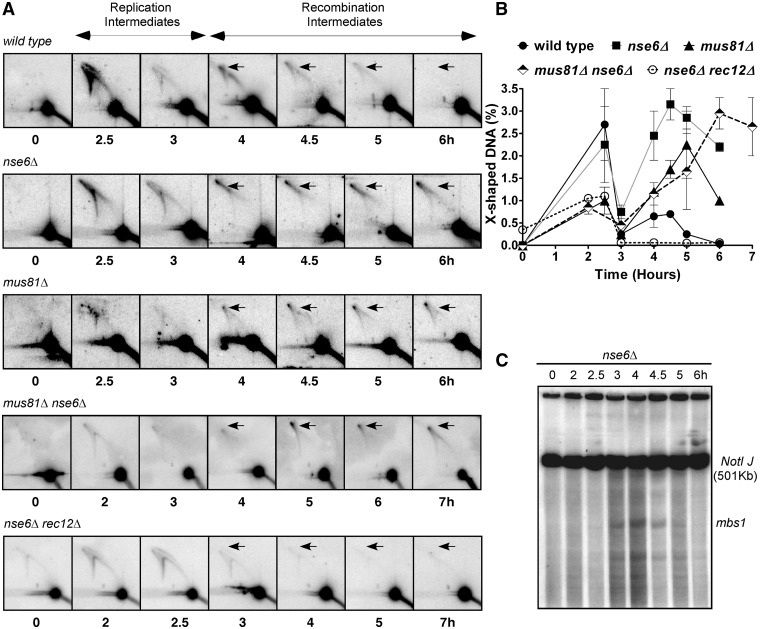
The similarities of the vegetative and meiotic phenotypes of mus81Δ and nse6Δ led us to investigate whether DNA JMs accumulate during meiosis in nse6Δ, as they do in mus81Δ (8). Synchronous meiosis was induced in pat1-114 nse6Δ and nse6Δ mus81Δ diploids, and the formation of branched DNA structures, indicative of both replication and recombination intermediates, was analyzed via 2D gel electrophoresis as previously performed for mus81Δ (8). Branched DNA structures produced during replication were visible at 2.5 h and decreased toward 3 h (Figure 4A and B), the time at which DNA content shifts from 2n to 4n as measured by flow cytometry ((8); our unpublished data). After replication was complete and DSB formation had begun (Figure 4C), the amount of X-shaped DNA increased again at 4 h, persisted at 4.5 h and disappeared after 5 h in the wild-type strain (Figure 4A and B), as expected for the formation and resolution of HJs. In the nse6Δ strain, the recombination intermediates were formed with the same timing as in wild-type, but they did not disappear; the same was true for mus81Δ and nse6Δ mus81Δ mutants, although the double mutant showed a slight delay in JM formation (Figure 4A and B; (8)). As expected, early but not late appearing JMs were detected in an nse6Δ rec12Δ haploid double mutant (haploid and diploid analyses are indistinguishable (74) and our unpublished data). This result indicates that, as in mus81Δ mutants (8), the early arising JMs are replication derived, and the late arising JMs are recombination intermediates and are consistent with the meiotic phenotype of nse6Δ depending on DSB formation by Rec12 (Figure 2B). Thus, both nse6Δ and mus81Δ mutants accumulate recombination intermediates with a similar timing during meiosis.
Figure 4.
Joint molecules accumulate during meiosis in the nse6Δ mutant. (A) Diploid pat1-114 wild-type (GP6656), nse6Δ (GP6234), mus81Δ (GP6657) and nse6Δ mus81Δ (GP7765) and haploid pat1-114 nse6Δ rec12Δ (GP7773) strains were induced for meiosis. DNA was extracted at the time indicated and digested with PvuII to generate 11.2 kb DNA fragments containing mbs1 (Figure 3B). Branched DNA molecules were assayed by two-dimensional gel electrophoresis (8,88) in which the first dimension slowly separates DNA primarily by mass and the second dimension primarily by shape, as branched structures have less mobility than linear DNA, and subsequent Southern blot hybridization. DNA from 2.5–3 h shows branched DNA structures arising from replication, while at 4 h, the structures are primarily recombination intermediates (8). Note that X-shaped DNA (arrows) forms in the wild-type at 4 h and disappears by 6 h, as expected for HJs that form and are resolved during the repair of DSBs, but form and persist in nse6Δ, mus81Δ and nse6Δ mus81Δ mutants. (B) Quantification of the X-shaped DNA observed in (A). Each datum is the mean of two independent experiments, with the error bars indicating the range. (C) Meiotic DSBs arise and disappear with wild-type timing in a diploid nse6Δ strain GP6234. DNA prepared at the indicated time after induction of meiosis was digested with NotI, separated by pulsed-field gel electrophoresis and analyzed by Southern blot hybridization with a probe specific to the left end of the 501 kb NotI fragment J, which contains mbs1 near its middle. Note that DSBs appear after replication at 3 h, reach a maximum at 4 h and are mostly repaired by 5 h, consistent with previous results from wild-type and mus81Δ strains (8,9,68,69) and with the timing of HJ formation observed in (A).
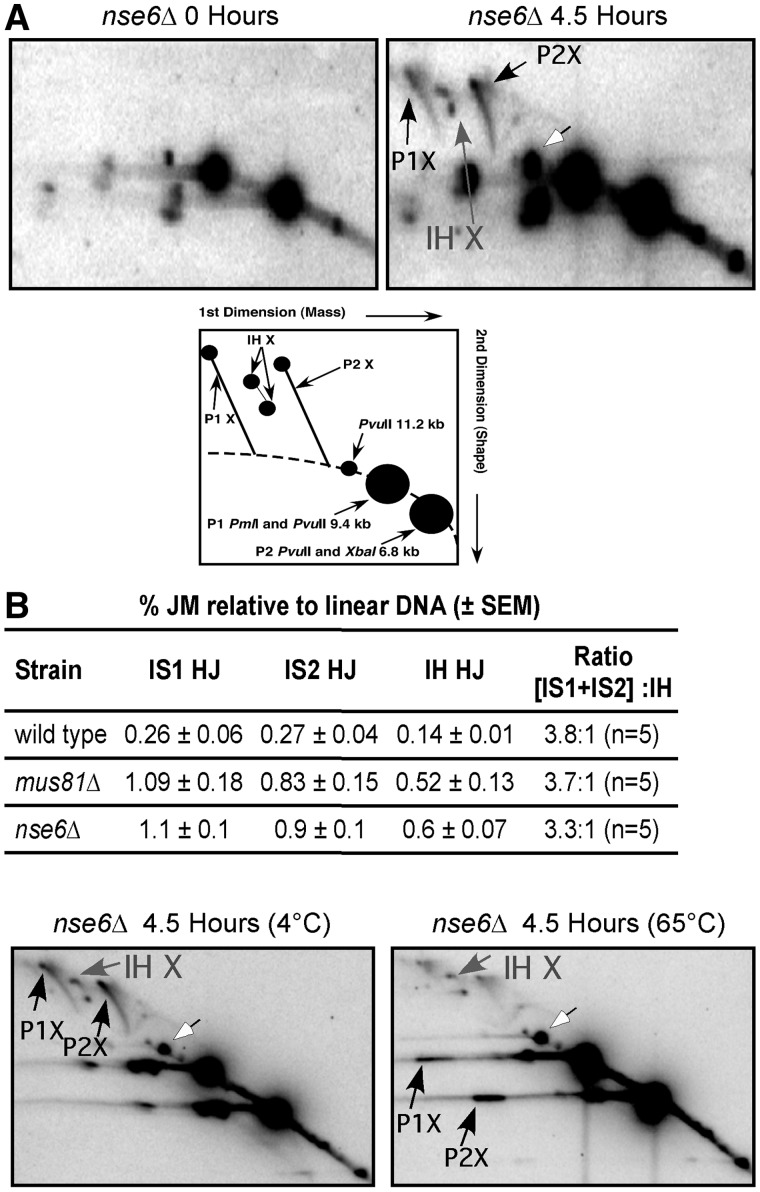
Next, we determined whether the JMs that accumulate in the nse6Δ mutant specifically form between sister chromatids or between homologs, and whether they are single HJs, as they are in a mus81Δ mutant (8), and not double HJs, a plausible alternative. A strain with heterozygous restriction site mutations that flank the DSB hotspot mbs1 was used to assay the relative interhomolog (IH) and intersister (IS) HJs formed during DSB repair. As was the case in the mus81Δ strain (8,9), IS HJs outnumbered IH HJs by ∼3.5:1 (Figure 5A). To distinguish single versus double HJs, the DNA in the agarose gel after the first dimension of electrophoresis was heated to 65°C to promote branch migration. The unequal length arms of homologous DNA in single HJs, which have two recombinant DNA strands, prevent the junction from migrating past the ends of the arms; the absence of recombinant DNA strands in double HJs and the equal length homologous arms of IS HJs allow their junctions to readily migrate off the end (8). When the DNA was heated, the IS HJs readily branch migrated and disassociated into the linear forms, whereas the IH HJs were resistant and remained intact (Figure 5B). This stability indicates that the IH-branched DNA structures are single HJs. This is the same behavior observed previously for branched DNA that accumulates in the mus81Δ mutant (8). These physical analyses further support a role for Nse6 in the resolution of DNA JMs during meiosis.
Figure 5.
IS and IH JMs accumulate in nse6Δ cells. (A) DNA prepared before (0 h) or 4.5 h after meiotic induction was digested with PvuII, PmlI and XbaI and separated by 2D-gel electrophoresis and Southern blot hybridized to assay for IS and IH JMs at mbs1 (8). DNA fragments with different masses are created with heterozygous restriction site mutations (depicted in cartoon). Parental fragments of 9.2 or 6.8 kb give rise to IS JMs (black arrows) that contain 18.4 or 13.6 kb of DNA, respectively. IH JMs (gray arrows) have an intermediate mass of 16 kb. Both IS and IH JMs accumulate, and the IS JMs outnumber the IH JMs 3.3 to 1, consistent with the 3.8:1 ratio previously observed in a mus81Δ mutant (8). The mean ± standard error of the mean of five experiments is indicated. (B) Branch migration of the JMs from the nse6Δ mutant, like those from mus81Δ, suggests single, not double, HJs. DNA prepared at 4.5 h after meiotic induction of an nse6Δ strain (GP6234) was digested as in (A). After the first dimension of electrophoresis, the DNA, in agarose, was incubated at either 4 or 65°C for 6 h, and the DNA was then subjected to the second dimension of electrophoresis. As expected, no detectable branch migration of JMs occurred at 4°C, but at 65°C, both IS JM species almost completely dissociated into parental length DNAs (black arrows). The IH JMs (gray arrows), however, remained intact, which is the characteristic of single HJs, as previously observed in a mus81Δ mutant (8). White arrows in (A) and (B) indicate partial digestion by either PmlI or XbaI, which results in the full-length 11.2 kb PvuII restriction fragment (the same as R1 in Figure 3B).
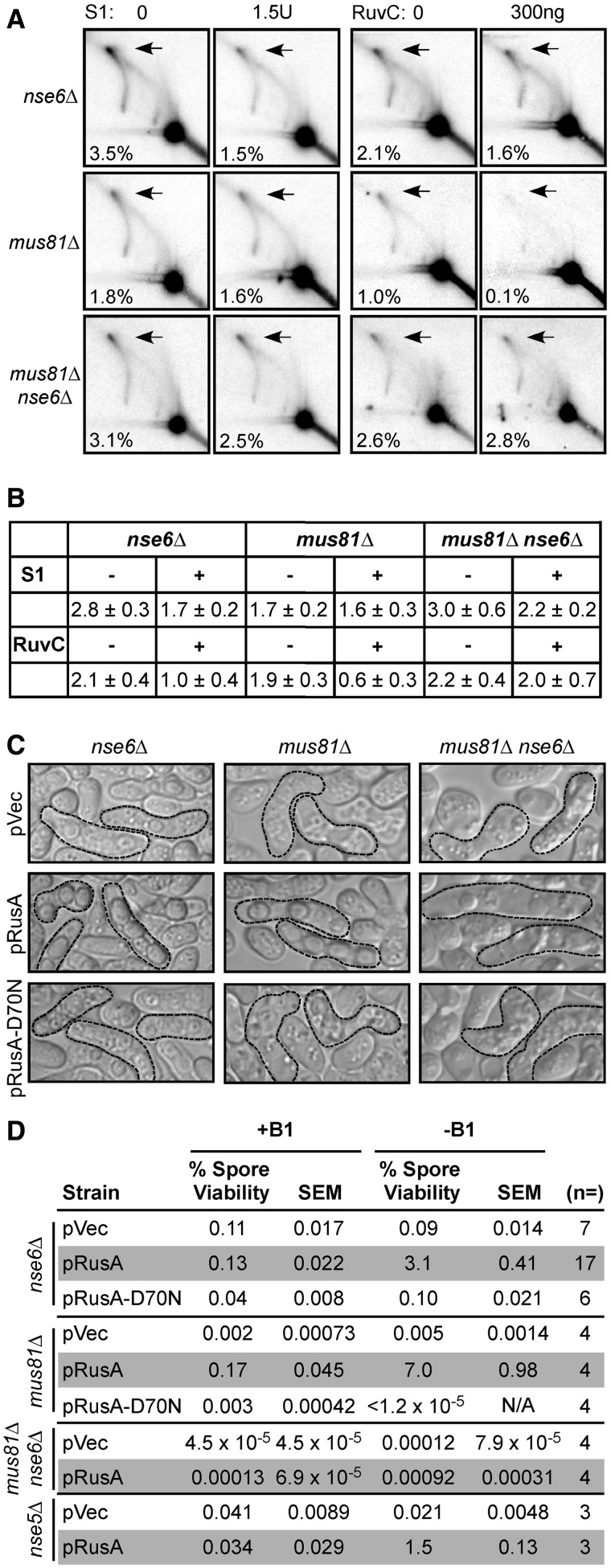
The X-shaped DNA structures seen in the 2D gel assays could be either HJs or a related four-stranded DNA structure, a hemicatenane, with two single DNA strands, or two pairs of strands, interwound. We first tested the sensitivity of these structures to the E. coli HJ resolvase RuvC (75). The JMs of the nse6Δ and mus81Δ strains were similarly sensitive to RuvC (Figure 6A and B), indicating that at least some of the X-shaped structures formed in both mus81Δ and nse6Δ strains are the same and are HJs. However, the JMs of the nse6Δ mus81Δ strain were largely resistant, suggesting that these X-shaped structures are not HJs but might be related structures such as hemicatenanes.
Figure 6.
nse6Δ Mutants fail in meiosis because of DNA joint molecule accumulation. (A) DNA prepared at 4.5 or 5 h (nse6Δ and mus81Δ) or 6 h (nse6Δ mus81Δ) after meiotic induction was digested with PvuII and treated with the indicated amounts of either the ss DNA-specific S1 nuclease or the purified RuvC and assayed by two-dimensional gel electrophoresis and Southern blot hybridization using the probe shown in Figure 3B. The percentage given is the amount of JMs, indicated by arrows, divided by the amount of total DNA. All experiments shown were performed concurrently. (B) The mean of two to five experiments performed as in (A), with either the range or standard error of the mean indicated. (C) nse6Δ, mus81Δ or mus81Δ nse6Δ mutants carrying an empty vector (pVec), pRusA or catalytically inactive pRusA-D70N were mated and sporulated, and the resultant asci photographed. Mature asci, outlined with a dashed line, arose from a meiotic cross of the indicated cells in medium lacking thiamine (B1) to allow vector-borne gene expression. (D) Spore viabilities were determined for nse5Δ, nse6Δ, mus81Δ or mus81Δ nse6Δ mutants bearing the indicated vectors and grown in media that either repressed (+B1) or derepressed (-B1) plasmid-borne gene expression. Spore viability data are the means from 3–17 independent experiments.
Although structurally similar to HJs, hemicatenanes have exposed ss DNA that would be susceptible to S1, a nuclease more active on ss DNA than on double-stranded DNA (76). To test this possibility, meiotic DNA prepared at 4.5 or 5 h after induction of mus81Δ or nse6Δ strains was treated with S1 before 2D gel electrophoresis. The accumulated branched DNA structures from the mus81Δ strain were insensitive to S1 treatment, but those from the nse6Δ and nse6Δ mus81Δ strains were partially sensitive (Figure 6A and B). We propose that both HJs and an S1-sensitive form of DNA, such as hemicatenanes, accumulate in nse6Δ mutants but only HJs accumulate in mus81Δ mutants during meiosis (Figure 6A and B).
Suppression of nse5Δ and nse6Δ meiotic defects by bacterial resolvase RusA
Our biochemical analyses provide evidence that nse6Δ mutants accumulate meiotic HJs, as observed in mus81Δ mutants (8,15). For in vivo validation of this conclusion, we used the RusA resolvase to probe the nature of the meiotic impediment in nse5Δ and nse6Δ cells. Our laboratories and others have shown that heterologous expression of E. coli RusA reduces the level of HJs that accumulate either on treatment with various DNA damaging agents or during meiosis in various DNA repair mutants (15,26,30–32). We expressed RusA and its catalytically inactive mutant, D70N, in nse6Δ cells and analyzed both spore formation and viability. Strikingly, RusA expression significantly rescued the meiotic defects of nse6Δ cells, yielding asci that often contained a wild-type complement of four spores (Figure 6C). This rescue required RusA endonuclease activity, as the RusA-D70N catalytically inactive mutant provided no benefits (Figure 6C). The spore viability was increased more than 20-fold when RusA was expressed in nse6Δ cells compared to an empty vector control (Figure 6D). Spore viability was also increased in nse5Δ cells (Figure 6D). Likewise, although more dramatic due to their initial lower viability, mus81Δ spore viability was increased more than 800-fold by RusA expression. Interestingly, spore viability was around 1.5% in nse5Δ and 3% in nse6Δ mutants expressing RusA, which was significantly lower (P < 0.005) than in mus81Δ-expressing RusA (7%; Figure 6D). This result is consistent with the accumulation of both HJs and an S1-sensitive form of DNA, such as hemicatenanes, in nse5Δ and nse6Δ meiosis, but only HJs in mus81Δ meiosis (Figure 6A and B). RusA expression did not so strongly improve the ascus morphology or spore viability in the nse6Δ mus81Δ double mutant (Figure 6C and D), which is consistent with the RuvC insensitivity of DNA isolated from these cells (Figure 6A and B). Overall, the remarkably similar suppression of both the ascus morphology and spore viability defects in nse5Δ or nse6Δ and mus81Δ by RusA endonuclease suggests that Nse5–Nse6 plays a role in facilitating the HJ resolvase activity of Mus81–Eme1.
DISCUSSION
Failure of the HR pathways underlies many human diseases including cancer and can cause birth defects through aberrant meiotic chromosome segregation. Here, we have identified a novel function for Nse5–Nse6 of the Smc5–Smc6 complex in processing mitotic and meiotic HR intermediates. Genetic and physical analyses indicate that nse6Δ mutants accumulate JMs resembling the HJs that accumulate in mus81Δ during meiosis. Thus, to our knowledge, Nse5–Nse6 is only the second factor in fission yeast required for the endonucleolytic processing of HJs, besides Mus81–Eme1, which has the catalytically active site for HJ resolution.
The key data supporting a function for Nse5–Nse6 in the resolution of JMs are as follows: (i) like mus81Δ mutants (15), nse6Δ mutants form few viable spores (Figure 2), due to the failure of chromosome segregation; (ii) like mus81Δ (15), rec12Δ is epistatic to nse6Δ in meiosis, demonstrating that Nse5–Nse6 acts after DSB formation (Figure 2); (iii) the double nse6Δ mus81Δ mutant also forms few viable spores and is suppressed, at least partially, by rec12Δ, indicating that Mus81 and Nse6 act in the same or closely related steps of meiosis (Figure 2); (iv) the rad51Δ dmc1Δ combination is epistatic to nse6Δ, demonstrating that Nse5–Nse6 acts after the formation of JMs (Figure 2); (v) JMs accumulate in meiotic nse6Δ and nse6Δ mus81Δ cells, and these JMs are temporally, genetically and electrophoretically indistinguishable from the HJs that accumulate in mus81Δ cells (Figures 4 and 5, (8)); (vi) at least some of these JMs are sensitive to E. coli RuvC HJ resolvase (Figure 6) and to RusA HJ resolvase, because, as for mus81Δ cells (15), expression of RusA partially rescues the meiotic defects of nse6Δ cells (Figure 6C and D) and (vii) crossovers are modestly reduced in nse6Δ cells, indicating that Nse5–Nse6 is important but not absolutely essential for HJ resolution, which requires Mus81–Eme1 (Figure 3).
In addition to HJ resolution, Nse5–Nse6 appears to have a second role because (i) the double nse6Δ mus81Δ mutant grows more slowly and is more CPT sensitive than either single mutant (Figure 1D); (ii) the double mutant is not quite as well suppressed to CPT resistance by expression of RusA as the single mutants (Figure 1D); (iii) unlike mus81Δ, nse6Δ is defective for mitotic DSB repair, as is rad51Δ (Figure 1); (iv) crossovers are formed, although at reduced level compared to wild-type, in nse6Δ but not in mus81Δ or nse6Δ mus81Δ mutants (Figure 3) and (v) the JMs that accumulate in nse6Δ and nse6Δ mus81Δ meiotic cells are at least partially sensitive to S1 nuclease, whereas those that accumulate in mus81Δ meiotic cells are not (Figure 6A and B), suggesting that Mus81–Eme1 and Nse6 have distinct, and overlapping, roles.
To reconcile these observations, we propose that in meiotic cells Nse5–Nse6 stimulates the resolution of HJs by Mus81–Eme1 and the resolution of other structures, such as hemicatenanes, which may arise during mitotic growth or during meiosis in the absence of Mus81–Eme1. These structures may arise during mitotic replication, as suggested by others (45,77), thereby partially accounting for the mitotic phenotypes of nse6Δ mutants. The structure of the JMs that accumulate in nse6Δ and nse6Δ mus81Δ mutants is not entirely clear, although those that accumulate in mus81Δ mutants are clearly single HJs (8). The JMs that accumulate in nse6Δ mutants are sensitive to both S1 nuclease and RuvC HJ resolvase, whereas those that accumulate in mus81Δ mutants are sensitive to RuvC and those in nse6Δ mus81Δ mutants are sensitive to S1 nuclease (Figure 6A and B). The stimulation of Mus81 HJ resolvase activity by the Nse5–Nse6 complex may be direct or indirect.
We propose that in nse6Δ mutants, Mus81–Eme1 slowly resolves HJs while some are converted into another structure and that in nse6Δ mus81Δ mutants, most or all of the HJs are converted into this structure. This proposal is consistent with the slight reduction in meiotic crossover frequency, among the few viable spores that arise, in nse6Δ mutants but strong reduction in mus81Δ mutants (Figure 3A (8,23,26)); with the slight reduction in total crossover DNA in nse6Δ mutants but strong reduction in mus81Δ and nse6Δ mus81Δ mutants (Figure 3B (8)); and with the suppression of nse6Δ, but not nse6Δ mus81Δ, by expression of the RusA HJ resolvase (Figure 6C and D).
The resistance to branch migration of the IH JMs in nse6Δ mutants suggests that these JMs, between heterozygous restriction sites, are single HJs with recombinant length strands (Figure 5B). This is because either double HJs, which have parental length strands, or hemicatenanes with either recombinant or parental length strands should dissociate into separate duplexes on heating (e.g. (8,76,78)). IS JMs would dissociate in any case, as observed. IH and IS JMs may differ in structure, but their low level has precluded our determining their sensitivity to S1 nuclease and RuvC. Although the S1 nuclease sensitivity is consistent with some JMs being hemicatenanes, to our knowledge, this structure has not been clearly demonstrated to arise in cells, for example by electron microscopy or comparison with synthetic DNA molecules. Further investigation is required to establish the structure of the population of non-HJ containing JMs in the absence of Nse6.
As Nse5–Nse6 acts as part of the Smc5–Smc6 complex, which has multiple roles in chromosome metabolism (44), it is no surprise that Nse5–Nse6 has multiple roles. One tempting hypothesis is that during meiosis, the primary role of Nse5–Nse6 is to stimulate the Mus81–Eme1 HJ resolvase and that during mitotic growth it plays both this and a second role. During both stages of the life cycle, expression of RusA suppresses at least partially the nse6Δ phenotype (Figures 1D and 6C and D), indicating that HJ resolution is stimulated by Nse5–Nse6 in both stages. The second role of Nse5–Nse6 might be to regulate any of the multiple functions of the Smc5–Smc6 complex (44). Further investigation is required to elucidate this function, but the requirement for Rad51 and Nse6 but not Mus81–Eme1 in mitotic DSB repair (Figure 1) suggests that Nse5–Nse6 is also important for the synapsis phase of mitotic DSB repair.
Although the Nse1 and Nse2 E3 ligase activities, like Nse5–Nse6, facilitate mitotic DNA repair (79–81), neither of these E3 ligases is required for meiotic nuclear division and recombination [(82) our unpublished data]. Thus, although the mitotic functions of Nse1, Nse2 and Nse5–Nse6 have not been dissected, in meiosis, Nse5–Nse6 acts independently of posttranslational modifications catalyzed by Nse1 and Nse2. Nevertheless, on the basis of our previous analyses showing that hypomorphic mutants of the essential Smc5–Smc6 subunits exhibit catastrophic meioses (83), we propose that Nse5–Nse6 acts in conjunction with the Smc5–Smc6 holocomplex to execute its meiotic HR role.
Budding yeast smc5–smc6 mutations also disrupt meiotic nuclear division (84), but the underlying defects appear strikingly different from those of nse6Δ fission yeast. Key differences are that a spo11 (rec12 homolog) mutation is not epistatic to an smc5–smc6 mutation, and crossovers are not affected in smc6 temperature-sensitive (Ts) mutant cells (84). Thus, the authors concluded that the crucial role of Smc5–Smc6 is executed during premeiotic S phase in budding yeast and not after the initiation of meiotic recombination. It is unclear whether this reflects real differences in the functions of Smc5–Smc6 between species or if the unavoidable disruption of both the essential and repair roles in smc6 (Ts) budding yeast masks functions analogous to those of fission yeast Nse5–Nse6.
HJ resolution must be carefully controlled, both during mitotic growth and in meiosis. When a DNA strand lesion blocks mitotic replication, the fork can regress and form a structure whose center is identical to that of an HJ. Were it resolved by Mus81–Eme1, a DSB would be formed and require further processing to allow completion of replication. Alternatively, the regressed fork can migrate back to the original position and allow immediate continuation of replication. Thus, Mus81–Eme1 may be kept inactive during this time. If strand exchange between sisters or homologs forms an HJ that is present at the time of mitosis, HJ resolution would appear to be the most expedient means to allow chromosome segregation, and Mus81–Eme1 may be activated at this time [e.g. (19)]. Similarly, during meiosis, many dozens of HJs must arise to account for the ∼45 crossovers in a fission yeast cell (85), and Mus81–Eme1 is clearly highly active in meiotic cells. We surmise that Nse5–Nse6, likely as part of the Smc5–Smc6 complex, regulates directly or indirectly the activity of Mus81–Eme1 allowing it to function at the proper time and place to maintain chromosome integrity and cell viability.
FUNDING
Scholar Award from the Leukemia and Lymphoma Society (to M.N.B.); National Institutes of Health (NIH) of the United States of America [GM068608 and GM081840 to M.N.B. and GM032194 to G.R.S.]. Funding for open access charge: NIH, United States.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ken Marians for a generous supply of RuvC protein, Tim Humphrey for his kind gift of the minichromosome assay strain, Sue Amundsen for helpful comments on the article and Jay Patel for media preparation.
REFERENCES
- 1.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 2.Mimitou EP, Symington LS. DNA end resection—unraveling the tail. DNA Repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 4.Fukushima K, Tanaka Y, Nabeshima K, Yoneki T, Tougan T, Tanaka S, Nojima H. Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 2000;28:2709–2716. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 6.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 7.Davis L, Smith GR. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickson ID, Mankouri HW. Processing of homologous recombination repair Intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle. 2011;10:3078–3085. doi: 10.4161/cc.10.18.16919. [DOI] [PubMed] [Google Scholar]
- 13.Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;39:7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, III, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 16.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz A, West SC, Whitby MC. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 2010;38:1866–1873. doi: 10.1093/nar/gkp1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, III, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 24.Blais V, Gao H, Elwell CA, Boddy MN, Gaillard PH, Russell P, McGowan CH. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell. 2004;15:552–562. doi: 10.1091/mbc.E03-08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hope JC, Cruzata LD, Duvshani A, Mitsumoto J, Maftahi M, Freyer GA. Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe. Mol. Cell Biol. 2007;27:3828–3838. doi: 10.1128/MCB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81·Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair (Amst) 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 31.Bolt EL, Sharples GJ, Lloyd RG. Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol. 1999;286:403–415. doi: 10.1006/jmbi.1998.2499. [DOI] [PubMed] [Google Scholar]
- 32.Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc. Natl. Acad. Sci. USA. 2011;108:4944–4949. doi: 10.1073/pnas.1014240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Piccoli G, Torres-Rosell J, Aragon L. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- 34.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, III, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 37.De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 2006;8:1032–1034. doi: 10.1038/ncb1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell. 1999;10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvey SH, Sheedy DM, Cuddihy AR, O'Connell MJ. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell Biol. 2004;24:662–674. doi: 10.1128/MCB.24.2.662-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann AR. The role of SMC proteins in the responses to DNA damage. DNA Repair (Amst) 2005;4:309–314. doi: 10.1016/j.dnarep.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 42.McDonald WH, Pavlova Y, Yates JR, III, Boddy MN. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- 43.Morikawa H, Morishita T, Kawane S, Iwasaki H, Carr AM, Shinagawa H. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell Biol. 2004;24:9401–9413. doi: 10.1128/MCB.24.21.9401-9413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kegel A, Sjögren C. The Smc5/6 complex: more than repair? Cold Spring Harb. Symp. Quant. Biol. 2011;75:179–187. doi: 10.1101/sqb.2010.75.047. [DOI] [PubMed] [Google Scholar]
- 45.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 46.Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bermudez-Lopez M, Ceschia A, de Piccoli G, Colomina N, Pasero P, Aragon L, Torres-Rosell J. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 2010;38:6502–6512. doi: 10.1093/nar/gkq546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chavez A, Agrawal V, Johnson FB. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 2011;286:5119–5125. doi: 10.1074/jbc.M110.201608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chavez A, George V, Agrawal V, Johnson FB. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J. Biol. Chem. 2010;285:11922–11930. doi: 10.1074/jbc.M109.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi K, Szakal B, Chen YH, Branzei D, Zhao X. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2010;21:2306–2314. doi: 10.1091/mbc.E10-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyabe I, Morishita T, Hishida T, Yonei S, Shinagawa H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell Biol. 2006;26:343–353. doi: 10.1128/MCB.26.1.343-353.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, Dalgaard JZ, Aragon L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- 55.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 56.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 57.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cullen JK, Hussey SP, Walker C, Prudden J, Wee BY, Dave A, Findlay JS, Savory AP, Humphrey TC. Break-induced loss of heterozygosity in fission yeast: dual roles for homologous recombination in promoting translocations and preventing de novo telomere addition. Mol. Cell Biol. 2007;27:7745–7757. doi: 10.1128/MCB.00462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khasanov FK, Savchenko GV, Bashkirova EV, Korolev VG, Heyer W-D, Bashkirov VI. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics. 1999;152:1557–1572. doi: 10.1093/genetics/152.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Y, Smith GR. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatebe H, Goshima G, Takeda K, Nakagawa T, Kinoshita K, Yanagida M. Fission yeast living mitosis visualized by GFP-tagged gene products. Micron. 2001;32:67–74. doi: 10.1016/s0968-4328(00)00023-8. [DOI] [PubMed] [Google Scholar]
- 63.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 64.Bahler J, Wu J, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Phileppsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targetting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 65.Davis L, Rozalen AE, Moreno S, Smith GR, Martin-Castellanos C. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr. Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutz H, Heslot H, Leupold U, Lopreno N. In: Schizosaccharomyces pombe Handbook of Genetics. King RC, editor. Vol. 1. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 67.De Veaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 69.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 70.Tinline-Purvis H, Savory AP, Cullen JK, Dave A, Moss J, Bridge WL, Marguerat S, Bahler J, Ragoussis J, Mott R, et al. Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast. EMBO J. 2009;28:3400–3412. doi: 10.1038/emboj.2009.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Cloning, overexpression, purification, and characterization of the Escherichia coli RuvC Holliday junction resolvase. J. Biol. Chem. 1994;269:5187–5194. [PubMed] [Google Scholar]
- 76.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bustard DE, Menolfi D, Jeppsson K, Ball LG, Dewey SC, Shirahige K, Sjogren C, Branzei D, Cobb JA. During replication stress Non-Smc-Element 5 is required for Smc5/6 complex functionality at stalled forks. J. Biol. Chem. 2012;287:11374–11383. doi: 10.1074/jbc.M111.336263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Strom L, Katou Y, Itoh T, Shirahige K, Sjogren C. Chromosome length influences replication-induced topological stress. Nature. 2011;471:392–396. doi: 10.1038/nature09791. [DOI] [PubMed] [Google Scholar]
- 80.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pebernard S, Perry JJ, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol. Biol. Cell. 2008;19:4099–4109. doi: 10.1091/mbc.E08-02-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watts FZ, Skilton A, Ho JC, Boyd LK, Trickey MA, Gardner L, Ogi FX, Outwin EA. The role of Schizosaccharomyces pombe SUMO ligases in genome stability. Biochem. Soc. Trans. 2007;35:1379–1384. doi: 10.1042/BST0351379. [DOI] [PubMed] [Google Scholar]
- 83.Pebernard S, McDonald WH, Pavlova Y, Yates JR, III, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farmer S, San-Segundo PA, Aragon L. The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS One. 2011;6:e20948. doi: 10.1371/journal.pone.0020948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leupold, Gutz Genetic fine structure in Schizosaccharomyces pombe. Proc IX Intl Congr Genet. 1964;2:31–35. [Google Scholar]
- 87.McLeod M, Stein M, Beach DH. The product of the mei3+ gene, expressed under the control of the mating type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]