Abstract
Binding of the DnaA protein to oriC leads to DNA melting within the DNA unwinding element (DUE) and initiates replication of the bacterial chromosome. Helicobacter pylori oriC was previously identified as a region localized upstream of dnaA and containing a cluster of DnaA boxes bound by DnaA protein with a high affinity. However, no unwinding within the oriC sequence has been detected. Comprehensive in silico analysis presented in this work allowed us to identify an additional region (oriC2), separated from the original one (oriC1) by the dnaA gene. DnaA specifically binds both regions, but DnaA-dependent DNA unwinding occurs only within oriC2. Surprisingly, oriC2 is bound exclusively as supercoiled DNA, which directly shows the importance of the DNA topology in DnaA-oriC interactions, similarly as previously presented only for initiator-origin interactions in Archaea and some Eukaryota. We conclude that H. pylori oriC exhibits bipartite structure, being the first such origin discovered in a Gram-negative bacterium. The H. pylori mode of initiator-oriC interactions, with the loop formation between the subcomplexes of the discontinuous origin, resembles those discovered in Bacillus subtilis chromosome and in many plasmids, which might suggest a similar way of controlling initiation of replication.
INTRODUCTION
Initiation is the first and strictly regulated step in chromosome replication (1,2). The basic mechanism of initiation is conserved in bacteria, Archaea and Eukaryota: a multiprotein complex (i.e. initiator) recognizes and binds a specific chromosomal region (or multiple regions in Archaea and Eukaryota) known as the origin of replication (ori) (3–5). The formation of an initiator-ori complex leads to DNA unwinding within the helically unstable AT-rich region. In bacteria, Archaea and lower Eukaryota, the ori regions are characterized by the presence of specific initiator binding sequences (5). However, no conserved initiator binding sequences have been identified in higher eukaryotes (metazoans), whose ori regions are featured by less-specific markers including CpG islands, DNA topology (especially negative supercoiling, loop formation) or nucleosome-free regions (5,6).
Most of the information about bacterial chromosome replication comes from studies on Escherichia coli, whose key initiation elements, oriC and DnaA, have been thoroughly characterized (reviewed in (7–9)). DnaA is composed of four functional domains, which are specialized in DnaA oligomerization, interaction with other proteins (e.g. DnaB, DiaA, Hda, HU) or cofactors (ATP/ADP), and DNA binding (9). The E. coli oriC consists of high- and low-affinity DnaA binding sites (DnaA boxes), an AT-rich region with a DNA unwinding element (DUE), and the binding sites for regulatory proteins IHF, Fis, IciA and SeqA (7,8). Sequential, cell-cycle coordinated DnaA binding to the DnaA boxes leads to formation of a highly ordered nucleoprotein complex (orisome) resulting in DNA unwinding at the DUE (7,8). Once the open complex is formed, DnaB, DnaG and finally DNA Pol III are loaded, forming replication forks which bi-directionally synthesize nascent DNA strands.
The initiation of chromosome replication is much less understood in bacteria other than E. coli. Almost all bacteria encode DnaA homologs (10,11). In all Gram-negative and in many Gram-positive bacteria, oriC is composed of a single DnaA-box cluster (DBC) and the DUE, which are localized in the intergenic region, usually, with the notable exception of E. coli, upstream or downstream of dnaA (12). However, in a few bacteria such as Gram-positive Bacillus subtilis and mollicute Mycoplasma pulmonis, the oriC is composed of two or three clusters indispensible for oriC activity (13–15). In such cases, DnaA binds and oligomerizes onto individual clusters, but usually also interacts with DnaA molecules bound to neighboring clusters, often forming a DNA loop (16,17). The AT-rich region with a DUE, a second conserved modular element of oriCs, is composed of a few tandem AT-rich repeats (e.g. 13-mers in E. coli) or is a stretch of an AT-rich sequence (27-mer in B. subtilis or 19-mer in Mycobacterium tuberculosis (16,18)).
Advanced in silico genome analyses have allowed many putative bacterial oriC regions to be identified. The methods are mainly based on the cumulative analysis of the genome skews, localization of DBC and an AT-rich region in the vicinity of the dnaA gene (10,19,20). However, the reliability of the in silico identification is limited and the results need to be confirmed experimentally. The DnaA binding sites might be localized outside oriC and serve as negative regulators of chromosome replication (datA in E. coli (21), D78 cluster in Streptomyces coelicolor (22) and DBCs in B. subtilis (23)) or regulatory sites in the promoters of genes controlled by DnaA, including dnaA autoregulation (24–26). Similarly, the in silico predicted helically unstable DNA sequences might be connected with gene transcription; thus their melting upon initiator binding should be experimentally proved. Indeed, out of many predicted bacterial oriC regions only a small number have been shown to be functional in vivo, whereas the DUE regions have been precisely localized only in a few of them (20).
Helicobacter pylori oriC has been identified in silico and, subsequently, DnaA-oriC interactions were characterized by a number of in vitro experiments (27–29). It is localized upstream of dnaA and contains five DnaA boxes bound with different affinities by DnaA (28,29). The binding of DnaA to oriC is enhanced in the presence of HobA—a protein interacting with DnaA, which is a structural homolog of DiaA from E. coli (30,31). The oriC region does not contain any other sequences related to known protein binding sites such as IHF or Fis, and genes encoding proteins homologous to known oriC-interacting proteins are not present on the H. pylori chromosome. Despite many attempts, no unwinding of the oriC region has been detected in vitro so far.
In this study, we used combined computational and experimental analyses to identify and characterize the DUE site within H. pylori oriC. In contradiction to our previous assumptions, this region was predicted downstream of the dnaA gene (here called oriC2). However, both upstream (oriC1) and downstream (oriC2) dnaA regions are required in vivo for the initiation of H. pylori chromosome replication, which indicates a bipartite structure of H. pylori oriC. Interestingly, oriC2 is bound by DnaA exclusively as supercoiled, resembling some archaeal and eukaryotic initiators, whose DNA binding activity also relies on local DNA topology.
MATERIALS AND METHODS
Materials, strains and culture conditions
The plasmids, proteins and bacterial strains used in this work are listed in Table 1. The oligonucleotide sequences are presented in Table 2. H. pylori 26 695 genomic DNA was used as a template to amplify DNA fragments for electron microscopy (EM), surface plasmon resonance (SPR) and cloning; H. pylori N6 ΔdnaAH(L) was used to prepare pTZ57R/TX plasmids. E. coli was grown at 37°C on solid or in liquid Luria-Bertani medium, supplemented with 100 µg/ml ampicillin when necessary. H. pylori was cultivated as described previously (32).
Table 1.
Strains, plasmids and proteins used in this work
| Strain/plasmid/protein | Relevant genotype/feature | Reference/source |
|---|---|---|
| H. pylori | ||
| 26695 | Parental strain | (68) |
| N6 ΔdnaAL | N6 ΔdnaAΩaphA-3, pILL2282, underproducing DnaA when compared with wild type 26695. | (32) |
| N6 ΔdnaAH | N6 ΔdnaAΩaphA-3, pILL2157dnaA, overproducing DnaA when compared with wild type 26695. | This work |
| Plasmids | ||
| GHPAQ41 | DNA fragment of H. pylori 26695 genome (1607422-1609162 bp) cloned into SmaIl site of pUC18, AmpR. The H. pylori DNA encodes dnaA flanked by 164 bp upstream (oriC1) and 202 bp downstream (oriC2) of the gene. | TIGR/ATCC microbial genome special collection |
| pOC170 | Plasmid carrying the E. coli oriC sequence, the replication origin of pBR322 on the NotI cassette and the bla gene of pBR322. | (69) |
| pILL2157 | IPTG-inducible E. coli and H. pylori expression vector, ChlR | (35) |
| pTZ57R/T | Cloning vector, AmpR | Fermentas |
| pori1ori2 | A pOC170 derivative, lacking E. coli oriC, containing DNA fragment of H. pylori 26695 genome (1607422–1609162 bp), restricted from GHPAQ41 with EcoRI and PstI. | This work |
| pori1 | A pOC170 derivative, lacking E. coli oriC, containing oriC1 region amplified with primers P-1 and P-2 and cloned into EcoRI site. | This work |
| pori2 | A pOC170 derivative, lacking E. coli oriC, containing oriC2 region amplified with primers P-3 and P-4 and cloned between EcoRI and PstI sites. | This work |
| pILL2157dnaA | pILL2157 derivative containing dnaA, constructed similarly as pILL2282 (32) | This work |
| pTZ57R/T1 | pTZ57R/T derivative carrying gentamicin resistance cassette used for homologous recombination with H. pylori N6 ΔdnaAL or N6 ΔdnaAH in order to delete oriC1. | This work |
| pTZ57R/T2 | pTZ57R/T derivative carrying gentamicin resistance cassette used for homologous recombination with H. pylori N6 ΔdnaAL or N6 ΔdnaAH in order to delete oriC2. | This work |
| pTZ57R/T3 | pTZ57R/T derivative carrying gentamicin resistance cassette used for homologous recombination with H. pylori N6 ΔdnaAL or N6 ΔdnaAH in order to delete both oriC1–oriC2 subregions. | This work |
| pTZ57R/T4 | pTZ57R/T derivative carrying gentamicin resistance cassette used for homologous recombination with H. pylori N6 ΔdnaAL or N6 ΔdnaAH in order to exchange aph-3 for gentamicin cassette. | This work |
| Proteins | ||
| DnaA | Recombinant, untagged H. pylori DnaA protein | (28,30) |
Table 2.
Oligonucleotides used in this work
| 5′ – 3′ sequence | |
|---|---|
| P-1 | GCGAATTCCGCAAAGCAGCATGAAAATCC |
| P-2 | GCGAATTCTTCAATATTGTTGTTGGTATCCAT |
| P-3 | GCCTGCAGGGTTTAGTAAAAGTCATAAATA |
| P-4 | GCGAATTCCCACAACCCCCCTAAAAAC |
| P-5 | CCAGCGCAAAGCAGCATGAAAATC |
| P-6 | CAATATTGTTGTTGGTATCCATGG |
| P-7 | CCCGCTTTCAATTCAAGTGAATG |
| P-8 | AAAGGGATTTTTTCATGCTTATT |
| P-9 | CCACAACCCCCCTAAAAACG |
| P-10 | CATGTTTGACAGCTTATCATCG |
| P-11 | Bio-CCAGCGCAAAGCAGCATGAAAATC |
| P-12 | Bio-CCGCTTGAACGAATTGAACGAC |
| P-13 | Bio-CTCTATTTTGAAAACCCCTATTTC |
| P-14 | CTATATTTTTTCAATGGTTTAGTGC |
| P-15 | GATGAGTTCCCTGAATTCCC |
| P-16 | CCCATCAATGAGTTTGGTG |
| P-17 | GGCTAACACATTAGAGAGC |
| P-18 | CGCTACCCCCCTATCGTCATT |
| P-19 | CAGTTTCTATGTGAATGAAATTAT |
| P-20 | TTTTGCTGATGGAGCTGCACGCTTTATAATAAGCTAATGGATG |
| P-21 | TCGCCAGTCGATTGGCTGAGGATCTGGTACCCGGGTG |
| P-22 | GCAAAGGACGCCATCGGC |
| P-23 | CCATTTAAAGATCCGCGCGA |
| P-24 | TTTTGCTGATGGAGCTGCACCACATTATTCCCTCCAGGTA |
| P-25 | TCGCCAGTCGATTGGCTGACTATAACCTATTTATGACTTTTAC |
| P-26 | GTTGTTTCTAAAGAAAGTTTTTCA |
| P-27 | GCTCCCTATAAAAATAAGGCTT |
| P-28 | TTTTGCTGATGGAGCTGCACCACCCGGGTACCAGATCC |
| P-29 | TCGCCAGTCGATTGGCTGACCTGGAGGGAATAATGTGAA |
| P-30 | GCTCGATCACAAGGGCTTG |
| P-31 | GTGCAGCTCCATCAGCAAAA |
| P-32 | TCAGCCAATCGACTGGCGA |
| P-33 | CTTATTTTGCTAGGAATTGCTAAAG |
| P-34 | CGCACCCTTTCAAAAAGAGCC |
| P-35 | CGCAAAGCAGCATGAAAATCC |
Bio - biotin.
In silico methods
WebSIDD (33) was used for the prediction of putative DUE(s) (34) in the H. pylori 26 695 dnaA region. The dnaA (HP1529)—punB (HP1530) upstream intergenic region (pos. 1 608 816–1 609 315) and the dnaA (HP1529)—HP1527 downstream intergenic region (pos. 1 607 325–1 607 824) were subjected to WebSIDD predictions as 2.5 kb DNA fragments with the intergenic regions located approximately in the middle (http://benham.genomecenter.ucdavis.edu/sibz/). Default values (37°C, 0.1 M salt, circular DNA, copolymeric) were chosen for the predictions, and negative superhelicity values were tested in the range of σ = –0.040 (low) to σ = – 0.060 (high) in increments of 0.005 (33). The prediction output data were obtained as raw text files and further processed with Microsoft Excel v97SR-1 and Corel Draw v.11.
Construction of H. pylori N6 ΔdnaAH mutant
H. pylori N6 ΔdnaAH mutant was constructed exactly as described for H. pylori N6 ΔdnaAL (32) using pILL2157 instead of pILL2150 (35).
Surface plasmon resonance
SPR analysis was done as previously described (30). Biotinylated DNA fragments were obtained by polymerase chain reaction (PCR) with the following primer pairs: oriC1 (P-6 and P-11; 191 bp), oriC2 (P-8 and P-12; 296 bp), non-box DNA (P-13 and P-14; 191 bp).
P1 nuclease assay and PE analysis
P1 nuclease assay was performed as described (36) with several modifications. The reaction mixture (15 μl) contained 25 mM Hepes-KOH (pH 7.6), 12% (v/v) glycerol, 1 mM CaCl2, 0.2 mM EDTA, 5 mM ATP, 0.1 mg/ml BSA, 200 ng of pori1ori2, pori2 or pori1 (50, 80 or 76 fmol, respectively) and H. pylori DnaA (50, 100, 200 ng; 1, 2, 4 pmol). After incubation at 30°C for 15 min, P1 nuclease (Sigma) was added (0.75 unit in 0.01 M sodium acetate, pH 7.6) and the reaction was continued for 5 min at 30°C. The P1 digestion was stopped by the addition of 85 μl of water and 300 μl of QG buffer (Qiagen) followed by immediate DNA purification using QIAquick spin columns (Qiagen). The P1 digestion was monitored either by restriction analysis or primer extension (PE) analyses. In the case of agarose gels analyses, the whole purified DNA was digested by ScaI restriction enzyme, loaded on a 1% agarose gel and separated. After ethidium bromide staining, the gels were analyzed with the Typhoon 8600 Variable Mode Imager (GE Healthcare). For a single PE reaction (37), 0.3 unit of Taq DNA polymerase (Fermentas), 20 fmol of digested DNA and 350 fmol 32P-labeled primer were used. After PE (30 s at 95°C, 30 s at 55°C and 60 s at 72°C, 30 cycles) samples were separated on an 8% polyacrylamide gel under denaturing conditions and analyzed with the Typhoon 8600 Variable Mode Imager (GE Healthcare).
RIP mapping
Replication initiation point (RIP) mapping was performed similarly as previously described (38–40). H. pylori cells were grown in 400 ml of brain heart infusion (32) to OD600 = 1.0 and pelleted. The bacterial pellet was resuspended in 30 ml of TEN buffer (50 mM Tris-HCl, pH 8.0, 50 mM EDTA, 100 mM NaCl) and disrupted by the addition of sodium dodecyl sulphate (SDS) and sodium sarkosyl to 1% concentration of each. After one-step extraction with phenol:chloroform (1:1), 1.1 g/ml CsCl and 100 μg/ml Hoechst-33 258 were added to the aqueous phase and the refractive index was adjusted to 1.400 with 5 M CsCl. Then genomic DNA was purified by CsCl gradient ultracentrifugation. To enrich for replicating intermediates, total isolated DNA (400 μg) was passed through a BND-cellulose column pre-equilibrated with NET buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA and 1 M NaCl). After washing with 5 column volumes of NET buffer, DNA was eluted at 50°C with NET buffer containing 1.8% caffeine. In order to remove nicked DNA, the recovered DNA (ca 40 μg) was subjected to phosphorylation by T4 kinase (Fermentas) followed by λ-exonuclease (Fermentas) digestion. For PE reaction 1 unit of vent (exo-) DNA polymerase (New England Biolabs), 0.3 or 1.2 μg of prepared DNA and 350 fmol of 32P-labeled primer were used. After 35 cycles of reaction (30 s at 95°C, 30 s at 55°C and 60 s at 72°C) amplification products were separated on an 8% polyacrylamide gel under denaturing conditions and analyzed with the Typhoon 8600 Variable Mode Imager (GE Healthcare).
Electron microscopy
60 ng of plasmid DNA was mixed with 30 ng of the H. pylori DnaA protein in 20 μl of buffer containing 25 mM Hepes-KOH (pH 7.6), 12% (v/v) glycerol, 1 mM CaCl2, 0.2 mM EDTA and 5 mM ATP. Samples were incubated for 15 min at 30°C followed by fixation in 0.2% glutaraldehyde for 10 min at 30°C. DNA was purified by gel filtration using Sephacryl S500 (GE Healthcare) and spin columns in buffer containing 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2 (41). The purified nucleoprotein complexes were adsorbed to mica as described (42). In the case of samples subjected to restriction enzyme digestion, glutaraldehyde was titrated by the addition of 50 mM glycylglycine (10 min at 30°C) and subsequently the sample volume was increased to 60 μl by the addition of 24.5 μl of water, 5 μl of MgCl2 (0.1 M), 5 μl of NaCl (1 M) and the appropriate restriction enzyme (5 units, 0.5 μl). Digestion was carried out for 1 h at 37°C. DNA was treated afterwards as described for undigested samples. The formed complexes were analyzed using a Philips CM100 electron microscope (FEI, Hillsboro, USA) with a Fastscan CCD camera (TVIPS, Gauting, Germany). The positions of the proteins bound to the DNA were measured on 35 mm negatives using an LM4 digitizer (Brühl, Nuremberg, Germany).
Immunoprecipitation assay
The immunoprecipitation assay was done as described elsewhere (43). H. pylori cells were grown in 30 ml of brain heart infusion (32) to OD600 = 1.0. The affinity-purified anti-DnaA rabbit antibody was used to precipitate DnaA–DNA nucleoprotein complexes. The following primers were used to amplify regions of interest: P-5 and P-6—oriC1, P-7 and P-8—oriC2, P-15 and P-16—dnaN gene fragment, P-17 and P-18—dnaB gene fragment. The PCR fragments were resolved in 1% agarose gel and analyzed on a Pharos FX Plus imager (Bio Rad).
Deletion of oriC1 and oriC2 regions in vivo
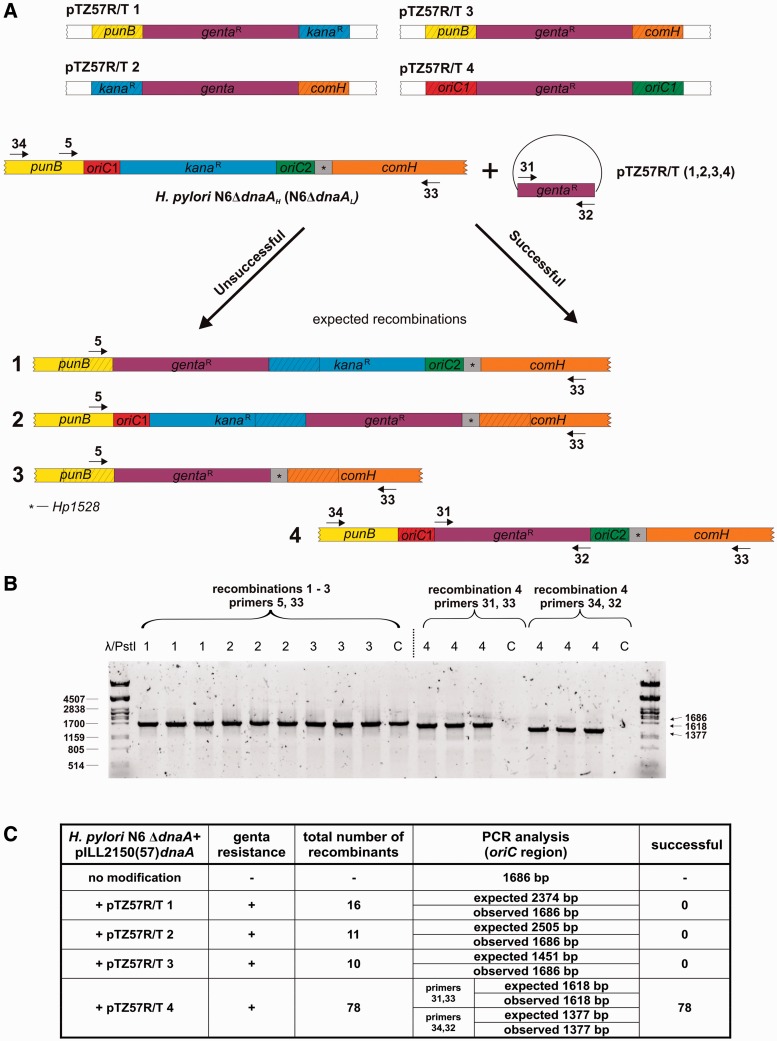
pTZ57R/TX (X: 1, 2, 3, 4) plasmids (Table 1 and Figure 8), bearing DNA fragments which allowed for homologous recombination with desired H. pylori chromosomal regions and the resistance cassette cloned into pTZ57R/T plasmid (Fermentas) were used to delete the H. pylori oriC (sub)regions. The fusion amplicons of H. pylori DNA and gentamicin cassette were prepared by the three-fragment PCR fusion method (44). N6ΔdnaAL was used as a template to amplify the H. pylori DNA. The gentamicin cassette was amplified from pUC1813Gm (45) using primers P-31 and P-32. Primer pairs P-19, P-20 and P-21, P-22 were used to amplify ca 300 bp regions upstream and downstream of oriC1, while the external P-19 and P-22 primers were used to amplify the final fused DNA used to delete oriC1. The same approach was used to construct cassettes to delete oriC2 (primers P-23, P-24 and P-25, P-26), the whole oriC region (P-19, P-20 and P-25, P-26) or to exchange aph-3 for gentamicin (P-27, P-28 and P-29, P-30). Resulting fusion PCRs were cloned into pTZ57R/T plasmid. The obtained plasmid constructs were used to transform H. pylori N6 ΔdnaAH or N6ΔdnaAL strains. Mutant selection was done on BA plates supplemented with 20 μg/ml apramycin and 1 mM IPTG.
Figure 8.
Comparative analysis of the DnaA interaction with oriC1 and oriC2 regions. (A) Schematic representation of the strategy used to delete chromosomal oriC1 and/or oriC2 regions. Fully viable H. pylori N6 ΔdnaAH and N6 ΔdnaAL strains with a plasmid-borne dnaA gene (Table 1) were used to ensure that dnaA expression was not changed in case of oriC1 deletion. H. pylori cells were transformed with pTZ57R/TX (X: 1, 2, 3, 4) suicide vectors in order to introduce the gentamicin resistance gene by recombination between homologous fragments. Successful recombinations should lead to deletion of oriC1 (pTZ57R/T 1), oriC2 (pTZ57R/T 2), oriC1 and oriC2 (pTZ57R/T 3) or to exchange of the aph-3 cassette for the gentamicin cassette (pTZ57R/T 4). The first three attempts were lethal for the cells, however, in a small number of transformants the insertion of the gentamicin resistance gene into an unknown chromosomal locus was observed leading to acquisition of the antibiotic resistance without detrimental changes within oriC (sub)regions. Control transformation with pTZ57R/T 4 resulted in desired recombination in case of all obtained clones. The primers used PCR analysis are indicated by arrows. (B) PCR analysis of a representative set of acquired recombinants. Primers used: P-5, P-33 for pTZ57R/TX (X: 1, 2, 3) transformations; P-31, P-33 and P-34, P-32 for pTZ57R/T 4 transformation; primers P33 and P34 hybridized to the chromosomal regions flanking the designed 3′ and 5′ homology arms, respectively. Symbols above lanes correspond to a recombination type (numbers 1–4) and control reactions with a DNA template of the N6 ΔdnaAH unmodified strain (C letter). (C) Table summary of data relevant to this experiment.
RESULTS
In silico prediction of the DUE in the H. pylori replication origin
In previous experiments we showed that in silico predicted H. pylori oriC was specifically bound by DnaA (27,28), but we could not show any DNA unwinding within the identified sequence (data not shown). Therefore we aimed for in silico predictions and employed the WebSIDD tool to localize the DNA-unwinding site within H. pylori oriC. WebSIDD was developed by Bi and Benham to predict SIDD (stress-induced DNA duplex destabilization) sites, i.e. short DNA sequences that are prone to strands opening under negative superhelical stress (33). Although designed for the analysis of superhelicity-responsive promoters in prokaryotes (46), we thought it likely that an origin-specific DUE (34) might be detectable by this prediction method.
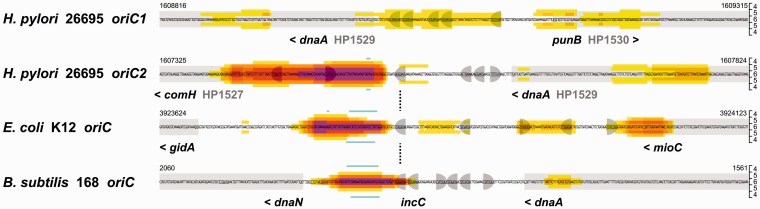
The results of the WebSIDD predictions are shown in Figure 1 (upper 2 panels, see also size-adjustable Supplementary Figure S1). A single strong SIDD site is predicted for the dnaA downstream region (oriC2) with strand-opening at high negative superhelicity in a particularly AT-rich stretch of 64 bp (AT: 81% versus 61% of overall AT content) (Figure 1, see also Figure 4). No significant prediction for an SIDD site was obtained for the dnaA upstream region (oriC1). To address the question whether the predicted SIDD site in oriC2 coincides with the DUE of H. pylori oriC, we performed WebSIDD prediction analyses for oriC of E. coli and the incC (47) region of B. subtilis oriC. The predicted strong SIDD sites in both cases agree well with the sites of DNA unwinding in vitro (16,48) (Figure 1, lower 2 panels). This supports the notion that the WebSIDD prediction is meaningful also for H. pylori oriC2 and suggests that the method can be used to predict the DUE sites of other bacterial chromosomes (CW, manuscript in preparation).
Figure 1.
In silico prediction of the DUE in H. pylori oriC. The heatmaps visualize the WebSIDD predictions for the central 500 bp of the 2.5 kb DNA sequences analyzed. Energy input values (kcal·mol−1) required for strand separation as predicted basewise are shown by the following color code: no color >5, yellow <5, light orange <4, orange <3, dark orange <2, red <1, pink <0 above and below the sequence. Superhelicity values tested from σ = –0.040 (4) to σ = –0.060 (6) in increments of 0.005 are shown as the y-axis on the right of the heatmaps, mirrored on the sequence. Genome position numbering is according to GenBank entries for H. pylori 26695 [AE000511], E. coli K12 W3110 [AP009048] and B. subtilis 168 [AL009126]. Open reading frames are shown as light gray boxes with the assigned gene names (and/or IDs); arrowheads indicate the direction of transcription. DnaA-binding sites are shown within the DNA sequences as gray half-circles, rightward-bound for the consensus TTWTNCACA and leftward-bound for the reverse orientation TGTGNAWAA, respectively, according to Schaper and Messer (1995) (51). Light blue lines above and below the heatmaps indicate the experimentally determined unwound regions; data are from this study for H. pylori oriC2 and taken from Krause et al. (1997) for E. coli oriC and B. subtilis oriC (incC) (16).
Figure 4.
H. pylori oriC1 and oriC2 regions. Coding strand sequence (with respect to the dnaA gene) is presented. The most important features are indicated as follows: genes are distinguished by open boxes, DnaA boxes are shaded in gray, in silico identified DUE is indicated by a dotted line, and the position of the RIP (Figure 5) is marked by a thick triangle. Single and double lines refer to the areas unwound by DnaA in vitro and susceptible to P1 digestion (as determined by comparison of PE results on primers P-9 and P-10, Figure 3) on pori2 and pori1ori2 plasmids, respectively. The position of the region of the highest P1 nuclease sensitivity is similar for both tested plasmids and is indicated as a major cut region.
DnaA-dependent unwinding takes place at oriC2 in vitro and in vivo
In order to experimentally verify the in silico identified H. pylori DUE, two experimental methods were applied: P1 nuclease assay, and RIP mapping. The first allows localization of the DNA sequences unwound upon DnaA binding in vitro, whereas the second maps the start site for chromosome replication at the nucleotide level directly in the cell.
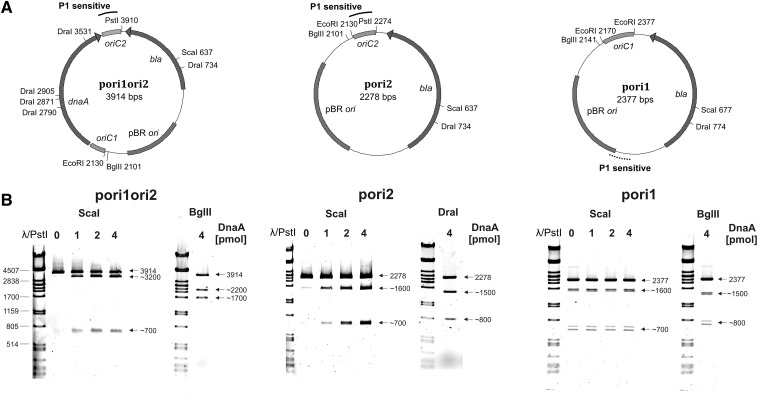
For the P1 nuclease assay we used plasmids containing single (pori1 or pori2) or the two oriC regions separated by the dnaA gene, exactly as on H. pylori 26 695 chromosome (pori1ori2). The supercoiled plasmids were incubated with the increasing amounts of H. pylori DnaA protein and subsequently treated with P1 nuclease, which hydrolyzes single-stranded DNA at the opened helix and hence linearizes the unwound plasmid. The further digestion by ScaI or BglII (pori1ori2 and pori1) and ScaI or DraI (pori2), which cut the plasmids once, excised the DNA fragment from the plasmid and allowed us to approximately determine the region unwound by DnaA (Figure 2). In the case of pori2 and pori1ori2, a DNA fragment of about 0.7 kb was excised by P1 and ScaI, whereas 0.8 and 1.7 kb was excised by P1-DraI and P1-BglII, respectively. The excision was observed only in the presence of DnaA and never occurred outside of oriC2 (Figure 2B). The presence of the two regions, oriC1 and oriC2, probably favors the orisome formation and unwinding at oriC2, because pori1ori2 is melted and the reaction reaches the saturation point at a lower DnaA concentration than pori2 (see also Figure 3). In contrast to oriC2, we could not detect any unwinding within oriC1, in the case of either pori1 or pori1ori2. The size of ScaI- or BglII-excised DNA fragments from pori1 indicated that melting was DnaA-independent and took place within a vector sequence mapping around the plasmid origin (i.e. helically unstable region, Fig. 2). This is consistent with the known phenomena that plasmids may contain helically unstable regions, usually related to origins of replication or transcription units, which, when negatively supercoiled, might undergo strand separation under certain conditions (49).
Figure 2.
In vitro identification of the DUE in H. pylori oriC. (A) Maps of the plasmids used in the P1 nuclease assay. The oriC regions, dnaA, bla, plasmid origin of replication and the positions of the most important restriction sites are marked. The P1 sensitive sites are indicated; the DnaA-dependent unwinding is distinguished from DnaA-independent by solid and dotted lines, respectively. (B) P1 nuclease assay localizing the region unwound by H. pylori DnaA. Supercoiled plasmids were incubated with the indicated amounts of the HpDnaA protein, treated with P1 nuclease and restricted by ScaI. The unwinding site was additionally verified by BglII or DraI digestion. The DNA fragments were analyzed by separation in 1% agarose gel and ethidium bromide staining.
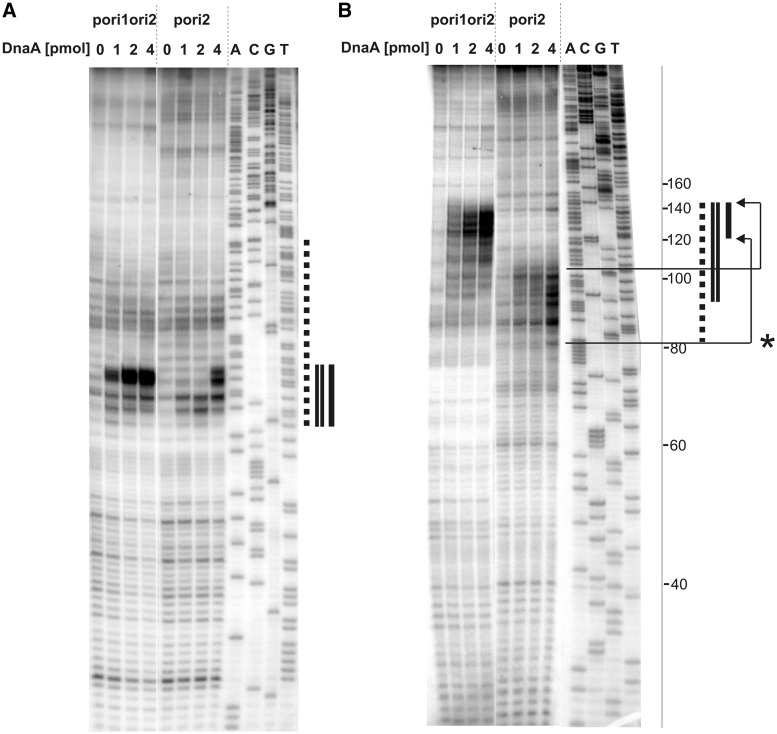
Figure 3.
Determination of the H. pylori oriC sequence unwound by DnaA in vitro. Plasmid DNA, after incubation with the indicated amounts of the DnaA protein and P1 nuclease treatment, was used as a substrate for PE analysis. 32P labeled primers P-9 and P-10 were complementary to the coding strand (with respect to the dnaA gene) (A) and non-coding strand (B), respectively. Dotted line corresponds to the AT-rich region identified in silico as a DUE. Single and double lines refer to the areas susceptible to P1 nuclease digestion in pori2 and pori1ori2 plasmids, respectively. A, C, G, T sequencing reactions were carried out with 32P labeled primers P-9 (A), P-10 (B) and the pori1ori2 plasmid DNA. The ladder on the right side of the figure corresponds to the distance from P-10 primer annealing site on pori1ori2. The P-10 PE product on pori2 is 40 bps shorter than that on pori1ori2 (Supplementary Figure S4). Thus to determine the position of the unwound region on pori2 it is necessary to add 40 bp to the observed PE bands positions; * - arrows correspond to the actual position of the pori2 unwound region within oriC2.
The above experiments proved that H. pylori DnaA unwinds DNA exclusively within oriC2. To precisely determine the unwound region, the P1-digested plasmids were subjected to PE using 32P labeled primers P-9, P-10 and P-35 (Figure 3, Supplementary Figures S2–S4 and Table 2). The primers, which hybridize to the template DNA 90 bp (P-9 on pori1ori2 and pori2, P-10 on pori2) and 130 bp (P-10 on pori1ori2) away from the in silico predicted DUE region or 57 bp from box1 of oriC1 (P-35 on pori1ori2), are extended by Taq polymerase until they reach P1 digested DNA. The observed extension products (Figure 3 and Supplementary Figure S3) confirm that DNA unwinding occurs at oriC2, which corresponds well with the predicted DUE sequence and allow one to estimate the unwound region of pori1ori2 and pori2 for about 20 bp (Figures 3 and 4). In the case of pori1ori2, the unwound region is broader, extending in the 3′ direction and reaching up to 52 bp. The fewer unwound base pairs in the case of pori2 might suggest that the interaction with the two oriC subregions, when compared with the single oriC2 origin binding, changes the oligomerization mode of DnaA, and leads to formation of a larger open complex possibly meeting the requirements for loading of other initiation proteins. The increased intensity of the extension products corresponds with the increasing concentration of DnaA and confirms that the reaction is strictly DnaA dependent. Moreover, the intensity of the extension products on pori1ori2 is higher than that on pori2, suggesting that the presence of the two oriC sites on one plasmid favors the orisome formation and the unwinding reaction.
To prove that the unwinding in vivo occurs at the identified DUE, we performed RIP analysis, which allows determination of the initiation sites in non-synchronized cultures (50). The chromosomal DNA was extracted from exponentially growing H. pylori cells and the replication intermediates were enriched on BND-cellulose (see Materials and Methods). Series of subsequent enzymatic reactions selectively separated nascent strands from nicked DNA and prepared the template for the PE reaction. The shortest extension product should represent the DNA fragment amplified on a leading strand starting at the unwinding site. The comparison of the lengths of the obtained PE products with the control sequencing reactions indicated that the replication initiation starts within oriC2 and co-localizes with the identified in vitro DUE region (Figure 5, see also Figure 4 and Supplementary Figure S4).
Figure 5.
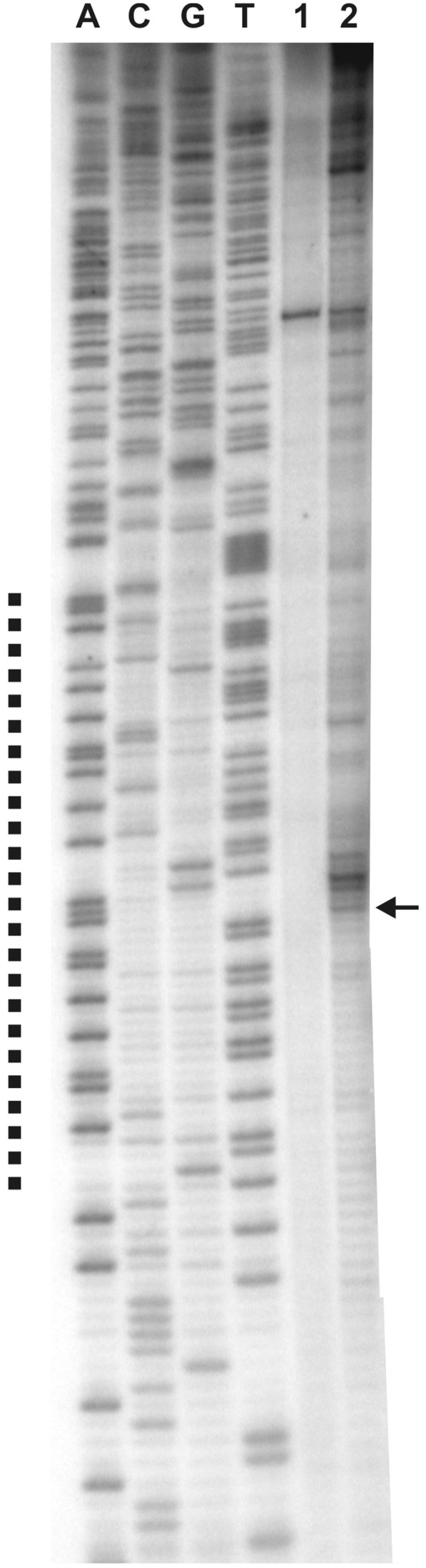
Mapping of the RIP in vivo. PE was performed with enriched replication intermediates and 32P labeled oligonucleotide P-9 complementary to the coding strand (with respect to the dnaA gene). Dotted line corresponds to the AT-rich region, identified in silico as a DUE. Arrow indicates the transition point between continuous and discontinuous DNA synthesis. 0.3 μg (lane 1) and 1.2 μg (lane 2) of a template DNA was used; A, C, G, T sequencing reactions were carried out with 32P labeled primer P-9 and the pori1ori2 plasmid DNA.
The results obtained by independent experimental methods clearly demonstrated that the DNA is unwound in the predicted downstream dnaA region; thus we postulate that the H. pylori chromosome replication starts in oriC2.
DnaA binds only supercoiled oriC2
It has been shown previously that HpDnaA exhibits the highest affinity for ‘strong’ TCATTCACA DnaA box and especially for ‘twin’ DnaA boxes in oriC1 (boxes 2–3 and 4–5, Figure 4) (27–29). The oriC2 region is characterized by the presence of a few putative DnaA boxes matching the consensus E. coli TTWTNCACA sequence (51) (Figures 1 and 4); however, only one TCATTCACT resembles a strong H. pylori DnaA box (28). In order to analyze DnaA binding to oriC2 and compare the results with DnaA-oriC1 interactions, we performed SPR, gel shift and EM analyses.
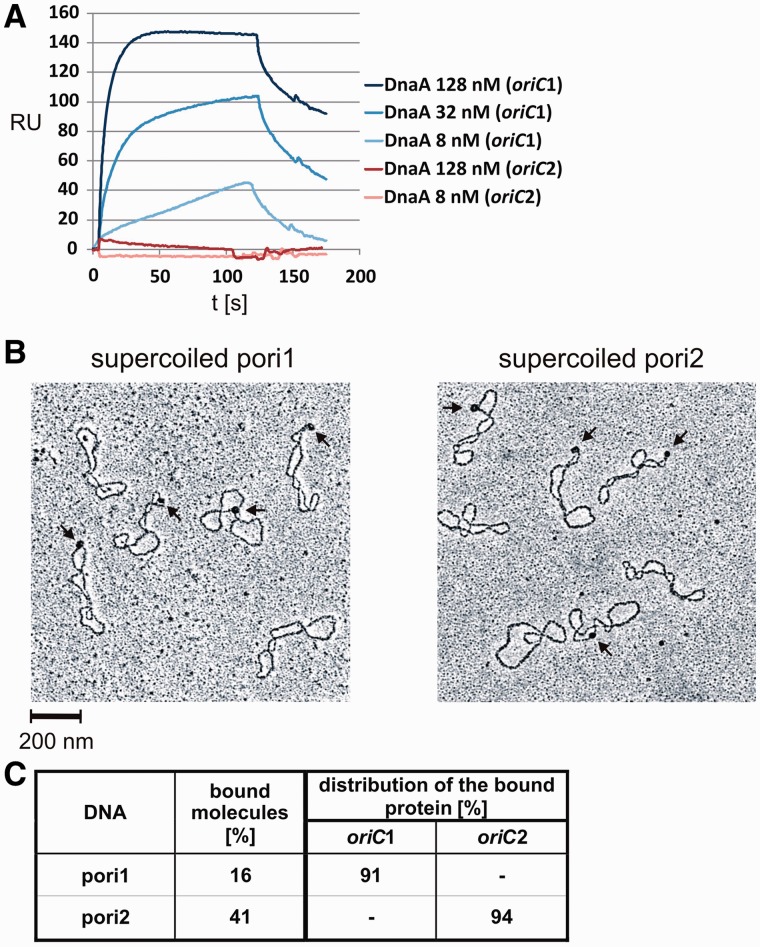
The SPR excluded interactions between DnaA and oriC2, while it confirmed previously characterized DnaA binding to oriC1 (Figure 6A and (30)). Similar results were obtained by gel shifts (data not shown). Moreover, no oriC2 binding was observed with the E. coli DnaA protein, which interacted with H. pylori oriC1 region (data not shown and (29)). The results were contradictory to the unwinding experiments in which we observed DnaA-dependent DNA melting. However, it has to be noted that, in contrast to SPR and gel shift, in all unwinding experiments supercoiled oriC plasmids were used. Therefore, we applied EM to visualize the interaction between supercoiled oriC2 and DnaA. The supercoiled pori1 and pori2 plasmids (Table 1) were incubated with DnaA under similar conditions as in the unwinding assay. The complexes were fixed by glutaraldehyde and then visualized by EM. The analysis revealed the interactions between DnaA and each of the plasmids (Figure 6B). Only one DnaA complex was observed on one supercoiled plasmid molecule. The binding of DnaA to oriC1 and oriC2 was further confirmed by ScaI digestion and measuring the distance between the bound protein and one of the plasmid ends, similarly as described by Krause et al. (16) (data not shown, see also below). The complexes were visible at about 500 nm from the proximal plasmid end which corresponds to the loci of oriC1 or oriC2 sites on the respective plasmids (700 bp from ScaI site, see Figure 2). When similar analyses were performed with linear DNA (PCR product or linearized plasmids) the HpDnaA binding to oriC1 was confirmed while no complexes were visible on oriC2 (data not shown and Figure 7C). This clearly shows that oriC2 is bound only when supercoiled, while DnaA binds to linear and supercoiled oriC1. Interestingly, the affinity of DnaA for supercoiled oriC1 was lower than for oriC2, since about 16% of pori1 and 41% of pori2 molecules were bound by DnaA (Figure 6C). Thus, these experiments show that superhelicity is the crucial factor enabling specific and high-affinity DnaA-oriC2 interactions (Figure 6C) leading to DNA unwinding (Supplementary Figure S2).
Figure 6.
Comparative analysis of the DnaA interaction with oriC1 and oriC2 regions. (A) SPR analysis. Protein concentrations and the types of DNA fragments used in the analysis are given in the legend of the sensogram. (B) EM study of the DnaA binding to the supercoiled pori1 and pori2 plasmids. (C) Statistical distribution and the level of DnaA binding to DNA molecules used in EM. The percentage of bound molecules was calculated on the basis of 250 molecules analyzed. Distribution of complexes on the DNA was evaluated by measuring 120 bound DNA molecules for each plasmid; data refer only to the molecules bound within oriC1 and/or oriC2.
Figure 7.
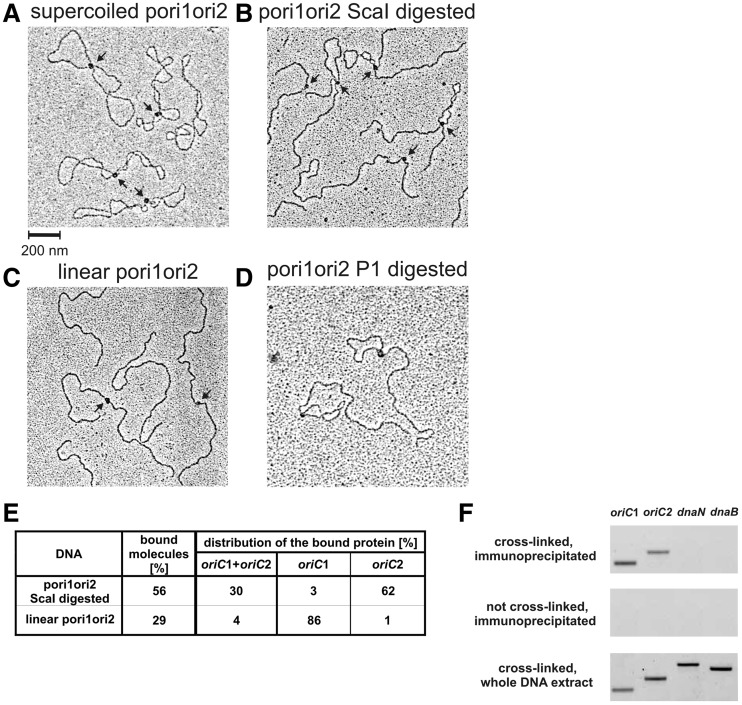
In vitro and in vivo analyses of DnaA binding to oriC1–oriC2 region. (A) DnaA interaction with supercoiled pori1ori2. The putative interactions between DnaA-oriC1 and DnaA-oriC2 subcomplexes are visible as DNA loops. (B) The localization of the complexes at oriC1 and oriC2 as well as the interactions between DnaA bound to oriC1 and oriC2 were confirmed by ScaI digestion and subsequent measurements. (C) DnaA interaction with linear pori1ori2. No loop structures were visible and the DnaA interacted almost exclusively with oriC1. (D) DnaA interaction with oriC1 and oriC2 regions on pori1ori2 and subsequent P1 nuclease digestion. The DnaA-oriC2 complex is formed between dnaA and the unwound DUE region, thus the P1 digestion allows to maintain the loop between oriC1-DnaA and oriC2-DnaA subcomplexes. (E) Statistical distribution and the level of DnaA binding to DNA molecules observed by EM. The percentage of bound molecules was calculated on the basis of 250 molecules analyzed. Distribution of complexes on the DNA was evaluated by measuring 120 bound DNA molecules for each plasmid; data refer only to the molecules bound within oriC1 and/or oriC2. (F) In vivo immunoprecipitation of H. pylori genomic DNA with a purified anti-DnaA antibody followed by PCR analysis. Amplified regions (dnaN and dnaB served as negative controls) and DNA templates used are indicated on the picture.
oriC1 and oriC2 are both important for initiation
The presented results revealed that H. pylori DnaA binds and unwinds the DNA within supercoiled oriC2. DnaA also interacts with linear and supercoiled oriC1 (Figures 6A and 6B and (29)); however, the presence of oriC1 is not required for DNA unwinding within oriC2 (Figure 2B). Additionally, the incidence of DnaA binding to supercoiled pori1 is lower compared with pori2 (Figure 6C), suggesting higher DnaA affinity for supercoiled oriC2 than for supercoiled oriC1. The question arose if and why in H. pylori both regions are required and what is the exact structure of H. pylori origin.
To analyze DnaA binding to oriC1 and oriC2, EM and immunoprecipitation were performed. As observed by EM, the incubation of DnaA with supercoiled pori1ori2 led to formation of a single-nucleoprotein complex per plasmid molecule (Figure 7A). The distance measurements between the plasmid ends and the protein core on ScaI digested nucleoprotein complexes confirmed the binding of DnaA to oriC1 and/or oriC2 (Figures 7B and 7E). By P1 digestion of the pori1ori2-DnaA complex we additionally showed, that the protein core of the DnaA-oriC2 subcomplex is localized upstream from the unwound DUE site (Figure 7D). The pori1ori2 was bound 1.4 and 3.5 times more frequently than pori2 and pori1 plasmids, respectively (Figures 6C and 7E), which shows that the presence of both regions enhanced DnaA binding to plasmid molecules. 65% of the complexes were formed only at one of the sites: 3% at oriC1 and 62% at oriC2, which confirmed higher DnaA affinity for supercoiled oriC2 than for oriC1. 30% of the complexes were visible as protein cores joining the two DNA strands (Figure 7A and B). The looped DNA, preserved after ScaI digestion, indicated that the two strands of a plasmid were indeed joined via DnaA-oriC1 and DnaA-oriC2 sub-complexes, similarly as in B. subtilis (16). However, unlike in B. subtilis, the loop formation was observed only when oriC1 and oriC2 were located on a supercoiled plasmid (Figure 7).
In order to analyze whether the two regions were also bound by DnaA in H. pylori cells, immunoprecipitation was performed. Exponentially growing 26 695 H. pylori cells were crosslinked with formaldehyde, sonicated and the DnaA–DNA complexes were immunoprecipitated with purified anti-DnaA antibodies. The immunoprecipitated DNA was identified by PCR reactions with pairs of primers amplifying oriC1, oriC2 and the intergenic regions of dnaB and dnaN genes as negative controls (Figure 7F). The PCR products indicated that oriC1 and oriC2 were specifically bound by DnaA, while comparable intensity of the oriC1 and oriC2 amplicons suggested equal binding by the initiator protein. Since the oriC1 DnaA box cluster is located in the promoter region of dnaA, the question arose whether oriC1 is indeed necessary for replication or is used rather for autoregulation of dnaA expression. To analyze whether both oriCs are important for initiation, a comprehensive oriC deletion analysis was performed. To distinguish the role of oriC1 in initiation and/or in transcription regulation, we used H. pylori N6 ΔdnaAH and N6 ΔdnaAL strains (Table 1), in which DnaA was ectopically synthesized at different expression levels (L, low, H, high) from the pILL2157 or pILL2150 vectors, respectively, and therefore independent from its own promoter (35,32). The pTZ57R/TX (X: 1, 2, 3) plasmids were constructed (Table 1) to allow deletion of oriC1, oriC2 or both regions simultaneously. In three independent transformations we were not able to delete any of the oriC regions (Figure 8). The control transformations with pTZ57R/T4 plasmid allowed exchange of kanamycin resistance cassette for gentamicin resistance cassette located between the two oriC sites, which confirmed that the inability to delete oriC1, oriC2 or oriC1–oriC2 regions was not due to transformation problems but was connected with the oriC function in H. pylori cells.
The performed experiments allowed us to conclude that DnaA binds oriC1 and oriC2 in vitro and in vivo, and that both regions are important for initiation of H. pylori chromosome replication.
DISCUSSION
The replication of a bacterial chromosome starts at a single chromosomal region (oriC) and is initiated by DnaA protein. Though the principle of initiation is similar in almost all bacteria, the oriC structure as well as the composition and architecture of nucleoprotein initiation complexes usually differ between unrelated species. In this work, we present new data concerning the initiation of H. pylori chromosome replication. Our experiments revealed the bipartite structure of H. pylori oriC; besides previously characterized oriC1, localized in the upstream region of dnaA, we identified the second oriC2, situated downstream of dnaA, which is subjected to DnaA-dependent unwinding in vitro (Figures 2B and 3) and was proven to be the replication initiation site in vivo (Figure 5). Neither oriC1 nor oriC2 can be deleted from H. pylori cells and both of them are bound by DnaA in vivo, which presumably leads to the loop formation (Figure 7B). Surprisingly, oriC2 is bound exclusively as a supercoiled DNA, indicating the importance of the DNA topology in the replication initiation. The recognition of DNA with respect to the particular sequence and topology revealed a new, so far hardly characterized feature of DnaA protein.
oriC2 supercoiling determines DnaA binding and subsequent unwinding
Our previous analysis identified the DnaA box cluster localized on the H. pylori chromosome, which, on the basis of the in vitro and in silico analyses, was assumed to be the H. pylori oriC (here named oriC1) (28,27). Despite many attempts, we were unable to detect DNA unwinding within oriC1. Here we showed that the DNA is in fact unwound within the neighboring region oriC2. Surprisingly, we discovered that oriC2 was bound by DnaA exclusively when present on a supercoiled plasmid, which proved that direct interaction between supercoiled oriC2 and DnaA leads to the DNA unwinding. In contrast, the E. coli DnaA binds oriC regardless of its topology (52). The recent data indicate that the E. coli initiator can unwind not only supercoiled but also the linear oriC (53). Thus, our studies for the first time directly showed that eubacterial DnaA is dependent on DNA topology for binding to oriC—a phenomenon reported previously only for initiator-ori binding in Archaea and higher eukaryotes (metazoans). In Archaea it was recently shown that apart from the ORB sequence, the Orc1/Cdc6 initiator also recognizes the local DNA structure (54). Metazoan ORC exhibit higher affinity for the supercoiled origin than for similar DNA sequences in linear form, and the clear lack of sequence conservation between known origins was already demonstrated (55,56). The H. pylori DnaA-oriC2 binding resembles that of archaeal Orc1/Cdc6-ori binding—it is both sequence- (i.e. localized within oriC2) and topology-specific. Thus, we conclude that topology-dependent recognition of DNA is not only restricted to archaeal and metazoan initiators, as suggested by Dueber et al. (54), but might be common in all three domains of life. All the initiators share the AAA+ domain with the Initiator-Specific Motif (ISM), which was suggested to participate in DNA structure recognition (7). In bacteria the ISM is engaged in ssDNA binding (57,53), but it cannot be excluded that it also recognizes dsDNA and its structure. In the future, we plan to identify a motif in the H. pylori DnaA that is responsible for topology-dependent recognition of DNA sequences within oriC2. More generally, our study demonstrated the need to reevaluate DnaA sensitivity toward DNA topology for orisome formation and function in other bacteria.
Bipartite structure of H. pylori oriC
Bacterial oriC regions, characterized by the presence of the DBC and the DUE, are often situated at the 3′ or 5′ regions of dnaA. However, in some Gram-positive bacteria, the two DBCs flank the dnaA gene. Both clusters can be indispensible for oriC activity (B. subtilis (13,14) and Mycoplasma pulmonis (15)) or one, the 5′ box cluster, is involved in autoregulation of dnaA expression whereas the second, the 3′ box cluster, serves as the oriC region (Streptomyces (58), Spiroplasma citri (13), Mycobacterium (18)). The two DnaA-box clusters were also reported to act as origins in Gram-negative Pseudomonas sp., however no mutual relationship between them have been established and only one of the origins (oriCI) was finally shown to be indispensible in vivo (59,60). We identified the two DnaA interaction regions on the H. pylori chromosome, oriC1 and oriC2, located in the 5′ and 3′ regions of the dnaA gene, respectively. Both are bound by DnaA in vitro and in vivo and neither of them can be deleted from the H. pylori chromosome, which suggested the discontinuous structure of the H. pylori oriC—the first bipartite origin discovered in a Gram-negative bacterium. The DUE is located within oriC2 and in vitro it does not require oriC1 for DnaA-dependent unwinding. The question arose then why H. pylori requires oriC1. Since the oriC1 DnaA cluster is located in the dnaA promoter region, it could be involved in regulation of dnaA expression. However, first, the DnaA level is invariant in growing and not growing H. pylori, and second, in contrast to some other bacteria (61,62), the alterations in cellular level of the DnaA protein have no effect on frequency of initiation replication in H. pylori strains expressing dnaA from inducible plasmids ((35,32), Table 1 and data not shown). Moreover, the oriC1 region cannot be deleted in N6 ΔdnaAH strain ectopically expressing dnaA from a plasmid (Figure 8, see also Table 1). Thus, we postulate that the oriC1 region exerts presumably only a marginal influence on dnaA expression but is indispensable for the origin activity and that both regions, oriC1 and oriC2, constitute the integral origin of H. pylori chromosome replication. The reason why some of the bacterial and plasmid origins are split into two parts is still not fully understood, but it is probably a way to control the initiation events, possibly through loop formation. Loop formation is important for many cellular processes, including initiation of DNA replication (63,64). However, the significance of this phenomenon is still unclear. It may generate cooperativity in the binding of the initiator protein, help to stabilize the nucleoprotein complex or provide the entry site for proteins regulating initiation of DNA replication (64). In Streptomyces lividans the observed loop formation between the two DBCs separated by a non-coding 134 bp region was suggested to be important for proper orisome formation (65). In B. subtilis the DNA looping between incA/B and incC DBCs was suggested either to be used for a coupled control for dnaA expression and the initiation of replication or, similarly as in P1, F factor and R6K plasmid or Epstein–Barr virus (EBV) viral origins constitute regulatory structures for initiation control by association of normally physically separated DnaA box regions ((16,14) and references herein). The initiator-mediated interaction between the two clusters of the initiator binding sites (DnaA boxes or iterons), either by looping (if positioned in cis) or by handcuffing (when placed in trans), are suggested to be the mechanism of negative regulation of chromosomal and plasmid replication. Interestingly, we have previously shown that HobA, the DiaA-related protein (30), can bridge the interaction between the two DnaA-oriC1 complexes (32). Particular χ structures have been observed in EM in which HobA mediated the cross-interaction. Our preliminary results suggest that HobA also increases the bridging between DnaA-oriC1 and DnaA-oriC2 subcomplexes, but we did not observe any significant influence on DNA unwinding (data not shown). On the other hand, HobA stimulates DnaA-oriC1 binding, and was suggested to be an activator of the initiation process (30). It is possible that HobA rearranges the orisome structure, but further studies are necessary to elucidate its role in the initiation of H. pylori chromosome replication.
The bipartite structure of the bacterial oriC region raises an intriguing question of whether and how the self-assembly of DnaA into filaments (66,57) is required for the formation of a functional orisome. Recent studies indicated that in B. subtilis inhibition of DnaA helix formation by Soj (an ortholog of ParA) stalls initiation of replication (67). Whether at the certain stage(s) of replication initiation the H. pylori DnaA protein forms filament, and more generally, what is the topology of such complexes with the bipartite oriC, is still not known and need to be elucidated in future.
In summary, our analyses showed that H. pylori oriC exhibits bipartite structure, being the first such origin discovered in a Gram-negative bacterium. Together with previously identified bipartite oriCs in Gram-positive bacteria and plasmids, it might represent a larger group of origins controlled by similar regulatory strategies. Our work also showed for the first time the direct relationship between DnaA initiator and the topology of its target sequence. This suggests that the DNA structure might be an important factor controlling DNA replication in three domains of life.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Funding for open access charge: Ministry of Science and Higher Education [project N N301 029 334]. J.Z.C. and R.D. acknowledge support by MISTRZ and A.Z.P. by the PARENT-Bridge programs of the Foundation for Polish Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Magdalena Felczak for helpful comments about the P1 nuclease test, Hilde de Reuse for pUC1813Gm and Kerstin Stingl for critical remarks concerning the manuscript. C.W. thanks David Ussery for suggesting use of the WebSIDD server for the prediction of DNA-unwinding elements in replication origins.
REFERENCES
- 1.Nielsen O, Løbner-Olesen A. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 2008;9:151–156. doi: 10.1038/sj.embor.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diffley JFX. The many faces of redundancy in DNA replication control. Cold Spring Harb. Symp. Quant. Biol. 2010;75:135–142. doi: 10.1101/sqb.2010.75.062. [DOI] [PubMed] [Google Scholar]
- 3.Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami H, Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem. Cell Biol. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- 6.Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 7.Ozaki S, Katayama T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009;62:71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 2010;13:766–772. doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 10.Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakrzewska-Czerwińska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 12.Jakimowicz D, Majka J, Messer W, Speck C, Fernandez M, Martin MC, Sanchez J, Schauwecker F, Keller U, Schrempf H, et al. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology (Reading, Engl.) 1998;144(Pt. 5):1281–1290. doi: 10.1099/00221287-144-5-1281. [DOI] [PubMed] [Google Scholar]
- 13.Lartigue C, Blanchard A, Renaudin J, Thiaucourt F, Sirand-Pugnet P. Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res. 2003;31:6610–6618. doi: 10.1093/nar/gkg848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriya S, Imai Y, Hassan AK, Ogasawara N. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid. 1999;41:17–29. doi: 10.1006/plas.1998.1381. [DOI] [PubMed] [Google Scholar]
- 15.Cordova CMM, Lartigue C, Sirand-Pugnet P, Renaudin J, Cunha RAF, Blanchard A. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J. Bacteriol. 2002;184:5426–5435. doi: 10.1128/JB.184.19.5426-5435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause M, Rückert B, Lurz R, Messer W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 1997;274:365–380. doi: 10.1006/jmbi.1997.1404. [DOI] [PubMed] [Google Scholar]
- 17.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Farhana A, Hasnain SE. In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS ONE. 2009;4:e4139. doi: 10.1371/journal.pone.0004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sernova NV, Gelfand MS. Identification of replication origins in prokaryotic genomes. Brief. Bioinformatics. 2008;9:376–391. doi: 10.1093/bib/bbn031. [DOI] [PubMed] [Google Scholar]
- 20.Rajewska M, Wegrzyn K, Konieczny I. AT-rich region and repeated sequences – the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 2011 doi: 10.1111/j.1574-6976.2011.00300.x. 10, 1111/j.1574-6976.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- 21.Nozaki S, Yamada Y, Ogawa T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells. 2009;14:329–341. doi: 10.1111/j.1365-2443.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 22.Smulczyk-Krawczyszyn A, Jakimowicz D, Ruban-Osmialowska B, Zawilak-Pawlik A, Majka J, Chater K, Zakrzewska-Czerwinska J. Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: from in silico to in vivo studies. J. Bacteriol. 2006;188:6184–6194. doi: 10.1128/JB.00528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okumura H, Yoshimura M, Ueki M, Oshima T, Ogasawara N, Ishikawa S. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 2011;40:220–234. doi: 10.1093/nar/gkr716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakimowicz D, Majka J, Lis B, Konopa G, Wegrzyn G, Messer W, Schrempf H, Zakrzewska-Czerwinska J. Structure and regulation of the dnaA promoter region in three Streptomyces species. Mol. Gen. Genet. 2000;262:1093–1102. doi: 10.1007/pl00008652. [DOI] [PubMed] [Google Scholar]
- 25.Salazar L, Guerrero E, Casart Y, Turcios L, Bartoli F. Transcription analysis of the dnaA gene and oriC region of the chromosome of Mycobacterium smegmatis and Mycobacterium bovis BCG, and its regulation by the DnaA protein. Microbiology (Reading, Engl.) 2003;149:773–784. doi: 10.1099/mic.0.25832-0. [DOI] [PubMed] [Google Scholar]
- 26.Messer W, Weigel C. DnaA as a transcription regulator. Meth. Enzymol. 2003;370:338–349. doi: 10.1016/S0076-6879(03)70030-5. [DOI] [PubMed] [Google Scholar]
- 27.Zawilak A, Cebrat S, Mackiewicz P, Król-Hulewicz A, Jakimowicz D, Messer W, Gosciniak G, Zakrzewska-Czerwinska J. Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 2001;29:2251–2259. doi: 10.1093/nar/29.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zawilak A, Durrant MC, Jakimowicz P, Backert S, Zakrzewska-Czerwińska J. DNA binding specificity of the replication initiator protein, DnaA from Helicobacter pylori. J. Mol. Biol. 2003;334:933–947. doi: 10.1016/j.jmb.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Zawilak-Pawlik A, Kois A, Majka J, Jakimowicz D, Smulczyk-Krawczyszyn A, Messer W, Zakrzewska-Czerwińska J. Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 2005;389:471–481. doi: 10.1042/BJ20050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zawilak-Pawlik A, Donczew R, Szafrański S, Mackiewicz P, Terradot L, Zakrzewska-Czerwińska J. DiaA/HobA and DnaA: a pair of proteins co-evolved to cooperate during bacterial orisome assembly. J. Mol. Biol. 2011;408:238–251. doi: 10.1016/j.jmb.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 31.Natrajan G, Noirot-Gros MF, Zawilak-Pawlik A, Kapp U, Terradot L. The structure of a DnaA/HobA complex from Helicobacter pylori provides insight into regulation of DNA replication in bacteria. Proc. Natl Acad. Sci. USA. 2009;106:21115–21120. doi: 10.1073/pnas.0908966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zawilak-Pawlik A, Kois A, Stingl K, Boneca IG, Skrobuk P, Piotr J, Lurz R, Zakrzewska-Czerwińska J, Labigne A. HobA – a novel protein involved in initiation of chromosomal replication in Helicobacter pylori. Mol. Microbiol. 2007;65:979–994. doi: 10.1111/j.1365-2958.2007.05853.x. [DOI] [PubMed] [Google Scholar]
- 33.Bi C, Benham CJ. WebSIDD: server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinformatics. 2004;20:1477–1479. doi: 10.1093/bioinformatics/bth304. [DOI] [PubMed] [Google Scholar]
- 34.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boneca IG, Ecobichon C, Chaput C, Mathieu A, Guadagnini S, Prévost M-C, Colland F, Labigne A, de Reuse H. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 2008;74:2095–2102. doi: 10.1128/AEM.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 37.Sasse-Dwight S, Gralla JD. Footprinting protein-DNA complexes in vivo. Meth. Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 38.Bielinsky AK, Gerbi SA. Chromosomal ARS1 has a single leading strand start site. Mol. Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 39.Gerbi SA, Bielinsky AK. Replication initiation point mapping. Methods. 1997;13:271–280. doi: 10.1006/meth.1997.0526. [DOI] [PubMed] [Google Scholar]
- 40.Matsunaga F, Norais C, Forterre P, Myllykallio H. Identification of short “eukaryotic” Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl Acad. Sci. USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiess E, Lurz R. 13 Electron Microscopic Analysis of Nucleic Acids and Nucleic Acid-Protein Complexes. Vol. 20. Academic Press; 1988. pp. 293–323. [Google Scholar]
- 43.Jakimowicz D, Chater K, Zakrzewska-Czerwínska J. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 2002;45:1365–1377. doi: 10.1046/j.1365-2958.2002.03102.x. [DOI] [PubMed] [Google Scholar]
- 44.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 2003;38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 45.Bury-Moné S, Skouloubris S, Dauga C, Thiberge J-M, Dailidiene D, Berg DE, Labigne A, De Reuse H. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 2003;71:5613–5622. doi: 10.1128/IAI.71.10.5613-5622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opel ML, Aeling KA, Holmes WM, Johnson RC, Benham CJ, Hatfield GW. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 2004;53:665–674. doi: 10.1111/j.1365-2958.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- 47.Fukuoka T, Moriya S, Yoshikawa H, Ogasawara N. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 1990;107:732–739. doi: 10.1093/oxfordjournals.jbchem.a123117. [DOI] [PubMed] [Google Scholar]
- 48.Bramhill D, Kornberg A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 49.Kowalski D, Natale DA, Eddy MJ. Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc. Natl Acad. Sci. USA. 1988;85:9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bielinsky AK, Gerbi SA. Where it all starts: eukaryotic origins of DNA replication. J. Cell. Sci. 2001;114: 643–651. doi: 10.1242/jcs.114.4.643. [DOI] [PubMed] [Google Scholar]
- 51.Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995;270:17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 52.Weigel C, Schmidt A, Rückert B, Lurz R, Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16:6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2011;40:1648–1665. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dueber EC, Costa A, Corn JE, Bell SD, Berger JM. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 2011;39:3621–3631. doi: 10.1093/nar/gkq1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houchens CR, Lu W, Chuang R-Y, Frattini MG, Fuller A, Simancek P, Kelly TJ. Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex-DNA interactions. J. Biol. Chem. 2008;283:30216–30224. doi: 10.1074/jbc.M802649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zakrzewska-Czerwińska J, Majka J, Schrempf H. Minimal requirements of the Streptomyces lividans 66 oriC region and its transcriptional and translational activities. J. Bacteriol. 1995;177:4765–4771. doi: 10.1128/jb.177.16.4765-4771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yee TW, Smith DW. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl Acad. Sci. USA. 1990;87:1278–1282. doi: 10.1073/pnas.87.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y, Yao S, Helinski D, Toukdarian A. Functional analysis of two putative chromosomal replication origins from Pseudomonas aeruginosa. Plasmid. 2006;55:194–200. doi: 10.1016/j.plasmid.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Løbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- 62.Greendyke R, Rajagopalan M, Parish T, Madiraju MVVS. Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology (Reading, Engl.) 2002;148:3887–3900. doi: 10.1099/00221287-148-12-3887. [DOI] [PubMed] [Google Scholar]
- 63.Matthews KS. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 65.Jakimowicz D, Majka J, Konopa G, Wegrzyn G, Messer W, Schrempf H, Zakrzewska-Czerwińska J. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 2000;298:351–364. doi: 10.1006/jmbi.2000.3686. [DOI] [PubMed] [Google Scholar]
- 66.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 67.Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31:1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388: 539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 69.Messer W, Hartmann-Kühlein H, Langer U, Mahlow E, Roth A, Schaper S, Urmoneit B, Woelker B. The complex for replication initiation of Escherichia coli. Chromosoma. 1992;102:S1–S6. doi: 10.1007/BF02451779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.