Abstract
Bacteriophage DNA packaging motors translocate their genomic DNA into viral heads, compacting it to near-crystalline density. The Bacillus subtilis phage ϕ29 has a unique ring of RNA (pRNA) that is an essential component of its motor, serving as a scaffold for the packaging ATPase. Previously, deletion of a three-base bulge (18-CCA-20) in the pRNA A-helix was shown to abolish packaging activity. Here, we solved the structure of this crucial bulge by nuclear magnetic resonance (NMR) using a 27mer RNA fragment containing the bulge (27b). The bulge actually involves five nucleotides (17-UCCA-20 and A100), as U17 and A100 are not base paired as predicted. Mutational analysis showed these newly identified bulge residues are important for DNA packaging. The bulge introduces a 33–35° bend in the helical axis, and inter-helical motion around this bend appears to be restricted. A model of the functional 120b pRNA was generated using a 27b NMR structure and the crystal structure of the 66b prohead-binding domain. Fitting this model into a cryo-EM map generated a pentameric pRNA structure; five helices projecting from the pRNA ring resemble an RNA claw. Biochemical analysis suggested that this shape is important for coordinated motor action required for DNA translocation.
INTRODUCTION
The Bacillus subtilis bacteriophage ϕ29 is a member of the double-stranded DNA phages that, during assembly, package their genome into a preformed head (prohead) to near-crystalline density (1,2). This adenosine triphosphate (ATP)-dependent process is driven by a molecular motor situated at a unique vertex of the head and is one of the most powerful macromolecular motors, capable of generating forces in excess of 57 picoNewtons (3). The motor is comprised of a head–tail connector embedded in a unique vertex of the virus capsid, a pentameric ring of prohead RNA (termed pRNA) and the multimeric ring-ATPase gp16 (Figure 1A) (4–6). pRNA is an essential component of the packaging motor (7) and provides the scaffold to which the packaging ATPase binds during motor assembly (5,8,9). The ATPase and pRNA motor components are transiently assembled for packaging, as they are not part of the mature virion (7).
Figure 1.
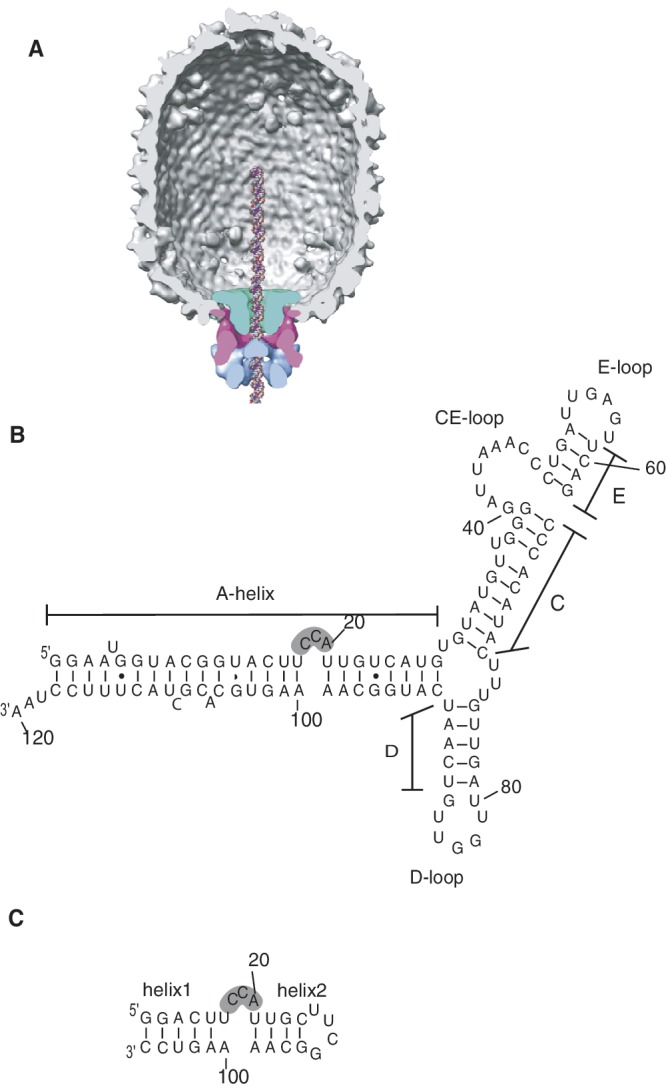
ϕ29 pRNA. (A) Cross-section of cryoEM 3D reconstruction of prohead-ATPase motor complex, with molecular envelopes identified as connector (green), pRNA (pink) and gp16 ATPase (blue). DNA is modeled in the channel. Reprinted from Journal of Molecular Biolology, 410, Grimes,S., Ma,S., Gao,J., Atz,R. and Jardine,P.J., Role of phi29 Connector Channel Loops in Late-Stage DNA Packaging, 50–59, (2011), with permission from Elsevier. (B) Predicted secondary structure of the 120 base (120b) pRNA. The CCA-bulge region is shadowed. (C) 27 base (27b) construct containing the CCA-bulge.
Although the pRNA is a viral-encoded 174 base transcript, a truncated 120 base form lacking the terminal 54 bases is fully functional in vitro and is widely used in biological assays (e.g. (10)) (Figure 1B). Phylogenetic analysis of pRNAs from ϕ29 and related phages predicted a secondary structure consisting of a long helical region (A-helix) connected to several stem–loop structures (11). Mutagenesis studies (4,12), combined with structural studies (5,13–15), showed that the loops of the pRNA are responsible for pRNA multimerization forming a ring structure (CE- and D-loops), and for binding pRNA to the prohead (CE-, D- and E-loops). Recently, the crystal structure of the prohead-binding domain (bases 25–95) that encompasses these loops was determined (13). In contrast to this stem–loop region, mutations in the A-helix do not affect prohead binding or oligomerization, but rather impact other aspects of DNA packaging (4,12). In this helix, maintenance of secondary structure is important for function (16) and a key bulge, the CCA bulge that includes bases 18–20, is essential for DNA packaging (17,18). Deletion of two or three bases from this bulge renders the DNA packaging motor completely inactive, although motor assembly with these mutants is similar to assembly with wild-type (wt) pRNA (18). Secondary structure predictions for prohead RNAs of related phages suggested that a bulge structure near this position in the A-helix is conserved, despite low overall conservation of primary sequence (30–50%) (11).
Here, we report the structure of this crucial bulge region of pRNA as determined by nuclear magnetic resonance (NMR) spectroscopy. We show that the bulge, rather than containing the predicted three bases, is comprised of four nucleotides with a single base bulge on the opposite strand. Together, these structural features introduce a bend in the A-helix. Residual dipolar coupling (RDC) analysis suggests that inter-helical motion around this bend is restricted. We generated a model of the packaging competent 120b pRNA by combining our 27b NMR structure with the 66b crystal structure of the prohead-binding domain and a segment of ideal A-form RNA. Fitting this 120b structure into the pRNA density of a prohead–pRNA cryo-EM 3D reconstruction generated a complete pseudoatomic model of the pentameric pRNA ring. Finally, DNA packaging and ATPase activity experiments using bulge mutants suggest that this bulge structure plays a critical scaffolding role in determining the functional conformation of the packaging ATPase.
MATERIALS AND METHODS
NMR sample preparation and spectroscopy
All RNAs were synthesized enzymatically from synthetic DNA templates using the AmpliScribe T7 transcription kit (Epicentre). The RNAs were purified by denaturing 15–20% polyacrylamide gel electrophoresis (PAGE), identified by ultraviolet absorbance and excised from the gel. RNAs were eluted from the gel slice in water and desalted using SepPak columns (Waters). The RNAs were concentrated using Centricon YM-3 and exchanged into 50 mM Na-phosphate (pH 6.5), 10 mM NaCl and 5 mM MgCl2. Samples for exchangeable proton detection were lyophilized, and then H2O was added to achieve a final solution of 95% H2O/5% D2O. Samples for non-exchangeable proton detection were lyophilized and then D2O was added. Before NMR experiments, the RNAs were heated to 80°C for 2 min and cooled in ice water. Unlabeled, uniformly 13C, 15N-labeled and base-specific 13C, 15N-labeled RNA samples were synthesized as described. NMR experiments were performed on Varian Inova 600, 800 or 900 MHz equipped with a cryogenic probe. Exchangeable protons were observed at 283 K in H2O/D2O (95:5) buffer and non-exchangeable protons were at 298 K in D2O buffer. One-bond C–H and N–H RDCs were measured on uniformly 13C, 15N-labeled 27b sample in 20 mg/ml Pf1 phage (ASLA Biotech Ltd) on 800 MHz spectrometer using rnaCtrosy (19) and gNhsqc2fb (20) experiments at 298 and 283 K. All spectra were processed with NMRPipe/NMRDraw (21) and analyzed with NMRView (22). The exchangeable protons were assigned using 2D NOESY (23), 2D DQF-COSY (24) on the unlabeled 27b sample, 2D HCN-H1′, HCN-H6/H8 (25) 1H–13C HSQC and a suite of 3D-filtered/edited NOESY (26) experiments on the uniformly labeled and base-specific 13C, 15N-labeled samples. The non-exchangeable protons were assigned using 2D ssNOSY (23) experiments on the unlabeled sample (a NOE-walk of sequential assignments are shown in Supplementary Figure S1) and 1H–15N HSQC experiment on uniformly 15N-labeled sample. J couplings were measured using 2D DQF-COSY experiment. Large JH1′-H2′ were assigned to C2′-endo and small JH1′-H2′ were assigned to C3′-endo dihedral angles, respectively, and used for structure calculation.
Structure calculation
The structure of the 27b was calculated using a standard CNS protocol (27) as described in (28). 65 C–H and N–H RDCs collected by two temperature sets were used for structure calculation along with distance and dihedral angles. Planarity restraints and hydrogen bonds were incorporated into the calculation for eight Watson–Crick base pairs (G12–C105, G13–C104, A14–U103, C15–G102, U16–A101, U22–A98, G23–C97 and C24–G96). The best values for the magnitude and asymmetry of alignment tensors are Da (C–H, 25°C) = −17 Hz, Da (C–H, 10°C) = −22 Hz, Da (N–H, 25°C) = −8.18 Hz, Da (N–H, 10°C) = −10.59 Hz, R (25°C) = 0.2 and R (10°C) = 0.078. Two standard pseudo-molecules representing alignment orientations for 10°C and 25°C were introduced to facilitate use of RDC constraints. The force constants for RDCs were gradually increased from 0.001 to 0.25 kcal·mol−1·Hz−2 for all RDCs in the structure calculation. The RDC incorporation generated conformations with anti-parallel helical orientation (180° rotation about Sxx and Syy components of alignment tensor in PAS of one helix), and these conformations were discarded from further analyses. The 10 lowest energy structures with no anti-parallel helical orientation are reported in this article. Structures were viewed and analyzed with PYMOL (DeLano Scientific; www.pymol.org). The 3DNA program (http://rutchem.rutgers.edu/∼xiangjun/3DNA/) was used to calculate the angle between the two helical segments. Helical regions of 20 calculated structures were extracted, and helical axes were assigned to each part. The angle between these axes was calculated using MATLAB (http://www.mathworks.com/) as described in the 3DNA homepage.
RDC analysis to elucidate order parameters
The bestfit algorithm of PALES (29) was used to elucidate order parameters. Ideal helices and calculated structures were fitted to experimental RDC values of the two helical parts separately, as described above. Calculations with ideal helices were included as part of the RDC data analysis in order to see dynamics parameters independent of calculated structures. The scaled axial component Da and rhombicity R of alignment tensor, generalized degree of order (GDO) and directionality (η) parameters were reported. In order to estimate the order parameters uncertainty, the structural noise Monte–Carlo method implemented in the bestfit algorithm was used, generating 1000 similar structures and fitting all structures to the observed RDCs. Average value and standard deviations for Da and Dr were directly reported by PALES. From these values, averages and errors for neta (η) and R were calculated using equations η = 3/2Dr/Da and R = Dr/Da. for 1000 steps. Alignment tensor components Sxx, Szz and Syy in PAS of the molecule were reported for the 1000 Monte–Carlo steps. Average value and standard deviation in 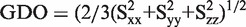 were calculated using MATLAB.
were calculated using MATLAB.
Production of packaging components
Proheads, the packaging ATPase gp16 and DNA-gp3 were purified as described previously (18,30). Proheads were purified from B. subtilis 12A infected with the mutant 900-16-14- (defective in the packaging ATPase and non-essential head fibers) as described previously (18). To create RNA-free proheads, purified proheads were treated with RNase A and re-purified as previously described (18). RNA-free proheads were then reconstituted with the desired form of pRNA by incubating proheads (83 nM) with an excess of pRNA (830 nM) in 0.5× TMS buffer (25 mM Tris, pH 7.6, 5 mM MgCl2 and 50 mM NaCl) in a total volume of 10 µl for 10 min at room temperature. For ATPase assays, the reconstituted proheads were further purified by sedimentation through a 5 ml 5% sucrose cushion in 0.5× TMS buffer in the SW55 rotor at 35 000 rpm for 2.5 h. The pellets containing the reconstituted proheads were re-suspended in 0.5× TMS buffer at 4°C overnight. The packaging ATPase was expressed from the plasmid pSACB-gp16 in B. subtilis and purified as described previously (18). DNA-gp3 was extracted and purified from phage as described previously (30).
Production of pRNA and mutant pRNAs
Mutant pRNAs were produced by inverse PCR (31,32) using the plasmid pRT72 that encodes the wt ϕ29 pRNA gene as a template (33). Plasmid DNA was subsequently sequenced to verify the presence of the mutation. Wt and mutant pRNAs were made by in vitro transcription using T7 RNA polymerase and purified by denaturing urea PAGE as described previously (33).
In vitro DNA packaging assay
The in vitro DNA packaging assay was performed as described previously (18). Briefly, DNA-gp3 (4.2 nM), proheads reconstituted with wt or mutant pRNA (8.4 nM) and the packaging ATPase (100–125 nM) were mixed in 0.5× TMS buffer in 20 µl and incubated at room temperature for 5 min. ATP was then added to the reaction (0.5 mM final) to initiate packaging and the reaction incubated for an additional 15 min. DNase I was added to 1 µg/ml to digest the unpackaged DNA and incubated for 10 min. Ethylenediaminetetraacetic acid (EDTA) (25 mM final) and Proteinase K (500 µg/ml final) were added and the mixture incubated at 65°C to extract the packaged DNA. Packaging was then analyzed by agarose gel electrophoresis.
Generation of a structural model of 120b pRNA
The structure of 27b construct and the recently published 66b crystal structure of the prohead-binding domain of pRNA (pdb: 3R4F) (13) were combined to generate the structural model of 120b pRNA (Supplementary Figure S6 and Supplementary Methods).
Generation of a pentameric pRNA structural model
Five copies of the 120b structural model were fitted as rigid bodies into the 10 Å, 5-fold symmetric cryo-EM reconstruction of prohead [(34), Morais,M.C., unpublished data] using the program EMFIT (35). A very good overall fit was obtained with very few structural clashes between neighboring molecules (Supplementary Figure S5). A virtually identical fit was obtained using SITUS program suite [(36), data not shown] indicating that the fit is not algorithm dependent.
ATPase assay
ATPase activity of prohead/gp16/DNA-gp3 packaging complexes or prohead/gp16 motor complexes was assessed by measuring production of inorganic phosphate by the EnzChek Phosphate Assay Kit (Invitrogen). TMS buffer (50 mM Tris, pH 7.8, 10 mM MgCl2 and 100 mM NaCl) was used to suppress ATPase activity that is unrelated to packaging (i.e. DNA-free prohead/ATPase motor complex activity), while TM buffer (25 mM Tris, pH 7.6 and 5 mM MgCl2) stimulates activity of prohead/motor ATPase complexes (37). Briefly, a reaction mixture containing reaction buffer (either TMS or TM) with 0.2 mM of 2-amino-6-mercapto-7-methylpurine riboside was pre-incubated with DNA-gp3 (4.2 nM), reconstituted proheads (4.2 nM) and gp16 (125 nM) in 100 µl at room temperature for 10 min in the presence of purine nucleoside phosphorylase (0.1 unit). ATP was added to 1 mM to initiate the reaction and production of Pi measured in the spectrophotometer at 360 nM at 15 s intervals for 10 min.
RESULTS
Overview of the solution structure of the 27 base construct
To assess the structural role of the CCA bulge, we determined the structure of a 27b RNA construct that contained the CCA bulge and a portion of the adjacent helices (Figure 1c). In order to facilitate NMR analysis, we introduced a UUCG tetra-loop at one end and two G–C base pairs at the other end to stabilize the secondary structure.
After resonance assignment, a total of 426 NOEs (∼16 per nucleotide), 206 dihedral angle restraints and 65 dipolar couplings (RDCs) were used for structure calculations. Since an order tensor analysis showed no significant difference between the degrees of alignment of the two helical elements, we used one axial (Da) and one rhombic (R) value to calculate the structure of the full molecule. Ten structures with the lowest energy were chosen as a representative ensemble. Excluding residues 17–20 and 100, which are not base paired (see below), this ensemble yielded an RMSD of 0.85 ± 0.14 Å for heavy atoms (Figure 2A). The RMSD of heavy atoms including all nucleotides was 1.31 ± 0.25 Å. Helix 1 and helix 2 are well-defined A-form structures (Figure 2B and D) with RMSDs of 0.54 ± 0.21 and 0.48±0.07 Å, respectively. As expected, the tetra-loop adopts a standard tetra-loop conformation (38) with C2′-endo puckering for the U and C residues in the middle of the loop (Supplementary Figure S3).
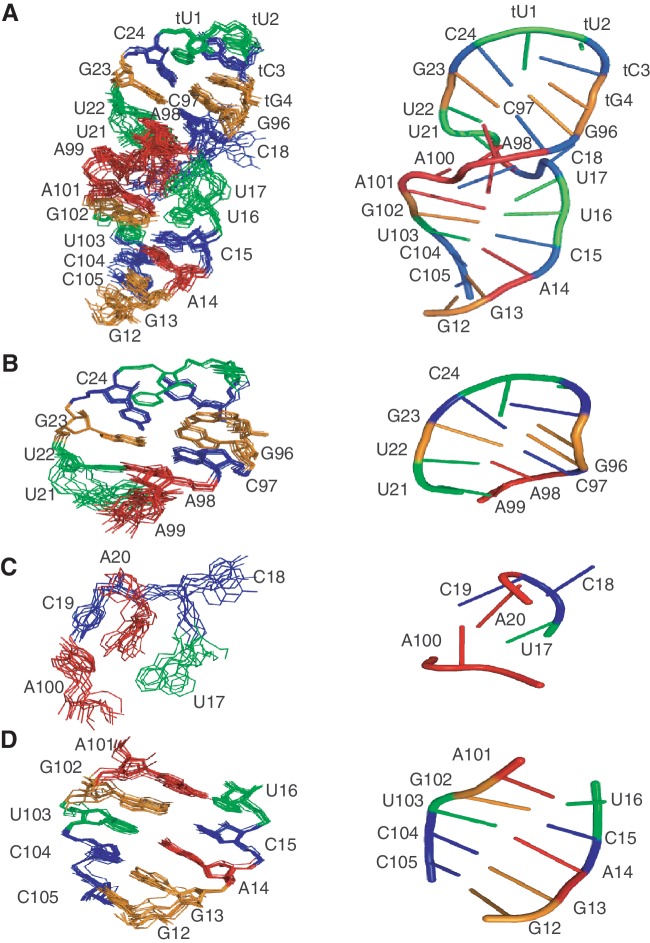
Figure 2.
Structure of the 27b RNA. Ensembles of 10 lowest energy structures (left) and cartoon diagrams of one representative structure (right) are shown. (A) 27b RNA, (B) helix2 with tetra-loop, (C) bulge region and (D) helix1. In (C), structures were rotated 60° about the x- and y-axes and 40° about the z-axis to see all nucleotides of the bulge. Nucleotides are coloured blue (C), green (U), red (A) and orange (G). Images created in PyMol (54).
Secondary structure prediction suggested that the CCA bulge would have contiguous base paring on either side of this three base feature (18-CCA-20) (11). Although most of the base pairing is evident in our ensemble structure of the 27b construct, we found that U17 and A100 are not paired as predicted. These residues have a minimum distance of 7.6 Å between bases, far too distant to form hydrogen bonds (Figure 2C). Instead, U17 is partly stacked over U16 which forms the last base pair of helix 1 prior to the bulge (Supplementary Figure S2). Rather than being base paired with U17, A100 is closer to the last two residues of the bulge, C19 and A20, which are positioned 3.4 and 3.0 Å away, respectively (Figure 2C). In our ensemble, the base of C18 is exposed to solvent and the position of C18 is less well defined relative to the rest of the bulge nucleotides. U16 and A101 form a Watson–Crick base pair, while A20 N6 amino is able to form a hydrogen bond with U16 O2 carbonyl, which enables U16-A101-A20 to form a base triple (Supplementary Figure S8). Additionally, A20 has a hydrophobic interaction with A100 involving C2H from each base (Supplementary Figure S8).
Furthermore, the N3 of A100 is within hydrogen bonding distance of O2′ of U21 (3.5 ± 0.3 Å), which could be important for stabilizing the overall conformation of the 27b construct as seen in the A-minor II motif (39,40). Similar to other known RNA structures containing bulges and loops determined by NMR (e.g. (41,42)), the 18-CCA-20 bulge appears to weaken neighboring base pairing, as evidenced by the very small imino signal from base-paired U16 and no detectable imino signal from U21. In addition, the bulge appears to have widened the major groove from the ideal A-type helix from 10.3 to 16 Å (Supplementary Figure S4). The first unpaired nucleotide in the bulge, U17, adopts a C2′-endo puckering conformation in 7 of 10 low-energy structures in our ensemble. An H1′–H2′ crosspeak was observed in DQF-COSY, although the measured H1′–H2′ scalar coupling constant was intermediate and therefore not assigned to either C2′-endo or C3′-endo for the structure calculation. C2′-endo conformations are rare in nucleotides located in helical structures of RNA, but are found in bulged regions such as the 5 nucleotide bulge in a Group I intron (43) and the one nucleotide bulge in the B. subtilis RNase P ribozyme (44).
RDC analysis indicated absence of significant inter-helical motion
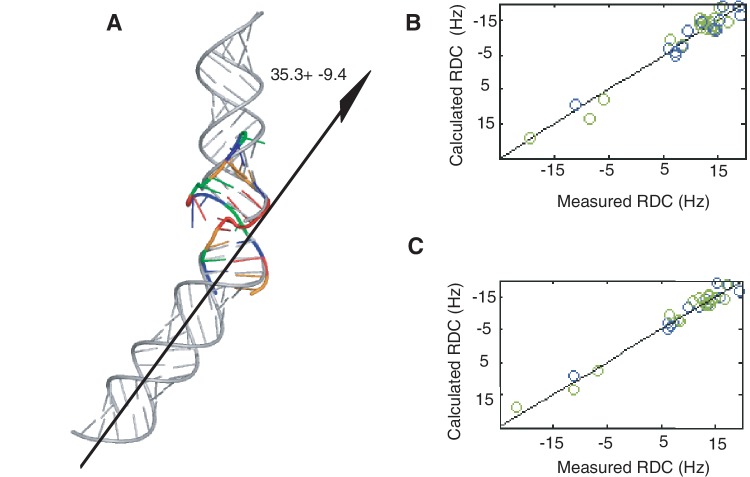
The CCA bulge introduces a bend between helix 1 and helix 2 with an average inter-helical angle of 35 ± 9°, calculated from 20 low-energy structures (Figure 3). The analysis using experimental RDC values fit to ideal helices yielded an estimated 33° between helices, which was similar to the angle observed in the structures. No significant difference in maximal RDC values between helix 1 and helix 2 was detected in RDCs collected at two temperatures (Supplementary Figure S5), which suggests a similar alignment for both helix 1 and helix 2. As expected, RDCs of the bulged residues are smaller than RDCs of the other residues, indicating a lower degree of alignment. Order tensor analysis performed for RDCs of helix 1 and helix 2 yielded very similar order parameters for both helical domains if fitted to ideal helices or calculated structures (Table 1). The ratio of GDO values of helix 1 and helix 2 are 0.98 and 0.86 for an ideal helix and 27b structure, respectively, suggesting that inter-helical motions are small (Table 1). For a comparison, the ratio of GDO values of HIV-1 TAR RNA, which has significant inter-helical motions, was 0.59 (45). Fitting both helical domains separately to RDCs did not significantly improve the fit compared with fitting all RDCs together (Figure 3B and C). Although, theoretically the similarity between order parameters could be due to independent movements of the two helices, this case is very unlikely for 27b because even the bulged nucleotides showed many NOE signals (Supplementary Table S2).
Figure 3.
The bulge-induced bend in the RNA helical axis. (A) 27b structure (in colour) is overlaid with two ideal helices (gray) aligned with the two helical segments of the 27b RNA (helix1 and helix2). The bend angle of the helical axis was calculated from the 20 lowest energy structures using 3DNA (55). The two helical segments of the 27b construct were fitted to experimentally measured RDC values as one molecule (B) and as two separate molecules (C). Fitting parameters are summarized in Table 1. (Complete list of RDC values are shown in Supplementary Figure S5).
Table 1.
Order tensor analysis of the 27b RNA construct
| Helix 1 ideal | Helix 2 ideal | Helix 1 27b | Helix 2 27b | |
|---|---|---|---|---|
| −Da × e−4 | 3.8 ± 0.4 | 3.7 ± 0.3 | 3.6 ± 0.4 | 4.0 ± 0.3 |
| R | 0.13 ± 0.07 | 0.11 ± 0.06 | 0.12 ± 0.05 | 0.17 ± 0.06 |
| GDO × e−4 | 7.2 ± 0.7 | 7.3 ± 0.5 | 7.1 ± 0.4 | 8.2 ± 0.5 |
| Neta | 0.20 ± 0.11 | 0.17 ± 0.09 | 0.18 ± 0.08 | 0.26 ± 0.09 |
Helix1 and helix2 were analyzed separately using ideal helices or calculated structures using the PALES program (29). Da is the axial component of the alignment tensor and neta is the directionality parameter.
Our analysis of RDC data argues that flexibility is limited between the two helical parts, suggesting that the inter-helical angle observed in the calculated structures is stable in solution and likely important for DNA packaging. To test this, and other predictions generated by our structure, we performed mutagenesis of pRNA in the bulge region and assessed the ability of modified pRNAs to support DNA packaging.
Mutational analysis revealed key residues of the bulge for DNA packaging
Since the conformation of the unpaired bulge residues introduced a bend between the two helical parts of the 27b RNA construct, we hypothesized that this bending angle is important for packaging activity. Although previous mutagenesis studies focused on the CCA residues known to be part of the bulge (18), here we investigated the influence of newly identified unpaired residues revealed by the NMR structure on DNA packaging. NMR analysis of the 27b construct showed that, whereas U21/A99 were base paired, U17 and A100 were not base paired. To assess the importance of this feature, G/C residues were substituted in order to force base pairing at this position. While a G/C substitution at the 21/99 position supported DNA packaging at a level similar to wt pRNA (Figure 4A), G/C substitution at the 17/100 position led to a ∼10× decrease in DNA packaging efficiency. This loss of activity from the introduction of a G/C base pair is likely due to modification of the structure, and supported our findings that residues 17 and 100 are not part of a helix, but rather have another role that is critical for pRNA function. Furthermore, mutation of U17 resulted in packaging effects similar to that observed for mutation of other nucleotides in the bulge (18,46). Although base substitutions were tolerated (46), deletion of U17 led to a ∼10× decrease in packaging, a result similar to that of deleting a C residue from the bulge (Figure 4A (18)). Deletion of both U17 and C18 led to no detectable packaging, again in agreement with previous results where deletion of any two bulge bases destroys packaging activity (18). Deletion of the unpaired A100 yields very low packaging activity, consistent with the NMR structure where A100 has multiple interactions with other residues. One of those interactions is the hydrophobic interaction with A20 involving C2H of each other. Interestingly, introducing a potential G/C base pair (A20 to G and A100 to C) preserved wt packaging activity. Taken together, the mutational analysis supports the structural observations from NMR and highlights the functional importance of the bulge conformation, and consequently helical bending, for DNA packaging.
Figure 4.
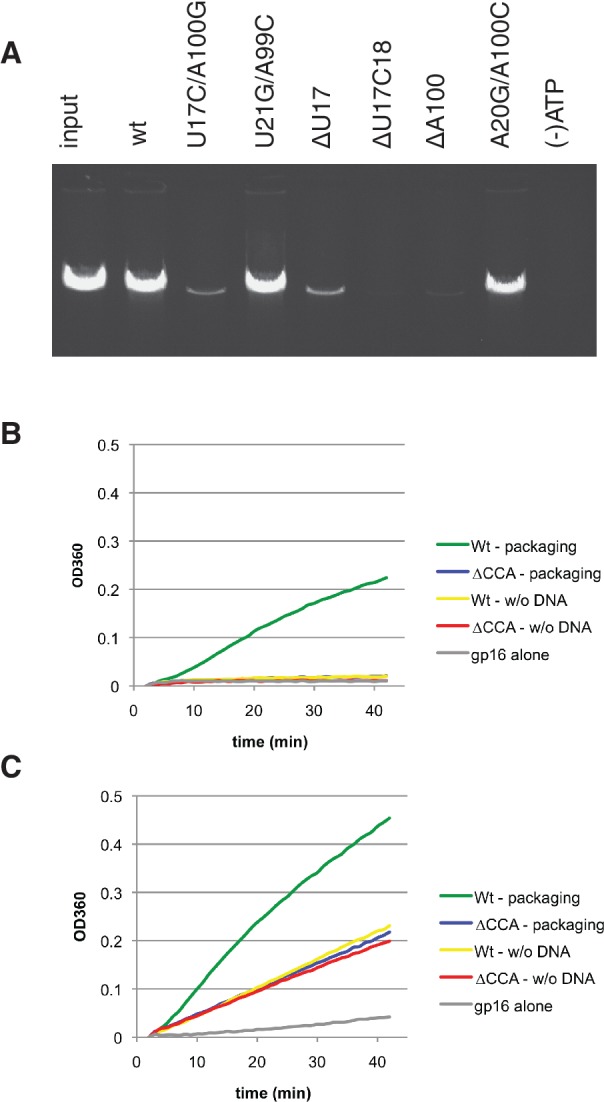
DNA packaging and ATPase activity of pRNA bulge mutants. (A) In vitro DNA packaging of proheads with wt or bulge-mutant 120b pRNA. Packaged DNA is protected from DNase digestion, and the packaged DNA is then extracted from the head and analyzed on an agarose gel. Input is amount of DNA added to the reaction and (−) ATP omits ATP from the packaging reaction. (B and C) ATPase activity of packaging complexes (packaging) or prohead/gp16 motor complexes (w/o DNA) using proheads with wt or ΔCCA pRNA. (B) High salt buffer conditions suppress non-packaging-related ATPase activity and (C) no salt buffer conditions promote non-packaging prohead/gp16 ATPase activity. Pi production is monitored spectrophotometrically (OD360) using the EnzChek assay. Representative traces from n = 3 experiments (Table 2).
The bulge did not affect ATP hydrolysis
pRNA is the scaffold upon which the ATPase assembles to the head. Previously, cryo-EM 3D reconstruction of proheads with ΔCCA pRNA showed that the appearance of the A-helix is altered in the absence of the bulge, with the distal portion of the A-helix appearing displaced and rotated compared with wt (18). Since the motor still assembles on this altered scaffold, we sought to further characterize the nature of the defect created by deleting this bulge.
The ATPase activity of the gp16 by itself is weak, but activity is strongly stimulated by pRNA bound to proheads (9,47). ATPase activity was assessed using proheads with wt or ΔCCA pRNA in both DNA-free (prohead/gp16) and DNA packaging conditions. Under packaging conditions that suppress non-packaging-related ATPase activity (high salt buffer) (37), the wt pRNA supported both ATPase activity and DNA packaging, while the ΔCCA mutant could not (Table 2 and Figure 4B). Under these conditions, ATPase activity of the prohead/gp16 complexes is at background levels similar to that of gp16 alone. In contrast, in the absence of DNA and under buffer conditions that promote DNA-free prohead/gp16 ATPase activity (no salt buffer), the ΔCCA mutant and wt pRNA supported similar levels of ATPase activity (Table 2 and Figure 4C). In packaging reactions, the activity level for ΔCCA remained the same as in the DNA-free sample, indicative of its inability to support DNA packaging, while the Pi production for wt increased, representing both packaging-related and prohead/motor ATPase activity. Since the ΔCCA mutant can support wt levels of ATPase activity in the absence of DNA but is defective in packaging, this suggests that the defect in motors with ΔCCA pRNA lies not in ATP hydrolysis per se, but in the coordinated action of the motor to drive DNA movement.
Table 2.
Effect of pRNA CCA bulge on ATPase activity related to DNA packaging and in the absence of DNA
| Packaging specific ATPase activity (slope × 10−5) | ATPase activity (slope × 10−5) | |
|---|---|---|
| Wt—packaging | 34.67 ± 4.99 | 68.33 ± 6.18 |
| ΔCCA—packaging | 2.33 ± 0.17 | 29 ± 4.55 |
| Wt—w/o DNA | 1.87 ± 0.25 | 38 ± 2.16 |
| ΔCCA—w/o DNA | 1.13 ± 0.21 | 30 ± 2.45 |
| Gp16 alone | 0.73 ± 0.33 | 5.33 ± 1.25 |
Values reflect relative rate of ATP hydrolysis determined by the generation of Pi. (n = 3, results are mean ± SD).
Complete model of the pentameric pRNA
Here, we have determined the structure of the CCA bulge, a key functional feature of the pRNA A-helix, given that all other single-base bulges can be deleted from the A-helix without abolishing function (46). By combining the recently published crystal structure of the prohead-binding domain of pRNA (13), which contains the elements for prohead attachment and oligomerization, with the 27b NMR structure, and a 10 base pair model A-form helix representing the distal portion of the pRNA A-helix, we created a complete model of the functional 120 nucleotide pRNA (Figure 5A).
Figure 5.
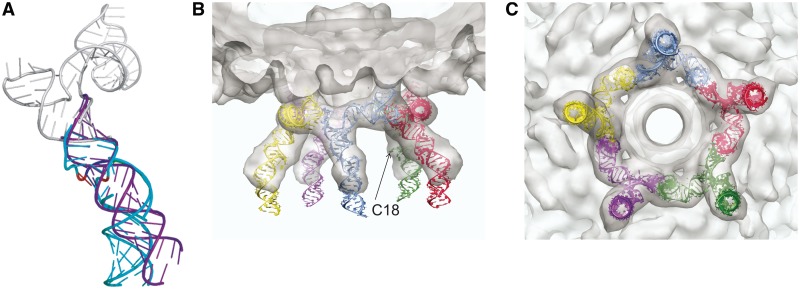
Structural models of the 120b pRNA. (A) Model of the 120b monomer. The prohead-binding region and intermolecular interaction region (13) are coloured gray. The A-helix containing the UCCA bulge is shown in blue. An ideal helix representing the A-helix lacking the bulge is shown in purple. (B and C) A pentameric pRNA model fit into the cryoEM density map is shown from the side (B) and from the ATPase gp16-binding side (C). Each pRNA monomer is coloured differently. The position of the bulged nucleotide C18 is indicated by the arrow in one monomer (B).
To create a pentameric model, hybrid 120b monomers were fit into the 10 Å cryo-EM 3D map of the 120b pRNA bound to proheads ((5), Morais et al., unpublished data; Supplementary Methods). The resulting docked structure showed very good fitting of these structures to density (Figure 5B and C). The pentameric model has the following geometry: the inner diameters before the bulge and after the bulge are 128 and 144 Å, respectively, and the diameter at the end of the A-helix is ∼150 Å. Therefore, the ATPase (gp16) binding region has a diameter between 144 and 150 Å.
DISCUSSION
We have determined the NMR structure of the 27mer RNA containing the CCA bulge region of the ϕ29 pRNA. The 27mer structure revealed that the bulge structure is more complex than suggested by secondary structure predictions and phylogenetic analysis. The bulge region is formed by four nucleotides, with the addition of residue U17 to the previously predicted three-base 18-CCA-20, thus comprising a UCCA bulge (Figure 2C). Since U17 is part of the bulge structure, the absence of the putative base pairing with A100 leaves this residue as a single base bulge on the opposite strand. Mutational analysis revealed the importance of these newly assigned structural features provided by the NMR structure. Substitution of the unpaired U17 and A100 with a G/C base pair to create a strong interaction at this site diminished DNA packaging (Figure 4A). In contrast, substituting a G/C base pair at positions A20 and A100, which were shown to interact in the NMR structure, resulted in retention of activity similar to wt pRNA. The A20G mutation would lose the hydrogen bond between A20 N6 amino and U16 O2 carbonyl, but a hydrogen bond between A20G N2 amino and A101 N3 would be newly able to form, which would keep the coordination of the U16-A101-A20 base triple. In addition, substitutions of A20G and A100C could replace the A100 C2H to A20 C2H interaction with a hydrogen bond between A100C O2 carbonyl and A20G N2 amino, resulting in keeping the base orientation of A20 and A100. Therefore, the A20G/A100C double mutation could maintain the structure of the bulge as well as the DNA-packaging activity of wt pRNA.
Of note, other members of the ϕ29 phage family, including M2, NF, SF5 and GA1, were all predicted to have pRNAs with four nucleotide bulges based on their phylogeny (Figure 6B, (11)). Thus, the presence of a four-nucleotide bulge in the 27b structure suggests that this is a conserved structural feature of prohead RNAs, even though the nucleotide sequence is not fully conserved between phages. Furthermore, since the 27b RNA also has a bulged adenine nucleotide on the opposite side of the helix (A100) similar to that predicted for M2, NF and SF5 pRNAs, ϕ29 and these other phages likely share similar 3D bulge structures (Figure 6B).
Figure 6.
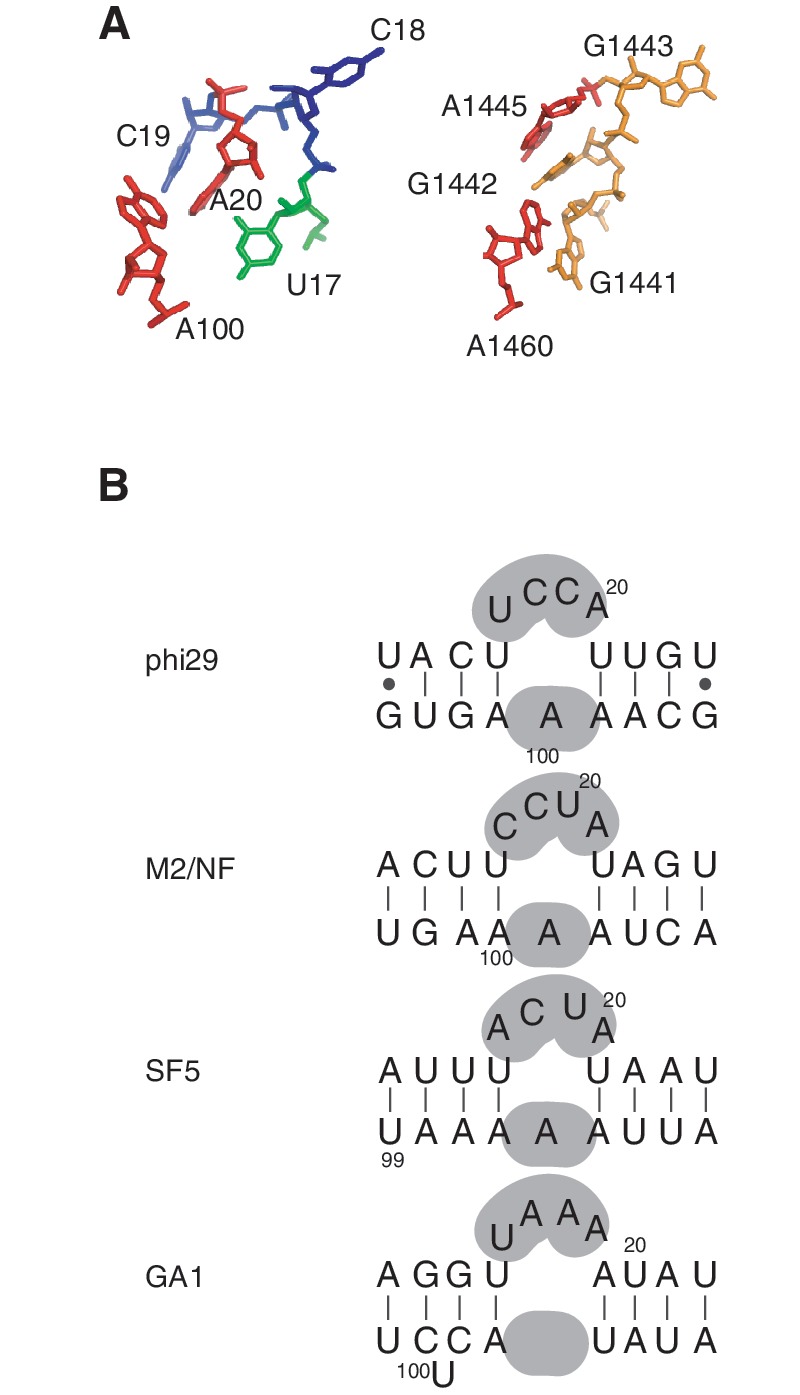
The four residue bulge of pRNAs. (A) Bulge regions of the 27b and 16S ribosomal RNA of Thermus thermophilus (PDB code 1FJG) are shown. Nucleotides of 27b are colored according to Figure 2. (Note that the numbering of nucleotides in 16S rRNA of T. thermophilus is irregular as the sequence was transformed to map them onto the Escherichia 16S rRNA.) (B) Secondary structure predictions of the bulged regions of other phage pRNAs, with bulge nucleotides highlighted in gray.
In the 27b structure, the bulge creates an ∼35° bend in the helical axis (Figure 3A). This bend is consistent with cryo-EM reconstructions comparing wt and ΔCCA pRNA bound to proheads. Superimposition of these structures showed that the distal portion of the A-helix of the ΔCCA pRNA is displaced by ∼30° compared with wt pRNA (18). Order tensor analysis using RDC data suggests that this angle is stable. The structure and dynamics of a bulge-containing HIV-1 TAR RNA is strongly dependent on Mg2+ concentration (48,49). Therefore, we tested the Mg2+ dependence of the 27b structure by folding the RNA without Mg2+ in NMR buffer. Chemical shifts of base, sugar and imino protons of 27b did not change with and without Mg2+, indicating that the bulge does not bind Mg2+ (Supplementary Figure S7).
We compared the 27b structure with other known bulge-containing RNAs using the RNA FRABASE (http://rnafrabase.cs.put.poznan.pl). Since the non-base-paired U17 is partly stacking over helix 1, we included both three- and four-nucleotide bulge structures in our search. A search for three-nucleotide bulges yielded 20 hits, but was comprised of only four RNA molecules other than ϕ29 pRNA. Ten structures were HIV-1 TAR RNA with different ligands, and another five were cryo-EM 3D reconstructions of the ϕ29 pRNA pentamer. The remaining five structures included the complexes of tRNA with pseudouridine synthase TruB, an endonuclease from Archaeglobus fulgidus with its substrate, and the substrate for the archael pre-tRNA splicing endonucleases; none of these three-nucleotide bulge structures were similar to the bulge structure of our 27b RNA. A search for four nucleotide bulges with an unpaired nucleotide in the opposite strand yielded 209 hits. Of these, a fragment of the 30S ribosomal subunit of Thermus thermophilus containing 16S ribosomal RNA has a bulge structure (PDB code 1FJG; 1441-GGGA-1445:A1460) similar to our 27b RNA (Figure 6A). This bulge is a part of the 3′-minor domain of 16S rRNA and introduces a 26.4° angle between the two helical parts of rRNA that flank this bulge. Since the 27b structure showed 35 ± 9° bend, it is likely that a similar bulge conformation is responsible for introducing similar angles in RNA structures.
Cryo-EM 3D reconstruction of the ϕ29 prohead–ATPase motor complex demonstrated that the gp16 pentamer is attached to the prohead solely through interaction with pRNA (5). RNase footprinting experiments showed that portions of the A-helix, including the bulge nucleotides U17, C18 and C19, are part of the region that is protected from RNase digestion by gp16 (8). Widening of the major groove by non-canonical base pairs, triples and bulges is frequently found in RNA–protein interactions (50,51). Since the UCCA bulge induces widening of the major groove (Supplementary Figure S4), this may also provide an opportunity for sequence-specific recognition of residues in the bulge.
A key function of pRNA is to act as a scaffold for binding the gp16 packaging ATPase during motor assembly. gp16 is, by itself, a weak ATPase, and its catalytic function is strongly stimulated by binding to pRNA on the head (9,52). Although the gp16 attached to ΔCCA pRNA showed ATP hydrolysis activity comparable to wt under DNA-free, low salt conditions, ΔCCA pRNA cannot support DNA packaging (Figure 4C). Some variants of pRNA containing mutations in the bulge region support DNA packaging, albeit with much less efficiency compared with wt pRNA (18). Interestingly, no partially packaged intermediates were found in these mutant pRNAs experiments, suggesting that once packaging has started it goes to completion (18). Therefore, this suggests that pRNA plays an important role in positioning gp16 for the initiation of the DNA packaging. The claw shape of the pRNA induced by the UCCA bulge appears key for proper alignment of gp16 in the motor. In the absence of the bulge, the ends of pRNA would be more splayed relative to wt (Figure 5). Alteration of the scaffold by deletion of the bulge may cause misalignment in the ATPase ring, possibly affecting the coordinated action of the ATPases required for packaging or altering the ATPase interaction with the DNA. Future studies focusing on the initiation of DNA packaging will reveal further details of the pRNA scaffold function and shed additional light on this ATPase-driven DNA translocation process.
ACCESSION NUMBERS
Atomic coordinates, constraints and chemical shifts were deposited to protein data bank under accession code 2LQZ. NMR assignments were deposited to Biomagnetic Resonance Bank under accession number 18336.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–8, Supplementary Methods and Supplementary References [56,57].
FUNDING
National Institutes of Health (NIH) [GM-059604 and GM-095516]. Funding for NMR instrumentation was provided by the Office of the Vice President for Research, the Medical School, the College of Biological Science, NIH, NSF and the Minnesota Medical Foundation; NIH [P41RR02301, P41GM66326, RR02781 and RR08438 to the National Magnetic Resonance Facility at Madison]; National Science Foundation [DMB-8415048, OIA-9977486 and BIR-9214394 to the National Magnetic Resonance Facility at Madison]. Funding for open access charge: NIH [GM-059604].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Key instrumentation was provided by the University of Minnesota NMR Facility, Supercomputing Institute, the University of Wisconsin NMRfam and University of Texas Medical Branch SCSB Cryo-electron Microscopy Laboratory.
REFERENCES
- 1.Casjens SR. The DNA-packaging motor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 2.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 3.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage ø29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 4.Grimes S, Jardine PJ, Anderson D. Bacteriophage ø29 DNA packaging. Adv. Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 5.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage ϕ29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage ø29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 8.Koti JS, Morais MC, Rajagopal R, Owen BAL, McMurray CT, Anderson DL. DNA packaging motor assembly intermediate of bacteriophage phi29. J. Mol. Biol. 2008;381:1114–1132. doi: 10.1016/j.jmb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TJ, Zhang H, Lianga D, Guo P. Strand and nucleotide-dependent ATPase activity of gp16 of bacterial virus phi29 DNA packaging motor. Virology. 2008;380:69–74. doi: 10.1016/j.virol.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wichitwechkarn J, Bailey S, Bodley JW, Anderson D. Prohead RNA of bacteriophage ø29: size, stoichiometry and biological activity. Nucleic Acids Res. 1989;17:3459–3468. doi: 10.1093/nar/17.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J. Biol. Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- 12.Guo P. Structure and function of ø29 hexameric RNA that drives the viral DNA packaging motor. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:415–472. doi: 10.1016/s0079-6603(02)72076-x. [DOI] [PubMed] [Google Scholar]
- 13.Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc. Natl Acad. Sci. USA. 2011;108:7357–7362. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X, Schroeder SJ. Different sequences show similar quaternary interaction stabilities in prohead viral RNA self-assembly. J. Biol. Chem. 2011;286:14419–14426. doi: 10.1074/jbc.M110.191064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris S, Schroeder SJ. Nuclear magnetic resonance structure of the prohead RNA E-loop hairpin. Biochemistry. 2010;49:5989–5997. doi: 10.1021/bi100393r. [DOI] [PubMed] [Google Scholar]
- 16.Zhang CL, Lee CS, Guo P. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage phi29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 17.Reid RJD, Bodley JW, Anderson D. Identification of bacteriophage phi29 RNA domains necessary for in vitro DNA-gp3 packaging. J. Biol. Chem. 1994;269:9084–9089. [PubMed] [Google Scholar]
- 18.Zhao W, Morais MC, Anderson DL, Jardine PJ, Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage ø29 in DNA packaging. J. Mol. Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigelt J. A single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 1998;120:10778. [Google Scholar]
- 20.Lewis Kay E, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BA, Blevins RA. NMRview: a computer program for the visualization and analysis for NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 23.Smallcombe SH. Solvent suppression with symmetrically-shifted pulses. J. Am. Chem. Soc. 1993;115:4776–4785. [Google Scholar]
- 24.Range M, Wright PE. Analysis of 1H NMR spectra of proteins using multiple-quantum coherence. J. Magn. Res. 1986;66:372–378. [Google Scholar]
- 25.Marino JP, Diener JL, Moore PB, Griesinger C. Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly 13C-labeled RNA. J. Am. Chem. Soc. 1997;119:7361–7366. [Google Scholar]
- 26.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Staple D, Butcher S. Solution structure of the HIV-1 frameshift inducing stem–loop RNA. Nucleic Acid Res. 2003;31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 30.Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Wilkinson MF. Site-directed mutagenesis of large (13-kb) plasmids in a single-PCR procedure. Biotechniques. 2000;29:976–978. doi: 10.2144/00295bm09. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Wilkinson MF. Deletion mutagenesis of large (12-kb) plasmids by a one-step PCR protocol. Biotechniques. 2001;31:722–724. doi: 10.2144/01314bm01. [DOI] [PubMed] [Google Scholar]
- 33.Reid RJD, Bodley JW, Anderson D. Characterization of the prohead-pRNA interaction of bacteriophage ø29. J. Biol. Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 34.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol. Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with x-ray crystallographic structures. J. Struct. Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 36.Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 37.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 38.Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 39.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulyanov NB, James TL. RNA structural motifs that entail hydrogen bonds involving sugar-phosphate backbone atoms of RNA. New J. Chem. 2010;34:910–917. doi: 10.1039/b9nj00754g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura A, Jardine PJ, Anderson DL, Grimes S, Matsuo H. Analysis of intermolecular base pair formation of prohead RNA of the phage phi29 DNA packaging motor using NMR spectroscopy. Nucleic Acid Res. 2008;36:839–848. doi: 10.1093/nar/gkm874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Kima N-K, Peterson RD, Wanga Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc. Natl Acad. Sci. USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luebke KJ, Landry SM, Tinoco I. Solution conformation of a five-nucleotide RNA bulge loop from a group I intron. Biochemistry. 1997;36:10246–10255. doi: 10.1021/bi9701540. [DOI] [PubMed] [Google Scholar]
- 44.Mortimer SA, Weeks KM. C2′-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc. Natl Acad. Sci. USA. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C, Tellinghuisen T, Guo P. Use of circular permutation to assess six bulges and four loops of DNA-packaging pRNA of bacteriophage phi29. RNA. 1997;3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 47.Grimes S, Anderson D. Cleaving the prohead RNA of bacteriophage phi 29 alters the in vitro packaging of restriction fragments of DNA-gp3. J. Mol. Biol. 1989;209:101–108. doi: 10.1016/0022-2836(89)90173-3. [DOI] [PubMed] [Google Scholar]
- 48.Al-Hashimi H, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casiano-Negroni A, Sun X, Al-Hashimi HM. Probing Na+-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry. 2007;46:6525–6535. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hermann T, Westhof E. Non-Watson–Crickbase pairs in RNA–protein recognition. Chem. Biol. 1999;6:R335–R343. doi: 10.1016/s1074-5521(00)80003-4. [DOI] [PubMed] [Google Scholar]
- 51.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 52.Grimes S, Anderson DL. RNA dependence of the bacteriophage phi 29 DNA packaging ATPase. J. Mol. Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 53.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of phi29 Connector Channel Loops in Late-Stage DNA Packaging. J. Mol. Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. [Google Scholar]
- 55.Lu X-J, Olson1 WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Nozinovic S, Fürtig B, Jonker HRA, Richter C, Schwalbe H. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acid Res. 2010;38:683–694. doi: 10.1093/nar/gkp956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.