Abstract
In eukaryotes, multiple genes encode histone proteins that package genomic deoxyribonucleic acid (DNA) and regulate its accessibility. Because of their positive charge, ‘free’ (non-chromatin associated) histones can bind non-specifically to the negatively charged DNA and affect its metabolism, including DNA repair. We have investigated the effect of altering histone dosage on DNA repair in budding yeast. An increase in histone gene dosage resulted in enhanced DNA damage sensitivity, whereas deletion of a H3–H4 gene pair resulted in reduced levels of free H3 and H4 concomitant with resistance to DNA damaging agents, even in mutants defective in the DNA damage checkpoint. Studies involving the repair of a HO endonuclease-mediated DNA double-strand break (DSB) at the MAT locus show enhanced repair efficiency by the homologous recombination (HR) pathway on a reduction in histone dosage. Cells with reduced histone dosage experience greater histone loss around a DSB, whereas the recruitment of HR factors is concomitantly enhanced. Further, free histones compete with the HR machinery for binding to DNA and associate with certain HR factors, potentially interfering with HR-mediated repair. Our findings may have important implications for DNA repair, genomic stability, carcinogenesis and aging in human cells that have dozens of histone genes.
INTRODUCTION
Histones are essential eukaryotic proteins encoded by multiple genes (1,2). They help to package the lengthy genomic deoxyribonucleic acid (DNA) into the relatively small nucleus, thereby regulating DNA accessibility. However, as histones are positively charged, when present in excess they can bind non-specifically to negatively charged DNA and affect all aspects of DNA metabolism, including DNA repair (3–5). Not surprisingly, the accumulation of excess histones is harmful for the cells, resulting in genomic instability and enhanced sensitivity of the budding yeast Saccharomyces cerevisiae to DNA damaging agents (4,6). All eukaryotes carry multiple genes encoding each core histone protein, ranging from two genes for each core histone in the budding yeast to several hundred in other popular model systems such as Drosophila and Xenopus (2). Each diploid human cell has 28 copies of histone H4 gene alone that encode identical H4 proteins (7). Of the multiple genes encoding each histone protein in eukaryotes, some are non-allelic variants that may have specialized functions (1,2). However, the rationale behind the existence of multiple genes encoding the same histone protein is unclear. As histones are essential for viability, one possibility is that the multiple histone genes simply serve as a backup in case of inactivating mutations in one or more genes. However, this is unlikely as the majority of genes essential for viability in eukaryotes do not have multiple gene copies to serve as backup. A second possibility is that multiple histone genes may be required to synthesize the enormous quantities of histones that are required for chromatin assembly during DNA replication. However, several studies over the past two decades suggest that the full complement of the multitude of histone genes is not required for maintaining cell viability in several species, including the budding yeast (8,9), fission yeast (10), fruit fly (11), chicken (12–14) and mice (15–18). Further, in several cases, the levels of chromatin associated histones or the gross chromatin structure appeared to be unaffected on histone gene deletions, although it is possible that subtle alterations in the fine structure of chromatin do occur and may be exacerbated when combined with other mutations, as exemplified in the budding yeast where histone gene deletions exhibit numerous genetic interactions (19,20), several of which would be indicative of an underlying chromatin structure defect. On the other hand, a potential problem that may arise with the presence of multiple copies of histone genes being driven by very strong promoters is that more histones may end up being synthesized than what is required for chromatin assembly and maintenance (5). To protect the cells from the deleterious effects of excess histone accumulation, histone levels are tightly regulated transcriptionally (21,22), posttranscriptionally (23–25), translationally (26,27) and posttranslationally (3,4). These mechanisms ensure a tight coupling between the levels of DNA and histone synthesis and downregulate histones in response to replication arrest or DNA damage during S-phase (3–5,22,28,29).
To preserve genomic integrity, cells have evolved a number of highly efficient DNA surveillance and repair mechanisms to detect and repair DNA damage caused by both external and endogenous genotoxic agents. Defects in these mechanisms increase the incidence of mutations and genome instability, which are implicated in oncogenesis (30). Upon DNA damage or replication stress, checkpoint responses arrest the cell cycle to provide additional time for efficient repair (31). In the budding yeast, two essential protein kinases, Mec1 and Rad53, play multiple roles in the DNA damage and replication arrest response (32). DNA damage in the budding yeast leads to the Mec1/Tel1 dependent hyperphosphorylation of Rad53 and a dramatic increase in its kinase activity (33). Activation of Rad53 triggers a phosphorylation-mediated cascade of events that bring about all the known responses to DNA damage. Mec1 and Rad53 are also required to prevent both spontaneous and DNA damage-induced collapse of replication forks (34,35) and block the firing of late origins in response to DNA damage in early S-phase (36). As a result, DNA damage leads to an abrupt decrease in DNA synthesis (37). Hence, it is not surprising that both mec1 and rad53 mutants are exquisitely sensitive to DNA damaging agents and strong genetic suppressors of their DNA damage sensitivity are not known.
The role of posttranslational histone modifications in DNA repair has been intensively studied for over a decade (38–42). However, the contribution of histone proteins themselves in regulating DNA repair has only been investigated sporadically over the past few years. The budding yeast linker histone Hho1 was reported to increase the DNA damage sensitivity by suppressing homologous recombination (HR) in a manner dependent on its DNA-binding activity, though the exact mechanism by which Hho1 affects HR is unclear (43). Despite significant differences between the yeast and vertebrate linker histone proteins, murine embryonic stem cells with reduced histone H1 levels were also reported to be resistant to DNA damaging agents, although the DNA repair pathways mediating these effects were not evaluated (44). Using a tetracycline regulated histone H4 gene, it was reported that a reduction in the level of histone H4 results in replication fork collapse, elevated recombination and genomic instability in budding yeast cells (45,46). However, the relevance of these findings to wild-type cells is complicated by the fact that the substantial depletion of histones in these studies resulted in pleiotropic effects on cell physiology including chromatin structure alterations, slow growth, spontaneous replication fork collapse and DNA damage. In fact, the results obtained on the depletion of histone H4 in yeast cells are very reminiscent of the spontaneous DNA damage and S-phase arrest observed in human cells on inhibition of chromatin assembly (47). Nevertheless, given the high degree of conservation of chromatin and DNA repair pathways in all eukaryotes, overall these studies suggest that any effect of histones on DNA damage and repair pathways may also be similarly conserved among all eukaryotes. In recent years, we have uncovered a novel role for the budding yeast checkpoint kinase Rad53 in histone metabolism (3,4). Rad53, but not Mec1, is required for degradation of excess histones that are not packaged into chromatin. As a consequence, rad53 mutants accumulate abnormally high amounts of soluble histones and are sensitive to histone overexpression. The DNA damage sensitivity, slow growth and chromosome loss phenotypes of rad53 mutants can be significantly suppressed by disrupting one of the two loci encoding histones H3/H4, arguing that these phenotypes are partly due to the presence of excess histones. Taken together, these data strongly suggest that there may be an intimate connection between DNA damage sensitivity, DNA repair processes and histone dosage.
In this study, we have investigated the mechanism by which changes in core histone gene dosage affect the DNA damage sensitivity of budding yeast cells. We discovered that a reduction in histone H3 and H4 gene dosage results in substantial resistance to all common DNA damaging agents tested. This appears to be largely due to an increase in the efficiency of repair by the HR pathway (48) on a reduction in histone gene dosage, whereas non-homologous end joining (NHEJ) remains unaffected. Further, the observed effects on DNA damage sensitivity on changes in histone gene dosage are not associated with global changes in the expression of DNA repair genes, alterations in the gross chromatin structure or the DNA damage checkpoint. In fact, we found that a reduction in histone H3 and H4 gene dosage results in a specific reduction in the free pool of histones, whereas the levels of histones associated with chromatin remained unaffected. Using the galactose-inducible HO endonuclease-mediated double-strand break (DSB) formation and repair system (GAL-HO system) (49,50) in budding yeast strains with intact silent mating type loci, we found that cells with reduced histone gene dosage experience greater histone loss around the lesion, whereas the recruitment of several key repair and HR factors to the lesion was concomitantly enhanced. Additionally, free histones were found to associate with several factors involved in HR. On the basis of these data, we propose that excess histones interfere with the HR machinery and prevent it from accessing DNA lesions, thus reducing the efficiency of repair by HR. As such, even a moderate reduction in histone dosage enhances the efficiency of DNA repair and makes cells more resistant to DNA damaging agents. Further, although the reasons behind the existence of multiple histone genes in eukaryotes are unclear, our results suggest the possibility that they may be required to generate high levels of histones to potentially suppress HR, thereby protecting cells against the deleterious effects of excessive recombination (51), particularly during S-phase. Overall, our findings may have major implications for DNA repair, genomic stability and carcinogenesis in human cells that have dozens of histone genes (7).
MATERIALS AND METHODS
Yeast strains, plasmids and media
The yeast strains used are listed in Supplementary Table S2. Standard techniques for growing and transforming yeast were used (52). Plasmids used for histone H3 overexpression have been described in detail elsewhere (3,4). For the galactose-induced expression of HO endonuclease, cells were routinely grown in minimal media supplemented with ‘yeast synthetic drop-out medium supplement’ (Sigma) containing 2% raffinose before the addition of 2% galactose. Galactose inducible genes were repressed by addition of 2% glucose. α-factor was used at a concentration of 5 μg/ml for 1.5–2 h to arrest cells in G1. The hht1-hhf1Δ and hht2-hhf2Δ gene pairs were deleted from various strains using a polymerase chain reaction (PCR)-based HR-dependent strategy described previously (53). Similar plasmid construct-based HR strategies were used to generate the strains carrying a 3XFLAG epitope (consisting of the artificial peptide sequence DYKDDDDK) on the 3′-end of the chromosomal ASF1 gene (3), as well as for the FLAG epitope tagging of the MRE11 gene. Strains carrying NAP1 expression vectors (54) were generated by transforming the cells either with the low-copy plasmid pRS316-FLAG-yNap1 (for Nap1 IP experiment in Figure 2d) or with the high-copy number plasmid pYES2-Nap1 (for the genetic analysis shown in Supplementary Figure S5). The tandem affinity purification (TAP)-tagged strains were obtained commercially (Open Biosystems).
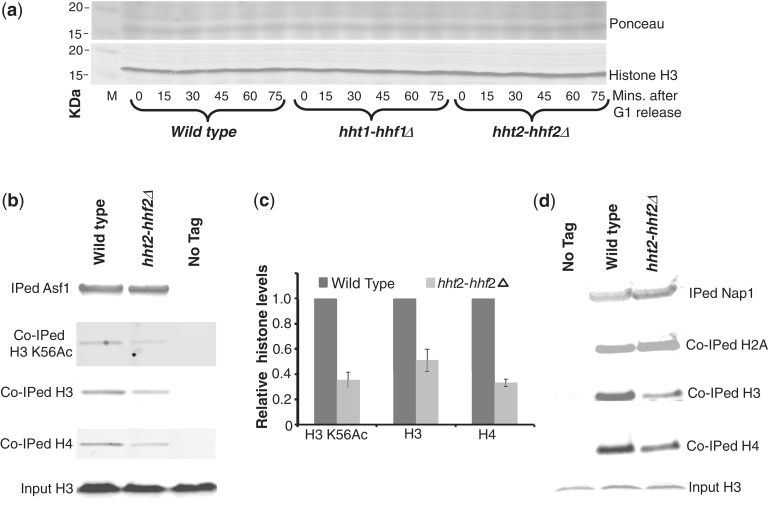
Figure 2.
Deletion of the histone gene pair hht2-hhf2 does not alter the levels of chromatin associated histones but reduces free histone pools. (a) The levels of chromatin associated histones are unchanged on deletion of the hht2-hhf2 gene pair. Cells from the indicated strains were grown overnight in rich media and arrested in G1 by treating with α-factor for 90 min. Cells were then released from the G1 arrest in fresh media and allowed to progress through S-phase. Cells (1 × 107) were harvested at the indicated time points following release from G1 arrest, and whole cell lysates were prepared as described previously (107). The lysates were resolved on a sodium dodecyl sulphate 18% polyacrylamide gel and processed for western blotting using the H3-C antibody as described previously (3,4) to measure the total endogenous histone H3 levels (which mainly represent the chromatin bound histones). The upper panel shows the Ponceau S staining of the total proteins on the membrane to demonstrate equal loading. (b) Histones associated with Asf1 are significantly reduced in the hht2-hhf2 deletion strain. Asf1-FLAG was immunoprecipitated (IPed) from soluble S-phase whole cell extracts (WCEs) corresponding to the wild-type and hht2-hhf2 deletion strains as described in ‘Materials and Methods’ section. The IPed material was resolved on gradient gels and processed for western blotting with different antibodies to measure the levels of histone H3, H4 and H3 K56Ac (a marker for newly synthesized histones) co-immunoprecipitated (co-IPed) with Asf1-FLAG. The bottom panel shows the total amount of histone H3 in 0.5% of the input material used for the IP reactions to demonstrate that roughly equal amounts of proteins were present in the WCEs. (c) Quantitation of the data shown in (b) from three independent experiments. The signals obtained for histones associated with Asf1-FLAG were normalized to the signals obtained for IPed Asf1-FLAG on western blots. Error bars depict standard error of the mean. (d) Nap1-associated histones H3 and H4, but not H2A, are significantly reduced in the hht2-hhf2 deletion strain. Histones co-IPed with Nap1-FLAG were measured as described earlier for Asf1-FLAG-associated histones in (b).
Immunoprecipitations and western blotting
For routine immunoprecipitation (IP) of tagged proteins, exponentially growing cells were harvested from 1 liter cultures at ∼2.5 × 107 cells/ml. For the IP of Asf1-FLAG and Nap1-FLAG (Figure 2b–d), as well as Rad52-TAP (Figure 7d and e) from S-phase cells, the cell cultures were first arrested in G1 by treating with α-factor for 90 min and then released for 40 min in fresh media lacking α-factor to allow them to progress to mid S-phase. Cells were harvested and processed essentially as described previously (3,4,55). Briefly, whole-cell extracts (WCEs) were prepared for IP by grinding cells in liquid nitrogen in a SPEX CertiPrep 6850 Freezer Mill in 20 ml lysis buffer (per liter of yeast culture) containing protease, phosphatase, deacetylase and proteasome inhibitors (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH pH 7.5, 110 mM potassium acetate, 10% glycerol, 0.1% Tween-20, 1 mM sodium vanadate, 50 mM sodium fluoride, 50 mM sodium β-glycerophosphate, 10 mM sodium butyrate, 10 mM 2-mercaptoethanol, 2X Roche protease inhibitor cocktail and 10 µM MG-132 proteasome inhibitor). Following clarification of the extracts by centrifugation at 18 000g for 20 min at 4°C to remove insoluble material and precipitate chromatin fragments, soluble extracts from equal amounts of cells were incubated overnight at 4°C with FLAG M2 antibody resin (Sigma) or IgG sepharose (Amersham Biosciences) to IP proteins tagged with the FLAG and TAP epitopes, respectively. The immunoprecipitated (IPed) material was resolved on a pre-cast 4–12% polyacrylamide gradient gel (BioRad) and processed for western blotting as described previously using specific antibodies to detect the presence of FLAG (M2 from Sigma) or TAP (TAP from Open Biosystems) tagged IPed and co-immunoprecipitated (co-IPed) proteins. The H3-C and the polyclonal H4 antibodies used to detect histone H3 and H4, respectively, have been described previously (3,4). The affinity purified antibody used to specifically detect H3 K56 Ac has also been described previously (56). Antibodies used to detect histone H2A were obtained commercially (Millipore).
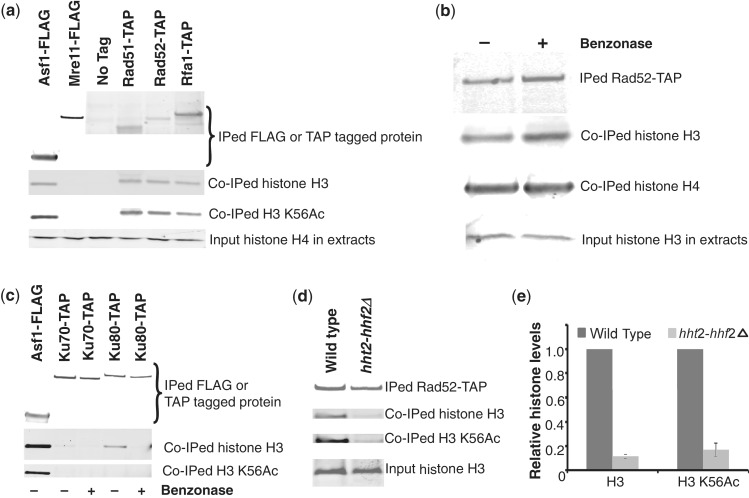
Figure 7.
Free histones associate physically with several HR factors but not with NHEJ factors. (a) Histones associate with Rad51, Rad52 and Rfa1 in vivo. The indicated epitope-tagged proteins were IPed from soluble extracts prepared from exponentially growing cultures and processed for western blotting using specific antibodies to detect co-IPed histones as described in ‘Materials and Methods’ section. As indicated by the presence of H3 K56Ac in the immunoprecipitates from TAP-tagged Rad51, Rad52 and Rfa1, substantial amounts of newly synthesized histone H3 appear to associate with these HR factors. No histones were found to associate with Mre11. The association of histones with Asf1 is used as a positive control and to allow a rough estimate of the relative amounts of histones associated with the recombination factors. The bottom panel shows the total amount of histone H4 in 0.5% of the input material used for the IP reactions to demonstrate that roughly equal amounts of proteins were present in the WCEs. (b) Free histones rather than chromatin-associated histones interact with Rad52 in soluble cell extracts. Rad52-TAP was IPed as described above in (a) except that the IP reactions were treated with or without Benzonase, a potent synthetic nuclease, for 30 min at 37°C to degrade any DNA. The IPed material was then processed for western blotting as described above in (a). No difference in the association of histones with Rad52-TAP was observed on Benzonase treatment, strongly suggesting that these histones arise from the soluble free histone pool rather than chromatin-associated histones. (c) NHEJ factors Ku70 and Ku80 associate with small amounts of chromatin bound histones. The indicated proteins were IPed as described above in (a) and treated with or without Benzonase as described above in (b). Co-IPed histones were detected as in (a). (d) The association of free histones with Rad52 is drastically reduced in the hht2-hhf2 deletion strain. Rad52-TAP was IPed from S-phase wild-type and hht2-hhf2Δ strains and the co-IPed histones were detected by western blotting as in (a). (e) Quantitation of the data shown in (d). The signals obtained for histones associated with Rad52-TAP were normalized to the signals obtained for IPed Rad52-TAP on western blots from multiple independent experiments. Error bars depict standard error of the mean.
Plasmid recircularization assay for NHEJ efficiency
The plasmid recircularization assay to study NHEJ was carried out as described previously (39). In this assay, a plasmid containing Kanr marker is first digested using the restriction endonuclease HindIII in the middle of the marker to mimic a DSB. Then 0.2 µg of the digested (linearized) or undigested (supercoiled) plasmid was transformed into 3 × 107 cells from each strain, and the transformants were selected on G418 plates. The Kanr marker encodes resistance to the antibiotic G418 and lacks any significant homology within the budding yeast genome, so the Hind III generated DSB can only be repaired by NHEJ. Only those cells that have successfully repaired the linearized plasmid and generated a functional Kanr gene will survive on media containing G418. NHEJ repair efficiency was calculated by comparing the number of colonies formed by strains transformed with the digested plasmid versus undigested plasmid.
Chromatin immunoprecipitation
Cells were grown overnight to reach a concentration 1–2 × 107 cell/ml in YP-Raffinose media. Galactose (2%) was added to the culture at the 0’ time point, and then 10 ml aliquots of the cultures were collected every 30 min and fixed with 1% formaldehyde for 15 min. Fixed cells were disrupted using glass beads in FA-lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 µg/ml leupeptin and 1 µg/ml pepstatin), and cell lysate was sonicated on ice to shear the chromatin into 300–1000 bp fragments. Equal amounts of WCE were incubated at 4°C for 1.5 h with 20 µl of IgG-coupled sepharose beads (GE healthcare) to immunoprecipitate TAP-tagged proteins or histone H4 antibody-bound protein-A beads to pull down histone H4. The beads were washed sequentially with 2 ml FA-lysis buffer, FA-500 (50 mM HEPES–KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate), LiCl wash solution (10 mM Tris-HCl pH 8, 250 mM lithium chloride, 1 mM EDTA, 0.5% Nonidet P40 and 0.5% sodium deoxycholate) and TES (10 mM Tris–HCl pH 7.5, 1 mM EDTA and 100 mM NaCl). IPed material was eluted twice by incubating the beads in 100 µl elution buffer (100 mM Tris–HCl pH 7.8, 400 mM NaCl, 10 mM EDTA and 1% sodium dodecyl sulphate) for 10 min at 37°C. To degrade proteins and reverse crosslinks, samples were first incubated with proteinase K (20 µg) for 1 h at 42°C and then overnight at 65°C. The IPed DNA was purified by phenol–chloroform extraction and then quantitated by real-time PCR (Applied Biosystems) using primers and fluorescently labeled probes (TaqMan) to specifically amplify sequences near the HO cleavage site (70 bp downstream) at the MATa locus. The ACT1 locus was also assayed to serve as internal control. Real-time PCR reactions were performed in triplicate for each DNA sample, and the data from three independent experiments were averaged.
RESULTS
Histone gene dosage has a major influence on the sensitivity of budding yeast cells to DNA damaging agents
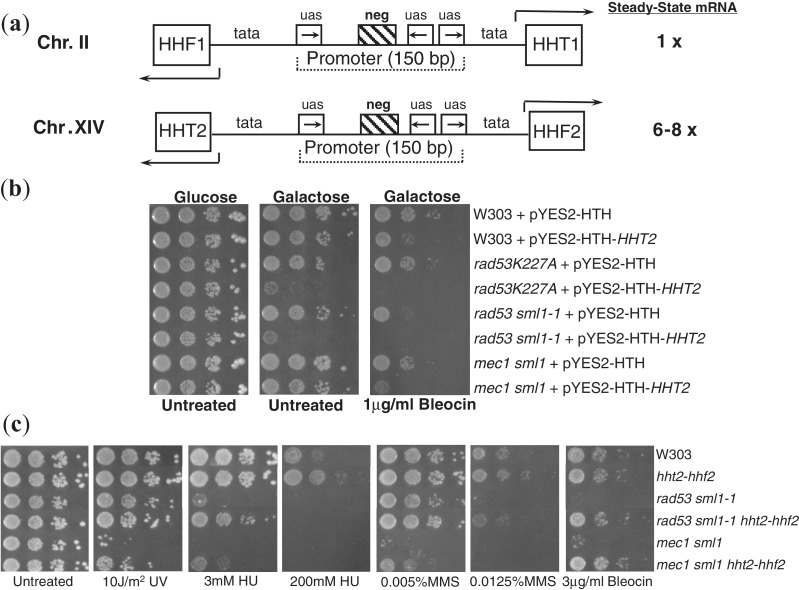
The budding yeast has two copies of each core histone gene (Figure 1a), and only one copy is required for survival (21). Of the two gene pairs (HHT1-HHF1 and HHT2-HHF2) encoding histone H3 and H4, the second gene pair HHT2-HHF2 contributes 6–8 times more histone messenger ribonucleic acid (mRNA) during S-phase than the first gene pair HHT1-HHF1 (57). However, any one of the two H3–H4 gene pairs is sufficient for viability, despite the lack of dosage compensation by the remaining gene pair. These characteristics of the budding yeast histone H3 and H4 genes make it an ideal system to study the relevance of multiple genes encoding the same histone protein in eukaryotes. We have previously reported that histone overexpression increases the DNA damage sensitivity of the checkpoint mutant rad53 (that is also defective in degrading excess histones), whereas a reduction in histone gene dosage results in resistance to DNA damaging agents (3,4). However, any potential influence of histone dosage on DNA repair in wild-type cells has not been evaluated previously. Hence, we first assayed the effect of histone overexpression on the DNA damage sensitivity of wild-type W303 and isogenic DNA damage checkpoint mutant yeast strains as controls (Figure 1b). Consistent with our previous findings (3,4), rad53 mutants were extremely sensitive to the strand-break reagent Bleocin (58), particularly in the presence of histone overexpression. Even the wild-type cells were more sensitive to DNA damage in the presence of histone overexpression, suggesting that excess histones can presumably block efficient DNA repair in wild-type cells as well. Although we have presented data for histone H3 overexpression only over here, we have previously demonstrated that this provides an excellent phenocopy of co-overexpression of histone gene pairs (4,59). As histone overexpression resulted in an increase in the DNA damage sensitivity of all strains tested, we wondered if a reduction in histone gene dosage could reduce DNA damage sensitivity of yeast strains. To test this idea, we next examined the effect of deletion of the hht2-hhf2 gene pair on the DNA damage sensitivity of wild type and the rad53 and mec1 DNA damage checkpoint mutant strains to a variety of common DNA damaging agents (Figure 1c). Compared with the wild-type strain, there is only 14∼20% histone H3–H4 mRNA in the strains carrying hht2-hhf2 deletions (57). Mec1 is the homolog of the essential human checkpoint kinase ATR (Ataxia Telangiectasia and Rad3 related) and regulates all aspects of the DNA damage response (DDR) in budding yeast (32). Surprisingly, deletion of the hht2-hhf2 gene pair resulted in a substantial decrease in the sensitivity of not just the rad53 mutant strains that accumulate excess histones but mec1 mutant strains as well that do not accumulate excess histones (3,4). In fact, the mec1 mutant carrying the hht2-hhf2 deletion was as resistant as the wild-type W303 cells to DNA damage with significantly high amounts of Bleocin (Figure 1c). Such a dramatic suppression of the DNA damage sensitivity of DNA checkpoint mutants such as mec1 toward a variety of different DNA damaging agents is unprecedented and highlights the enormous influence that histone levels can have on DNA damage sensitivity. Similar to the response of cells lacking the linker histone HHO1 to methylmethane sulfonate (MMS) (43), cells carrying the hht2-hhf2 deletion were more resistant to DNA damaging agents than their wild-type counterparts with the full complement of histone genes, and this effect was more prominent at higher doses of the damaging agent (Figure 1c, compare the sensitivity to 200 mM hydroxyurea [HU] and 0.0125% MMS). These results imply that histone gene dosage is a major influence on the DNA damage sensitivity of budding yeast cells and even normal wild-type levels of histones can potentially interfere with efficient repair.
Figure 1.
Histone gene dosage influences DNA damage sensitivity of wild type and DNA damage checkpoint deficient yeast strains. (a) Organization of histone H3 and H4 genes in the budding yeast. The paired histone genes are transcribed divergently from a common promoter. Regulatory elements in the promoter are indicated. uas, upstream activating sequence; neg, negative element; tata, TATA box; HHT, histone H3 gene; HHF, histone H4 gene. The relative levels of total histone mRNA for H3 and H4 contributed by each gene pair is indicated. (b) Overexpression of histone H3 increases DNA damage sensitivity of yeast strains. Indicated strains transformed with either a vector encoding galactose-inducible histone H3 (pYES2-HTH-HHT2) (3), or the empty vector (pYES2-HTH) was plated in 10-fold serial dilutions on glucose or galactose media with or without Bleocin. The plates were incubated for 4 days at 30°C before being photographed. (c) Deletion of one of the hht2-hhf2 histone gene pair encoding histone H3 and H4 decreases DNA damage sensitivity of yeast strains. Tenfold serial dilutions of exponentially growing yeast strains of the indicated genotypes were plated and treated with the indicated amounts of various DNA damaging agents. UV, ultraviolet; HU, hydroxyurea; MMS, methyl methane sulfonate. The plates were incubated for 4 days at 30°C before being photographed.
Changes in histone gene dosage have little effect on the bulk chromatin structure or global gene expression but significantly reduce free histone pools
We next sought to understand the mechanism/s by which histone gene dosage may influence the DNA damage sensitivity of yeast cells. Because we have recently published the effects of histone overexpression on yeast cells in extensive detail (59), we will focus mainly on the effects of histone gene deletion on yeast cells for the rest of the article. As histones are a key component of the chromatin, it is reasonable to hypothesize that an alteration of histone gene dosage may change chromatin structure. To determine whether global chromatin structure is altered in strains carrying histone gene deletions, we performed micrococcal nuclease (MNase) digestion analysis of chromatin from wild-type and hht2-hhf2 deletion strains (Supplementary Figure S1). Consistent with previous reports (8,60), MNase digestion patterns were identical between wild-type and hht2-hhf2Δ strains, suggesting that the bulk chromatin structure is not altered on histone gene deletion. However, this does not rule out fine structural changes in the chromatin of the hht2-hhf2Δ strain, which are in fact known to occur on histone H3 overexpression that does not alter the bulk chromatin structure either (59). Chromatin structure is generally considered as a barrier to gene expression by inhibiting the binding and function of the transcriptional machinery (61). We have recently reported changes in the fine structure of chromatin and in the expression patterns of ∼4% of the budding yeast transcriptome on an increase in histone dosage (59). Thus, another possible effect of a reduction in histone gene dosage could be changes in the expression of genes (60), especially those genes known to be involved in DDR (62). We investigated this possibility using microarray-based genome wide gene expression analysis (63). Compared with the wild-type cells, the expression of ∼50 genes was significantly affected (expression changes of more than 2-fold) in the hht2-hhf2 deletion strain (Supplementary Figure S2 and Supplementary Table S1), none of which are known to be involved in the DDR (62). These results suggest that yeast cells can form a largely intact and functional chromatin structure even in hht2-hhf2Δ strains that lack the main supplier of the histone H3/H4 mRNAs. This is not surprising given that yeast cells normally appear to produce more histone proteins than is necessary for packaging their genome into chromatin (5).
Cells typically have very small pools of free histones that are extremely hard to detect in a vast background of chromatin associated histones, but they accumulate to higher levels on replication arrest (64). Free histone pools constitute only ∼1% of the total histones in human cells (65) but appear to play important roles in cellular physiology (66). Our experience suggests that free histone levels in the budding yeast cells are similar or slightly lower than in human cells. The absence of chromatin structure changes in the budding yeast cells lacking the hht2-hhf2 (Supplementary Figure S1) suggests that these cells produce sufficient quantities of histones. We verified this directly by measuring the total levels of histone H3 in cells carrying hht1-hhf1 or hht2-hhf2 deletions. We did not detect any differences in the total amount of histone H3 (which is primarily a measure of chromatin bound histones) in these strains compared with the wild-type strain (Figure 2a). Next, we also measured the free pools of histones in the wild-type and hht2-hhf2 deletion strains by quantitating the histones associated with the H3–H4-specific histone chaperone Asf1 (3,4,55). In cells synchronized in S-phase (when histone genes are expressed), we observed a significant reduction in the levels of histones H3 and H4 associated with FLAG-tagged Asf1 in the hht2-hhf2Δ strain (Figure 2b and c), suggesting that free histone pools are reduced in this strain. To further confirm that the histones associated with Asf1 are indeed free histones, we measured the levels of acetylated lysine 56 on histone H3 (H3 K56Ac) bound to Asf1 and found its levels to mirror those of histone H3 itself. As the H3 K56Ac mark is present only on newly synthesized histone H3 and is rapidly removed following its deposition on chromatin (56), we conclude that the histones associated with Asf1 represent newly synthesized histones. We further confirmed these findings by measuring histones associated with another histone chaperone, Nap1, which binds all four core histones (67). Once again, the levels of histone H3 and H4 associated with Nap1-FLAG were specifically reduced on the deletion of hht2-hhf2, whereas the levels of H2B were unaffected (Figure 2d). Hence, overall we conclude that free histone H3 and H4 pools are considerably reduced in the hht2-hhf2Δ strain and could have a significant effect on cell physiology.
The DNA damage checkpoint is efficiently activated in cells with reduced histone dosage
The observed effect of a reduction in histone gene dosage on the DNA damage sensitivity of yeast cells (Figure 1c) could potentially be due to differences in the DNA damage checkpoint activation. Hence, we also investigated whether there were any differences in DNA damage checkpoint activation between wild-type and hht2-hhf2Δ strains. First, we used pulsed field gel electrophoresis (68) to visualize the presence of ongoing chromosome replication in HU-treated cells (Supplementary Figure S3), which revealed that both wild-type and the hht2-hhf2 deletion strains have intact intra-S phase checkpoint. Then we used flow cytometry to follow the cell cycle progression in the presence of MMS, which showed a clear DNA damage checkpoint-mediated arrest in both wild-type and the hht2-hhf2 deletion strains (Supplementary Figure S4). Hence, we were unable to detect any significant differences in the activation of the DNA damage cell cycle checkpoints or in the rate of S-phase progression between wild-type and hht2-hhf2Δ using these assays.
Reduction in histone gene dosage enhances survival of yeast cells following a DNA DSB
Because a reduction in histone gene dosage results in resistance to a variety of DNA damaging agents that cause different kinds of DNA lesions and are often repaired by different repair pathways (Figure 1c), we next turned our attention to potential DNA repair pathway/s that may be used in common for the repair of a variety of different DNA lesions. Several different kinds of DNA lesions may be processed by different repair machineries to ultimately generate DSBs (69) that can be repaired by the two competing pathways of HR or NHEJ (48,70). A DSB is perhaps the most harmful kind of DNA damage that a cell could incur as this event could potentially lead to loss of the entire chromosome arm. In fact, even a single unrepaired DSB can cause lethality (71). An in vivo model system has been developed in budding yeast cells using galactose inducible-HO endonuclease (GAL-HO) expression (49,50,72,73) for the study of DSB repair. In this system, a single DSB at the naturally occurring HO cleavage site in the mating type (MAT) locus on chromosome III can be created on GAL-HO expression in budding yeast cells lacking a functional endogenous HO gene. This DSB at the MAT locus is predominantly repaired by the multi-step and multi-factorial process of HR (48), where the broken ends are first resected in a 5′–3′ direction, giving rise to 3′ overhangs with single-stranded DNA (74). Then, one of the two silent mating loci (HMLα and HMRa) adjacent to the MAT locus is used as a donor template and copied by new DNA synthesis to accurately repair the DSB at the MAT locus (48). NHEJ involves the simple ligation of the broken ends and normally repairs the DSB at MAT locus at a much lower frequency than HR in the presence of the silent mating loci but becomes the only way to repair the DSB at MAT locus when the silent mating type loci are deleted.
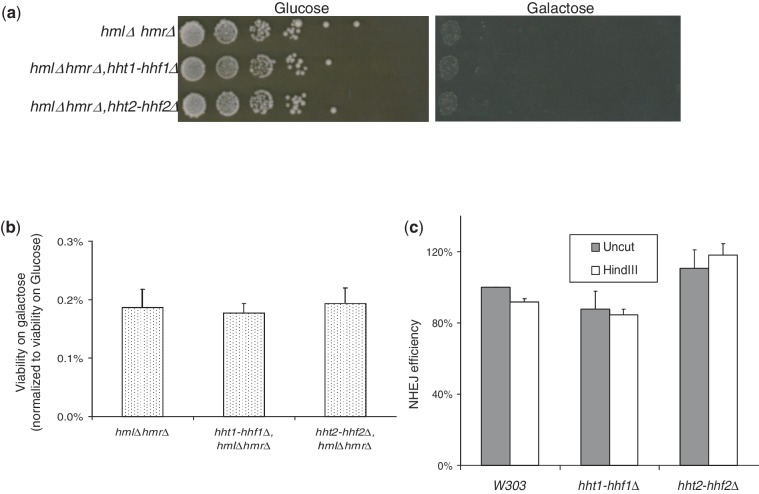
Next we decided to test whether DSB repair was indeed affected on deletion of the hht2-hhf2 gene pair by inducing a single defined DSB in the yeast genome using GAL-HO For this, we first stably integrated the GAL-HO construct at the ADE3 locus in wild-type, hht1-hhf1Δ and hht2-hhf2Δ strains with intact silent mating type loci. Fivefold serial dilutions of these strains were plated on glucose or galactose containing plates. Compared with wild type, the hht2-hhf2Δ strain showed dramatic improvement of viability on galactose containing plates, whereas hht1-hhf1Δ strain appeared to exhibit a small but reproducible improvement in survival on galactose media (Figure 3a). This result was confirmed by a viability assay to quantify the ratio of viable colonies formed on galactose versus glucose plates, which revealed that the survival of the hht2-hhf2Δ strain following the GAL-HO-mediated DSB at the MAT locus was ∼5000-fold better than the wild-type strain (Figure 3b). Using the same assay, yeast cells lacking the Hho1 linker histone have been reported to have a ∼2-fold better survival on GAL-HO induction compared with wild-type cells (43). The difference in the magnitude of the effects of hho1Δ and hht2-hhf2Δ in terms of surviving GAL-HO expression is likely to be a reflection of the much stronger affinity of core histones for DNA compared with linker histones. The effect on viability of the hht1-hhf1Δ and hht2-hhf2Δ strains following GAL-HO cleavage mirrors the relative transcriptional output of the HHT1-HHF1 and HHT2-HHF2 gene pairs. As the HHT2-HHF2 gene pair contributes 80–86% of the histone H3–H4 mRNAs in the cell (57), it strongly argues that the observed effect of the hht2-hhf2Δ strain on viability in response to the GAL-HO break is due to the differences in endogenous histone levels in the cells carrying histone gene deletions. These results also predict that overexpression of histone chaperones such as Nap1 (67) to sequester these excess endogenous histones should improve viability of cells following a DSB. Consistent with this idea, we find that Nap1 overexpression enhances the survival of wild-type cells following a GAL-HO-mediated DSB at the MAT locus (Supplementary Figure S5). Although the magnitude of the Nap1 effect was not as spectacular as the effect of hht2-hhf2 deletion on the survival of a DSB, it is possible that overexpression of different or multiple histone chaperones known in the budding yeast may be required to observe the full potential effect of histone chaperone overexpression on the survival of a DSB.
Figure 3.
A reduction in histone gene dosage enhances survival of budding yeast cells following an induced DSB in a checkpoint independent manner. (a) Survival of strains following induction of HO-mediated DSB. The indicated yeast strains carrying galactose inducible HO endonuclease (GAL-HO) were grown overnight in YP-raffinose media to reach a concentration of 1–2 × 107 cells/ml. Then 5-fold serial dilutions of the indicated strains were plated on glucose or galactose plates. The plates were incubated for 4 days at 30°C before being photographed. (b) Quantitation of the survival of yeast strains following induction of HO-mediated DSB. The viability of the strains was tested by plating 1000 cells from each strain in (a) on YP-glucose media or 100 000 cells on YP-galactose media in duplicate (only 10 000 cells were plated for the hht2-hhf2Δ strain due to its much higher viability on galactose media). The viability was quantitated by normalizing the number of colonies formed on galactose to the number of colonies formed on glucose media. The error bars represent standard error of the mean from three independent repeats of the experiments under identical conditions. (c) Survival of rad53Δ strains following induction of HO-mediated DSB. Cell growth was assayed as described above in (a). (d) Survival of mec1Δ strains following induction of HO-mediated DSB. Tenfold serial dilutions of the indicated strains were plated on glucose or galactose plates, and cell growth was assayed as described above in (a).
It is possible that the dramatic differences between wild-type and hht2-hhf2Δ cells in surviving the GAL-HO-mediated DSB is due to differences in the cleavage efficiency of HO at the MAT locus in these strains. To exclude this possibility, we directly assayed the cleavage efficiency at the MAT locus in these two strains at various time points following the addition of galactose by southern blotting using a probe specific to a region adjacent to the HO cleavage site as described previously (72). As judged by the appearance of the cleaved MATa product in both the strains with similar kinetics following galactose addition, we conclude that there is no significant difference between the rate and extent of HO cleavage in wild-type and hht2-hhf2Δ strains (Supplementary Figure S6).
Another potential reason for the dramatic increase in the survival of the hht2-hhf2Δ strain carrying the GAL-HO construct on galactose media compared with wild-type cells could be that this strain mutates the HO cleavage site at the MAT locus making it refractory to further cleavage. We investigated this directly by sequencing the HO cleavage site in 17 colonies of the hht2-hhf2Δ strain isolated from galactose plates and found that they all had identical wild-type HO cleavage sites. Further, the hht2-hhf2Δ strain did not show any increase in mutation frequency compared with wild-type cells as assayed by the reversion rates of the ade2-1 mutation in these strains. Hence, all our results are consistent with the hht2-hhf2Δ strain repairing the GAL-HO-mediated DSB at the MAT locus with high fidelity and efficiency.
Reduction in histone gene dosage enhances survival of yeast cells following a DSB in a checkpoint-independent manner
Lethality due to the GAL-HO-mediated DSB in the continuous presence of galactose has been linked to the persistent activation of the DNA damage checkpoint, resulting in the prolonged arrest of the cells in G2/M (75–77). However, we did not find any differences in checkpoint activation between the wild-type and hht2-hhf2Δ strains (Supplementary Figures S3 and S4). Consistent with these results, the rad53 deletion strain that is defective in the DNA damage checkpoint also showed much better survival of GAL-HO-mediated DSB on the deletion of hht2-hhf2 (Figure 3c). Because of the much milder effect of the hht1-hhf1 deletion on the DNA damage sensitivity compared with the dramatic effects observed on hht2-hhf2 deletion (Figure 3a–c), we decided to focus mainly on the hht2-hhf2Δ strains for the rest of our studies. We obtained similar results for the mec1 deletion strain that lacks a functional DNA damage checkpoint but nevertheless exhibited a remarkably improved survival of the GAL-HO-mediated DSB on the deletion of hht2-hhf2 (Figure 3d). These data strongly argue that the better survival of yeast strains carrying the GAL-HO construct on galactose media is independent of a functional DNA damage checkpoint.
Improved survival of strains carrying a reduced histone dosage following DSB induction depends on HR and not on NHEJ
The HO-mediated DSB at the MAT locus is predominantly repaired by the HR pathway in the presence of the homologous donor sequences, but NHEJ may also play a role (48–50,72,73). Hence, we next tested whether the improved survival of strains lacking hht2-hhf2 to a GAL-HO-mediated DSB was dependent on the HR or NHEJ-mediated repair. For this we used a strategy similar to the one used in Figure 3a and b, except that the GAL-HO carrying strains now had the HMRa and HMLα sequences deleted from the silent mating type loci to prevent HR-mediated repair of the HO-induced DSB at the MAT locus and force the repair to occur exclusively by the NHEJ pathway. Tenfold serial dilutions of these strains were plated on glucose or galactose containing media, and their growth was monitored over several days (Figure 4a). Although the growth on galactose media was very poor presumably due to the repeated cycles of DSB repair and cleavage by HO in the continuous presence of galactose, the improved survival of strains lacking the hht2-hhf2 gene pair was completely abolished on the deletion of HMRa and HMLα sequences, and no significant differences were observed between the different strains. To obtain more quantitative data, a survival assay was also carried out using these cells, and the repair efficiency was estimated by comparing the number of colonies formed on galactose versus glucose plates, but once again no significant differences were observed in the survival of the strains (Figure 4b). To further rule out any significant contribution of NHEJ, we also used a plasmid recircularization assay to study NHEJ as described previously (39). Consistent with the NHEJ-mediated repair of the DSB at the MAT locus in the absence of HMRa and HMLα sequences (Figure 4a and b), no significant differences in NHEJ repair efficiency were observed in strains with altered histone gene dosage using the plasmid recircularization assay (Figure 4c). Together, these results clearly show that NHEJ efficiency is not affected by a reduction in histone gene dosage and that the improved survival of a GAL-HO-mediated DSB in strains carrying reduced histone dosage is dependent on HR-mediated repair.
Figure 4.
Reduction in histone gene dosage affects the efficiency of DSB repair by the HR but not the NHEJ pathway. (a) Survival of strains lacking the HMRa and HMLα donor sequences on GAL-HO-mediated DSB induction. Tenfold serial dilutions of the indicated GAL-HO strains carrying deletions of hml and hmr sequences were plated to determine their relative viability on glucose and galactose media. The plates were incubated for 4 days at 30°C before being photographed. (b) Quantitation of the survival of strains lacking the HMRa and HMLα donor sequences following a DSB created by HO endonuclease. Quantitative representation of the result shown in (a) exactly as described for Figure 3b. (c) Plasmid recircularization assay for NHEJ. Equal amounts of HindIII linearized or uncut supercoiled plasmids carrying a KanR marker for G418 resistance were transformed into the same number of yeast cells from each strain and plated on media containing the selection antibiotic G418. Viable colonies were counted and normalized to wild-type cells transformed with uncut supercoiled plasmid. The error bars represent standard error of the mean from three independent repeats of the experiments under identical conditions.
Reduction in histone gene dosage enhances spontaneous recombination rates
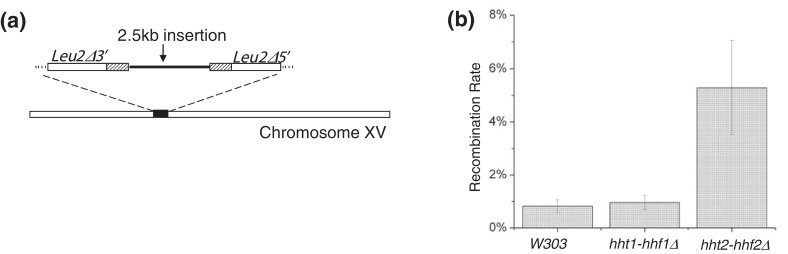
The recombination-mediated repair of a HO-mediated DSB at the MAT locus is a highly specialized, site-specific and natural event in the budding yeast life cycle, and the results obtained in Figure 3a and b may not be generally applicable to all HR-mediated events occurring in yeast cells (73). To ascertain whether the observed effect of hht2-hhf2 deletion on the repair of a DSB at the MAT locus was applicable to HR at other sites in the yeast genome, we next investigated the effect of hht2-hhf2 deletion on HR between direct repeats using the previously described LU system (Figure 5a) (78). Using the LU system integrated at the HIS3 locus, we found that spontaneous HR between direct repeats is significantly elevated in the hht2-hhf2 deletion strain (Figure 5b), and nearly identical results were obtained with the LU system maintained on a plasmid or a linear mini-chromosome. Hence, we conclude that a reduction in histone gene dosage results in a general increase in HR and may explain the lower sensitivity of hht2-hhf2 deletion strains to multiple DNA damaging agents.
Figure 5.
Reduction in histone gene dosage results in elevated rates of spontaneous homologous recombination (HR). (a) Scheme for the LU direct repeat recombination system. In the LU system (78), two 600 bp nonfunctional internal fragments of the LEU2 gene are interrupted by a 2.5 kb fragment. Successful HR will excise the 2.5 kb fragment and generate a functional LEU2 gene, allowing recombinants to be scored as Leu+ colonies on synthetic media lacking leucine. The LU recombination system can be integrated in different locations in the yeast genome or placed on a centromeric plasmid and as such it can be used to obtain a clear picture of a general effect on HR. (b) Reduction in histone gene dosage elevates HR rates between direct repeats. A LU system was inserted into the HIS3 locus on chromosome 15 in the indicated strains. Wild-type W303, hht1-hhf1Δ and hht2-hhf2Δ strains carrying the LU direct repeat system were plated on YPD or synthetic media lacking leucine (-LEU). The frequencies of Leu+ recombinants in the different strains were calculated as described previously (78) to calculate the recombination frequencies. The error bars represent standard error of the mean from three independent repeats of the experiments under identical conditions.
Histones compete with HR factors for binding to damaged DNA
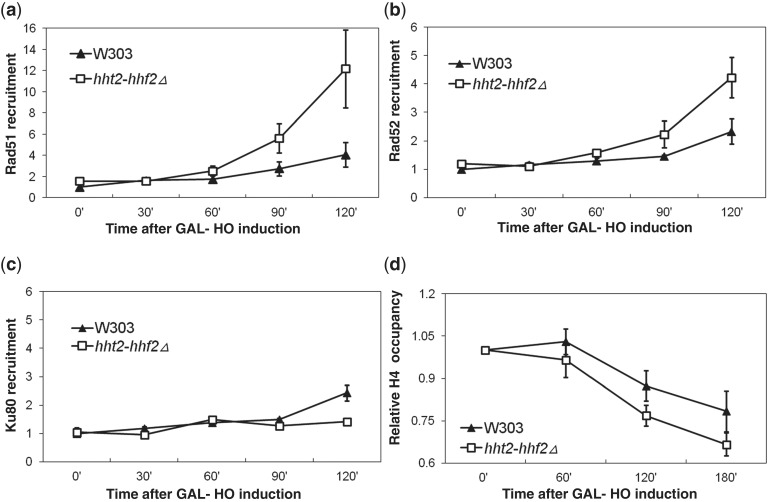
Both HR and NHEJ pathways require that a number of repair factors have direct access to the DNA at the site of the lesion (48). As the major DNA-binding proteins in eukaryotic cells are histones, accessibility to the DSB would be nearly guaranteed if the region around the lesion was largely histone free during DNA repair (79). This is indeed the case during repair of the HO-mediated DSB at the MAT locus from which histones are displaced presumably due to the actions of chromatin remodeling factors (80,81). However, if excess free histones are present in the vicinity of the DNA lesion, they can potentially compete with repair proteins for binding to DNA repair sites, thereby inhibiting or slowing down the repair process. The converse of this scenario is also possible, such that if free histone levels are low, the repair factors will not have to compete with these histones for binding to the DNA at the site of the lesion and thus carry out the repair more efficiently. To test whether this is true in vivo, we introduced the hht2-hhf2 deletion and the GAL-HO construct in commercially obtained strains carrying TAP-tagged HR factors (Rad51, Rad52 and Rfa1, the largest subunit of the replication protein A [RPA]) and the NHEJ factor Ku80. The association of the TAP-tagged proteins and histone H4 with the DNA in the vicinity of the DSB at the MAT locus on cleavage by the HO endonuclease was assayed using chromatin immunoprecipitation (ChIP) as described previously (82). The DNA co-IPed by the HR, NHEJ or histone H4 was quantitated by quantitative real-time PCR (83) using fluorescently labeled primers to specifically amplify sequences adjacent to the HO cutting site in the MATa locus. ChIP experiments that have been carried out on the MAT locus in the context of DSB repair so far have largely used strains lacking the HMRa and HMLα donor sequences that are incapable of repairing the HO break by HR, which is likely to result in the accumulation of long stretches of single-stranded DNA in the continuous presence of galactose (41,42,80,84). Our experiments have been carried out using strains that are more physiologically relevant for the study of HR as they have intact HMRa and HMLα sequences that allow high efficiency repair of the HO break by HR. Using these strains, we consistently observed a greater enrichment of the HR factors Rad51 (Figure 6a), Rad52 (Figure 6b) and RPA (Supplementary Figure S7) at the HO-mediated DSB in the MAT locus, particularly at the later time points. This result is consistent with the idea that a reduced histone dosage facilitates HR by increasing the probability of the association of the HR repair factors with DNA in the vicinity of the DSB. In contrast, no significant differences were observed in the recruitment of the NHEJ factor Ku80 between wild-type and hht2-hhf2Δ strains (Figure 6c), consistent with our finding that NHEJ is not affected by a reduction in histone dosage. Moreover, we also consistently observed a greater loss of histone H4 from the vicinity of the DSB in the hht2-hhf2 deletion strain compared with the wild-type cells (Figure 6d). No significant differences were observed in histone occupancy at the MAT locus between the wild-type and hht2-hhf2 deletion strains before HO cleavage (0’ time point in Figure 6d). Together, these results strongly suggest that there is competition between DNA repair factors and histones for binding to damaged DNA in wild-type cells and a reduction in histone dosage alleviates this competition, thereby facilitating DNA repair.
Figure 6.
Histones compete with HR factors for binding to DNA repair sites. (a) Recruitment of Rad51-TAP to a HO-mediated DSB at the MAT locus. Galactose was added to induce a HO-mediated DSB at the MATa locus in wild-type or hht2-hhf2Δ yeast strains carrying TAP-tagged Rad51. Cells were harvested every 30 min following the addition of galactose and subjected to ChIP analysis as described in the ‘Materials and Methods’ section. Real-time PCR signal obtained from the MATa locus was normalized to the background signal obtained from the ACT1 locus, which serves as an internal control to ensure that equal amounts of DNA was used in each reaction. All data are shown relative to the Ct (cycle threshold) of the 0 time point, which was given an arbitrary value of 1 for the wild-type sample. The error bars represent standard error of the mean from three independent repeats of the experiments under identical conditions. (b) The recruitment of Rad52-TAP was studied as described above in (a). (c) The recruitment of Ku80-TAP was studied as described above in (a). (d) Loss of histone H4 from a HO-mediated DSB at the MAT locus which was studied as described above in (a).
Histones associate with HR factors
Numerous reports have suggested genetic interactions between HR factors, histones and histone chaperones (40,43,46,81,85–90). Further, certain HR factors such as RPA have been reported to physically interact with histones (91,92). Hence, excess histones may not only be interfering with the activities of HR factors by competing for binding to the DNA but also by physically associating with HR factors and inhibiting their activities. We tested this idea directly by immunoprecipitating FLAG-tagged Mre11 or TAP-tagged Rad50, Rad51 and Rfa1 from soluble cell extracts and checking for the presence of co-IPed histones. Consistent with published reports (91,92), we clearly observed the association of histone H3 with TAP-tagged Rfa1, which served as a good positive control in our experiments. Interestingly, histones were also found to associate with both Rad51 and Rad52 but not with Mre11 (Figure 7a). Further, the relative amounts of histone H3 and H3K56Ac co-IPed with the HR factors was similar to the levels of these histones co-IPed with the histone chaperone Asf1. This suggested that histones bound to the HR factors originate from the newly synthesized pool of free histones, rather than chromatin bound histones. However, it is still possible that the histones associated with HR factors in fact represent histones associated with nascent chromatin that is bound by the HR factors. Hence, to categorically rule out the contribution of histones associated with DNA in our IP reactions, we treated them with a potent nuclease Benzonase (Novagen). Treatment with this nuclease did not alter the association of histones with the Rad52 protein, thereby strongly suggesting that these histones arise from the free histone pool rather than the chromatin bound histones (Figure 7b). We also investigated whether NHEJ factors such as Ku70 and Ku80 associated with free histones. Although barely detectable amounts of histone H3 was found in association with Ku70 and somewhat higher levels of H3 were bound to Ku80, in both cases this binding was abolished on Benzonase treatment (Figure 7c), suggesting that this interaction was with chromatin bound histones rather than free histones. Consistent with this, no H3 K56Ac signal was detectable from H3 co-IPed with Ku70/80.
As deletion of hht2-hhf2 results in a reduction in free histone levels (Figure 2b–d) concomitant with improved recruitment of HR factors to a DSB (Figure 6), next we investigated whether the levels of free histones associated with HR factors were also altered in the hht2-hhf2Δ strain. For this, we IPed Rad52-TAP from WT and hht2-hhf2Δ strains and measured the levels of co-IPed histone H3. We observed a dramatic drop in the levels of both histone H3 and H3 K56Ac associated with Rad52-TAP in the hht2-hhf2Δ strain (Figure 7c–d), which is consistent with the increased efficiency of HR in this strain (Figure 5 and 6). Taken together, our data suggest that the physical association of free histones with HR factors can potentially interfere with their function, thereby reducing the efficiency of HR-mediated repair.
DISCUSSION
In this study, we have shown that the histone gene dosage has a dramatic influence on the DNA damage sensitivity of budding yeast cells (Figure 1). Our results suggest that even the normal histone gene dosage in wild-type yeast cells appears to generate an excess of free histones, which in turn appears to interfere with efficient DNA repair by competing with the repair factors for binding to the DNA lesion (Figures 3–5) and also by physically associating with HR factors (Figure 7). Further, deletion of the hht2-hhf2 gene pair that contributes most of the histone H3 and H4 mRNA in the cell results in a significant reduction in the free histone pools of histone H3 and H4 (Figure 2b and c), although these cells still synthesize enough histones to assemble and maintain the bulk chromatin structure (Figure 2a; Supplementary Figure S1). A reduction in free histone pools following the deletion of the hht2-hhf2 gene pair results in a reduction in the DNA damage sensitivity of wild type and DNA damage checkpoint mutant yeast strains (Figures 1 and 3). Similar to the effect of linker histone deletion in yeast (43), the effect of a reduced core histone gene dosage on the DNA damage sensitivity of yeast cells is primarily mediated by an increase in the efficiency of DNA repair by the HR pathway (Figures 3, 5 and 6), whereas the NHEJ pathway is unaffected (Figure 4). This is not surprising as repair by the HR pathway involves intimate DNA and protein transactions over long stretches of chromatin, whereas repair by the NHEJ pathway essentially involves the religation of the broken ends with limited amount of end processing (48) (Figure 8). In fact, the resection of the broken ends is the first step in HR-mediated DSB repair (74) and involves the generation of single-stranded DNA with concomitant loss of histones (80). Additionally, the donor loci also exhibit substantial chromatin remodeling that facilitates repair of the DSB at the MAT locus by HR (81). As histones can interact efficiently with single-stranded DNA and may even form nucleosome-like structures (93–95), the presence of appreciable quantities of free histones in the vicinity of such HR repair intermediates is likely to present a significant competition for the HR factors for binding to the exposed DNA (Figure 8). Consistent with this mechanism, better recruitment of HR factors at a DSB is observed concomitant with an accelerated loss of histones from this region in the hht2-hhf2 deletion strain (Figure 6). Given the dependence of Hho1 on its predicted DNA-binding residues to exert an effect on DNA damage sensitivity (43), it is very likely that this effect is also mediated by competition with HR factors for binding damaged DNA. Additionally, our data suggest that free histones may physically associate with HR factors such as Rad51 and Rad52 (Figure 7a), thereby potentially interfering with their activities. Not surprisingly, reduction in histone dosage results in diminished binding of histones to Rad52 (Figure 7d–e) concomitant with improved kinetics of HR factor recruitment to a DSB (Figure 6), as well as improved survival following a DSB (Figure 3) in the hht2-hhf2Δ strain. DSBs in haploid yeast are repaired by HR primarily during the S and G2 phases of the cell cycle when homologous sequences are readily available for HR-mediated repair. As the yeast histone genes are predominantly expressed during early S-phase (21), free histones are also likely to be present at their highest concentrations in the cell during S and G2 phases of the cell cycle and thus have their maximal effect on HR-mediated repair, rather than NHEJ.
Figure 8.
Model to illustrate the potential effect of excess histones on HR or NHEJ-mediated repair of a DSB. (a) Effect of free histones on HR-mediated DSB repair. Because of the presence of significant stretches of nucleosome deficient single-stranded DNA as intermediates at both the recipient and donor loci during HR-mediated DSB repair, the availability of significant amounts of free (non-chromatin associated) histones even in wild-type cells could result in competition between the HR factors and histones for binding to the exposed DNA (Figure 6). Additionally, free histones can physically interact with certain HR factors such as Rad51, Rad52 and RPA (Figure 7a), potentially interfering with their activities (depicted in the cartoon within the dashed box). Both these situations are likely to lead to a reduction in the efficiency of HR-mediated repair. For simplicity, only a few HR factors and one nuclease complex (MRX) are indicated. (b) Effect of free histones on NHEJ-mediated DSB repair. Significant stretches of DNA are not exposed at the DSB site during NHEJ-mediated repair, and as such this repair process is unlikely to be affected by the presence of significant quantities of excess histones. Further, unlike recombination factors that associate with free histones, NHEJ factors appear to associate with chromatin bound histones on binding to DNA (Figure 7c). NHEJ factors are indicated.
Our results raise the important question of why eukaryotes have evolved with multiple copies of histone genes when they are clearly not required for viability (10,11,14,21). Despite the tight transcriptional and posttranscriptional control of histone genes (21), calculations based on the published estimates of histone proteins arising from each core histone gene suggest that yeast cells make an excess of histone proteins over what is required for chromatin assembly (5,96), which can potentially have deleterious consequences for the cells (59). On the other hand, it is also known that delays between DNA replication and chromatin assembly due to histone insufficiency during S-phase results in spontaneous DNA damage, genomic instability and inviability (45–47,97–99). Hence, to avoid such deleterious consequences, one possibility is that high levels of histones are synthesized during S-phase to ensure a sufficiently high histone concentration for rapid chromatin assembly by a relatively limited number of histone chaperone molecules. For the assembly of 300 000 new histone H3 and H4 molecules during each S-phase in the budding yeast, the estimated number of all the known histone H3 and H4 chaperone molecules available are as follows: Asf1, 6230; Cac1, 1590; Hir1, 846 and Nap1, 8070 (note: histone chaperones that work as multi-subunit complexes have similar or lower number of molecules per cell for their other subunits than shown here) (96). So, essentially just over 15 000 histone chaperone molecules have the challenging task of assembling 300 000 histone H3 and H4 molecules during the budding yeast S-phase, which typically lasts around 20 min at 30°C. We speculate that this task can only be achieved at high effective histone protein concentrations that would allow the histone chaperones to have a quick turn-around time after each round of histone deposition, such that they do not have to spend too much time looking for the next histone molecule to deposit.
Mutant yeast strains such as those lacking the sgs1, srs2 or rrm3 helicases exhibit greatly elevated rates of mitotic recombination that are highly deleterious for the cells as they result in genomic instability (100–102). On the basis of our current results, we speculate on the possibility that the high levels of histones produced by multiple histone genes may in fact play a role in keeping the HR machinery in check by potentially preventing it from initiating spurious recombination events using DNA replication intermediates as templates. As such, an intriguing possibility is that eukaryotic cells may have evolved multiple histone genes for high levels of histone production not only to facilitate efficient chromatin assembly but also to restrain HR activity during the S and G2 phases of the cell cycle, which in turn would contribute to the overall genomic stability of the cells. This idea would also be consistent with the finding that partial depletion of histone H4 results in genomic instability due to elevated recombination (45). More importantly, combined with the reported requirement of Hho1 for normal lifespan in yeast (43), as well as the exciting recent finding that aging yeast cells have reduced levels of histones (103), our results could help explain in part the hyper-recombinational state of aging yeast cells (51). Not surprisingly, histone overexpression increases the life span of yeast cells (103), and our findings suggest that this may be in part due to the dampening of HR activity, thereby limiting genomic instability that would otherwise accelerate aging in these cells. However, much more work needs to be carried out in the future to test this idea.
Irrespective of what the underlying reasons for the presence of multiple histone genes in all eukaryotes prove to be eventually, due to the high degree of conservation of DNA repair and chromatin processes, the presence of multiple histone genes is likely to have major ramifications for these processes in all eukaryotes, including humans. For example, many current anti-cancer drugs rely on their ability to cause DNA damage for their effectiveness, and based on our studies in yeast, it is possible that the loss of histone genes from genetically unstable cancer cells may make it relatively easy for them to develop resistance to such drugs. Hence, our results could have serious implications for genomic stability of human cells with their dozens of histone genes (7) that can significantly influence numerous aspects of human health and disease. Future studies will reveal if histone gene copies are indeed lost from histone gene clusters in human cancer cells that are refractory to DNA damaging chemotherapeutic drugs, and if the mechanism of resistance is dependent on elevated HR, then the HR machinery could be targeted in an attempt to destroy such refractory cancers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–7 and Supplementary References [3,4,104–106].
FUNDING
A Bankhead-Coley Cancer Research Program, Florida Department of Health [07BN-02]. Funding for open access charge: Departmental ‘start up’ funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Drs. Andres Aguilera, John Diffley, Muriel Grenon, Noel Lowndes, Rohinton Kamakaka, Kyosuke Nagata, Mary Ann Osley, Rodney Rothstein, Jasper Rine, Alain Verreault and Yanchang Wang for strains and reagents. They thank Drs. David Brown, Johanna Paik and Yanchang Wang for critical reading of this manuscript.
REFERENCES
- 1.Wolffe A. Chromatin: Structure and Function. 2nd edn. London: Academic Press; 1995. [Google Scholar]
- 2.Stein GS, Stein JL, Marzluff WF. Histone Genes: Structure, Organization, and Regulation. New York: Wiley; 1984. [Google Scholar]
- 3.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 5.Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front. Biosci. 2009;14:3145–3158. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 6.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 7.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 8.Norris D, Dunn B, Osley MA. The effect of histone gene deletions on chromatin structure in Saccharomyces cerevisiae. Science. 1988;242:759–761. doi: 10.1126/science.2847314. [DOI] [PubMed] [Google Scholar]
- 9.Norris D, Osley MA. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 1987;7:3473–3481. doi: 10.1128/mcb.7.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Gunesdogan U, Jackle H, Herzig A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 2010;11:772–776. doi: 10.1038/embor.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takami Y, Nakayama T. A single copy of linker H1 genes is enough for proliferation of the DT40 chicken B cell line, and linker H1 variants participate in regulation of gene expression. Genes Cells. 1997;2:711–723. doi: 10.1046/j.1365-2443.1997.1550353.x. [DOI] [PubMed] [Google Scholar]
- 13.Takami Y, Nakayama T. One allele of the major histone gene cluster is enough for cell proliferation of the DT40 chicken B cell line. Biochim. Biophys. Acta. 1997;1354:105–115. doi: 10.1016/s0167-4781(97)00077-8. [DOI] [PubMed] [Google Scholar]
- 14.Takami Y, Takeda S, Nakayama T. An approximately half set of histone genes is enough for cell proliferation and a lack of several histone variants causes protein pattern changes in the DT40 chicken B cell line. J. Mol. Biol. 1997;265:394–408. doi: 10.1006/jmbi.1996.0733. [DOI] [PubMed] [Google Scholar]
- 15.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 16.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell. Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Q, Sirotkin A, Skoultchi AI. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol. Cell. Biol. 2000;20:2122–2128. doi: 10.1128/mcb.20.6.2122-2128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirotkin AM, Edelmann W, Cheng G, Klein-Szanto A, Kucherlapati R, Skoultchi AI. Mice develop normally without the H1(0) linker histone. Proc. Natl Acad. Sci. USA. 1995;92:6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 20.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 22.Su C, Gao G, Schneider S, Helt C, Weiss C, O'Reilly MA, Bohmann D, Zhao J. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 25.Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 26.Borun TW, Gabrielli F, Ajiro K, Zweidler A, Baglioni C. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975;4:59–67. doi: 10.1016/0092-8674(75)90134-8. [DOI] [PubMed] [Google Scholar]
- 27.Graves RA, Pandey NB, Chodchoy N, Marzluff WF. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 28.Sittman DB, Graves RA, Marzluff WF. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl Acad. Sci. USA. 1983;80:1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lycan DE, Osley MA, Hereford LM. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:614–621. doi: 10.1128/mcb.7.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 31.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 32.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 34.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 36.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 37.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 38.Qin S, Parthun MR. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 40.Evans ML, Bostelman LJ, Albrecht AM, Keller AM, Strande NT, Thompson JS. UV sensitive mutations in histone H3 in Saccharomyces cerevisiae that alter specific K79 methylation states genetically act through distinct DNA repair pathways. Curr. Genet. 2008;53:259–274. doi: 10.1007/s00294-008-0182-1. [DOI] [PubMed] [Google Scholar]
- 41.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 44.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prado F, Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol. Cell. Biol. 2005;25:1526–1536. doi: 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clemente-Ruiz M, Prado F. Chromatin assembly controls replication fork stability. EMBO Rep. 2009;10:790–796. doi: 10.1038/embor.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 48.Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair. 2004;3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Haber JE. Uses and abuses of HO endonuclease. Methods Enzymol. 2002;350:141–164. doi: 10.1016/s0076-6879(02)50961-7. [DOI] [PubMed] [Google Scholar]
- 50.Holmes A, Haber JE. Physical monitoring of HO-induced homologous recombination. Methods Mol. Biol. 1999;113:403–415. doi: 10.1385/1-59259-675-4:403. [DOI] [PubMed] [Google Scholar]
- 51.McMurray MA, Gottschling DE. An age-induced switch to a hyper–recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 52.Burke D, Dawson D, Stearns T Cold Spring Harbor Laboratory. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2000 edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 53.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 54.Miyaji-Yamaguchi M, Kato K, Nakano R, Akashi T, Kikuchi A, Nagata K. Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol. Cell. Biol. 2003;23:6672–6684. doi: 10.1128/MCB.23.18.6672-6684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morillo-Huesca M, Maya D, Munoz-Centeno MC, Singh RK, Oreal V, Reddy GU, Liang D, Geli V, Gunjan A, Chavez S. FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 2010;6:e1000964. doi: 10.1371/journal.pgen.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 57.Cross SL, Smith MM. Comparison of the structure and cell cycle expression of mRNAs encoded by two histone H3-H4 loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:945–954. doi: 10.1128/mcb.8.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, Stubbe J. Bleomycins: towards better therapeutics. Nat. Rev. Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 59.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 61.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 62.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl Acad. Sci. USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horak CE, Snyder M. Global analysis of gene expression in yeast. Funct. Integr. Genomics. 2002;2:171–180. doi: 10.1007/s10142-002-0065-3. [DOI] [PubMed] [Google Scholar]
- 64.Bonner WM, Wu RS, Panusz HT, Muneses C. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry. 1988;27:6542–6550. doi: 10.1021/bi00417a052. [DOI] [PubMed] [Google Scholar]
- 65.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 66.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 67.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. J. Biol. Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 69.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 70.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 71.Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc. Natl. Acad. Sci. USA. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 74.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 76.Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 77.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 78.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Linger JG, Tyler JK. Chromatin disassembly and reassembly during DNA repair. Mutat. Res. 2007;618:52–64. doi: 10.1016/j.mrfmmm.2006.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsukuda T, Lo YC, Krishna S, Sterk R, Osley MA, Nickoloff JA. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair. 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 82.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 83.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 84.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 86.Game JC, Kaufman PD. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Linger J, Tyler JK. The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics. 2005;171:1513–1522. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martini EM, Keeney S, Osley MA. A role for histone H2B during repair of UV-induced DNA damage in Saccharomyces cerevisiae. Genetics. 2002;160:1375–1387. doi: 10.1093/genetics/160.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 93.Palter KB, Alberts BM. The use of DNA-cellulose for analyzing histone-DNA interactions. Discovery of nucleosome-like histone binding to single-stranded DNA. J. Biol. Chem. 1979;254:11160–11169. [PubMed] [Google Scholar]
- 94.Palter KB, Foe VE, Alberts BM. Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell. 1979;18:451–467. doi: 10.1016/0092-8674(79)90064-3. [DOI] [PubMed] [Google Scholar]
- 95.Caffarelli E, De Santis P, Leoni L, Savino M, Trotta E. Interactions of the histone octamer with single-stranded DNA. Sedimentation analysis and low-angle X-ray diffraction. Biochim. Biophys. Acta. 1983;739:235–243. doi: 10.1016/0167-4781(83)90034-9. [DOI] [PubMed] [Google Scholar]
- 96.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 97.Prado F, Cortes-Ledesma F, Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han M, Chang M, Kim UJ, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 99.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl Acad. Sci. USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kerrest A, Anand RP, Sundararajan R, Bermejo R, Liberi G, Dujon B, Freudenreich CH, Richard GF. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat. Struct. Mol. Biol. 2009;16:159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmidt KH, Kolodner RD. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl Acad. Sci. USA. 2006;103:18196–18201. doi: 10.1073/pnas.0608566103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol. Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 105.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 107.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]