Abstract
The CRISPR arrays found in many bacteria and most archaea are transcribed into a long precursor RNA that is processed into small clustered regularly interspaced short palindromic repeats (CRISPR) RNAs (crRNAs). These RNA molecules can contain fragments of viral genomes and mediate, together with a set of CRISPR-associated (Cas) proteins, the prokaryotic immunity against viral attacks. CRISPR/Cas systems are diverse and the Cas6 enzymes that process crRNAs vary between different subtypes. We analysed CRISPR/Cas subtype I-B and present the identification of novel Cas6 enzymes from the bacterial and archaeal model organisms Clostridium thermocellum and Methanococcus maripaludis C5. Methanococcus maripaludis Cas6b in vitro activity and specificity was determined. Two complementary catalytic histidine residues were identified. RNA-Seq analyses revealed in vivo crRNA processing sites, crRNA abundance and orientation of CRISPR transcription within these two organisms. Individual spacer sequences were identified with strong effects on transcription and processing patterns of a CRISPR cluster. These effects will need to be considered for the application of CRISPR clusters that are designed to produce synthetic crRNAs.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (cas) genes define an anti-viral defence system in Archaea and Bacteria. CRISPR loci are composed of repeat sequences with an average length of 24–47 nt, which alternate with unique spacer sequences derived from previous encounters with foreign nucleic acids (i.e. viruses, plasmids) (1–4). CRISPR loci are transcribed and processed to generate the small interfering crRNAs. Diverse sets of cas genes are often found adjacent to a CRISPR locus and encode proteins that are involved in the three phases of CRISPR/Cas activity: acquisition of new spacers, processing of crRNAs and interference with foreign nucleic acid (5–9). Although there is little information available for the process of new spacer acquisition, recent progress has led to a better understanding of the other two phases. The maturation of precursor crRNA into small crRNAs is performed by diverse Cas endonucleases that belong to a protein family termed Cas6 (10–16). In CRISPR/Cas Type-I the interference step is mediated by a complex of different Cas proteins (Cas complex for antiviral defence: Cascade) bound to crRNAs that target the invading nucleic acid through base complementarity which ultimately results in the inactivation or degradation of foreign DNA by Cas3 (17–24). Type-II CRISPR/Cas systems use the single Cas9 protein for interference (25) and Type-III systems use a multi Cas protein complex that is distinct from Cascade (26,27).
Computational analyses of these defence systems identified a surprising diversity of different CRISPR/Cas types and subtypes, which are spread throughout archaeal and bacterial kingdoms.
This classification has defined three major types which can be further divided into at least 10 CRISPR/Cas subtypes (28). The subtype I-B, found, e.g. in Clostridia, methanogens and halophiles, is defined by the subtype-specific protein Cas8b. In Clostridium thermocellum and Methanococcus maripaludis the minimal subtype I-B Cas protein organization consists of the universal Cas1, Cas2 and Cas4 proteins that are proposed to mediate the integration of spacers as well as Cas3, Cas5, Cas7 and Cas8b which are proposed to form the Cascade complex of this subtype. Finally, a Cas6 protein is required for the processing of crRNA (10–16).
A Cas6 protein was first described for CRISPR/Cas subtype III-B in Pyrococcus furiosus as a metal-independent endonuclease involved in the processing of precursor crRNA into mature crRNA (10,11,14,15). Cas6 enzymes were also characterized for CRISPR/Cas subtype I-F in Pseudomonas aeruginosa (Cas6f, also termed Csy4) (13) and CRISPR/Cas subtype I-E in Thermus thermophilus and Escherichia coli (Cas6e, also termed Cse3) (12,16). The amino acid sequence similarity of these Cas6 proteins is limited, yet they share ferredoxin-like folds and perform analogous reactions in the different CRISPR/Cas systems. These Cas6 proteins do not only differ in substrate specificity, but also in the composition of their active sites. For example P. furiosus Cas6 (Pf Cas6) interacts with single-stranded RNA while Cas6e and Cas6f seem to specifically bind to hairpin structures formed by the repeats (10–16). Further differences can be found in the catalytic site of the Cas6 proteins. Pf Cas6 uses a catalytic triad composed of tyrosine, histidine and lysine residues (10,14), while in Cas6f a catalytic dyad of a histidine and a serine residue proved to be important for protein activity (13,29). Activity of Cas6e relies on a tyrosine and a histidine residue (12,16). Although there are variations in their active site composition and the recognition of RNA substrates, the different Cas6 cleavage reactions always generate crRNAs that consist of a spacer unit that is flanked by 8 nt of the repeat sequence as a 5′-terminal tag and a 3′-terminal repeat tag (11–13). Finally, Cas6 was shown to deliver the mature crRNA to the Cascade complex (18,30).
In this study, we provide the first analysis of crRNA processing for CRISPR/Cas subtype I-B for one bacterial model organism, C. thermocellum and one archaeal model organism, M. maripaludis (detailed information of CRISPR loci and gene organization can be found in Supplementary Figure S1). The abundance and processing of crRNAs were analysed in vivo by RNA-Seq methodology. In addition, the Cas6 enzymes of this CRISPR/Cas subtype (termed Cas6b) were identified and M. maripaludis Cas6b (Mm Cas6b) was analysed for crRNA processing in vitro.
MATERIALS AND METHODS
Growth of E. coli, M. maripaludis C5 and C. thermocellum cells
Methanococcus maripaludis C5 cells were a kind gift of W.B. Whitman (Georgia). Clostridium thermocellum (DSM1237) cells were obtained from DSMZ (German collection of micro-organisms and cell cultures). All E. coli cells were grown in LB-media with appropriate antibiotics at 37°C and shaking at 200 rpm.
Methanococcus maripaludis C5 was grown at 37°C in complex medium for methanococci (McC) (31) with H2/CO2 atmosphere (80%/20%) and one bar (15 psi) overpressure. Clostridium thermocellum cells were incubated in complex medium (32) at 60°C with an anaerobic atmosphere (N2).
Production of Cas6 and mutants
The cas6 genes MmarC5_0767, Cthe_3205 and Cthe_2303 were amplified from genomic DNA of M. maripaludis C5 or C. thermocellum ATCC 27 405 and cloned into the vector pET-20b to facilitate protein expression with a C-terminal His-tag. Oligonucleotides for site-directed mutagenesis were designed using Agilents QuickChange Primer Design tool and cas6 mutants were created using the QuickChange site-directed mutagenesis (Stratagene) according to the manufacturer’s instructions. Mutations were confirmed by sequencing (MWG Eurofins).
All Cas6 variants were produced in E. coli (Rosetta2 DE3) cells. Induction of protein expression was performed by addition of isopropylthio-β-d-galactoside (IPTG) to a final concentration of 0.5 mM after growing the cells to an OD578 of 0.6. Four hours after induction the cells were harvested, the pelleted cells re-suspended in lysis buffer (10 mM Tris–HCl [pH8.0], 300 mM NaCl, 10% glycerol and 0.5 mM DTT) and incubated on ice with lysozyme (1 mg/g cell pellet) for 30 min. Cell disruption was performed using sonication (8 × 30 s; Branson Sonifier 250). Clearing of the lysate was achieved by centrifugation (20 000 rpm, 30 min, 4°C) and the supernatant was applied to a Ni–NTA–Sepharose Column (GE-Healthcare) and purified using a FPLC Äkta-Purification system (GE-Healthcare). Elution of the proteins was performed by a linear imidazole gradient (0–500 mM). Purity of the proteins was determined by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie Blue staining. The protein was dialysed into lysis buffer and the protein concentration was determined by Bradford Assay (BioRad).
Generation of RNA substrates
The spacer2–repeat–spacer3 and repeat–spacer27–repeat RNA substrates were generated by in vitro run-off transcription using T7 RNA polymerase and internally labelled using [α-32P] adenosine triphosphate (ATP) (5000 ci/mmol, Hartman Analytic) (33). The repeat RNAs and repeat RNAs with a substitution of the first unprocessed nucleotide against a dexoy nucleotide were synthesized by Eurofins MWG Operon. End labelling of these substrates was performed using T4 polynucleotide kinase (Ambion) and [γ-32P] ATP (5000 ci/mmol) according to the manufacturer’s instructions.
Templates for in vitro transcription were obtained by cloning of the pre-crRNA sequences with an upstream T7 RNA polymerase promotor sequence into pUC19 vector. After linearization of the plasmid with HindIII, in vitro transcription was performed in a final volume of 20 μl [40 mM HEPES–KOH (pH8.0); 22 mM MgCl2; 5 mM DTT; 1 mM spermidine; 4 mM UTP, CTP, GTP and 2 mM ATP; 20 U RNase Inhibitor; 1 µg T7 RNA polymerase; 1 μg linearized plasmid] at 37°C for 1 h. End labelling of synthesized RNA was done in a 20 µl reaction volume: 10 µl of the RNA was labelled using 2 µl T4 Polynucleotide Kinase (PNK) buffer (New England Biolabs (NEB)) and 25 U T4 PNK (Ambion) at 37°C for 30 min.
The RNAs were separated by denaturing PAGE (8 M urea; 1× TBE; 10% polyacrylamide), and afterwards respective bands were cut out using sterile scalpels in reference to brief autoradiographic exposure. The RNA was eluted from the gel piece using 500 μl RNA elution buffer [250 mM NaOAc, 20 mM Tris–HCl (pH 7.5), 1 mM ethylenediaminetetraacetic acid (EDTA) (pH8.0), 0.25% SDS] and overnight incubation on ice. Precipitation of RNA was performed by adding two volumes EtOH (100%; ice cold) and 1/100 glycogen for 1 h at −20°C and subsequent washing with 70% EtOH of pelleted RNA.
Endonuclease assay
Different indicated concentrations of purified Cas6 enzyme were incubated with radio labelled RNA substrates and buffer [250 mM KCl, 1.875 mM MgCl2, 1 mM DTT, 20 mM HEPES–KOH (pH 8.0)]. The reaction mix was incubated for 10 min at 37°C and then immediately mixed with 2× formamide buffer [95% formamide; 5 mM EDTA (pH 8.0), 2.5 mg bromophenol blue, 2.5 mg xylene cyanol) and incubated at 95°C for 5 min to stop the cleavage reaction. The reaction was applied to a denaturing 12–15% polyacrylamide gel running in 1× TBE with 12 W for 1.5 h. Visualization was achieved by phosphorimaging.
RNA-sequencing
RNA and DNA were extracted from cell lysates with phenol/chloroform (1:1; phenol pH5 for RNA and pH 8 for DNA) (34). A Proteinase K and 55°C heat shock treatment preceded the phenol/chloroform step. Small RNAs (<200 nt) were purified from total RNA using the mirVana RNA extraction kit (Ambion). Three micrograms of isolated small RNA from either M. maripaludis C5 or C. thermocellum were treated with T4 PNK to ensure proper termini for ligation. A protocol for the dephosphorylation of 2′-, 3′-cyclic phosphate termini was modified from (35): 1 μg of RNA was incubated at 37°C for 6 h with 10 U T4 PNK and 10 μl 5× T4PNK buffer (NEB) in a total volume of 50 μl. Subsequently, 1 mM ATP was added and the reaction mixture was incubated for 1 h at 37°C to generate monophosphorylated 5′-termini. RNA libraries were prepared with an Illumina TruSeq RNA Sample Prep Kit and sequencing on an Illumina HiSeq2000 sequencer was performed at the Max-Planck Genomecentre Cologne.
Identification of crRNA abundance
Sequencing reads were trimmed [(i) removal of Illumina TruSeq linkers and poly-A tails and (ii) removal of sequences using a quality score limit of 0.05] and mapped to the reference genomes (GenBank: CP000568 and CP000609) with CLC Genomics Workbench 5.0 (CLC Bio, Aarhus, Denmark). The following mapping parameters were used (mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.5, similarity: 0.8). Reads <15 nt were removed. Initial crRNA identification was obtained from crisprdb (36) and gene annotations were obtained from Genbank.
Modelling of M. maripaludis Cas6b
A model of the Mm Cas6b (MmarC5_0767) protein structure was built with the I-TASSER platform (37). The program identified P. furiosus Cas6 (pdb ID 3PKM) as the top template for structure prediction. The protein model was compared with the Pf Cas6 crystal structure using the program DaliLite (38) and their alignment revealed two homologous structures (Z-score 19.7, RMSD 2.5 Å). Cas6b sequences were aligned with ClustalW2 (39).
RESULTS
crRNA processing for CRISPR/Cas subtype I-B
The processing of crRNAs of the CRISPR/Cas subtype I-B was analysed by RNA-Seq for M. maripaludis C5 and for C. thermocellum ATCC 27 405. The isolated total small RNAs were modified with T4 polynucleotide kinase to allow proper adapter ligation and were sequenced through Illumina HiSeq2000 RNA-Seq methodology. Over 14 million individual sequence reads were mapped to the corresponding reference genomes and elucidated the abundance and processing patterns of the CRISPR arrays of these two organisms. M. maripaludis C5 possesses a single CRISPR cluster with 28 repeats of 37-nt length that are interspersed by 27 unique spacers. The CRISPR region is constitutively transcribed and processed into small crRNAs (Figure 1). The crRNAs contain a clearly defined 5′-terminal 8-nt tag with the sequence 5′-AUUGAAAC-3′. The 3′-termini are gradually shortened and most often contain a minimal 2-nt tag with the repeat nucleotides 5′-CU-3′. The abundance of crRNAs declines gradually from the leader proximal to the leader distant region with the crRNA containing the highly AT-rich (30 A or T out of 34 nt) spacer 3 being underrepresented.
Figure 1.
crRNA processing in M. maripaludis. Illumina HiSeq2000 sequencing reads mapped to the M. maripaludis C5 reference genome highlight the abundance of crRNAs. Processing occurs within the repeat elements, generating crRNAs with a 5′-terminal AUUGAAAC 8-nt tag (boxed) and more variably trimmed 3′-terminal tags. Cleavage sites are indicated and a possible hairpin structure was predicted by RNAfold (41).
crRNA processing patterns in C. thermocellum reveal long-range influence of spacer sequences
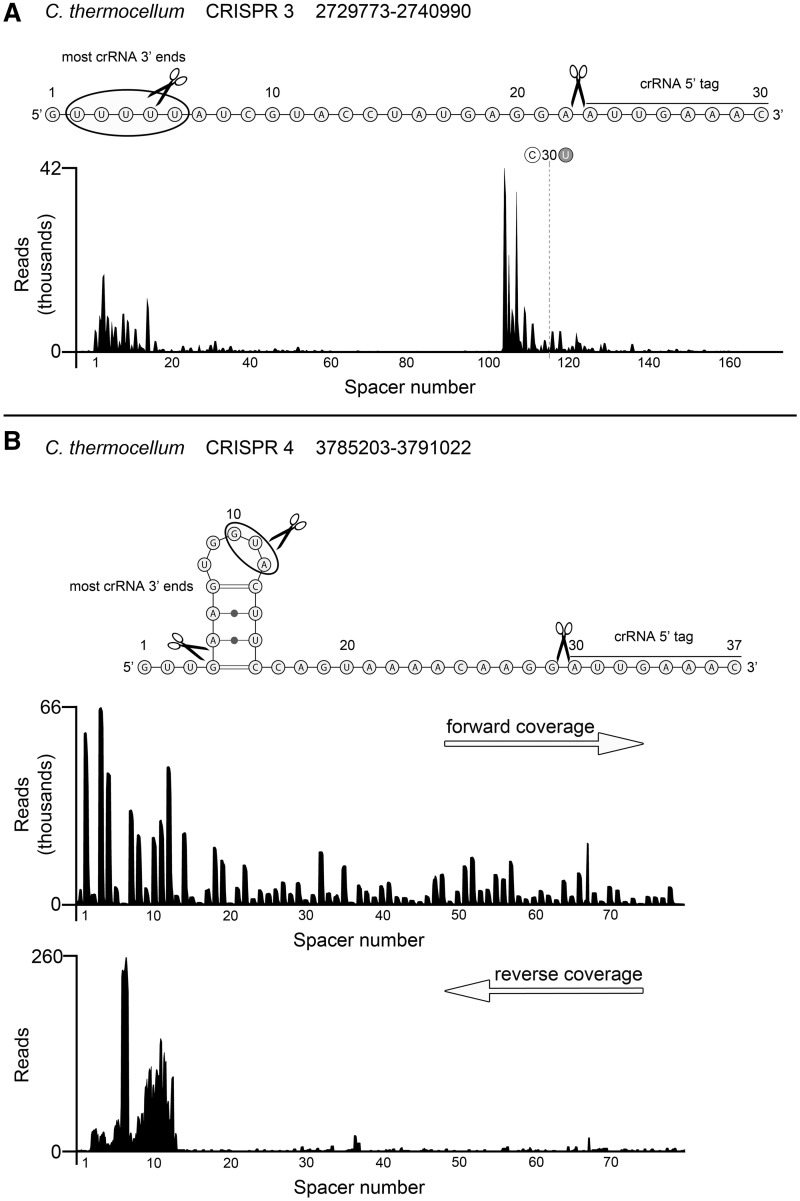
RNA-Seq analysis of the small RNAs of C. thermocellum revealed five constitutively transcribed and processed CRISPR clusters. Two of these CRISPR/Cas subtype I-B systems are very similar to the one found in M. maripaludis and contain 37-nt repeat elements. The other three CRISPR clusters have 30-nt repeat sequences. Processing of both C. thermocellum CRISPR repeat sequences into mature crRNAs yields the same 5′-terminal 8-nt (5′- AUUGAAAC-3′) tag that is also found for M. maripaludis crRNAs (Figure 2 and Supplementary Figure S2). The 3′-termini are trimmed leaving mostly short tags. The abundance of crRNAs follows the pattern found in M. maripaludis and described for other CRISPR/Cas subtypes with one notable exception. The CRISPR locus 3 contains an internal signal to promote crRNA transcription within the CRISPR array (Figure 2A and Supplementary Table S1). The overall crRNA abundance gradually declines from Spacer 1 to Spacer 103 before crRNA production peaks again starting with the crRNA containing spacer number 104. Interestingly, the 8-nt repeat tags are not identical for the crRNAs from this CRISPR locus 3 as at Spacer 116 the final U base of the 5′-terminal tag is changed to the commonly found base C (Figure 2A and Supplementary Table S1). Close analysis of this sudden spike of internal crRNA abundance revealed a transcription start site at the A residue at Position 29 within Spacer 103. Our data suggest that this spacer is sufficient to promote transcription within the CRISPR region and that the 28-nt upstream of the transcription start within the spacer provide the necessary promoter elements in the context of the flanking repeats. Although it is difficult to pinpoint the pribnow box, the extreme AT-richness of the spacer (26 out of 28 nt upstream of the transcription start site are A and T residues) suggests relaxed strand separation. DNA sequencing of the genomic region upstream of Spacer 104 excluded errors in the initial genome assembly during whole-genome sequencing.
Figure 2.
crRNA processing in C. thermocellum. Illumina HiSeq2000 sequencing reads were mapped to the C. thermocellum ATCC 27405 reference genome and selected CRISPR regions are displayed. Conserved 5′-terminal crRNA cleavage sites and variably trimmed 3′-termini are indicated within the repeat sequence. A possible hairpin structure was predicted by RNAfold (41). All C. thermocellum CRISPR mappings are found in Supplementary Figure S2. (A) CRISPR locus 3 reveals internal promotion of crRNA transcription at Spacer 104. (B) CRISPR locus 4 exemplifies bidirectional CRISPR transcription. Forward and reverse reads were separated to highlight the occurrence of processed anti-crRNAs that can correlate with reduced crRNA abundance.
In addition to internal promotion, we observed several cases of bidirectional transcript production for the CRISPR arrays. Anti-crRNA transcripts can start at the region opposite of the leader (CRISPR loci 1,2,5) or internally (CRISPR locus 4) (Figure 2B and Supplementary Figure S2). Although the amount of these anti-crRNA transcripts is usually very small in comparison to the abundance of crRNAs, individual anti-crRNAs show a conserved processing pattern within the repeats that yields RNAs with complete reverse complementary spacer sequences. These anti-crRNAs usually contain 18-nt 5′-tags and 15-nt 3′-tags for CRISPR loci 1 and 2 and 22-nt 5′-tags for CRISPR loci 4 and 5 (Supplementary Figure S3). The presence of processed anti-crRNAs can correlate with the reduced abundance of the respective sense crRNA (Figure 2B and Supplementary Figure S2).
Identification of Cas6 I-B enzymes that generate crRNAs
To identify the enzyme that generates crRNAs for CRISPR/Cas subtype I-B, we analysed the cas genes of M. maripaludis and C. thermocellum. A set of only eight cas genes was identified in the genome of M. maripaludis C5. One of these potential cas gene products (MmarC5_0767, Mm Cas6b) showed 12% amino acid identity to Pf Cas6 which identified it as a Cas6b candidate for CRISPR/Cas subtype I-B. As the protein shares limited sequence identity with Cas6 proteins of other CRISPR/Cas subtypes (11–13), the structure of Mm Cas6b was modelled with I-Tasser (40). Pf Cas6 was identified as the closest structural homologue and shares a very similar overall architecture [Dali-Lite Z-score 19.7, RMSD 2.5 Å, (38)] (Figure 3). The structural alignment of the Mm Cas6b model and Pf Cas6 also reveals a conserved histidine residue of Mm Cas6b in close proximity to the catalytic histidine of Pf Cas6. The comparison of different Cas6b homologues of CRISPR subtype I-B (Figure 3) indicates high sequence similarity and conserved residues. Clostridium thermocellum contains one Cas6b homologue (Cthe_3205) associated with the 37-nt repeat sequences and a potential second Cas6 enzyme (Cthe_2303) associated with the 30-nt repeat sequences. Classification of Cthe_2303 is not unambiguously possible as the neighbouring cas genes do not clearly fit into the commonly used 10 CRISPR/Cas subtypes.
Figure 3.
Structural model of Cas6b shows high similarity to Pf Cas6. (A) A structure model (I-Tasser) of M. maripaludis Cas6b was aligned to Pf Cas6 (DaliLite). The catalytic site of Pf Cas6 is indicated and a histdine at Position 38 in Cas6b is located in close proximity to the catalytic histdine in P. furiosus. (B) An alignment of Cas6b homologues (ClustalW) of subtype I-B shows conserved amino acid residues (black coloured residues) and reveals putative catalytic residues (black bars).
Repeat specific endonucleolytic activity of subtype I-B Cas6 homologues
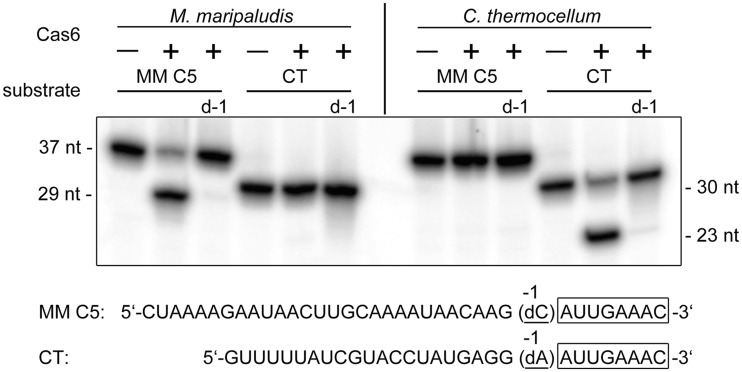
The genes for Cas6 enzymes from M. mariplaudis and C. thermocellum were cloned and the recombinant enzymes were produced. Clostridium thermocellum Cas6b (Cthe_3205) production yielded only insoluble protein, but the two other Cas6 proteins were obtained in soluble form and allowed the analysis of their involvement in crRNA processing. Size exclusion separation of Mm Cas6b revealed a monomeric structure of the protein. The purified Cas6 candidates (Mm Cas6b and Cthe_2303) showed in vitro endonuclease activity and processed repeat RNA sequences from M. maripaludis and C. thermocellum, respectively (Figure 4). Therefore, the unclassified Cthe_2303 provided a good control for crRNA processing specificity of Mm Cas6b, as both enzymes specifically recognized only the repeats from their associated CRISPR clusters. Addition of bivalent metal ions did not influence the activity of Mm Cas6b. To define the cleavage site, a repeat RNA was synthesized with a deoxy nucleotide substitution at the proposed processing position that generates the 8-nt crRNA 5′-tag observed in vivo. This substitution resulted in the loss of Mm Cas6b and Cthe_2303 processing.
Figure 4.
Cas6b of M. maripaludis (MM C5) and C. thermocellum Cthe_2303 (CT) cleave their specific repeat structure. Cas6b endonuclease assay were performed with 5′-terminal radioactively labelled repeat RNA and the respective deoxy variants (indicated at the bottom, −1 displaying the first base upstream of the 5′-tag) of M. maripaludis and C. thermocellum. Cas6b processes the 37-nt repeat into the smaller 29-nt fragment, while the deoxy variant (d-1) and the 30-nt repeat RNA of C. thermocellum are not cleaved. Cthe_2303 is specific for its 30-nt repeat RNA.
A single repeat structure is sufficient for Mm Cas6b in vitro processing
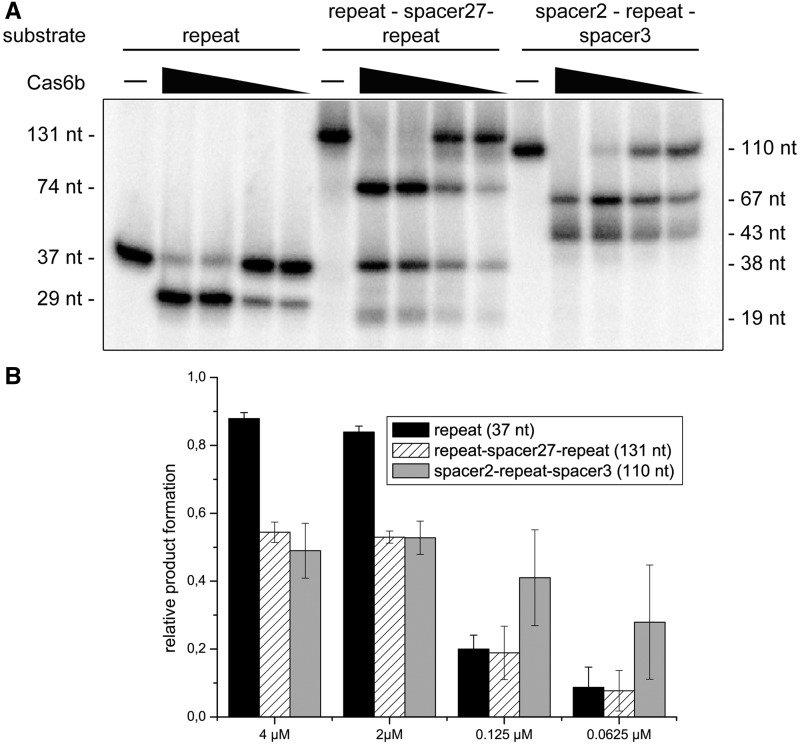
To test the influence of different RNA substrates (Supplementary Table S2) for Mm Cas6b activity, cleavage assays were performed with (i) repeat RNA, (ii) repeat–spacer–repeat RNA and (iii) spacer–repeat–spacer RNA using repeat and spacer sequences of the M. maripaludis CRISPR. For all three substrates, product formation was observed that corresponds with Mm Cas6b processing at the cleavage site determined by the deoxy nucleotide substitution within the repeat (Figure 4). This cleavage site determines that the conversion of the repeat (37 nt) results in a 29-nt fragment, while the repeat–spacer27–repeat structure (131 nt) is processed into three fragments (74, 38 and 19 nt) and the spacer2–repeat–spacer3 substrate (110 nt) is cleaved into two fragments (67 and 43 nt) (Figure 5). Since all used substrates were cleaved in similar efficiency, the repeat RNA was used for further analysis of the catalytic site of Mm Cas6b. In order to test the influence of the computationally predicted [RNAfold (41)] short hairpin structure of the repeat (Figure 1), the mutation G16C was introduced that disrupts a G–C base pair within this hairpin. The mutated repeat was cleaved less effectively than wild-type repeats (Supplementary Figure S4).
Figure 5.
RNA substrates for Cas6b processing. The two ‘repeat–spacer27–repeat’ and ‘spacer2–repeat–spacer3’ substrates were internally labelled by in vitro transcription and repeat RNA molecules were 5′-end labelled. The substrates were used in three independent cleavage assays using different concentrations of Cas6b (4, 2, 0.125 and 0.0625 µM). (A) A representative assay shows the ability of Cas6b to process all used substrates in similar manner and efficiency. (B) Product formation for three independent reactions was quantified.
Mm Cas6b contains two catalytic histidine residues
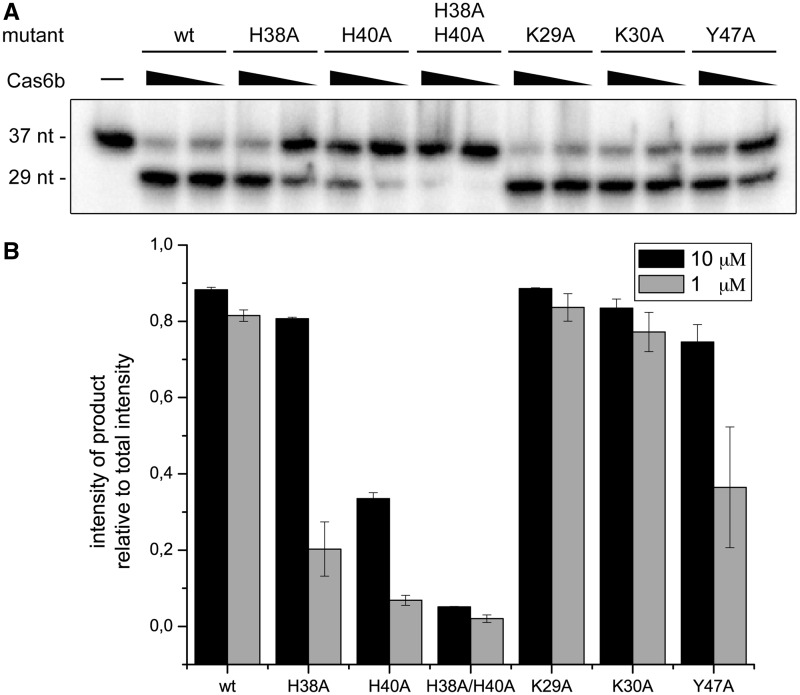
To deduce catalytic residues, potentially important amino acids were identified based on the structural model of Mm Cas6b and the observed conservation of amino acids in the alignment of Cas6b homologues (Figure 3). Cas6 proteins of other CRISPR/Cas subtypes contain a single catalytic histidine residue. In Cas6b, there are two conserved histidine residues (H38 and H40), separated only by a single amino acid that could potentially fulfil this role. A set of Mm Cas6b mutants was produced (Supplementary Figure S5) of which two mutants (Y49A and Y47A/Y49A) yielded insoluble proteins. The other mutants were used in endonucleolytic cleavage assays testing the processing of the repeat RNA substrate in comparison to wild-type Mm Cas6b (Figure 6). The single histidine mutant H38A and the tyrosine mutant Y47A showed reduced processing activity compared with wild-type Mm Cas6b. The H40A mutation reduced Mm Cas6b activity by >50%. Surprisingly, both single histidine mutants retained considerable cleavage activity. However, the mutation of both histidine residues into alanine (H38A/H40A) resulted in a drastic loss of substrate processing. Mutation of the lysines at Position 29 or 30 did not show any notable effect on endonucleolytic activity.
Figure 6.
Two histidine residues play a critical role for Cas6b activity. Cas6b endonuclease assays were performed with the indicated Cas6b variants and 5′-end-labelled repeat RNA substrates. (A) A representative assay shows the activities of the Cas6b variants. While mutation of two lysines at Position 29 and 30 did not show any influence on activity in comparison to wild-type (wt), a mutation of two histidines at Positions 38 and 40 as well as a mutation of tyrosine at Position 47 show reduced processing activity. (B) Product formation for three independent reactions was quantified.
DISCUSSION
The observed crRNA processing and abundance patterns of CRISPR/Cas subtype I-B are in good agreement with crRNA maturation previously analysed for other CRISPR/Cas subtypes and substantiate that 8-nt 5′-terminal and trimmed 3′-terminal crRNA tags are an universal feature of many CRISPR/Cas subtypes. The surprising observation of a spacer sequence that promotes crRNA production internally in C. thermocellum exemplifies the effect that an individual spacer can have for the abundance of mature crRNAs and subsequently the efficiency of entire CRISPR regions. The exchange of 1 nt of the otherwise universal 5′-terminal 8-nt tag of the crRNAs in the vicinity of the internal CRISPR transcription start site opens the possibility that two CRISPR elements might have been fused and subsequently portions of a leader element might have been incorporated into the repeat–spacer–repeat pattern. In addition, spacer elements were shown to promote transcripts of the reverse orientation. These anti-crRNAs were first described in Sulfolobus (42) and were also found in P. furiosus (43). However, they appear to be absent in most organisms. The occurrence of specific processing patterns for anti-crRNAs from different repeats of C. thermocellum CRISPR and the absence of anti-crRNAs in M. maripaludis indicate that this phenomenon might be specific for organisms with relaxed transcription start site definition rather than for the CRISPR/Cas subtype. Individual anti-crRNAs appear to be better suited for a conserved maturation process at their termini by a currently unknown mechanism. Reverse transcripts can form double-stranded RNA duplexes that might reduce the abundance and efficiency of crRNAs. Taken together these results highlight the effects that individual spacer sequences within a CRISPR region can have in both forward and reverse direction. These strong effects will need to be taken into consideration in the anticipated and proposed design of synthetic CRISPR regions for biotechnologically or medically important processes.
We identified the Cas6b endonuclease responsible for crRNA maturation in the CRISPR/Cas subtype I-B. Cas6 enzymes are among the most diverse members in the sets of Cas protein of the different CRISPR/Cas subtypes and can be used to classify CRISPR/Cas systems. The similarity of repeat sequences and Cas6b enzymes between Bacteria (i.e. Clostridia) and Archaea (i.e. methanogens) hints at a horizontal transfer event for these CRISPR/Cas systems. Cas6 proteins might show this remarkable degree of divergence due to their individual adaptation to the given repeat sequence and/or structure. Evidence for this can also be found in the different principles of recognition for Pf Cas6, Cas6e and Cas6f. While Pf Cas6 binds to unstructured RNA, both Cas6e and Cas6f need a secondary structured RNA to bind and process in vitro (13,14,16). For Type II CRISPR systems, no Cas6 activity was reported. In these systems, the presence of a guide RNA (tracrRNA) recruits RNase III for the processing of crRNAs (44). These pathways exemplify the differences in crRNA maturation among organisms and CRISPR subtypes.
The Cas6 enzymes Pf Cas6, Cas6e and Cas6f of different CRISPR/Cas subtypes were all shown to require a single conserved histidine residue for catalysis (10,12–14,16,29). In this study not one but two conserved histidine residues were identified for Mm Cas6b. Only the simultaneous mutation of both histidine residues resulted in a drastic loss of endonuclease activity. This implies that Cas6b exhibits the first example of a more flexible catalytic core in which both histidine residues are potentially representing the catalytic histidine and able to complement the loss of the other residue. Why did Cas6b evolve two catalytic histidine residues where this function can be fulfiled by a single histidine in other Cas6 enzymes? One possible explanation is the advantage such setup would have in coping with different substrates, e.g. with crRNA precursors that contain spacer of different length or structure. In Pf Cas6 a catalytic triad was described that provides a catalytic site for general acid–base catalysis (10,14). Mm Cas6b does not contain an identical catalytic triad but our observation of the importance of tyrosine 47 for Mm Cas6b activity and the occurrence of clustered amino acids that could provide general bases and acids might indicate more flexible active site architecture.
In conclusion, we provide the first description of crRNA processing in vivo and in vitro for CRISPR/Cas subtype I-B. These analyses of Cas6b in a bacterial and an archaeal model organism highlight the similarities between different CRISPR/Cas subtypes and the differences in crRNA processing. Two interchangeable catalytic histidine residues in Cas6b and internal promotion of crRNA production in C. thermocellum exemplify two new concepts that were found for CRISPR/Cas I-B systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
The Deutsche Forschungsgemeinschaft [DFG, FOR1680] and the Max Planck Society. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Andreas Su for technical help and André Plagens for advice and discussions The authors are very grateful to Michael Rother, Rolf Thauer and William B. Whitman for their help with the handling of Methanococcus maripaludis.
REFERENCES
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 3.Sorek R, Kunin V, Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 4.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou R, Horvath P. CRISPR: New Horizons in Phage Resistance and Strain Identification. Annu. Rev. Food Sci. Technol. 2011;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, Li Y, Gorge O, Platonov ME, Yan Y, Guo Z, Pourcel C, Dentovskaya SV, Balakhonov SV, Wang X, et al. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One. 2008;3:e2652. doi: 10.1371/journal.pone.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 8.Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol. Rep. 2009;1:95. doi: 10.3410/B1-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 13.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Zheng H, Preamplume G, Shao Y, Li H. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–417. doi: 10.1002/pro.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 17.Howard JA, Delmas S, Ivancic-Bace I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem. J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 18.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 19.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J. Biol. Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J. Biol. Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl Acad. Sci. USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl Acad. Sci. USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones WJ, Nagle DP, Jr, Whitman WB. Methanogens and the diversity of archaebacteria. Microbiol. Rev. 1987;51:135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynd LR, Grethlein HE. Hydrolysis of dilute acid pretreated mixed hardwood and purified microcrystalline cellulose by cell-free broth from Clostridium thermocellum. Biotechnol. Bioeng. 1987;29:92–100. doi: 10.1002/bit.260290114. [DOI] [PubMed] [Google Scholar]
- 33.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randau L, Münch R, Hohn MJ, Jahn D, Söll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 35.Schurer H, Lang K, Schuster J, Mörl M. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002;30:e56. doi: 10.1093/nar/gnf055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lillestol RK, Shah SA, Brugger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 43.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45: 292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.