Abstract
Malaria parasites have a complex life cycle, during which they undergo significant biological changes to adapt to different hosts and changing environments. Plasmodium falciparum, the species responsible for the deadliest form of human malaria, maintains this complex life cycle with a relatively small number of genes. Alternative splicing (AS) is an important post-transcriptional mechanisms that enables eukaryotic organisms to expand their protein repertoire out of relatively small number of genes. SR proteins are major regulators of AS in higher eukaryotes. Nevertheless, the regulation of splicing as well as the AS machinery in Plasmodium spp. are still elusive. Here, we show that PfSR1, a putative P. falciparum SR protein, can mediate RNA splicing in vitro. In addition, we show that PfSR1 functions as an AS factor in mini-gene in vivo systems similar to the mammalian SR protein SRSF1. Expression of PfSR1-myc in P. falciparum shows distinct patterns of cellular localization during intra erythrocytic development. Furthermore, we determine that the predicted RS domain of PfSR1 is essential for its localization to the nucleus. Finally, we demonstrate that proper regulation of pfsr1 is required for parasite proliferation in human RBCs and over-expression of pfsr1 influences AS activity of P. falciparum genes in vivo.
INTRODUCTION
Plasmodium falciparum is the causative agent of the deadliest form of human malaria infecting millions each year, resulting in over million deaths primarily of children and pregnant women (1). The malaria parasite has a complex life cycle, during which it undergoes significant morphological changes to be able to adapt to different hosts and changing environments. These parasites alternate between the mosquito and human hosts. Within each host they have extracellular motile forms that evolved specific molecular machinery to invade and propagate within either the mid-gut epithelial cells in the mosquito or hepatocytes and red blood cells (RBCs) in the human host. These changes are mediated by tight regulation of cell cycle-dependent gene-expression patterns (2). Thus, the complexity of P. falciparum biology is achieved with a rather small genome that contains ∼5700 genes (3) (http://plasmodb.org/plasmo/showXmlDataContent.do?name=XmlQuestions.GeneMetrics). It is striking that P. falciparum adapted to this complex life cycle with less number of genes than the yeast Saccharomyces cerevisiae, an organism that has a much simpler life cycle (http://www.yeastgenome.org/cache/genomeSnapshot.html). One way by which eukaryotic organisms can regulate gene expression and expand their protein repertoire out of a limited number of genes is through post-transcriptional regulation of alternative splicing (AS) of pre-mRNA that enable synthesis of functionally different protein isoforms from the same pre-mRNA transcripts. In mammals, it is estimated that >90% of genes undergo AS (4).
In recent years there have been an increasing number of studies reporting on different AS events in transcripts from numerous Plasmodium genes. To our knowledge, the first report of AS in plasmodium was of the blood stage antigen 41-3 precursor (PFL0385c) in P. falciparum (5). In addition, multiple spliced variants were found to encode for different functional isoforms of P. falciparum adenylyl cyclase, including a novel isoform that is expressed in sexual stages (6). AS also plays a key role in developmental regulation of different isoforms encoded by the maebl gene implicated in red cell invasion in several plasmodium species, demonstrating a high degree of conservation of AS mechanisms across Plasmodium evolution (7). Interestingly, distinct spliced variants were reported in the 5′-UTRs of transcripts from the yir multi-copy gene family, a large complement of genes implicated in antigenic variation in P. yoelii (8). Additional AS events have been reported in a small-scale analysis of cDNA libraries (9). Recently genome wide studies using RNA-seq of different stages during the intra erythrocytic development cycle (IDC) implied that over 300 AS events occur in ∼4% of the genes in the malaria genome (10,11). This evidence suggests that post-transcriptional regulation of gene expression through AS of pre-mRNAs is an important mechanism by which Plasmodium parasites regulate gene expression and expand their proteome diversity. However, despite the fact that ∼54% of the parasite’s genes contain introns (3) and that ∼30% of the genes contain at least two introns (Supplementary Figure S1), very little is known about splicing factors in Plasmodium and even less is known about mechanisms that regulate gene expression through AS.
The best characterized AS factors in eukaryotes belong to the family of SR proteins (12–14). These proteins have a modular structure consisting of a C-terminus SR domain as well as one or two RNA recognition motifs (RRMs). SR proteins function as a part of the spliceosome and are required for both constitutive and alternative splicing (15). SR proteins are functionally regulated through specific phosphorylation of their RS domain by several kinases, particularly by SR protein-specific kinases belonging to the SRPK family (13). The complex regulation of AS is only partly understood in higher eukaryotes and remains elusive in lower eukaryotes. Nevertheless, the involvement of SR proteins with AS activity was reported in Schizosaccharomyces pombe (16) and Trypanosoma cruzi (17). A recent study demonstrated that P. falciparum has a functional SRPK homolog (PfSRPK1), which is involved in mRNA splicing in this parasite (18). This elegant study also showed that PfSRPK1 is associated with a putative splicing factor candidate named PfSR1. PfSRPK1 could phosphorylate PfSR1 and thus affect its binding affinity to mRNA in vitro, implying that these two proteins are potentially regulatory components of the P. falciparum splicing machinery.
Using recombinant purified PfSR1 protein, we demonstrate that it is a functional SR protein that can complement splicing reactions in vitro, thus identifying the first validated splicing factor in P. falciparum. In addition, we show that PfSR1 regulates AS activity in mini-gene systems in mammalian cells similar to the human splicing factor SRSF1 (previously known as SF2/ASF) (19) indicating that it can also function as an AS factor. Using parasites transfected with PfSR1-myc, we determined its cellular localization during the IDC and by mutation analyses, we identified the RS domain as its nuclear localization signal (NLS). Finally, we show that proper regulation of the pfsr1 gene is essential for P. falciparum development in human RBCs.
MATERIALS AND METHODS
Bioinformatics search for putative SR proteins
In order to find putative SR proteins in P. falciparum, we initially performed BLAST searches against all predicted proteins from P. falciparum using a consensus RS domain sequence consisting of 30 tandem Ser/Arg dipeptides repeats as a query. This enabled us to identify sequences that are compositionally biased in alternating Ser/Arg residues as described (20). Further searches for protein motifs such as the SR domain of SRSF1 or dipeptide patterns such as RSRS or SRSR were performed against all predicted proteins in P. falciparum using the PlasmoDB (http://plasmodb.org/plasmo/). Candidate genes were further analyzed using Myhits (SIB) (http://myhits.isb-sib.ch/) in order to identify the genes that contain a predicted RRM as well as a putative RS domain. Multiple sequence alignments of the putative SR and SR-like proteins were performed with the MUSCLE program (21). Phylogenetic tree was built with Neighbor-Joining algorithm (22) and the evolutionary distances among candidate SR proteins were calculated using the Poisson correction method with MEGA5 software (23). Evolutionary distances are essentially the number of amino acids substitutions per aligned site.
Cell cultures
All parasites used were derivatives of the NF54 parasite line and were cultivated at 5% hematocrit in RPMI 1640 medium, 0.5% Albumax II (Invitrogen), 0.25% sodium bicarbonate and 0.1 mg/ml gentamicin. Parasites were incubated at 37°C in an atmosphere of 5% oxygen, 5% carbon dioxide and 90% nitrogen. Parasite cultures were synchronized using percoll/sorbitol gradient centrifugation as previously described (24,25). Briefly, infected RBCs were layered on a step gradient of 40/70% percoll containing 6% sorbitol. The gradients were then centrifuged at 12 000g for 20 min at room temperature. Highly synchronized, late stage parasites were recovered from the 40/70% interphase, washed twice with complete culture media and placed back in culture. The level of parasitemia was calculated for each time point by counting three independent blood smears stained with Giemsa under light microscope.
HEK 293 T cells were grown in Dulbecco’s modified Eagle’s medium (Biological Industries Inc., Israel) supplemented with 10% fetal bovine serum (FCS), penicillin and streptomycin at 37°C with 5% CO2.
Plasmid construction
In order to express PfSR1 in the mammalian cells a synthetic cDNA of pfsr1 with codons optimized for mammalian cells was synthesized by GeneArt, fused to a T7 tag. The synthetic pfsr1 was amplified using PfSR1PTTF-XbaI–PfSR1PTT-HisR primers (see Supplementary Materials) and cloned into the pTT3 plasmid (26) using XbaI and BamHI restriction sites, creating His-tagged PfSR1 similar to the His-tagged SRSF1 previously described (27).
The synthetic pfsr1 fused to a T7 tag was cloned into pcDNA3− plasmid (Invitrogen) using XhoI and KpnI restriction sites creating a similar plasmid to the pCDNA3-SRSF1 previously described (27).
PfSR1 was cloned into the expression vector pHBIRH (28) previously described and fused to a myc epitope tag to generate pHBISR1myc. The myc tag was amplified using the primers mycF–mycR (see Supplementary Materials) and cloned using SpeI and SacI restriction sites. pfsr1 was amplified using the primers PfSR1F–PfSR1R (see Supplementary Materials) and cloned into the pHBIRH plasmid using SalI and SpeI restriction sites.
To express PfSR1ΔRRM1myc the pfsr1 gene was amplified using the primers dRRM1F–PfSR1R (see Supplementary Materials) and cloned into the pHBIRH plasmid using SalI and SpeI restriction sites. Similarly, PfSR1ΔRSmyc was created using the primers PfSR1F–dRSR (see Supplementary Materials). PfSR1ΔRRM2myc was created using quick-change mutagenesis with the dRRM2F–dRRM2R primers (see Supplementary Materials) and using pHBIPfSR1myc as template for polymerase chain reaction (PCR) amplification.
Mammalian cell transfections
HEK 293 T cells were transfected using Fugene 6 (Roche Diagnostics), according to the manufacturer’s recommendations. In order to purify the PfSR1-His protein, cells were transfected using linear polyethylenimine (PEI 25000; Polysciences Inc.). A transfection mix consisting of 1 μg plasmid DNA, 2 μg PEI and 50 μl of JMEM was prepared for every 1 ml of cell culture to be transfected. The mix was incubated at room temperature for 10–15 min and was then added to the cell culture. Forty-eight hours post-transfection cells were collected and analyzed.
Parasite transfections
Parasites were transfected as previously described (29). Briefly, 0.2 cm electroporation cuvettes were loaded with 0.175 ml of erythrocytes and ∼100 µg of plasmid DNA in incomplete cytomix solution. Stable transfectants were initially selected on 2 µg/ml blasticidin-S-hydrochloride (Invitrogen, USA). In order to obtain parasites carrying high plasmid copy numbers, these cultures were then subjected to 6 µg/ml blasticidin.
Genomic DNA extraction, RNA extraction and cDNA synthesis
Genomic DNA was extracted as described (30). Briefly, iRBCs were pelleted and lysed with Saponin. RBC-free parasites from 20 ml cultures were then pelleted, washed with phosphate buffered saline (PBS), and taken up in 200 μl TSE buffer (100 mM NaCl, 50 mM EDTA, 20 mM Tris, pH 8), 40 μl of 10% sodium dodecyl sulfate (SDS) and 20 μl 6 M NaClO4. This suspension was rocked for 12 h and genomic DNA was extracted with phenol/chloroform. The resulting DNA was taken up in 100 μl dH2O.
RNA extraction and cDNA synthesis were performed as described (31). Briefly, RNA was extracted from synchronized parasite cultures at 36 h post-invasion. RNA was extracted with the TRIZOL LS Reagent® as described (32) and purified on PureLink column (Invitrogen) according to manufacturer’s protocol. Isolated RNA was then treated with recombinant Deoxyribonuclease I® (Takara) to degrade contaminating gDNA. cDNA synthesis was performed from 750 ng total RNA with Primescript RT reagent® (Takara) with oligo dT (Takara) as described by the manufacturer.
Real-time reverse transcriptase–qPCR
Transcript copy numbers were determined using the formula 2−ΔΔCT as described in the Applied Biosystems User Bulletin 2 using NF54 gDNA as the calibrator. Specifically, relative copy number was calculated as two exponential negative ((Ct target gene in cDNA − Ct reference gene in cDNA) − (Ct target gene in gDNA–Ct target gene in gDNA)). Relative copy numbers of pfsr1 and renilla luciferase were calculated comparing to the expression of the housekeeping gene arginyl–tRNA synthetase (PFL0900c). All reverse transcriptase (RT)–qPCR assays were performed at least in duplicate for each template with no apparent differences, and each experiment was completed three times in its entirety, again with no significant differences.
The housekeeping and other control primers used for RT–qPCR were previously published (renilla in (28) and arginyl–tRNA synthetase in (31)). Ectopic expression of pfsr1 was measured using primers designated to specifically amplify the fusion of pfsr1 and the myc tag (see Supplementary Materials).
Protein purification
293T cells were transfected with human pTT3-SRSF1-His or pTT3-PfSR1-His and harvested 48 h after transfection. Cells were re-suspended in lysis buffer (50 mM Tris–HCl [pH 8.0], 1 M NaCl, 10 mM imidazole, 20 mM β-mercaptoethanol and 0.1% [vol/vol] Triton X-100) and centrifuged after sonication to remove insoluble material. Most cellular proteins were precipitated by using ammonium sulfate at 40% saturation at 4°C and were separated by centrifugation. His-tagged proteins were purified from the supernatant by nickel–agarose affinity chromatography.
In-vitro splicing assay
In vitro splicing assays were done as described previously (33,34). Briefly, radiolabeled Ftz reporter pre-mRNAs was prepared by the in vitro transcription of plasmids by SP6 RNA polymerase, in the presence of [α-32P]UTP. Pre-mRNAs were incubated under splicing conditions in HeLa cells nuclear or cytoplasmic S100 extracts, as described previously. RNAs were extracted and analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) and phosphorimager analysis on a Fujifilm FLA-5100 instrument (Fuji Medical Systems Inc., USA).
Immuno-fluorescence assay
Immuno-fluorescence assays (IFA) were performed as described (http://www.mr4.org/Portals/3/Pdfs/ProtocolBook/Methods_in_malaria_research.pdf) with slightly modifications. Briefly, RBCs were lysed, washed twice in PBS and fixed in 4% paraformaldehyde, followed by three washes in PBS. Parasites were blocked using 2% bovine serum albumin (BSA) in PBS + 0.1% tween20 (PBST). Mouse-anti-myc (Santa-Cruz) antibodies were incubated at 1:1000 dilution in blocking solution, washed three times in PBST and incubated with 1:250 dilution of 488-conjugated rabbit anti-mouse anitbodies (Invitrogen). After final washes, slides were sealed with ProLong Gold Reagent (Invitrogen).
Western blot analysis
In order to collect proteins from the 293 T cells, transfected cells were washed twice with PBS and collected in sample buffer (63 mM Tris–HCl, 10% glycerol, 2% SDS and 0.0025% Bromophenol Blue). To collect parasite proteins, iRBCs were lysed in saponin, parasites were washed twice in PBS and collected in sample buffer. Proteins were subjected to SDS–PAGE (10% polyacrylamide) and electroblotted to nitrocellulose membrane. Immunodetection was carried out using mouse-anti-myc antibody (Santa-Cruz) or mouse-anti-T7 antibody (Novagen; 1:5000), followed by rabbit anti-mouse antibody conjugated to Horseradish Peroxidase (HRP) (Jackson, ImmunnoResearch Laboratories, Inc.) and developed in EZ/ECL solution (Israel Biological Industries Ltd).
RESULTS
PfSR1 is a functional SR protein that can mediate pre-mRNA splicing
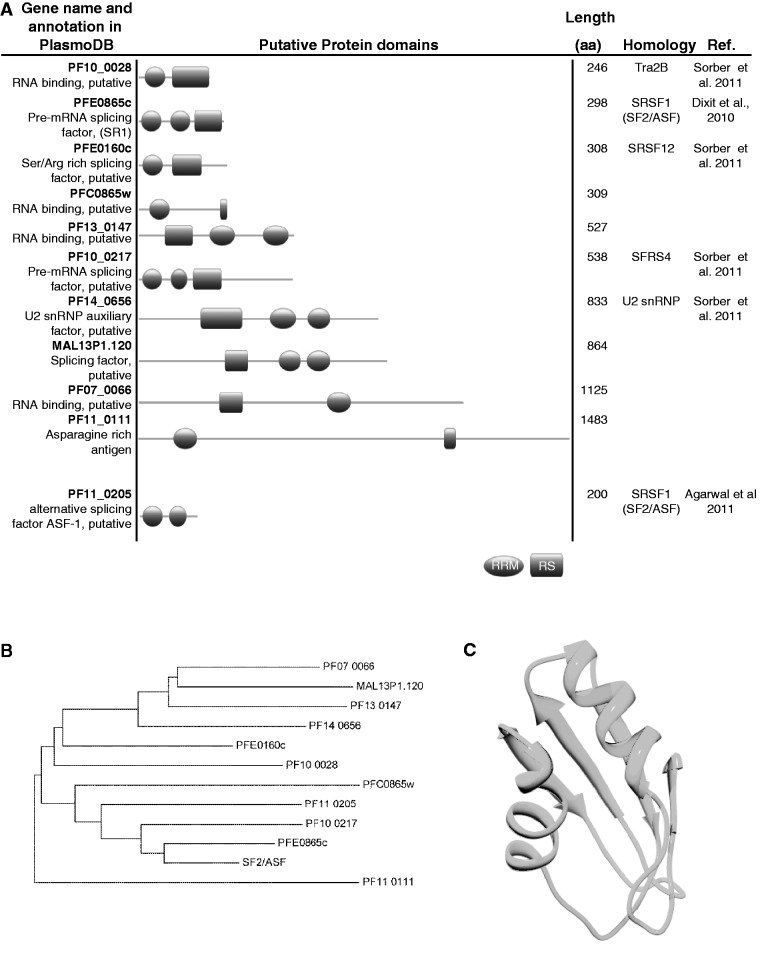
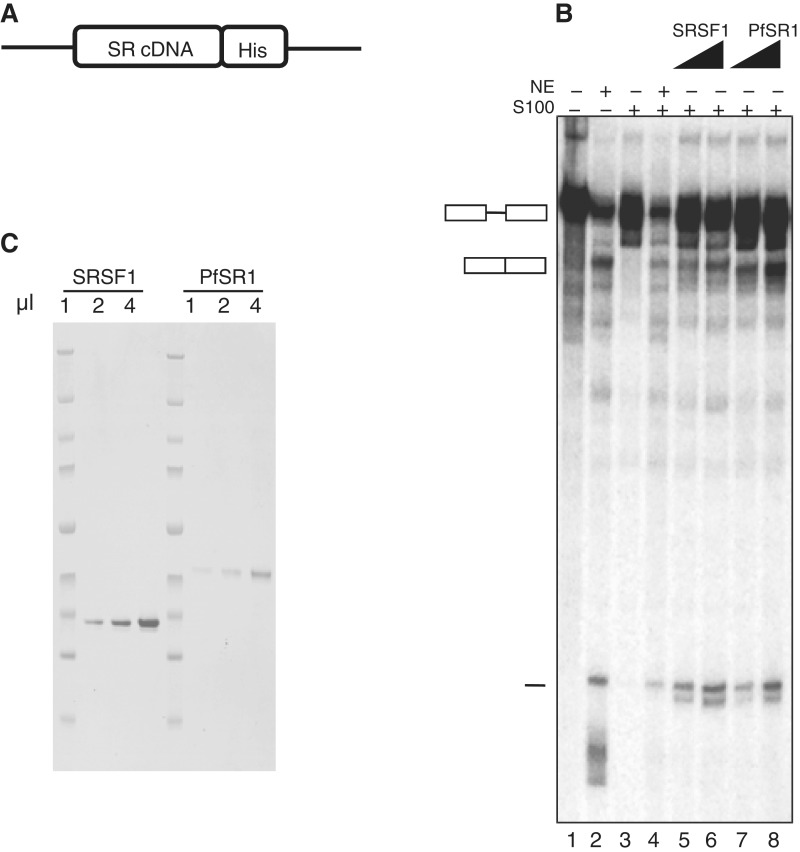
Very little is known about splicing factors in Plasmodium spp., however, it is clear that the splicing machinery as well as the factors involved in regulation of AS functions in these pathogens. We used BLAST search in the Plasmodium genome database http://plasmodb.org/plasmo/ to look for candidate genes in P. falciparum that encode potential SR proteins (Figure 1A). We found that P. falciparum may have more SR proteins than were previously identified (9,11,18,35). Nevertheless, to our knowledge their involvement in RNA metabolism had not been demonstrated. As a first step to investigate whether P. falciparum has functional SR proteins, we chose to begin our analysis with PfSR1, a protein that shares the highest sequence homology to the mammalian SR protein SRSF1 (SF2/ASF) (Figure 1B). PfSR1 was recently shown to be phosphorylated by PfSRPK1 at its predicted SR domain, and its ability to bind RNA in vitro was also demonstrated (18). We compared the first RRM domain of PfSR1 and the human SRSF1 (PDB ID 1x4a), using homology modeling. Overall the RRMs are highly conserved with 51% sequence identity over a total length of 80 amino acids (Figure 1C). To test whether PfSR1 has the same splicing activity as metazoan SR proteins that can complement an in vitro splicing reaction, we purified a recombinant version of PfSR1 fused to a 6× His-tag (Figure 2A). We then applied an in vitro S100 complementation assay to determine whether the recombinant PfSR1 by itself can restore the splicing of pre-mRNA in splicing deficient S100 extract. The S100 extracts lack SR proteins and this assay has been previously used to characterize several recombinant mammalian SR proteins including SRSF1 that could complement splicing when added to this extract (33,34). We therefore compared the complementation activity of PfSR1 with the mammalian SRSF1 as a positive control. Incubation of increasing concentrations of recombinant PfSR1 to the S100 incomplete extracts was sufficient to complement splicing of the Ftz reporter pre-mRNA similar to the recombinant SRSF1 (Figure 2B, Lane 3 compared with Lanes 7 and 8). The appearance of splicing products was correlated with the increasing concentration of the recombinant PfSR1, similar to the effect observed with the recombinant SRSF1 (Figure 2B, Lanes 5 and 6). Similar results were obtained with the β-globin reporter pre-mRNA (Supplementary Figure S2). We concluded that PfSR1 by itself could mediate splicing of pre-mRNA and demonstrate that PfSR1 is a functional splicing factor.
Figure 1.
Putative SR and SR-like proteins in P. falciparum. (A) Schematic representation of the domain structure of P. falciparum putative proteins that are predicted to contain RNA Recognition Motif (RRM, ellipses) and SR domain (RS, rectangles). Pervious predictions of homologs are shown with references (B) A phylogenetic tree built with the Neighbor-Joining algorithm based on the entire amino acid sequence of the predicted P. falciparum SR proteins. As shown the closest homologs of SRSF1 are PfSR1 (PFE0865c) and PF10_0217, respectively. (C) Homology modeling of the first RRM of PfSR1 based on the human SRSF1 RRM1. The crystal structure of the human SRSF1 was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do, PDB-1x4aA). The two RRMs have 51% identity over 80 amino acids. The Z-score for the model quality calculated using ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) is 5.5.
Figure 2.
PfSR1 is a functional SR protein that can mediate pre-mRNA splicing. Recombinant PfSR1 can complement S100 extracts and rescue the splicing activity similar to SRSF1. (A) Schematic of the expression vector used to express the recombinant PfSR1 and SRSF1 in 293 T cells. cDNA of both genes was cloned into the expression vector fused to a His-tag. (B) SDS–PAGE presenting the purified SRSF1 and PfSR1 loaded on gel in increasing volumes (1, 2 and 4 µl, respectively). (C) Complementation of splicing activity of PfSR1 and SRSF1 in S100 extracts (in vitro splicing assay). Increasing amounts of purified SRSF1 or PfSR1 were added to S100 extracts in the presence of radiolabeled Ftz reporter pre-mRNA. Lane 1: labeled Ftz pre-mRNA only; Lane 2: nuclear extract (NE) + Ftz; Lane 3: S100 extract + Ftz; Lane 4: NE + S100 + Ftz; Lanes 5–6: S100 + Ftz with increasing amounts of SRSF1; Lanes 7 and 8: S100 + Ftz with increasing amounts of PfSR1. Schematics of pre-mRNA, spliced mRNA and the spliced intron are shown in the left.
PfSR1 is an AS factor with similar activity to SRSF1
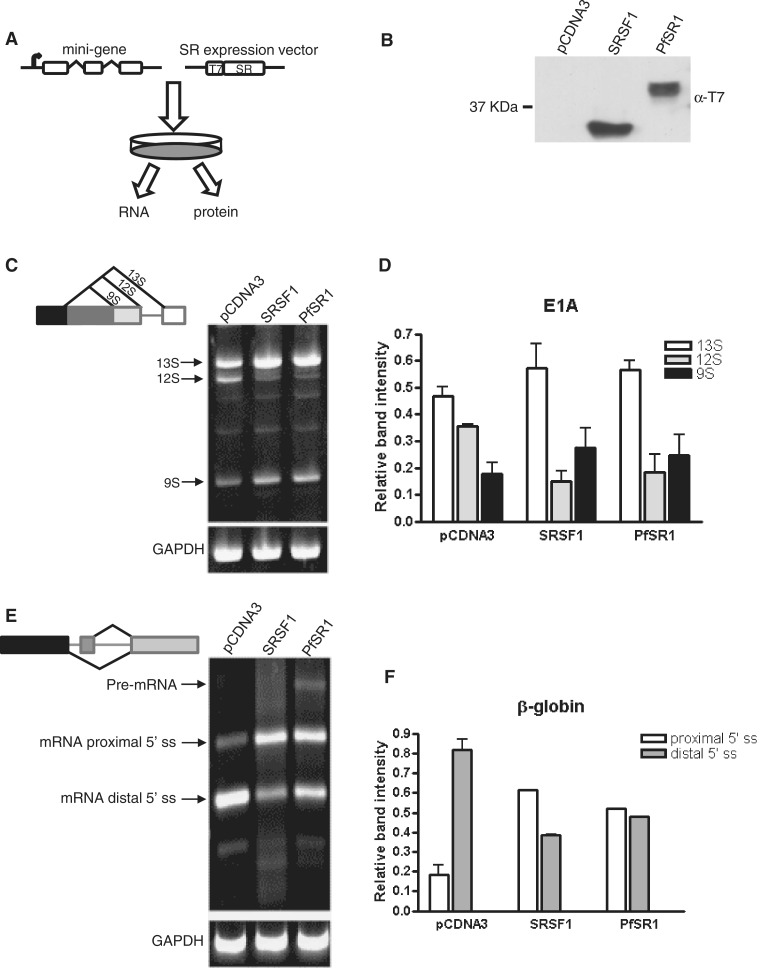
The fact that PfSR1 by itself could complement splicing of pre-mRNA in an in vitro S100 complementation assay encouraged us to further investigate whether PfSR1 can also function as AS factor in living cells. Toward this aim we applied two mini-gene constructs (E1A and β-globin duplicated 5′-splice sites [D5′-ss]) successfully used to demonstrate the AS activity of the mammalian SRSF1 (19,27,36). In these systems, HEK 293T cells are cotransfected with an expression vector that over-expresses a candidate AS factor together with another vector that carries an artificial mini-gene. The E1A mini-gene contains several competing 5′-ss and could potentially give rise to a few different alternative spliced mRNA products (19). Following transfection, the cotransfected cells are harvested for further analyses of protein expression and detection of differential AS isoforms of pre-mRNA of the mini-gene (Figure 3A). Our initial attempts to express PfSR1 in the HEK 293 T cells had very poor protein yield (data not shown). We reasoned that since the P. falciparum genome is AT rich, its codon usage might not be optimal for protein translation in mammalian cells. Therefore, we synthesized PfSR1 with a codon bias that was predicted to be optimal for expression in these mammalian cells (Supplementary Figure S3). The synthetic pfsr1 gene was cloned into the pcDNA3 expression vector fused to a T7 epitope tag (Figure 3A) and was found be well expressed in the HEK 293 T cells (Figure 3B). To determine whether PfSR1 can influence the choice of different 5′-ss and thus give rise to alternatively spliced isoforms we expressed PfSR1 in the presence of either the E1A or the β-globin mini-genes and found that for both mini-gene constructs PfSR1 acts as AS factor. In cells that were cotransfected with the adenovirus E1A mini-gene and an empty pcDNA3 vector, the putative AS machinery gave rise to three differentially spliced isoforms of E1A that are detected by RT–PCR. Remarkably, in cells in which either PfSR1 or SRSF1 are coexpressed with the mini-gene, there is a clear change in the ratio of the different mRNA isoforms compared with the cells that were transfected with the empty pcDNA3 vector. In both PfSR1 and SRSF1 over-expressing cells there is an increase in the amount of the distal and proximal isoforms S13 and S9 (600 and 150 bp, respectively) while the amount of the intermediate isoform S12 (450 bp) decreases (Figure 3C and D). These results show that ectopic expression PfSR1 promotes the proximal 5′-ss selection and modulate the AS regulation of the E1A mini-gene similar to SRSF1. Similarly, when PfSR1 is expressed in the presence of the β-globin D5′-ss mini-gene (27,36) the balance between the distal and proximal 5′-ss selection is affected, indicating that PfSR1 strongly promotes the proximal 5′-ss. In cells that were transfected with the empty pcDNA3 vector the dominant spliced mRNA product is the 586-bp isoform that corresponds with splicing at the distal ss. However, in cells in which PfSR1 is expressed the 810-bp isoform, which corresponds with the proximal ss, becomes significantly more abundant than in the cells containing the empty pcDNA3 vector (Figure 3E and F). This splicing pattern is similar to the results obtained in the SRSF1 expressing cells. Altogether, these results demonstrate that PfSR1 regulate ss selection resulting in AS patterns which are similar to those that are obtained with SRSF1 and thus indicate that PfSR1 functions as a bona fide SR protein.
Figure 3.
PfSR1 is an AS factor with similar activity to SRSF1. (A) Schematic illustration of the experimental procedure designed to demonstrate AS activity of PfSR1. HEK 293 T cells were cotransfected with plasmids that carried a mini-gene as well as an expression vector (pcDNA3−). Forty-eight hours post-transfection the cells were harvested and their RNA and protein content were analyzed. (B) Western blot analysis using anti-T7 antibody showing the expression of the tagged SRSF1 (Lane 2) and PfSR1 (Lane 3). An empty pcDNA3− vector was used as a negative control (Lane 1). (C) RT–PCR amplification of total RNA extracted from HEK 293 T cells that were cotransfected with the E1A mini-gene obtained differential splicing patterns mediated by PfSR1 and SRSF1 compared with the control. Schematic of the mini-gene and the differentially spliced isoforms is shown on the left. GAPDH was used as loading control and is presented in the lower panel. (D) Quantification of the changes in the proportion of each spliced isoform of E1A obtained after transfection with PfSR1 and SRSF1 compared with the control. The density of each band is presented as a proportion of the total signal obtained. The averages and standard errors of three different experiments are presented. (E) RT–PCR amplification of total RNA extracted from 293 T cells that were cotransfected with the β-globin mini-gene obtained differential splicing patterns mediated by PfSR1 and SRSF1 compared with the control. Schematic of the mini-gene and the differentially spliced isoforms is shown on the left. GAPDH was used as loading control and is presented in the lower panel. (F) Quantification of the changes in the proportion of each spliced isoform of β-globin obtained after transfection with PfSR1 and SRSF1 compared with the control. The density of each band is presented as a proportion of the total signal obtained. The averages and standard errors of two different experiments are presented.
Analyses of the dynamic cellular localization of PfSR1 during IDC indicates that its RS domain function as NLS
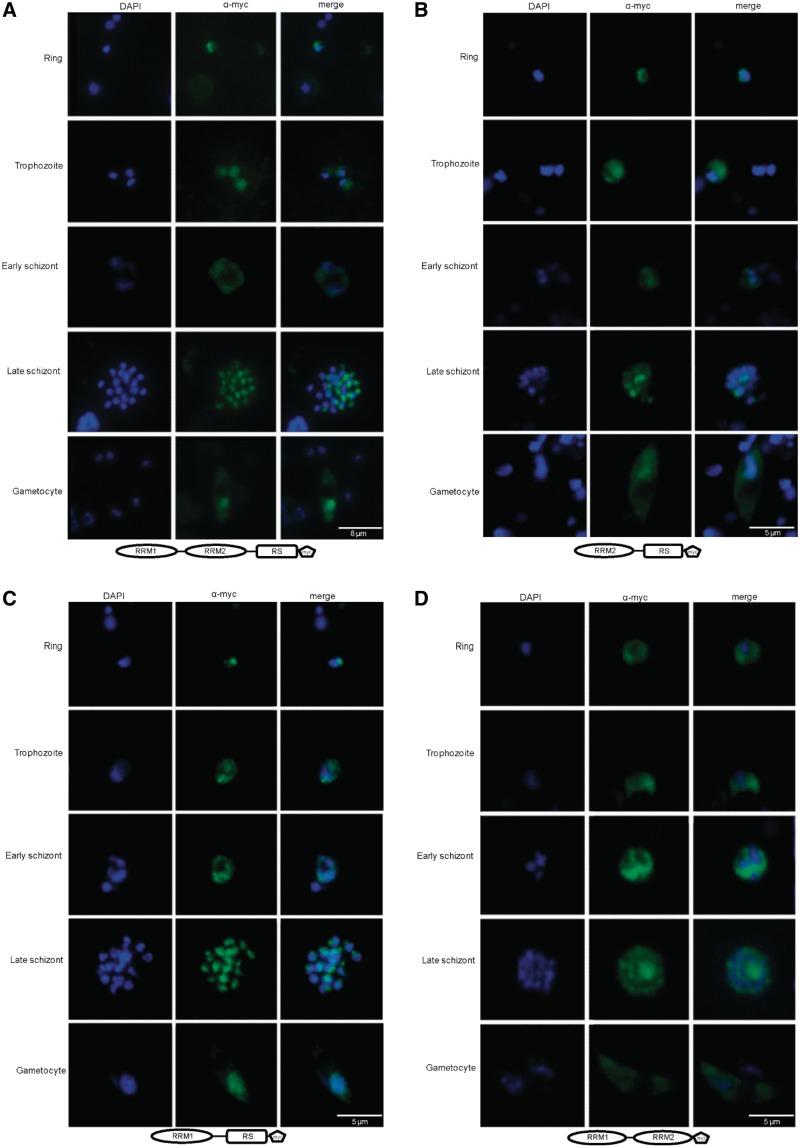
In higher eukaryotes it was shown that several SR protein shuttle between the cytoplasm and the nucleus and that specific protein domains are important for proper cellular localization (37,38). It was recently shown that the dynamic cellular localization of PfSR1 during the IDC correlates with the localization of PfSRPK1 (18). Therefore, we were interested to investigate whether the RRM domains and/or the RS domain have a role in determining the cellular localization of PfSR1. We ectopically expressed the entire PfSR1 protein as well as three mutants in which either RRM1, RRM2 or the RS domains had been deleted. These mutants were named ΔRRM1, ΔRRM2 and ΔRS, respectively. The entire pfsr1 as well as the mutated genes were cloned into the expression vector pHBISR1myc (see plasmid map in Figure 5A) fused to a myc epitope tag that enables specific immuno-fluorescence visualization. These constructs also express the bsd resistance gene that allows us to select for parasite populations that stably carry the episomes using drug selection with blasticidin S. Interestingly, we found that the ΔRS mutants (Figure 4D) had a different cellular localization than that of the entire PfSR1 protein or the ΔRRM mutants (Figure 4A–C). In early ring stages the complete PfSR1-myc as well as ΔRRM mutants localizes to the nucleus in foci that are mainly located at the nuclear periphery. This pattern changes later during the IDC in trophozoites, schizonts and in gametocytes, in which PfSR1-myc and all the mutated forms of PfSR1 are detected both in the nucleus and in the cytoplasm. It is also apparent that the cytoplasmic location of these different isoforms is restricted to regions that are outside the parasite’s food vacuole. Remarkably though, the ΔRS mutant proteins are located in the cytoplasm and not in the nucleus indicating that the RS domain is essential for nuclear targeting of PfSR1 (Figure 4D). Altogether these results indicate that the cellular location of PfSR1 in the parasite is dynamic during IDC and that nuclear localization of PfSR1 is determined by the RS domain.
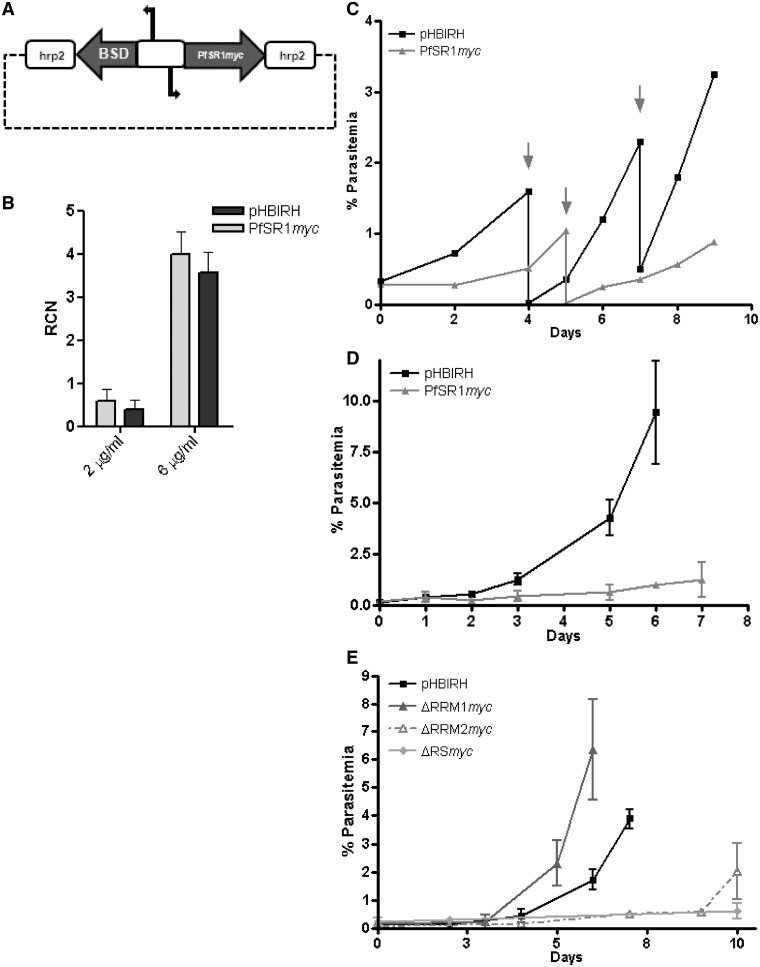
Figure 5.
Proper regulation of the pfsr1 gene is essential for parasites’ proliferation in human RBCs. Over-expression of pfsr1 gene inhibits parasite proliferation. (A) Schematic map of the plasmid (pHBIPfSR1myc) used for ectopically over-expression using increasing concentrations of blasticidin as described (28). Steady-state mRNA levels of pfsr1 and renilla luciferase genes were measured using qRT–PCR from tightly synchronized parasite cultures 36-h post-invasion (hpi). Values are presented as relative copy number to the housekeeping genes arginyl-tRNA synthetase (PFL0900c). (B) Transcription levels of pfsr1 and renilla luciferase expressed from the transfected episomes by parasites growing under 2 and 6 µg/ml blasticidin selection. (C) Growth curves of the parasite populations over-expressing pfsr1 (light gray) compared with those that similarly express renilla luciferase (black) from the pHBIPfSR1myc and the pHBIRH plasmids, respectively, under 2 μg/ml blasticidin. Arrows indicate days in which the culture was cut down to avoid over-parasitemia. Each curve represents the average of three different cultures grown in parallel. (D) Growth curves of the parasite populations over-expressing pfsr1 (light gray) compared with those that similarly express renilla luciferase (black) from the pHBIPfSR1myc and the pHBIRH plasmids, respectively, under 6 μg/ml blasticidin. Day 0 represents the day in which blasticidin concentration was increased. Each curve represents the average of three different parasitemia measurements. (E) Growth curves of the parasite populations over-expressing the different PfSR1 mutants (gray) compared with those that similarly express renilla luciferase (black) under 2 μg/ml blasticidin.
Figure 4.
The RS domain localizes PfSR1 to the nucleus. IFAs showing the localization of the entire PfSR1 as well as different PfSR1 mutants lacking either the RRM or the RS domains (ΔRRM1, ΔRRM2 and ΔRS, respectively) during IDC. The different forms of PfSR1 were ectopically express fused to a myc epitope tag and detected using anti-myc antibodies. PfSR1 (green); DAPI staining (blue); developmental stages are indicated on the left. (A) Cellular localization of PfSR1-myc. (B) Cellular localization of the PfSR1ΔRRM1-myc. (C) Cellular localization of the PfSR1ΔRRM2-myc. (D) Cellular localization of the PfSR1ΔRS-myc. The entire PfSR1 and two ΔRRM mutants shuttle between the nucleus and cytoplasm, whereas the ΔRS mutant localizes to the cytoplasm.
Proper expression of the pfsr1 gene is essential for proliferation of blood stages parasites
In mammals, it was possible to over-express SRSF1 only by ∼2-fold before the over-expressed cells underwent apoptosis (39). Interestingly, in our IF assays on parasites that were transfected with the pHBISR1myc constructs, we were able to detect fluorescent signal only from a portion of the parasite population while many parasites were not stained. This suggests that episomal expression of PfSR1 is not obtained from the entire parasite population. This assumption was also supported by the fact that the detection of PfSR1-myc by western blot analysis was very inefficient and required a large amount of parasites (Supplementary Figure S4). These results suggest that ectopic over-expression of the pfsr1 gene could interfere with the proper proliferation of P. falciparum in RBCs. The plasmid pHBISR1myc is a great transgenic tool that enables both ectopic protein expression as well as fine tuned and a precise over-expression in P. falciparum by forcing the parasite to carry increasing copy numbers of the over-expression episomes (28). Using increasing concentration of blasticidin S, we attempted to achieve fine tuned over-expression of the pfsr1 gene. In order to eliminate the possible effect of the drug itself, we transfected parasites with pHBIRH plasmid as a control. pHBIRH is an identical plasmid in which renilla luciferase was cloned into the same expression vector instead of pfsr1. Following transfections, these parasites were initially selected on a low dose of 2 µg/ml blasticidin until populations that stably carried these expression vectors were obtained. Using qPCR of gDNA with PfSR1-myc specific primers we determined that <2 µg/ml blasticidin both parasite populations carried a single copy of the expression vectors and transcribed endogenous pfsr1 at similar levels (data not shown). Interestingly, although under this dose of drug pressure pfsr1 and renilla luciferase are transcribed at similar levels we observed significant growth differences between the parasite populations (Figure 5B). Proliferation of the parasites cultures that over-expressed pfsr1 was slower than the population expressing renilla luciferase (Figure 5C). We then increased blasticidin concentrations to 6 µg/ml in attempt to increase the levels of ectopic over-expression of the pfsr1gene. By this increase in drug pressure we were able to achieve ∼8-fold increase in both pfsr1and renilla luciferase transcription levels (Figure 5B). However, while the renilla luciferase expressing parasites grew rather well under this drug pressure the parasite population over-expressing pfsr1 proliferated grew much slower (Figure 5D). Mutation analyses of SRSF1 pointed toward functional differences of the RRM and RS domains in splicing (40), translation (41) and oncogenic activity (42). We were therefore interested to test the influence of over-expression of the different PfSR1 mutants on parasites’ proliferation. We found that parasite over-expressing the ΔRRM1 mutant had normal growth rate while the proliferation of parasites over-expressing the ΔRRM2 and the ΔRS mutants was significantly slower (Figure 5E). Altogether, these results imply that ectopic over-expression of the pfsr1 gene had caused a distinct growth inhibition phenotype indicating that proper regulation of pfsr1 is essential for parasite proliferation in human RBCs.
Over-expression of pfsr1 influence AS activity in P. falciparum
In order to demonstrate that PfSR1 influences AS activity in P. falciparum, we performed large-scale screening for different splicing isoforms in genes in which exon skipping event has been previously reported in blood stages parasites (10,11,43). We used numerous primer sets designated to identify such events by RT–PCR and compared the different cDNA isoforms that were amplified from parasites over-expressing pfsr1 (pHBISR1myc) with those detected in wild-type NF54 parasites as well as the parasites transfected with the pHBIRH plasmid that express renilla luciferase. We have identified AS events in three different P. falciparum genes that are specifically associated with over-expression of pfsr1 (Figure 6). The first example is the influence of PfSR1 on the blood stage antigen 41–3 precursor (PFL0385c). For this gene, two mRNA isoforms are detected in NF54 parasites using primers designed to anneal to the third and the last exons (Figure 6A). Both mRNA isoforms are also detected in parasites that were transfected with pHBIRH and pHBISR1myc growing on low dose of 2 µg/ml blasticidin. However, when drug pressure was increased to 6 µg/ml blasticidin, the splicing pattern shifted only in parasite over-expressing pfsr1 that under these conditions favored the large (726 bp) isoform over the short (474 bp) isoform (Figure 6A). This change is specific to the parasite line that over-expressed pfsr1 on 6 µg/ml blasticidin while the control parasite populations showed no change in mRNA splicing. Similarly, over-expression of pfsr1 was associated with specific changes in mRNA splicing of putative kinesin (PFA0535c) and PFE0480c (conserved plasmodium protein with unknown function) in parasites carrying the pHBIRS1myc episome and selected on 6 µg/ml blasticidin (Figure 6B and C, respectively). Over-expression of PfSR1 caused a clear preference for the 394-bp mRNA isoform of the putative kinesin (Figure 6B) and for the 857 - and 322-bp mRNA isoforms in PFE0480c (Figure 6C). Here as well the observed effect on AS was specific to the high level of pfsr1 over-expression. Altogether, these data indicate that PfSR1 influences AS activity of endogenous P. falciparum genes in vivo.
Figure 6.
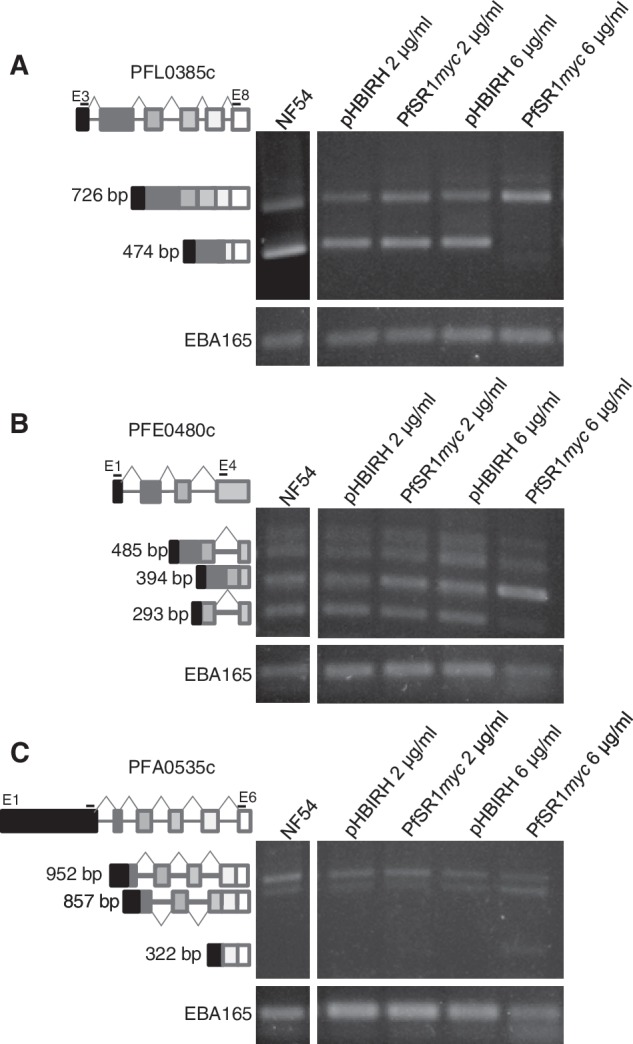
Over-expression of the pfsr1 influences AS activity in P. falciparum. RT–PCR amplifications on cDNA of three P. falciparum endogenous genes reveals specific changes in their splicing patterns mediated by PfSR1. cDNA from NF54, wile-type untrasfected parasites was used as control. For each gene RT–PCR amplifications of mRNA extracted from parasites transfected with either pHBIRH or pHBISR1myc and selected on 2 and 6 μg/ml blasticidin are presented. Primers used for these amplifications are listed in Supplementary Table S1. (A) The spliced isoforms obtained for PFL0385c. (B) The spliced isoforms obtained for PFE0480c and (C) The splicing isoforms obtained PFA0535c. Schematic of the genes and the differentially spliced isoforms is shown on the left. Exons are illustrated in boxes and introns in lines. Positions of the PCR primers that were used to amplify each gene are indicated. EBA165 (PFD1155w) was used as loading control and is presented in the lower panels. All of the detected isoforms were sub-cloned and sequenced.
DISCUSSION
Alternative pre-mRNA splicing is an important mechanism of post-transcriptional regulation of gene expression in eukaryotes. AS events were reported in Plasmodium many years ago; however, only recently with the establishment of whole genome approaches that allow genome-wide transcriptome analyses has it become clear that AS events are much more abundant in P. falciparum during the IDC than previously considered (10,11). The genome of P. falciparum seems to have a relatively low number of genes. The recent finding of genome-wide abundance of different mRNA isoforms that are encoded by the same genes (10,11) might indicate that AS in P. falciparum is an important mechanism by which the parasite can expand its proteome out of a limited number of genes to be able to adapt such a complex life cycle. To our knowledge, our findings demonstrate PfSR1 (PFE0865c) as the first functional AS factor found in the genome of Plasmodium or any other apicomplexan parasite. We show that purified recombinant PfSR1 added in vitro to splicing deficient HeLA S100 extracts is able to complement splicing activity similar to the mammalian SR protein SRSF1 indicating that it is a bona fide SR protein. Moreover, similar to SRSF1 we demonstrated that PfSR1 can affect the ss selection of two different mini-genes in mammalian cells. These results indicate that PfSR1 is able to cooperatively interact with the mammalian splicing machinery to function as an AS factor resulting in a preference for specific splicing isoforms. This might imply that the basic components that are required for alternative pre-mRNA splicing are conserved even in these evolutionarily distinct eukaryotes. However, it is clear that PfSR1 confers unique features that are distinct from other eukaryote SR proteins. A recent study that identified PfSRPK1 as the kinase that interacts with PfSR1 and phosphorylates its RS domain also showed that the phospho-epitopes generated by PfSRPK1 on the RS domain are not recognized by mAb104. This may imply that these phospho-epitopes are different than those of SRSF1, most likely due to variation in the RS domains of SRSF1 and PfSR1 (18). Our in silico search for putative SR proteins in P. falciparum indicated that some of the predicted proteins possess an unusual RS domain which is different than other eukaryotes and contain less Ser/Arg repeats. Whether these are indeed RS domains will need further functional validation.
The elegant study of Dixit et al. (18) demonstrated that PfSRPK1’s interaction with PfSR1 influences the ability of PfSR1 to bind RNA in vitro and showed that these two proteins colocalize during the IDC, thus supporting the hypothesis that PfSR1 is indeed an SR protein. However, while our results support the previous observation that PfSR1 shuttles between the nucleus and the cytoplasm during the IDC, we found some distinct differences in the patterns of cellular localization of PfSR1 compared with what was previously reported. First, we found that the nuclear location of PfSR1 at early ring stages is in a distinct punctuated pattern mainly at the nuclear periphery and is not equally spread all over the nucleus as previously shown. The pattern of sub-nuclear localization of PfSR1 in ring stages shown here is more similar to the sub-nuclear localization of SRSF1 which is found at distinct nuclear speckles (37). These speckles are believed to be sub-nuclear storage compartments for SR proteins and other splicing factors (12). In addition, while PfSR1 in the cytoplasm seems to be more evenly spread than in the nucleus, it is clear that its cytoplasmic location is restricted to region that are outside the food vacuole, while previously it was shown throughout the entire cytoplasm (18). These previous data were obtained using polyclonal antibodies raised against recombinant PfSR1. It is possible that IFAs using an epitope tagged PfSR1 have an advantage in getting better resolution for cellular localization as detected here.
Our deletion analyses of the PfSR1 mutants lacking different domains have indicated that the RS domain of PfSR1 acts as NLS similar to the mammalian SRSF1 (37). Interestingly, deletion of either of the RRM domains seems to have no effect on the cellular location of PfSR1. Some of the mammalian SR proteins, including SRSF1, shuttle continuously between the nucleus and the cytoplasm (44). This movement was found to be associated with phosphorylation of specific residues at the RS or RRM domains. One can hypothesize that phosphorylation of the RS domain of PfSR1 by PfSRPK1 influences it’s binding affinity for mRNA, and that this modification could also regulate its cellular localization and function in P. falciparum. The dynamic cellular localization of PfSR1 may reflect a functional transition of PfSR1 during the IDC, from mainly nuclear functions such as splicing and AS at early ring stages to additional roles that take place in cytoplasm in late stages and in gametocytes. In recent years there is mounting evidence that SR proteins in metazoans are involved in many other cellular processes. These include chromatin re-modeling, transcription, cell cycle progression, mRNA export, nonsense-mediated mRNA decay (NMD) and translation (12,13,36,45). One cannot exclude that in addition to its role in splicing and AS shown here, PfSR1 may have other functions as indicated by its localization both to the nucleus and cytoplasm. Ectopic over-expression of pfsr1 caused strong inhibition of culture growth even at relatively low levels of gene over-expression, indicating that tight regulation of pfsr1 is essential for parasite proliferation in human RBCs. These results are supported by experiments showing that only low levels (∼2-fold) of over-expression of several SR proteins could be achieved in mammalian cells before they underwent apoptosis (39). In addition, reduced expression of SRSF1 was found to elicit genomic instability and apoptosis (46,47). The involvement of SR proteins in several of the basic cellular processes in living cells can explain why P. falciparum parasites tightly regulate pfsr1 so that even the low levels of over-expression obtained here caused significant growth inhibition. Interestingly, over-expression of the ΔRRM2 and the ΔRS mutants had growth inhibition phenotypes while the parasites over-expressing ΔRRM1 had normal growth rate. These results might indicate that the role of PfSR1 in regulating parasite proliferation in human RBCs involves primarily RNA targets recognized by RRM1. Similarly, it has been recently shown that RRM1 of SRSF1 has an essential role in proliferation of mammary epithelial cells (42).
It is likely that P. falciparum harbors additional SR proteins that might be regulated by kinases other than PfSRPK1. It was recently shown that PfCLKs can phosphorylate recombinant PfASF-1 (PF11_0205) in vitro (35). However, even though this protein has an overall 42% amino acid identity with SRSF1 it lacks an RS domain. In addition, other putative SR proteins such as PfSR1 and PF10_0217 are closer orthologs of SRSF1. What role PfASF-1 or other putative P. falciparum SR proteins play in RNA metabolisms in Plasmodium and how they are regulated by PfCLKs is still an open question.
The identification of PfSR1 as the first functional AS factor in P. falciparum will hopefully set the platform toward investigating in depth the role of AS in the biology of this pathogen. We were able to show that PfSR1 influences AS activity of three endogenous genes in vivo including a gene that encodes for an antigen expressed on the surface of RBCs. Whole genome approaches are now available that will hopefully soon enable the identification of additional gene targets that are alternatively spliced by PfSR1 and thus point toward potential biological processes that are regulated by PfSR1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Israeli Academy for Science [660/09]; German Israeli Foundation [997/2008]; United States-Israel Binational Science Foundation [2007350]; Marie Curie International Reintegration Grant (IRG) [203675 to R.D.]; Jacob and Lena Joels Memorial Foundation Senior Lectureship for Excellence in the Life and Medical Sciences (to R.D.). Funding for open access charge: Jacobs and Lena Joels Memorial Foundation Senior Lectureship for Excellence in the Life and Medical Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.WHO. 2010 World Malaria Report (2010) [Google Scholar]
- 2.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu JC, Derisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PloS Biol. 2003;1:85–100. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 5.Knapp B, Nau U, Hundt E, Kupper HA. Demonstration of alternative splicing of a pre-mRNA expressed in the blood stage form of Plasmodium falciparum. J. Biol. Chem. 1991;266:7148–7154. [PubMed] [Google Scholar]
- 6.Muhia DK, Swales CA, Eckstein-Ludwig U, Saran S, Polley SD, Kelly JM, Schaap P, Krishna S, Baker DA. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 2003;278:22014–22022. doi: 10.1074/jbc.M301639200. [DOI] [PubMed] [Google Scholar]
- 7.Singh N, Preiser P, Renia L, Balu B, Barnwell J, Blair P, Jarra W, Voza T, Landau I, Adams JH. Conservation and developmental control of alternative splicing in maebl among malaria parasites. J. Mol. Biol. 2004;343:589–599. doi: 10.1016/j.jmb.2004.08.047. [DOI] [PubMed] [Google Scholar]
- 8.Fonager J, Cunningham D, Jarra W, Koernig S, Henneman AA, Langhorne J, Preiser P. Transcription and alternative splicing in the yir multigene family of the malaria parasite Plasmodium y. yoelii: identification of motifs suggesting epigenetic and post-transcriptional control of RNA expression. Mol. Biochem. Parasitol. 2007;156:1–11. doi: 10.1016/j.molbiopara.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Iriko H, Jin L, Kaneko O, Takeo S, Han ET, Tachibana M, Otsuki H, Torii M, Tsuboi T. A small-scale systematic analysis of alternative splicing in Plasmodium falciparum. Parasitol. Int. 2009;58:196–199. doi: 10.1016/j.parint.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, Barrell B, Pain A, Berriman M, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorber K, Dimon MT, DeRisi JL. RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res. 2011;39:3820–3835. doi: 10.1093/nar/gkq1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 16.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 17.Portal D, Espinosa JM, Lobo GS, Kadener S, Pereira CA, De La Mata M, Tang Z, Lin RJ, Kornblihtt AR, Baralle FE, et al. An early ancestor in the evolution of splicing: a Trypanosoma cruzi serine-arginine-rich protein (TcSR) is functional in cis-splicing. Mol. Biochem. Parasitol. 2003;127:37–46. doi: 10.1016/s0166-6851(02)00301-8. [DOI] [PubMed] [Google Scholar]
- 18.Dixit A, Singh PK, Sharma GP, Malhotra P, Sharma P. PfSRPK1, a novel splicing-related kinase from Plasmodium falciparum. J. Biol. Chem. 2010;285:38315–38323. doi: 10.1074/jbc.M110.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 20.Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. A genome-wide survey of RS domain proteins. RNA. 2001;7:1693–1701. [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aley SB, Sherwood JA, Howard RJ. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J. Exp. Med. 1984;160:1585–1590. doi: 10.1084/jem.160.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 26.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF. Mol. Cell Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epp C, Raskolnikov D, Deitsch KW. A regulatable transgene expression system for cultured Plasmodium falciparum parasites. Malar. J. 2008;7:86. doi: 10.1186/1475-2875-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deitsch KW, Driskill CL, Wellems TE. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzikowski R, Deitsch KW. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 2008;382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 33.Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 34.Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal S, Kern S, Halbert J, Przyborski JM, Baumeister S, Dandekar T, Doerig C, Pradel G. Two nucleus-localized CDK-like kinases with crucial roles for malaria parasite erythrocytic replication are involved in phosphorylation of splicing factor. J. Cell. Biochem. 2011;112:1295–1310. doi: 10.1002/jcb.23034. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 37.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell. Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allemand E, Gattoni R, Bourbon HM, Stevenin J, Caceres JF, Soret J, Tazi J. Distinctive features of Drosophila alternative splicing factor RS domain: implication for specific phosphorylation, shuttling, and splicing activation. Mol. Cell Biol. 2001;21:1345–1359. doi: 10.1128/MCB.21.4.1345-1359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl Acad. Sci. USA. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Manley JL. New talents for an old acquaintance: the SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle. 2005;4:1706–1708. doi: 10.4161/cc.4.12.2210. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.