Abstract
Nucleolin is a multi-functional nucleolar protein that is required for ribosomal RNA gene (rRNA) transcription in vivo, but the mechanism by which nucleolin modulates RNA polymerase I (RNAPI) transcription is not well understood. Nucleolin depletion results in an increase in the heterochromatin mark H3K9me2 and a decrease in H4K12Ac and H3K4me3 euchromatin histone marks in rRNA genes. ChIP-seq experiments identified an enrichment of nucleolin in the ribosomal DNA (rDNA) coding and promoter region. Nucleolin is preferentially associated with unmethylated rRNA genes and its depletion leads to the accumulation of RNAPI at the beginning of the transcription unit and a decrease in UBF along the coding and promoter regions. Nucleolin is able to affect the binding of transcription termination factor-1 on the promoter-proximal terminator T0, thus inhibiting the recruitment of TIP5 and HDAC1 and the establishment of a repressive heterochromatin state. These results reveal the importance of nucleolin for the maintenance of the euchromatin state and transcription elongation of rDNA.
INTRODUCTION
Nucleoli play essential roles in ribosome biogenesis and particularly in the transcription of ribosomal RNA (rRNA) genes. In a typical human cell, there are ∼400 copies of ribosomal DNA (rDNA) arranged in tandem repeats in nucleolar organizer regions (NORs). Each rDNA encodes a precursor transcript (45S pre-rRNA) that can be processed to generate 18S, 5.8S and 28S rRNA. The sequences that encode 45S pre-rRNA are separated by long intergenic spacers (IGSs).
rRNA genes are transcribed by RNA polymerase I (RNAPI) and require several associated factors for transcription specificity and efficiency. Recent works have highlighted the importance of the epigenetic control of rRNA transcription (1–5). In eukaryotic cells, only about half of the rRNA genes are transcriptionally active and can be distinguished by different epigenetic marks such as rDNA methylation, histone post-translational modifications and different chromatin-associated proteins (6). Actively transcribed rRNA genes possess euchromatin marks characterized by H4K12 acetylation, tri-methylation of H3K4 and hypo-methylated CpG dimers, while silent rRNA genes have heterochromatin features such as di-methylated H3K9, tri-methylated H4K20, tri-methylated H3K27 and hyper-methylated CpG dimers (5,6). Apart from the post-translational modifications of histones, specific incorporation of histone variants such as H2A.Z and macroH2A (7,8) may also contribute to the epigenetic control of rRNA transcription. Araya et al. (2010) recently observed the differential incorporation of macroH2A1 and two subtypes into the ribosomal cistron in fish could regulate gene expression during the acclimatization process (7).
A major mechanism for silencing active rRNA genes seems to be the recruitment of the nucleolar remodelling complex (NoRC) to the promoter by the transcription termination factor-1 (TTF-1), which binds to the promoter-proximal terminator T0 (1,9,10). NoRC, a member of ATP-dependent chromatin remodelling machines, comprises the ATPase SNF2H and TIP5. Once NoRC is recruited to the rDNA promoter by TTF-1, it mediates heterochromatin formation and silencing of rRNA transcription by recruiting histone-modifying enzymes histone deacetylases (HDAC), histone methyltransferases (HMT), DNA methyltransferase and by shifting the promoter-bound nucleosome into a silent position (1,2,10–12). Recent reports also showed that pRNA, a non-coding promoter-associated RNA, could form a triplex structure with T0, leading to displacement of TTF-1 from T0. The triplex could then recruit DNMT3b to the rDNA promoter, thus methylating CpG-133 and contributing to the repression of transcription (5,13). NoRC-dependent rDNA silencing and heterochromatin formation has been studied in detail. However, little is known about the mechanisms that counteract heterochromatin formation and promote the establishment and maintenance of the euchromatic state of active rDNA repeats.
Since it was first described as one of the main nucleolar proteins, nucleolin has been shown to be implicated in many steps of ribosome biogenesis including the synthesis of rRNA by RNAPI (14–20). Multiple functional domains allow the interaction of nucleolin with numerous proteins and nucleic acid sequences (RNAs and DNAs). Earlier experiments suggested a link between the proteolysis of nucleolin and RNAPI transcription elongation (21) and that only RNAPI (not Pol II or Pol III) transcription could be regulated by nucleolin, independent of the sequence of the transcribed RNA (15). Nucleolin depletion in different cell lines using small interfering RNA (siRNA) (14,19) or by conditional knockout in DT40 cells (20) results in the reduction of the accumulation of pre-rRNA. Metabolic labelling and analysis of the maturation or pre-rRNA produced in absence of nucleolin strongly suggest that nucleolin is required for efficient transcription of rRNA genes (20). Although the mechanism of nucleolin action on the synthesis of pre-rRNA remains unclear, several experiments indicate that this regulation may be achieved through chromatin. Nucleolin binds tightly to chromatin (22,23) and is able to modulate chromatin structure by interaction with histone H1 (24,25) or to stimulate the remodelling activities of the ATP-dependent remodelling complexes SWI/SNF and ACF on canonical or macroH2A1 containing nucleosomes (26). In vitro, nucleolin possesses a histone chaperone activity. It binds directly to H2A–H2B dimers and facilitates their assembly into nucleosomes on naked DNA (26). FRAP experiments on eGFP-tagged histones (H2B, H4 and macroH2A) in living cells depleted in nucleolin also confirmed a role for nucleolin in chromatin dynamics by its histone chaperone and co-remodelling activities (27).
In this study, we analysed the association of nucleolin with rDNA and the consequences of nucleolin silencing on the transcription of pre-rRNA. ChIP-Seq analysis shows that nucleolin distribution along the rDNA coding and promoter regions is very similar to that of RPA116 and UBF. The silencing of nucleolin expression modifies the distribution of RNAPI, UBF, TTF-1 and TIP5 on the rDNA together with an increase of heterochromatin marks on the rDNA promoter and coding region. We propose a model in which nucleolin plays an important role in maintaining the euchromatin state of rDNA, allowing the efficient transcription elongation of rDNA by RNAPI.
MATERIALS AND METHODS
Cell lines and siRNA transfection
HeLa cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% foetal bovine serum (Gibco) at 37°C in 5% CO2 incubator. To generate the cell line stably expressing Flag-tagged nucleolin, HeLa cells were transfected with pSG5–Flag–nucleolin (encoding full-length human nucleolin with an amino-terminal Flag tag) and pcDNATM3.1 plasmids. Clones stably expressing Flag-tagged nucleolin identified by immunofluorescence were selected with hygromycin. HeLa cells (6 × 105) were transfected twice with siRNA (Eurogentec) using Lipofectamine 2000 (Invitrogen) as described previously (14). The siRNA sequences used are listed in Supplementary Table S1. Control siRNA was BLOCK-iT™ Alexa Fluor Red Fluorescent Oligo (Cat.No.14750-100, Invitrogen). Proteins and RNAs were harvested at 96 h (for nucleolin siRNA) or 72 h after the first transfection (for TTF-1 siRNA) and analysed by western blot and reverse transcriptase–polymerase chain reaction (RT–QPCR), respectively.
Metabolic labelling and analysis of pre-rRNA transcription and processing
Cells (4 ×105) were incubated at 37°C in 3 ml of DMEM without phosphate (Invitrogen) supplemented with 10% foetal calf serum, 25 mM HEPES (Invitrogen). After 2 h, [32P]Pi (PerkinElmer) was added to the medium (125 µCi/ml) for various times. At the end of the labelling, total RNA was extracted with RNeasy kit (Qiagen), separated by denaturing electrophoresis and the gel was dried before exposure. The quantity of RNA corresponding to an equal number of cells was loaded for each sample. Radioactivity was detected with a Fujifilm FLA-5100 scanner and the pictures were analysed with the Multigauge V3.0 software (Fujifilm).
Transcription analysis by RT–QPCR
Total RNA was prepared by TRIzol (Invitrogen) extraction and digested with 20 U of RNase-free DNase I (Roche Diagnostics) for 1 h at 37°C. One hundred nanograms of total RNA was reverse-transcribed using hexamer random primers and first-strand cDNA synthesis kit (Fermentas) and the synthesized cDNA was used for RT–QPCR using FastStart Universal SYBR Green Master (ROX) (Roche). Primer sequences used are listed in Supplementary Table S2.
Immunofluorescence
HeLa cells grown on glass coverslips were washed with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, incubated on ice for 20 min and then permeabilized with 0.1% Triton X-100 in PBS for 2 × 5 min. After two washes in PBS, non-specific binding of antibodies was blocked with 10% foetal calf serum in PBS. After three washes with PBS, coverslips were incubated in primary antibodies at 37°C for 1 h. After two washes in PBS with 0.1% Triton X-100, coverslips were incubated with secondary antibodies coupled with Alexa dyes (A555 or A647). After two more washes in PBS with 0.1% Triton X-100, the coverslips were washed in PBS, rinsed in ddH2O and briefly dipped in 100% ethanol. After a quick dry, coverslips were mounted with Fluoromount G containing 200 ng/ml DAPI.
Chromatin immunoprecipitation and DNA methylation assays
Chromatin immunoprecipitation (ChIP) assays were done as described earlier (28). HeLa cells were cross-linked for 10 min with 1% formaldehyde in the culture medium at room temperature. Glycine (0.125 M) was added, and the cells were rocked for 5 min to stop the reaction. The cells were suspended in 200 µl sodium dodecyl sulphate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.1) and submitted to sonication to produce DNA fragments of 200–1000 bp in length. Chromatin was diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8.1, 167 mM NaCl), pre-cleared and immunoprecipitated with the respective antibodies. Precipitated DNA and protein complexes were reverse cross-linked and purified through phenol/chloroform extraction and ethanol precipitation. For RNase treatment, permeabilized HeLa cells were treated with 1 mg/ml RNase A for 5 min, cross-linked with formaldehyde and subjected to ChIP with anti-nucleolin antibody.
For sequential ChIP (reChIP) assays, the specific DNA–protein complexes from the first immunoprecipitation were extracted by adding 25 μl of 10 mM dithiothreitol, followed by incubation for 20 min at 37°C with vortexing every 5 min. The supernatants were pooled and diluted 10 times with reChIP buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.1% Triton X-100), followed by the second immunoprecipitation.
The purified DNAs were amplified by real-time PCR using StepOne Plus (Applied Biosystems) and FastStart Universal SYBR Green Master (ROX) (Roche). Primers for real-time PCR were designed or as reported (29) and are listed in Supplementary Table S2.
For nucleolin ChIP-seq analysis, ∼100 ng of input DNA and DNA precipitated by nucleolin antibody were sequenced with the Illumina/Solexa 1G technique (GATC, Germany). The input DNA sequences (27 615 364 reads) were used for normalization of the nucleolin ChIP-seq data (22 381 018 reads). Unique reads from the nucleolin ChIP dataset (15 467 484 reads) were mapped onto human rDNA reference sequence (NCBI accession number: HSU13369) using BWA (30,31). We optimized the quality threshold (=30) and mismatch penalty (−M) as 7 and –d as 12 to obtain the mapping of unique reads. The mapped nucleolin data and the control input data were then processed and filtered using tools and software algorithms from BioCOS Life Sciences, which uses data map quality-based filtering to eliminate repeated and poorly mapped reads. Furthermore, the mapped and filtered nucleolin data is normalized by the mapped and filtered input control data and plotted as shown on Figure 3A. ChIP-seq data of UBF and RPA116 (32) were downloaded from NCBI (NCBI sra SRR087746.sra (UBF HEK293T), SRR087747.sra (RPA116 HEK293T), SRR087753.sra (input HEK293T), SRR087754.sra (UBF K562) and SRR087755.sra (input K562) and analysed using the same methods (Supplementary Figure S3).
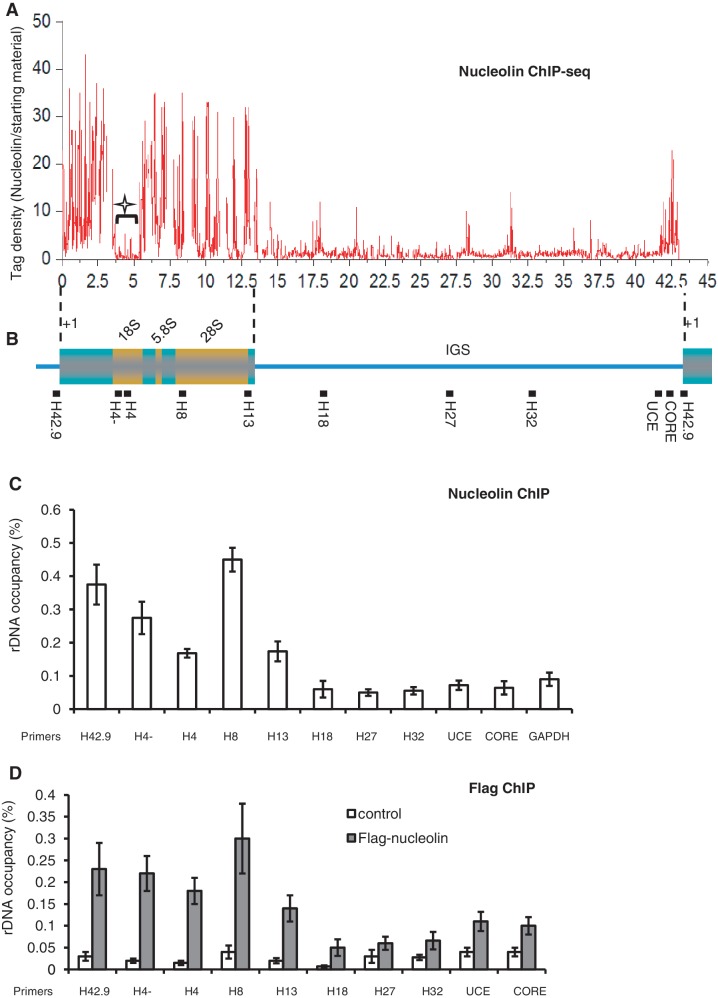
Figure 3.
Nucleolin is associated with rDNA. (A) ChIP-seq mapping of nucleolin binding sites throughout the rDNA locus. Data were analysed as explained in the ‘Materials and Methods’ section, and the nucleolin tag density normalized to the starting material was plotted. As reads that map to multiple locations were masked during the bioinformatic analysis, regions rich in repetitive sequences can leave gaps as shown in the 18S region (star). (B) Schematic representation of a human rDNA repeat. The positions of QPCR amplicons in ChIP assays are indicated with solid bars. (C) QPCR analysis of the enrichment of nucleolin on rDNA. Immuno-precipitated DNA was analysed by QPCR using sets of primers indicated in (B). The percentage of DNA immuno-precipitated with anti-nucleolin antibody was calculated relative to the ChIP input DNA. Values are means ± SD (standard deviation) derived from three independent ChIP experiments, each tested by at least three independent QPCRs. (D) HeLa cells stably transfected with control Flag-EV (white bars) or with Flag-tagged nucleolin (grey bars) were used for ChIP experiments using anti-flag antibody. The precipitated DNA was analysed by QPCR as previously. The percentage of DNA immuno-precipitated with anti-Flag antibody was calculated relative to the ChIP input DNA as in Panel C.
To monitor CpG methylation of rDNA, the immunoprecipitated as well as the input DNA were digested with HpaII or MspI, and the digested DNA along with an equal amount of the undigested DNA were amplified by real-time PCR using the H42.9 primers (Supplementary Table S2) specific for the human rDNA promoter region (29). The results are expressed as the ratio of methylated or unmethylated DNA to the respective input.
Psoralen cross-linking and Southern blot
HeLa cells grown in 60-mm Petri dishes (0.8 × 106 cells/Petri) were placed in serum-free media (1.5 ml) just before cross-linking began. A 1/20 volume (75 µl) of 200 µg/ml trioxsalen (4,5′,8-trimethylpsoralen; Sigma) in methanol was added with gentle mixing, and after 5 min incubation, cells were irradiated on ice for 5 min with a 366-nm UV lamp (BlackRay model B-100A, 100W) placed at a distance of 6–7 cm. Cross-linking was repeated three more times, each time adding fresh Trioxsalen. The cross-linking procedure was performed in a dark room. Subsequently, the cells were washed with PBS and lysed with 0.5 ml of lysis buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 25 mM EDTA, 2% SDS). Proteinase K (1 mg/ml; Fermentas) was added, and the combined lysate was incubated overnight at 50°C, adding fresh proteinase K after the first 2 h. The genomic DNA was then de-proteinized twice with phenol/chloroform, ethanol precipitated and re-suspended in 250 µl TE pH 7.5, 0.1% SDS. Twenty micrograms of RNase A (Fermentas) was added and after 30 min at 37°C, 0.5 mg/ml Proteinase K was also added and incubation continued for another 1 h at 50°C. After two phenol/chloroform extractions, the genomic DNA was ethanol precipitated, re-suspended in 50 µl TE and quantified. About 10 µg of genomic DNA was digested overnight with BamHI and resolved on a 1% TAE agarose gel at 2 V cm−1 for 22 h in the absence of ethidium bromide (EtBr). The gel was subsequently EtBr-stained, photographed, its cross-links reversed by UV irradiation at 254 nm (4000 mJ·cm−2) in a Stratalinker 1800 cross-linker (Agilent Technologies) and transferred onto a Biodyne B membrane (Pall). Hybridization was performed overnight at 65°C with labelled DNA probes in Southern Church Buffer (0.5 M sodium phosphate, pH 7.2, 1 mM EDTA pH 8.0, 7% SDS, 1% BSA). Subsequently, membranes were washed with 6× saline sodium citrate (SSC), 2× SSC and 0.1× SSC and 0.1% SDS. Data were analysed using a FUJI PhosphorImager and with MultiGauge software.
Antibodies
The following antibodies were used: a rabbit polyclonal antibody against human nucleolin (number 5567; developed by our laboratory), anti-acetyl H4K12 (ab1761; Abcam), anti-histone H3 (ab1791; Abcam), anti-trimethyl H3K4 (ab8580; Abcam), anti-G9a (ab40542; Abcam), anti-TTF-1 (A302-361A; Bethyl), anti-TIP5 (CS-090-100; Diagenode), anti-dimethyl H3K9 (ab1220; Abcam), anti-nucleolin monoclonal antibody (KAM-CP100; Stressgen), anti-RPA116 (a generous gift from Ingrid Grummt), anti-UBF (F-9) (sc-13125; Santa Cruz), anti-B23 antibody (ab10530; Abcam), anti-HDAC1 (ab46985; Abcam) and anti-Flag antibody (Sigma).
RESULTS
Depletion of nucleolin affects epigenetic marks of rDNA chromatin
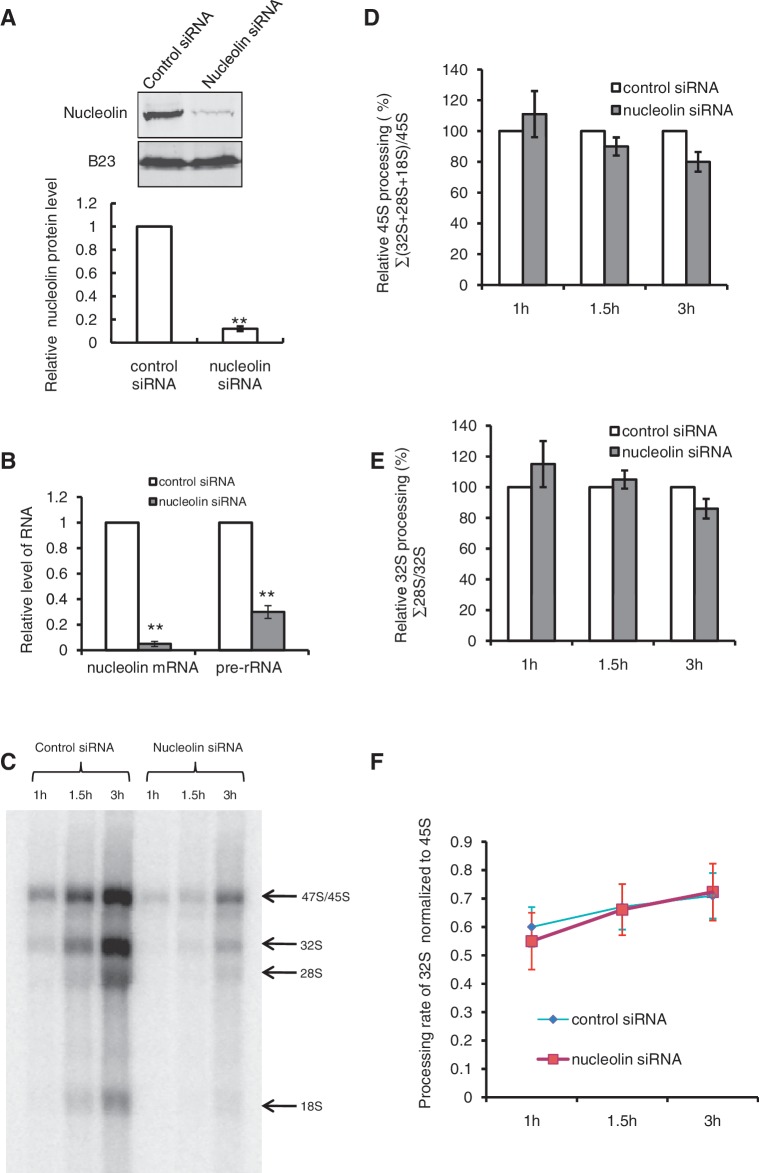
Previous experiments have shown that nucleolin is required for the efficient production of pre-rRNA by RNAPI, but the mechanisms implicated in this regulation have not been investigated (14,19,20). Transfection of HeLa cells with specific siRNAs against nucleolin efficiently reduced the levels of nucleolin protein (Figure 1A) and messenger RNA (Figure 1B) and this was associated with a low level of pre-rRNA accumulation (Figure 1B). Metabolic labelling with [32P]Pi and northern blot analysis of pre-rRNA transcribed in nucleolin depleted cells confirm that nucleolin depletion severely impairs rRNA production without affecting the global maturation of pre-rRNA (Figure 1C and Supplementary Figure S1). By summing the intensities of all four major products, the loss of nucleolin causes a reduction of ∼80% of rRNA accumulation compared to the control cells treated with a control siRNA. We then examined the maturation of the largest precursor (47S/45S) by comparing its intensity with the sum of all other maturation products and we observed that its maturation was only very moderately affected (decreased by ∼10%) (Figure 1D). The efficiency and rate of processing of the 32S species was not significantly affected (P > 0.05) in nucleolin-depleted cells (Figure 1E and F) showing that 45S pre-rRNA precursor is processed as in control cells. These data indicate that the low accumulation of pre-rRNA in nucleolin-depleted cells is not the consequence of an inhibition of rRNA processing that may affect ribosome biogenesis and transcription, but rather the result of a role of nucleolin on the transcription process itself. To better understand the mechanisms involved, we first investigated the consequences of nucleolin depletion on the epigenetic marks of rDNA chromatin (Figure 2).
Figure 1.
Effect of nucleolin on rRNA transcription and processing. (A) Inhibition of nucleolin expression by siRNA. HeLa cells were transfected with control (BLOCK-iT™) or nucleolin siRNA. Proteins (nucleolin and B23) were analysed by western blot 4 days after transfection. **P < 0.01, Student’s t-test was done between HeLa cells transfected with control siRNA and nucleolin siRNA. (B) Levels of nucleolin messenger RNA and pre-rRNA determined by RT–QPCR as described in the ‘Materials and Methods’ section. The data represent the average of three different independent experiments. **P < 0.01, Student’s t-test was done between HeLa cells transfected with control siRNA and nucleolin siRNA. (C) Metabolic labelling of total RNA from HeLa cells transfected with control siRNA or nucleolin siRNA. HeLa cells were incubated in phosphate-free DMEM for 2 h and [32P]orthophosphate was added to the medium (125 µCi/ml) for the indicated times. The 32P-labelled RNAs corresponding to an equal number of cells were resolved on 1% MOPS-formaldehyde gels and exposed on a Fujifilm FLA-5100 scanner. (D) Quantification of the relative 45S processing in HeLa cells transfected with control and nucleolin siRNA as in (A). For each pulse time, the 45S processing was calculated as the sum of the intensities of processed products 32S, 28S and 18S, divided by the intensity of the 45S precursor. Data were normalized to the control cells. Data were from two independent experiments. (E) Quantification of the relative 32S processing in HeLa cells transfected with control and nucleolin siRNA as in (A). For each pulse time, the 32S processing was calculated as the intensity of processed product 28S divided by the intensity of the 32S precursor. Data were normalized to the control cells. Data were from two independent experiments. (F) Quantification of the rate of 32S rRNA processing in HeLa cells transfected with control and nucleolin siRNA. The 32S rRNAs were normalized to the 45S rRNAs. Data were from two independent experiments. Student’s t-test was done between HeLa cells transfected with control siRNA and nucleolin siRNA, and no significant difference was detected (panels D, E and F).
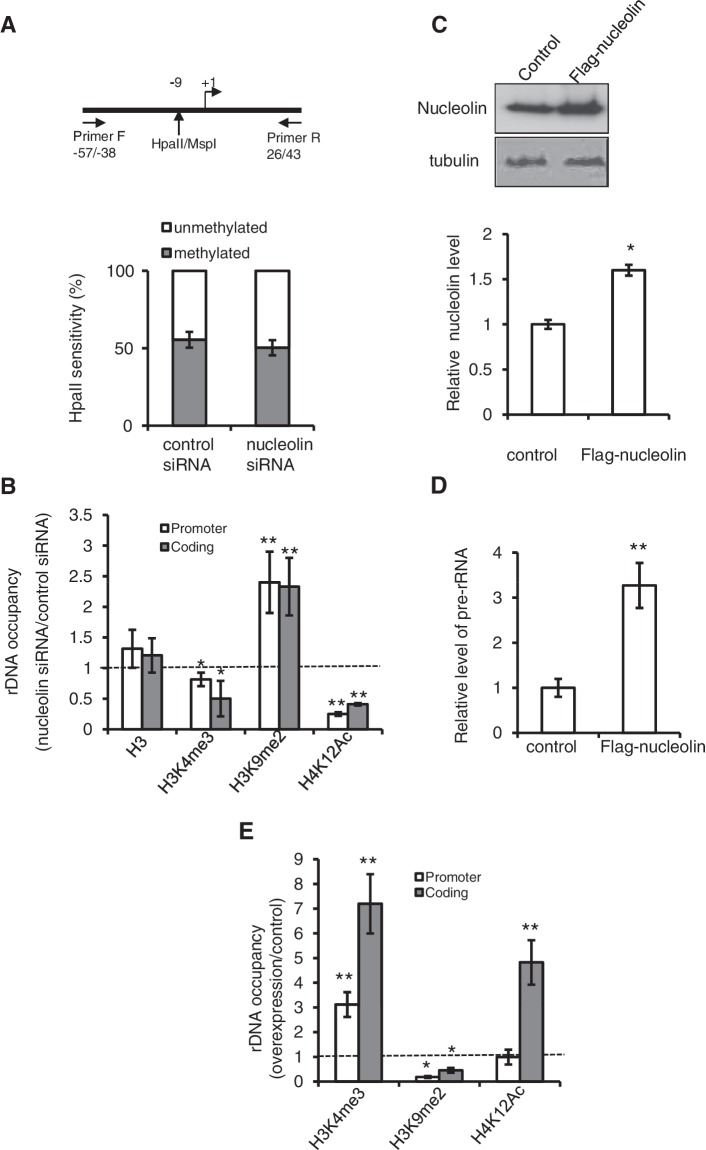
Figure 2.
Effect of nucleolin on epigenetic marks. (A) Depletion of nucleolin does not affect rDNA promoter methylation. DNA was isolated from control or siRNA-treated HeLa cells and digested with MspI (methylation-insensitive) or HpaII (methylation-sensitive). The human rDNA promoter was then amplified with primers flanking the CCGG site at −9. If the CpG dimer was methylated (resistant to cleavage with HpaII), amplification of rDNA from −57 to +43 yield a fragment of 100 nt, whereas unmethylated DNA would be cleaved and yield no PCR product. Student’s t-test was done between HeLa cells transfected with control and nucleolin siRNA, no significant difference was detected. (B) Histone post-translational modification marks of rDNA are affected after the depletion of nucleolin. QPCR amplification of either the 5′-end of the rRNA gene (H42.9) or the coding region (H8) after ChIP shows the level of H3, H3K4me3, H3K9me2 and H4K12Ac on different regions of rDNA. The ratio of rDNA occupancy between nucleolin and control siRNA transfected cells was calculated for each histone mark. (C) Over-expression of Flag-nucleolin. Total cell extract from the HeLa cell line stably expressing Flag-nucleolin was used for western blot analysis with an anti-nucleolin and anti-tubulin antibody, and the quantification was done using Image J. (D) The level of pre-rRNA in cells over-expressing nucleolin was determined by RT–QPCR with primers specific for the 5′-ETS. (E) Over-expression of nucleolin affects histone post-translational modifications on rDNA. ChIP experiments shows the level of H3K4me3, H3K9me2 and H4K12Ac on 5′-end of the gene (H42.9) and coding regions (H8) of rDNA in cells that stably over-express Flag-nucleolin. For all experiments, *0.01 < P < 0.05, **P < 0.01, Student’s t-test was done between control HeLa cells and HeLa cells transfected with nucleolin siRNA, or cells over-expressing Flag-tagged nucleolin.
To determine whether this reduced level was associated with an increase in silencing by rDNA methylation, the level of methylation of a critical CpG dimer in the core promoter element of the rDNA was determined in control and siRNA-treated HeLa cells (Figure 2A). Nucleolin depletion did not affect the level of rDNA methylation at this site, suggesting that the decrease of pre-rRNA level is most likely not associated with increased rDNA methylation. The level of active and inactive rRNA genes can be also determined by their susceptibility to the DNA cross-linking agent psoralen (33). To determine whether the decrease of pre-rRNA synthesis was associated with the alteration of the chromatin state of rDNA, we performed this psoralen cross-linking assay. We found that nucleolin depletion has no significant effect on the proportion of genes that are efficiently cross-linked by psoralen, suggesting that the ratio of active to inactive rRNA genes is not changed in nucleolin-depleted cells (Supplementary Figure S2). Then, we examined the histone post-translational modifications on rDNA promoter and coding regions (Figure 2B). In agreement with the lower accumulation of pre-rRNA molecules in nucleolin-depleted cells, we observed an important decrease in H4K12Ac and H3K4me3, two marks usually associated with transcriptionally active rDNA repeats, whereas the level of H3K9me2 increased significantly.
As nucleolin is required for rRNA transcription, we could also expect that over-expression of nucleolin affects the chromatin state of rDNA and pre-rRNA transcription. Indeed, in HeLa cells that stably over-express Flag-tagged nucleolin (Figure 2C), we observed a marked increase of pre-rRNA level (Figure 2D). This was associated with a strong increase in the activation marks H3K4me3 and H4K12Ac and a decrease in the silencing mark H3K9me2 in rRNA genes (Figure 2E). Altogether, these ChIP data indicate that the depletion and over-expression of nucleolin are associated with opposing alterations of histone modification states of rDNA but not with a change of the proportion of active genes.
Nucleolin is associated with rDNA
These alterations of post-translational modifications of rDNA chromatin upon nucleolin silencing or over-expression could be indirect or the result of nucleolin interaction with rDNA chromatin. To characterize these interactions, we performed ChIP-Seq experiments with nucleolin antibody. ChIP experiments from two independent experiments were analysed. The total number of reads from nucleolin ChIP-seq (22 381 018 reads) and from input DNA (27 615 364 reads) were mapped on the genome, and only unique reads (15 467 484) were mapped on rDNA (NCBI accession number: HSU13369). The mapped nucleolin data and the input starting material were then processed and filtered as described in the ‘Materials and Methods’ section, and the nucleolin dataset was then normalized by the starting input material and plotted (Figure 3A). These results show that nucleolin is particularly enriched in the coding region of rDNA, moderately enriched in the promoter region and insignificantly present in the IGS. Sequence reads that map to multiple locations were masked during bioinformatic analysis, which can leave gaps in regions of rDNA that are rich in repetitive sequences; this is particularly observed in the 18S region (star in Figure 3A). The distribution of nucleolin on rDNA was compared to the distribution of UBF and RNAPI (RPA116) that have recently been published (32) (Supplementary Figure S3). Interestingly, the global pattern of nucleolin distribution on rDNA is very similar to the distribution of UBF and PolI. To validate these ChIP-seq data, we performed several QPCR analyses using primers that span the human rDNA repeat (29) (Figure 3B and C). As expected from the ChIP-Seq data, nucleolin was preferentially found associated with the rDNA-coding regions, including the 18S rRNA, which does not show any unique reads in the ChIP-seq analysis (star in Figure 3A). The interaction of nucleolin with rDNA chromatin was independent of RNA since RNase treatment before the ChIP procedure does not abrogate the nucleolin with the rDNA locus (Supplementary Figure S4). Furthermore, when the ChIP experiment was performed with a cell line that stably expresses Flag-nucleolin (Figure 3D), we observed the similar enrichment of nucleolin in the rDNA-coding sequence.
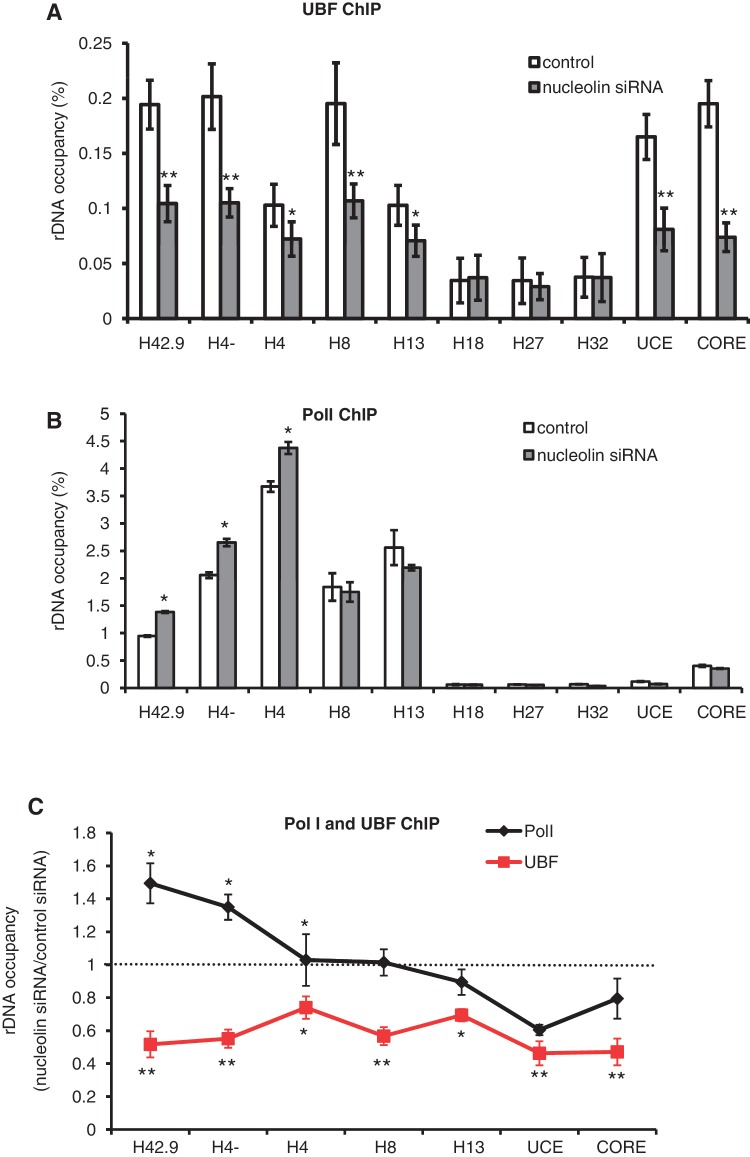
To determine if the decreased accumulation of pre-rRNA in nucleolin-depleted cells was linked to a modification of the interaction between RNAPI and UBF with the rRNA genes, we performed ChIP experiments using the antibodies to the RNAPI subunit (RPA116) and for UBF in nucleolin-depleted cells. Nucleolin depletion leads to a decrease in UBF on the promoter and coding regions of rDNA (Figure 4A and C), which is in agreement with the decrease in rRNA transcription in nucleolin-depleted cells. Interestingly, nucleolin depletion leads also to an increase of PolI loading at the beginning of coding region of rDNA but to a significant decrease at the 3′-end of the gene (Figure 4B and C). This pattern of RNAPI loading after nucleolin depletion is reminiscent of what happens after inhibition of transcription elongation by actinomycin D (34), suggesting that nucleolin could be required for efficient RNAPI elongation.
Figure 4.
Nucleolin depletion alters the distribution of RNAPI and UBF on rRNA. (A) Nucleolin depletion leads to a decrease in UBF in the promoter and coding regions of the rDNA repeat. ChIP experiments with UBF antibody were performed in HeLa control and nucleolin-depleted cells. The percentage of DNA immuno-precipitated with anti-UBF antibody was calculated relative to the ChIP input DNA. (B) Nucleolin depletion increases the level of RNAPI at the 5′-end of the coding region (H42.9 and H4−) of the rDNA repeat. ChIP experiments with RPA116 antibody were performed in HeLa control and nucleolin-depleted cells. The percentage of DNA immuno-precipitated with anti-PRA116 antibody was calculated relative to the ChIP input DNA. (C) Data from ChIP experiments with UBF antibody (panel A) and RPA116 antibody (panel B) were normalized to the level of occupancy in control siRNA transfected cells. *0.01 < P < 0.05, **P < 0.01, Student’s t-test was done between Hela cells transfected with control siRNA and nucleolin siRNA.
Nucleolin is associated with active rDNA repeats
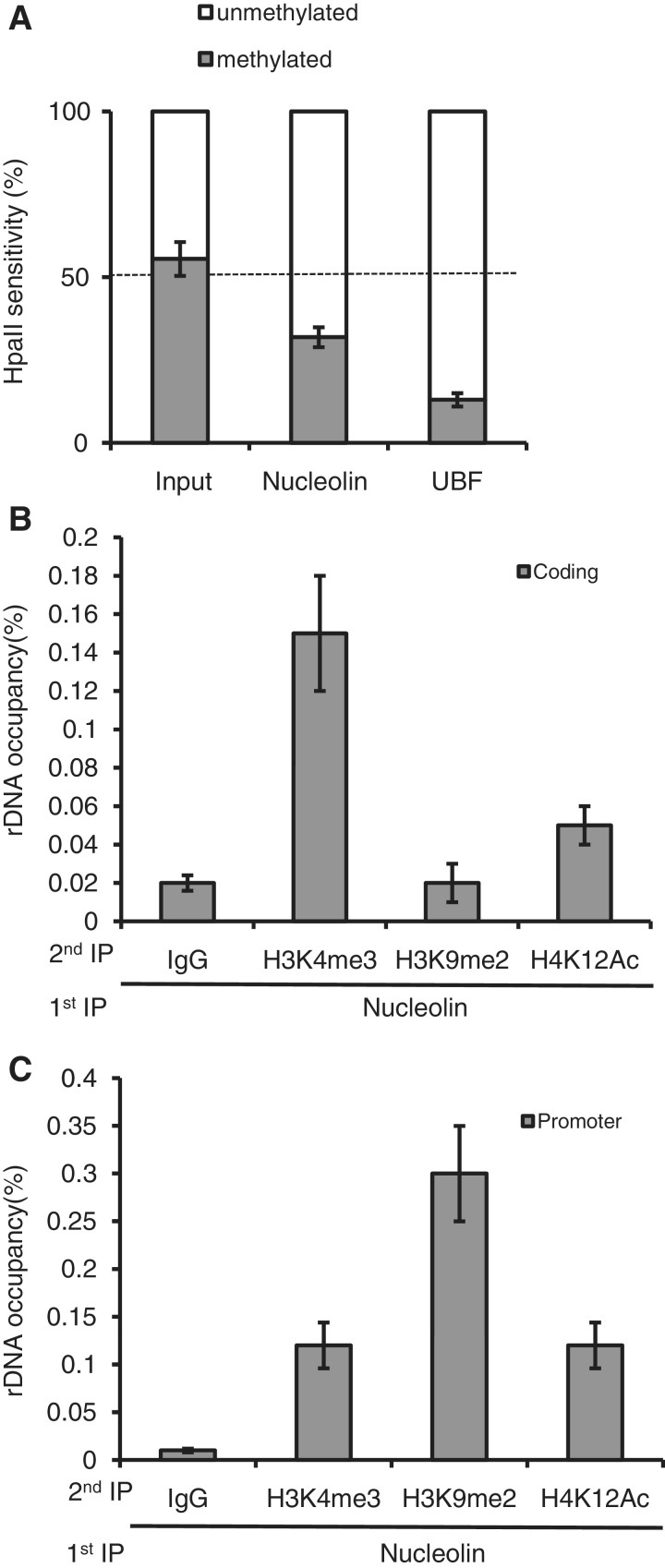
We then determined the epigenetic state of the rDNA chromatin associated with nucleolin (Figure 5). Active and silent rDNA can be distinguished by DNA methylation. Active rDNA is hypo-methylated, while silent rDNA is hyper-methylated (6). In human cells, methylation at a single site in the promoter of the rRNA gene correlates with the repression of the promoter activity (35). We performed a ChIP–CHOP experiment to look at rDNA methylation status at the −9 CpG site of the promoter sequences bound to nucleolin (Figure 5A). As a control, we did the same experiment with UBF, which is almost exclusively associated with unmethylated rDNA promoters (36). Interestingly, nucleolin was found to be also preferentially associated with unmethylated rDNA promoters, which correspond to the transcriptionally active rRNA genes. Re-ChIP experiments indicate that the heterochromatin mark H3K9me2 in the nucleolin-associated chromatin seems absent within the rDNA coding region (Figure 5B) but is highly enriched in the promoter sequence (Figure 5C). In contrast, H4K12Ac and H3K4me3 euchromatin marks are indeed associated with nucleolin in the rDNA coding sequence (Figure 5B) and at the 5′-end of the rRNA gene (H42.9 primers) (Figure 5C), which is in agreement with an association of nucleolin with the transcriptionally active copies of rDNA.
Figure 5.
Nucleolin is associated with active rDNA repeats. (A) ChIP input DNA and DNA precipitated by nucleolin or UBF antibody were digested with HpaII or MspI, or they were mock-digested. The relative levels of HpaII-resistant, methylated rDNA copies (grey bars) and unmethylated copies (white bars) were determined by QPCR with the pair of primers that flanks the HpaII/MspI site on the rDNA promoter as indicated in Figure 2A. (B) and (C) co-immunoprecipitation of endogenous nucleolin with active and repressive histone modification marks on coding (H8 primers) and 5′-end of the coding region (H42.9 primers) regions of rDNA, respectively. Re-ChIP experiments with anti-H3K4me3, H3K9me2 and H4K12Ac antibodies were performed after a first immuno-precipitation with anti-nucleolin antibody. Values are means ± SD (standard deviation) derived from three independent experiments, each tested by at least three independent QPCR reactions.
Nucleolin affects TTF-1 interaction with T0
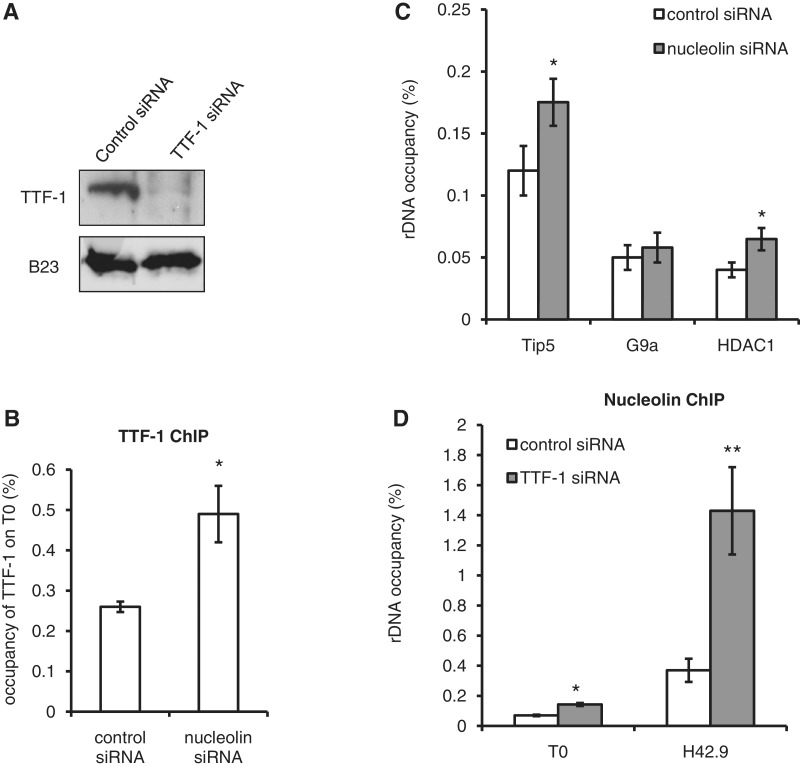
TTF-1, which binds the promoter-proximal terminator T0, plays a key role in the recruitment of the nucleolar remodelling complex NoRC and the silencing of rDNA (5), although earlier reports indicated that TTF-1 was required for the transcription by RNAPI of chromatinized DNA templates (37). In order to determine whether nucleolin plays any role in this process, we studied whether the interaction of nucleolin with ribosomal chromatin had any influence on the binding of TTF-1 to the T0 site (Figure 6). We found that nucleolin depletion leads to a two-fold increase in TTF-1 interaction with T0 (Figure 6B). As the binding of TTF-1 is required for the recruitment of NoRC and HDACs, we also performed ChIP experiments with TIP5 and HDAC1 after nucleolin depletion (Figure 6C). We also observed a slight increase in the recruitment of TIP5 and HDAC1, while the interaction of G9a was not changed. In a reverse experiment, we observed that the inhibition of expression of TTF1 leads to increased association of nucleolin with T0 (2-fold) and H42.9 (4-fold) (Figure 6D). Altogether, these data show that the presence of nucleolin on rDNA chromatin inhibits the binding of TTF-1 within the T0 region and vice versa and thus protects rRNA genes from TTF-1-mediated silencing of transcription.
Figure 6.
Nucleolin affects TTF-1 interaction with T0. (A) Western blot was performed 3 days after transfection of HeLa cells with TTF-1 specific siRNA. B23 antibody is used here as a control. (B) Depletion of nucleolin leads to an increase in TTF-1 on T0. TTF-1 ChIP experiments were performed after nucleolin depletion. Data were normalized to the TTF-1 occupancy in control siRNA transfected cells. (C) The rRNA occupancy of TIP5, HDAC1 and G9a was determined after siRNA specific for nucleolin were transfected in HeLa cells for 4 days. Data were normalized to the level of occupancy in control siRNA transfected cells. (D) Depletion of TTF-1 leads to an increase in nucleolin on T0, H42.9. ChIP experiments showed the level of nucleolin on different regions of the rDNA repeat T0 and H42.9. Data were normalized to nucleolin rDNA occupancy in control siRNA-transfected cells. *0.01 < P < 0.05, **P < 0.01, Student’s t-test was done between HeLa cells transfected with control siRNA and nucleolin siRNA or TTF-1 siRNA.
DISCUSSION
Over the past 20 years, many in vitro and in vivo studies have implicated nucleolin, one of the major nucleolar proteins, in the production of rRNAs by RNAPI transcription (14,15,19–21,38) without providing many mechanistic details on how nucleolin could participate in the production of rRNA. Previous works have shown that in HeLa cells the accumulation of 45S could be affected by the rate of pre-rRNA processing (39,40). Since nucleolin interacts specifically with pre-rRNA (41–47) and has been involved in the first processing step of pre-RNA in vitro (16), it was tempting to explain the low accumulation of pre-rRNA in nucleolin depleted cells by an indirect effect of nucleolin on pre-rRNA processing. However, by metabolic labelling or northern blot we could not detect major changes in the processing pathways of pre-rRNA or in the efficiency of this processing (Figure 1 and Supplementary Figure S1) that could explain the strong reduction of 45S accumulation. These data are also in agreement with our previous analysis of nucleolin knockout in chicken DT40 cells (14,20). One possible explanation is that the low level of nucleolin that remains in nucleolin-depleted HeLa cells is sufficient to support normal pre-rRNA processing, while it is affecting very strongly pre-rRNA accumulation through its transcription. Indeed, the accumulation of pre-rRNA is very sensitive to the level of expression of nucleolin (20). We have seen the same effect of nucleolin depletion on the level of pre-rRNA not only in HeLa cells but also in human primary fibroblast (14) and in chicken DT40 cells (20), showing that what we describe in this article is not a particularity of HeLa cells.
Recent reports indicate that nucleolin is a histone chaperone with a FACT-like (FAcilitates Chromatin Transcription) activity that facilitates chromatin remodelling and histone dynamics (26,27), suggesting that nucleolin could regulate rRNA transcription at the level of chromatin accessibility and transcription elongation.
In this report, we describe experiments that support the role of nucleolin in the maintenance of the euchromatin state of rRNA, which allows efficient RNAPI transcription elongation. We show that nucleolin is preferentially associated with unmethylated rDNA and with epigenetic marks characteristic of transcriptionally active genes, which is in agreement with earlier reports indicating that nucleolin is required for rRNA transcription. In Arabidopsis thaliana, disruption of the AtNUC-L1 gene induces the loss of DNA methylation without affecting histone epigenetic marks at rRNA genes (48). However, we found that, in human cells, the depletion of nucleolin strongly inhibits pre-rRNA transcription and affects the post-translational modifications of histones associated with the rDNA promoter (Figure 2B and E) but does not modify the level of CpG methylation (Figure 2A) in the promoter region. This suggests that in mammalian cells, increased CpG methylation is not required for the inhibition of rRNA transcription in response to nucleolin depletion. Of note, methylation-independent silencing of rRNA genes was also observed upon depletion of UBF (36). Interestingly, UBF depletion leads to an increase in the number of rRNA genes in an inactive condensed state without reducing the net rRNA synthesis as transcription from remaining active genes is increased (36). Upon nucleolin depletion, we observed a substantial decrease in UBF loading on rRNA gene (Figure 4); in contrast to Sanij et al. (2009) report, this is correlated with a strong decrease of rRNA accumulation, as nucleolin is probably required for the transcription activation of the genes that remained actives in UBF-depleted cells (36).
Nucleolin depletion does not affect the proportion of active rRNA genes detected by the psoralen cross-linking experiment (Supplementary Figure S2). It should be noted that there is not always a direct correlation between the level of psoralen cross-linking and the presence of heterochromatin marks or the level of RNAPI transcription. For instance, when DNA methylation is lost, 50% of rRNA genes still show an inactive state by psoralen (49). In addition, UBF depletion leads to a decrease in the active ribosomal genes in psoralen cross-linking experiments (36); however, rRNA synthesis does not decrease as the transcription from the active genes is increased. This result may suggest that, in nucleolin-depleted cells, the lower level of pre-rRNA accumulation is rather the consequence of a lower rate of transcription of the active genes rather than an increase in the number of inactive genes. Indeed, our data showed that nucleolin depletion leads to an increase in RNAPI at the 5′-end of the coding region of the rDNA (Figure 4B and C), as previously described for the effect of actinomycin D on transcription elongation (34), which is consistent with a role for nucleolin in RNAPI elongation.
In our ChIP-seq analysis, we found a strong enrichment of nucleolin at the rDNA coding region with little if any enrichment in the IGS. Interestingly, this distribution looks very similar to that of UBF, another factor required for rRNA transcription (32) and to that of RNAPI subunit RPA116. The association of nucleolin with rDNA does not result from its association with pre-rRNA precursor, as RNase treatment does not alter the distribution of nucleolin on rDNA (Supplementary Figure S4). The presence of nucleolin on the coding region is associated with the euchromatin marks H4K12Ac and H3K4me3 (Figure 5B and C). Although H3K9me2 is usually associated with heterochromatin formation, there are now several reports that demonstrate that components of heterochromatin such as H3K9me2 play additional role in establishing chromatin structure of actively transcribed genes. This is true for PolII genes (50) and also for Pol I genes (51) indicating that the association of nucleolin with H3K9me2 at the 5′-end of the gene may be another sign of the link between active genes and nucleolin. Upon nucleolin depletion, the level of H4K12Ac and H3K4me3 on rDNA decreased while the level of H3K9me2 increased. On the other hand, nucleolin over-expression caused an increase in H3K4me3 and H4K12Ac and a decrease in H3K9me2, suggesting that the presence of nucleolin is required for the modulation of histone modifications of rDNA associated with rRNA transcription.
The binding of TTF-1 to T0 is an important step for the silencing of rRNA as this allows the recruitment of the nucleolar remodelling complex NoRC (3,5,6), which then recruits HDAC and HMT to establish a heterochromatin structure of rRNA (1–3). Interestingly, we found that after nucleolin depletion, the interaction of TTF-1 with T0 increases, with a concomitant increase of TIP5 on rDNA promoter (Figure 6B and C). This is also accompanied by recruitment of HDAC1 (Figure 6C), in agreement with the lower level of H4K12Ac that we observed (Figure 2B). NoRC function requires the interaction of its TIP5 subunit with 150–250 nt pRNAs transcribed from a spacer promoter located ∼2 kb upstream of the major 45S pre-rRNA promoter (52). These pRNAs also seem to interact with T0 leading to displacement of TTF-1 and to the recruitment of DNA methyltransferase DNMT3b to the promoter (13). After nucleolin depletion, even if we observed higher recruitment of TIP5, we could not detect any difference in the −9 bp CpG methylation, suggesting that this mechanism for heterochromatin formation and rDNA silencing mediated by NoRC does not entirely take place in nucleolin-depleted cells.
In addition, nucleolin-depleted cells show a striking reorganization of nucleoli structure (14,20,53), and several reports show that nucleolin is implicated in chromatin condensation (54), chromatin loop organization (55,56) and histone dynamics in vivo (27). Therefore, nucleolin could also be implicated in the regulation of chromatin accessibility at a relatively large scale in addition to its effect on single nucleosome co-remodelling and destabilizing activity (26,57).
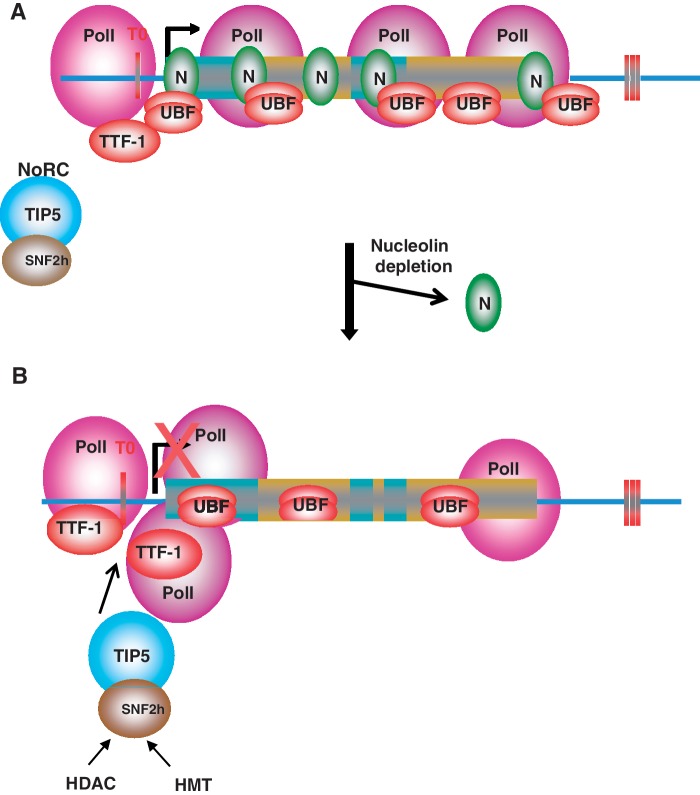
Altogether, this study provides new information to explain the role of nucleolin in regulation of RNAPI transcription (Figure 7). Nucleolin depletion results in an increase of RNAPI at the 5′-end of the coding region of the rDNA and a decrease of UBF along the rDNA. The association of nucleolin with the promoter region interferes with the binding of TTF-1 to T0, thereby inhibiting the recruitment of NoRC. Nucleolin depletion causes an increase of TTF-1 on T0, allowing a better recruitment of NoRC and then the establishment of a heterochromatin state contributing to the inhibition of RNAPI transcription.
Figure 7.
Model depicting the role of nucleolin in rRNA transcription. (A) Nucleolin is enriched in the promoter and coding regions of rDNA and protects rDNA from TTF-1. (B) Nucleolin depletion leads to an increase in TTF-1 on rDNA; TTF-1 could thereby recruit NoRC through its subunit TIP5, then recruits HDAC and HMT to the rDNA promoter to establish heterochromatin structure thus inhibit rRNA transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–4.
FUNDING
Agence Nationale de la Recherche [ANR-07-BLAN-0062-01]; Région Rhône-Alpes MIRA 2007, 2008, and 2010; Association pour la Recherche sur le Cancer n° ECL2010R01122, CEFIPRA n° 3803-1; CNRS and Ecole Normale Supérieure de Lyon. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank T. Moss for advices to perform the psoralen experiment. The authors also thank PLATIM (PLAteau Technique d’Imagerie et de Microscopie, UMS3444, Lyon, FRANCE).
REFERENCES
- 1.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 3.Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grummt I. Wisely chosen paths–regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden. FEBS J. 2010;277:4626–4639. doi: 10.1111/j.1742-4658.2010.07892.x. [DOI] [PubMed] [Google Scholar]
- 6.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 7.Araya I, Nardocci G, Morales J, Vera M, Molina A, Alvarez M. MacroH2A subtypes contribute antagonistically to the transcriptional regulation of the ribosomal cistron during seasonal acclimatization of the carp fish. Epigenet. Chromatin. 2010;3:14. doi: 10.1186/1756-8935-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008;27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strohner R, Nemeth A, Nightingale KP, Grummt I, Becker PB, Langst G. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 2004;24:1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Langst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeth A, Strohner R, Grummt I, Langst G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004;32:4091–4099. doi: 10.1093/nar/gkh732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiki H, Suganuma M, Nishiwaki S, Yoshizawa S, Winyar B, Sugimura T, Schmitz FJ. A new pathway of tumor promotion by the okadaic acid class compounds. Adv. Second Messenger Phosphoprotein Res. 1990;24:340–344. [PubMed] [Google Scholar]
- 14.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roger B, Moisand A, Amalric F, Bouvet P. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J. Biol. Chem. 2002;277:10209–10219. doi: 10.1074/jbc.M106412200. [DOI] [PubMed] [Google Scholar]
- 16.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roger B, Moisand A, Amalric F, Bouvet P. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma. 2003;111:399–407. doi: 10.1007/s00412-002-0221-5. [DOI] [PubMed] [Google Scholar]
- 18.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 19.Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storck S, Thiry M, Bouvet P. Conditional knockout of nucleolin in DT40 cells reveals the functional redundancy of its RNA-binding domains. Biol. Cell. 2009;101:153–167. doi: 10.1042/BC20080054. [DOI] [PubMed] [Google Scholar]
- 21.Bouche G, Caizergues-Ferrer M, Bugler B, Amalric F. Interrelations between the maturation of a 100 kDa nucleolar protein and pre rRNA synthesis in CHO cells. Nucleic Acids Res. 1984;12:3025–3035. doi: 10.1093/nar/12.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson MO, Ezrailson EG, Guetzow K, Busch H. Localization and phosphorylation of nuclear, nucleolar and extranucleolar non-histone proteins of Novikoff hepatoma ascites cells. J. Mol. Biol. 1975;97:611–619. doi: 10.1016/s0022-2836(75)80062-3. [DOI] [PubMed] [Google Scholar]
- 23.Olson MO, Rivers ZM, Thompson BA, Kao WY, Case ST. Interaction of nucleolar phosphoprotein C23 with cloned segments of rat ribosomal deoxyribonucleic acid. Biochemistry. 1983;22:3345–3351. doi: 10.1021/bi00283a007. [DOI] [PubMed] [Google Scholar]
- 24.Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur. J. Biochem. 1988;175:525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x. [DOI] [PubMed] [Google Scholar]
- 25.Erard M, Lakhdar-Ghazal F, Amalric F. Repeat peptide motifs which contain beta-turns and modulate DNA condensation in chromatin. Eur. J. Biochem. 1990;191:19–26. doi: 10.1111/j.1432-1033.1990.tb19088.x. [DOI] [PubMed] [Google Scholar]
- 26.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaume X, Monier K, Argoul F, Mongelard F, Bouvet P. In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem. Res. Int. 2011;2011:187624. doi: 10.1155/2011/187624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 29.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011;39:4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 34.Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 2004;279:6783–6793. doi: 10.1074/jbc.M309393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 1998;17:3135–3145. doi: 10.1093/emboj/17.11.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egyhazi E, Pigon A, Chang JH, Ghaffari SH, Dreesen TD, Wellman SE, Case ST, Olson MO. Effects of anti-C23 (nucleolin) antibody on transcription of ribosomal DNA in Chironomus salivary gland cells. Exp. Cell Res. 1988;178:264–272. doi: 10.1016/0014-4827(88)90397-7. [DOI] [PubMed] [Google Scholar]
- 39.Craig NC, Perry RP. Aberrant intranucleolar maturation of ribosomal precursors in the absence of protein synthesis. J. Cell. Biol. 1970;45:554–564. doi: 10.1083/jcb.45.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willems M, Penman M, Penman S. The regulation of RNA synthesis and processing in the nucleolus during inhibition of protein synthesis. J. Cell. Biol. 1969;41:177–187. doi: 10.1083/jcb.41.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouvet P, Allain FH, Finger LD, Dieckmann T, Feigon J. Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J. Mol. Biol. 2001;309:763–775. doi: 10.1006/jmbi.2001.4691. [DOI] [PubMed] [Google Scholar]
- 43.Bouvet P, Jain C, Belasco JG, Amalric F, Erard M. RNA recognition by the joint action of two nucleolin RNA-binding domains: genetic analysis and structural modeling. EMBO J. 1997;16:5235–5246. doi: 10.1093/emboj/16.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginisty H, Amalric F, Bouvet P. Two different combinations of RNA-binding domains determine the RNA binding specificity of nucleolin. J. Biol. Chem. 2001;276:14338–14343. doi: 10.1074/jbc.M011120200. [DOI] [PubMed] [Google Scholar]
- 45.Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J. Biol. Chem. 2000;275:18845–18850. doi: 10.1074/jbc.M002350200. [DOI] [PubMed] [Google Scholar]
- 46.Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- 47.Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J. Biol. Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- 48.Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell. 2009;35:414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 50.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 52.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 53.Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J. Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 54.Kharrat A, Derancourt J, Doree M, Amalric F, Erard M. Synergistic effect of histone H1 and nucleolin on chromatin condensation in mitosis: role of a phosphorylated heteromer. Biochemistry. 1991;30:10329–10336. doi: 10.1021/bi00106a034. [DOI] [PubMed] [Google Scholar]
- 55.Dickinson LA, Kohwi-Shigematsu T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell. Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.