Abstract
The majority of Cockayne syndrome (CS) patients carry a mutation in Cockayne Syndrome group B (CSB), a large nuclear protein implicated in DNA repair, transcription and chromatin remodeling. However, whether CSB may play a role in telomere metabolism has not yet been characterized. Here, we report that CSB physically interacts with TRF2, a duplex telomeric DNA binding protein essential for telomere protection. We find that CSB localizes at a small subset of human telomeres and that it is required for preventing the formation of telomere dysfunction-induced foci (TIF) in CS cells. We find that CS cells or CSB knockdown cells accumulate telomere doublets, the suppression of which requires CSB. We find that overexpression of CSB in CS cells promotes telomerase-dependent telomere lengthening, a phenotype that is associated with a decrease in the amount of telomere-bound TRF1, a negative mediator of telomere length maintenance. Furthermore, we show that CS cells or CSB knockdown cells exhibit misregulation of TERRA, a large non-coding telomere repeat-containing RNA important for telomere maintenance. Taken together, these results suggest that CSB is required for maintaining the homeostatic level of TERRA, telomere length and integrity. These results further imply that CS patients carrying CSB mutations may be defective in telomere maintenance.
INTRODUCTION
Telomeres are heterochromatic structures found at the ends of linear eukaryotic chromosomes. Mammalian telomeric DNA consists of tandem repeats of TTAGGG that are bound by a telomere-specific complex known as shelterin/telosome (1–3). Shelterin, composed of six protein subunits, including TRF1, TRF2, TIN2, hRap1, TPP1 and POT1, functions not only to regulate telomere length maintenance but also to protect natural chromosome ends from being recognized as damaged DNA (1,2,4). Telomeric DNA has been shown to be transcribed into a large non-coding telomere repeat-containing RNA (5), referred to as TERRA, which is implicated in maintaining the integrity of telomere heterochromatin (5,6). Disruption of the shelterin complex or the telomere heterochromatic state can lead to induction of telomere abnormalities, including telomere end-to-end fusions, telomere loss and telomere doublets/fragile telomeres (1,2,6). These dysfunctional telomeres have been shown to be associated with DNA damage response factors, such as γH2AX and 53BP1, resulting in the formation of nuclear structures that are referred to as telomere dysfunction-induced foci (TIF) (7–10).
TRF2 is one of the two shelterin subunits that bind specifically to duplex telomeric DNA (11,12), the other being TRF1 (13). Overexpression of TRF1 leads to telomere shortening, whereas removal of TRF1 from telomeres promotes telomerase-dependent telomere lengthening (14–16), implying that TRF1 may restrict the access of telomerase to the ends of telomeres.
While TRF1 has been implicated in telomere length maintenance, TRF2 is best known for its role in telomere protection. TRF2 contains a N-terminal basic domain, a central TRF homology (TRFH) domain and a C-terminal Myb-like DNA binding domain (11,12). The N-terminal basic domain is rich in glycine and arginine residues, also referred to as a GAR domain. The TRFH domain of TRF2 not only mediates homo-dimerization but also acts as a protein interaction platform at telomeres to recruit additional shelterin subunits and other accessory proteins (17,18). Removal of TRF2 from telomeres either by conditional knockout or overexpression of a dominant-negative allele of TRF2 lacking both the N-terminal basic/GAR domain and the C-terminal Myb-like DNA binding domain promotes telomere end-to-end fusions (19,20). Overexpression of TRF2 lacking its N-terminal basic/GAR domain promotes telomere loss (8), whereas overexpression of TRF2 carrying amino acid substitutions in the same basic/GAR domain induces the formation of telomere doublets (10).
Cockayne syndrome (CS) is a rare human hereditary disorder characterized by severe postnatal growth failure, progressive neurological degeneration and segmental premature aging, including sensorineural hearing loss, retinal degeneration and loss of subcutaneous fat (21,22). CS patients show hypersensitivity to UV light and the average life span of CS patients is ∼12 years (23–25). Although five genes have been identified to be responsible for the disease, including CSA, CSB, XPB, XPD and XPG, the majority of CS patients carry a defect in the CSB gene (21,22,25).
Cockayne Syndrome group B (CSB) protein, also known as ERCC6, is a nuclear protein of 1493 amino acids in length, containing several distinct domains, including an acidic domain, a glycine rich domain, a SWI/SNF-like ATPase domain, a nucleotide binding (NTB) domain and a ubiquitin binding domain (UBD) (Figure 1A) (21,26–28). CSB has been shown to play a key role in transcription-coupled repair (21,29), a subpathway of nucleotide excision repair (NER) responsible for removing bulky lesions such as UV-induced DNA damage (cyclobutane pyrimidine dimers and 6-pyrimidine-4-pyrimidone photoproducts). In addition to NER, CSB has also been implicated in base excision repair (30,31), transcription (32–35), chromatin maintenance and remodeling (36). However, whether CSB may play a role in telomere maintenance relevant to cancer and aging has not yet been characterized.
Figure 1.
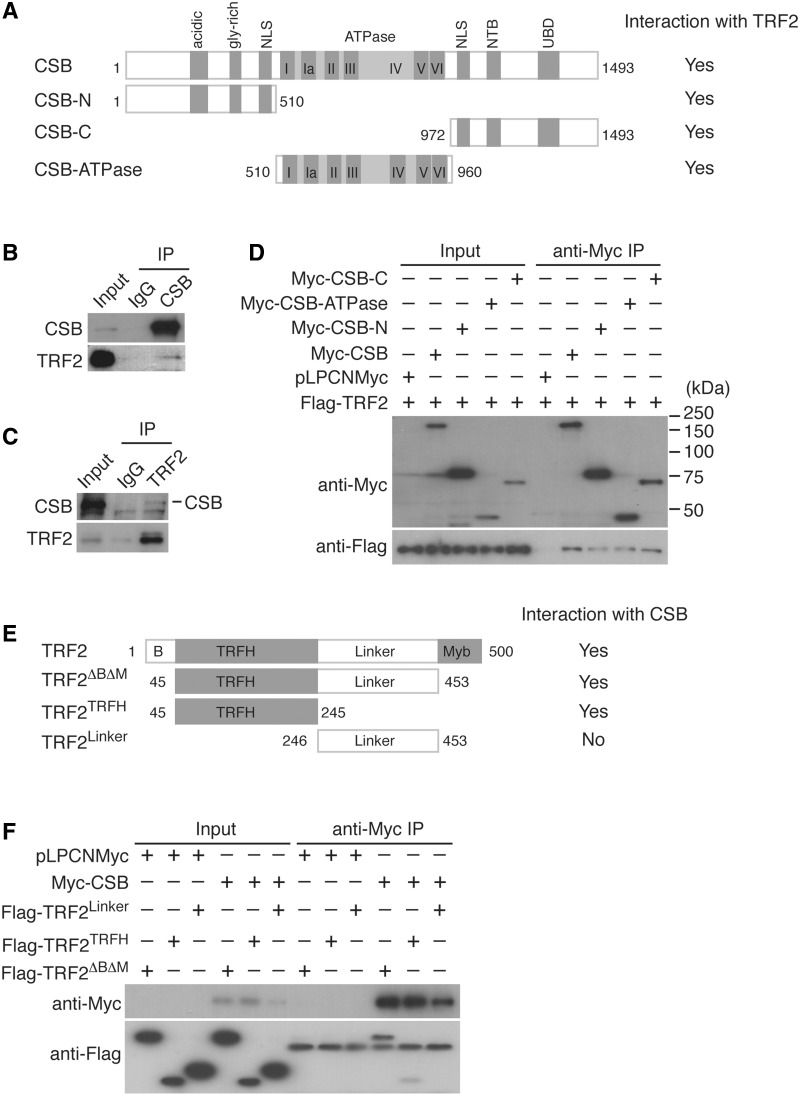
CSB interacts physically with TRF2. (A) Schematic diagram of CSB. NLS: nuclear localization sequence; NTB: nucleotide binding domain; and UBD: ubiquitin binding domain. (B) Co-immunoprecipitation with HeLa cell extracts and anti-CSB antibody. Anti-IgG IP was used as a negative control. Immunoblotting was carried out with anti-CSB or anti-TRF2 antibody. (C) Co-IP with HeLa nuclear extracts and anti-TRF2 antibody. Anti-IgG IP was used as a negative control. Immunoblotting was carried out with anti-CSB or anti-TRF2 antibody. (D) IP with anti-Myc antibody was carried out with protein extracts from 293T cells coexpressing Flag-TRF2 in conjunction with either the vector alone, Myc-CSB, Myc-CSB-N, Myc-CSB-ATPase or Myc-CSB-C. Immunoblotting was performed with anti-Myc or anti-Flag antibody. (E) Schematic diagram of TRF2. B stands for basic domain. (F) IP with anti-Myc antibody was carried out with protein extracts from 293T cells coexpressing the vector or Myc-CSB in conjunction with Flag-TRF2linker, Flag-TRF2TRFH or Flag-TRF2ΔBΔM. Immunoblotting was performed with anti-Myc or anti-Flag antibody.
Here, we report that CSB physically interacts with TRF2. While multiple domains of CSB are engaged in its interaction with TRF2, the TRFH domain of TRF2 is required and sufficient for binding CSB. We show that CS cells or CSB knockdown cells exhibit an accumulation of telomere doublets and an induction of TIF formation. Re-introduction of wild-type CSB into CS cells suppresses the formation of telomere doublets and TIFs, indicative of its role in telomere protection. In addition, we find that CS cells undergo telomere shortening, whereas overexpression of CSB into CS cells results in telomerase-dependent telomere lengthening. The latter is associated with a reduction in the amount of telomere-bound TRF1, a negative mediator of telomere length maintenance (14–16). Furthermore, we find that CS cells or CSB knockdown cells display misregulation of TERRA expression. Collectively, these results suggest that CSB is required for maintaining the homeostatic level of TERRA, telomere length and stability.
MATERIALS AND METHODS
DNA constructs and antibodies
The complementary DNA (cDNA) for CSB purchased from mammalian gene collection contained three missense mutations (C666, P1041 and P1294). The QuickChange site-directed mutagenesis kit (Strategene) was used to revert these mutations to wild-type. The corrected CSB cDNA was then subcloned into the retroviral vector pLPC-puro (37) or pLPC-N-Myc-puro (37). The pLPC-N-Myc-CSB plamid was used as a template for PCR to generate CSB truncation alleles CSB-N (amino acids 2–510), CSB-ATPase (amino acids 510–960) and CSB-C (amino acids 972–1493). The cDNA for TRF2 was a generous gift from Titia de Lange, Rockefeller University. The TRF2 truncation alleles TRF2ΔBΔM (amino acids 45–453), TRF2TRFH (amino acids 45–245) and TRF2linker (amino acids 246–453) were generated by PCR and cloned into pLPC-FH2 (38) (a kind gift from Titia de Lange, Rockefeller University). pBabe-neo-hTERT was kindly provided by Robert Weinberg, MIT.
The oligonucleotides encoding siRNA directed against CSB have been previously described (39). The annealed oligonucleotides were ligated into pRetroSuper vector (kindly provided by Titia de Lange, Rockefeller University), giving rise to pRetroSuper-shCSB.
Antibodies to TRF1 (13), TRF2 (40) and hRap1 (41) were kind gifts from Titia de Lange, Rockefeller University. Commercial antibodies used were rabbit anti-CSB (Bethyl A301–345A), mouse anti-CSB (Abcam Ab66598), anti-Myc (9E10, Calbiochem), anti-γ-H2AX (Upstate) and anti-γ-tubulin (GTU88, Sigma).
Cell culture and retroviral infection
HeLaI.211 cells were a gift from Titia de Lange, Rockefeller University. Primary fibroblast cell lines GM38 (normal), GM9503 (normal), GM8399 (normal), GM10901 (heterozygote), GM10905 (CS), GM739 (CS), GM1428 (CS) and a transformed CS cell line (GM16095) were obtained from the NIGMS Human Genetic Cell Repository (Coriell Institute for Medical Research, Camden, NJ, USA). GM16095 is a SV40-transformed cell line derived from GM739 (27). Supplementary Table S1 lists the nature of CSB mutations and the age of the individuals from whom biopsies were taken to establish the primary cell lines. Cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS) for transformed cell lines GM16095, HeLa and Phoenix cells, and 15% FBS for all primary fibroblasts, supplemented with non-essential amino acids, glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. Retroviral gene delivery was carried out as described (42,43). Phoenix amphotropic retroviral packaging cells were transfected with the desired DNA constructs. For hTERT-mediated immortalization, 3 days after the last infection, neomycin (600 µg/ml) was added to the medium to select for hTERT-expressing cells. Otherwise, 12 h after the last infection, puromycin (2 µg/ml) was added to the medium and the cells were maintained in the selection medium for the entirety of the experiments.
Immunoblotting and immunoprecipitation
Immunoblotting was carried out as previously described (10,40). Immunoprecipitation (IP) of endogenous TRF2 was performed essentially as described (10,40). For IP of endogenous CSB, HeLa cells were collected and resuspended in ice-cold NP-40 buffer (1% NP-40, 150 mM NaCl, 10 mM sodium phosphate, pH 7.2). Following incubation on ice for 20 min, the supernatant was recovered by micro-centrifugation at 13 000 rpm for 10 min. Protein extracts of 1.5 mg was mixed with 2 µl mouse anti-CSB antibody (Abcam) and the mixture was incubated overnight at 4°C. Protein G-beads (30 µl) was added to the mixture on the next day and the IP pellet was washed five times each with 1 ml of ice-cold NP-40 buffer containing 1 mM DTT, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 10 µg/ml pepstatin and 1 mM PMSF.
Co-immunoprecipitation from 293T cells was carried out essentially as described (38) except for the method of transfection used. Human 293T cells grown on 6-cm plates with 95% confluency were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For each co-transfection, a total of 8 µg DNA mixture was used. The ratio of CSB constructs to TRF2 constructs in each DNA mixture was 3:1.
Chromatin Immunoprecipitations
Chromatin immunoprecipitations (ChIPs) were carried out essentially as described (44–46). Cells were directly fixed with 1% formaldehyde in phosphate-buffered saline (PBS) for 1 h, followed by sonication (10 cycles of 20 s each, 50% duty and 5 output). For each ChIP, 200 µl cell lysate (equivalent to 2 × 106 cells) was used. For the total telomeric DNA, 50 µl supernatant (corresponding to one-quarter of the amount of lysate used for IP) were processed along with the IP samples at the step of reversing the crosslinks. Four-fifths of the immunoprecipitated DNA was loaded on the dot blots whereas two inputs each containing 5% of total DNA was included to assess the consistency of loading. The ratio of the signal from each ChIP relative to the signal from the input lane was multiplied by 5% (5% represents 5% of total DNA) and a factor of 1.25 (since four-fifths of the precipitated DNA was loaded for each ChIP), giving rise to the percentage of total telomeric DNA recovered from each ChIP.
Immunofluorescence and fluorescence in situ hybridizatioin
Immunofluoresence was performed essentially as described (40,43). Briefly, cells grown on coverslips were rinsed with PBS, incubated in Triton X-100 buffer [0.5% Triton X-100, 20 mM HEPES–KOH (pH 7.9), 50 mM NaCl, 3 mM MgCl2 and 300 mM sucrose] at room temperature (RT) for 5 min and then fixed for 10 min in PBS buffered 3% paraformaldehyde and 2% sucrose. Following permeablization at RT for 10 min in Triton X-100 buffer, fixed cells were blocked with 0.5% bovine serum ablumin (Sigma) and 0.2% gelatin (Sigma) in PBS and then incubated at RT for 2 h with both rabbit anti-hRap1 and mouse anti-γH2AX or mouse anti-CSB.
Immunofluorescence (IF)–fluorescence in situ hybridizatioin (FISH) analyses were conducted as described (9). Briefly, cells grown on coverslips were fixed at RT for 10 min in PBS buffered 2% para-formaldehyde, washed in PBS twice for 5 min each, followed by incubation at RT for 30 min in blocking buffer containing 1 mg/ml bovine serum albumin (BSA), 3% goat serum, 0.1% Triton X-100 and 1 mM ethylenediaminetetraacetic acid (EDTA) in PBS. Blocked coverslips were incubated with anti-Myc antibody in blocking buffer at RT for 1 h. After three washes in PBS, coverslips were incubated with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated donkey anti-mouse (1:100, Jackson Laboratories) at RT for 30 min. Subsequently, cells on coverslips were fixed again in PBS buffered 2% paraformaldehyde for 5 min and followed by dehydration in a series of 70, 85 and 100% ethanol. The air-dried coverslips were denatured at 80°C for 10 min and hybridized with 0.5 µg/ml fluorescein isothiocyanate (FITC)-conjugated-(CCCTAA)3 PNA probe (Biosynthesis Inc.) for 2 h in dark at RT. Following incubation, cover slips were washed with 70% formamide and 10 mM Tris–HCl (pH 7.2) twice for 15 min. After three washes in PBS, DNA was counter-stained with 4,6-diamidino-2-phenylindole (DAPI; 0.2 µg/ml) and embedded in 90% glycerol/10% PBS containing 1 mg/ml p-phenylene diamine (Sigma). All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 camera and processed in Open Lab.
Metaphase chromosome spreads
Metaphase chromosome spreads were essentially prepared as described (19,43). Cells were arrested in nocodazole (0.1 µg/ml) for 90–120 min. Following arrest, cells were harvested by trypsinization, incubated for 7 min at 37°C in 75 mM KCl and fixed in freshly made methanol/glacial acedic acid (3:1). Cells were stored overnight at 4°C, dropped onto slides and air-dried overnight in a chemical hood.
FISH analysis on metaphase chromosome spreads was carried out essentially as described (43,47). Slides with chromosome spreads were incubated with 0.5 µg/ml FITC-conjugated-(CCCTAA)3 PNA probe (Biosynthesis Inc.) for 2 h at room temperature. Following incubation, slides were washed, counter-stained with 0.2 µg/ml DAPI and embedded in 90% glycerol/10% PBS containing 1 mg/ml p-phenylene diamine (Sigma). All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 camera and processed in Open Lab.
Northern analysis of TERRA
Total RNA was isolated from cells using TRIzol® Reagent (Invitrogen) according to the manufacture’s instructions. Northern analysis was performed essentially as described with minor modifications (5). Briefly, 20 µg of the RNA was loaded onto 1.3% formaldehyde agarose gels and run at 60 V for 7 h. The gel was then stained with ethidium bromide to inspect the presence of the 28S and 18S ribosomal RNA, both of which were indicators of RNA quality. RNA was then transferred to a Nylon membrane (Hybond-N, GE) and was blocked in Church mix [0.5 M Na2PO4 (pH 7.2), 1 mM EDTA, 7% SDS and 1% BSA] for 1 h at 65°C. The membrane was then incubated overnight at 65°C with a radioactively labeled 800-bp TTAGGG repeat-containing fragment as previously described (44). For the GAPDH control, the membrane was incubated with a radioactively labeled DNA fragment containing the GAPDH gene. Following incubation, the membrane was washed once with 1× SSC, 0.1% SDS at room temperature, three times in 0.5× SSC at 65°C and then exposed to a PhosphorImager screen. The signals on the membrane were quantified by ImageQuant analysis.
Telomere length analysis and telomeric repeat amplification protocol (TRAP) assays
Genomic DNA isolated from cells was digested with RsaI and HinfI and loaded onto a 0.7% agarose gel in 0.5× TBE. Blotting for telomeric fragments was carried out according to standard protocols (48,49). The average telomeric restriction fragment length was determined by PhosphorImager analysis using ImageQuant and MS Excel as described (50).
The activity of telomerase in cells was determined using a Trapeze telomerase detection kit (Chemicon) according to the protocol provided by the manufacturer. PCR amplification was performed for 31 cycles. The products were separated on a 12.5% non-denaturing polyacrylamide gel in 0.5× TBE buffer and visualized using SYBR green (Invitrogen).
RESULTS
Physical interaction between CSB and TRF2
To investigate the role of CSB in telomere biology, we decided to ask whether CSB might interact with components of the shelterin complex essential for telomere maintenance. Co-immunoprecipitation with anti-CSB antibody brought down endogenous TRF2 (Figure 1B). CSB association with TRF2 was also detected in a reverse IP using anti-TRF2 antibody and HeLa nuclear extracts (Figure 1C). The interaction of CSB with TRF2 was further confirmed when Flag-tagged TRF2 was co-expressed with Myc-CSB in 293T cells (Figure 1D). Taken together, these results reveal that CSB interacts with TRF2 in vivo.
To gain further understanding of CSB interaction with TRF2, we examined the interaction between various CSB domains and TRF2. Flag-TRF2 was coexpressed with Myc-tagged CSB-N carrying the first 510 amino acids, including the acidic and the glycine-rich domains, Myc-tagged CSB-ATPase containing the central 450 amino acids or Myc-tagged CSB-C carrying the last 521 amino acids including the NTB domain and UBD in 293T cells. Co-immunoprecipitation studies with anti-Myc antibody revealed that all three CSB truncation mutants were able to pull down Flag-TRF2 (Figure 1D), suggesting that multiple domains of CSB may be engaged in its interaction with TRF2.
TRF2 contains an N-terminal basic/GAR domain, a central TRFH domain, a linker region and a C-terminal Myb-like DNA binding domain (Figure 1E). To investigate the domain of TRF2 important for its interaction with CSB, we coexpressed Myc-CSB with Flag-tagged TRF2ΔBΔM lacking both the basic domain and the Myb-like domain, Flag-tagged TRF2 carrying the TRFH dimerization domain alone (Flag-TRF2TRFH) or Flag-tagged TRF2 carrying the linker region alone (Flag-TRF2linker) in 293T cells. Co-immunoprecipitation with anti-Myc antibody showed that both Flag-tagged TRF2ΔBΔM and Flag-tagged TRF2TRFH were able to interact with Myc-CSB (Figure 1F). In contrast, no interaction between Myc-CSB and Flag-TRF2linker was detected despite a high level of expression of Flag-TRF2linker (Figure 1F). These results suggest that the TRFH domain is required and sufficient for TRF2 interaction with CSB.
CSB localizes at a fraction of human telomeres and is required to suppress the formation of TIFs in CS cells
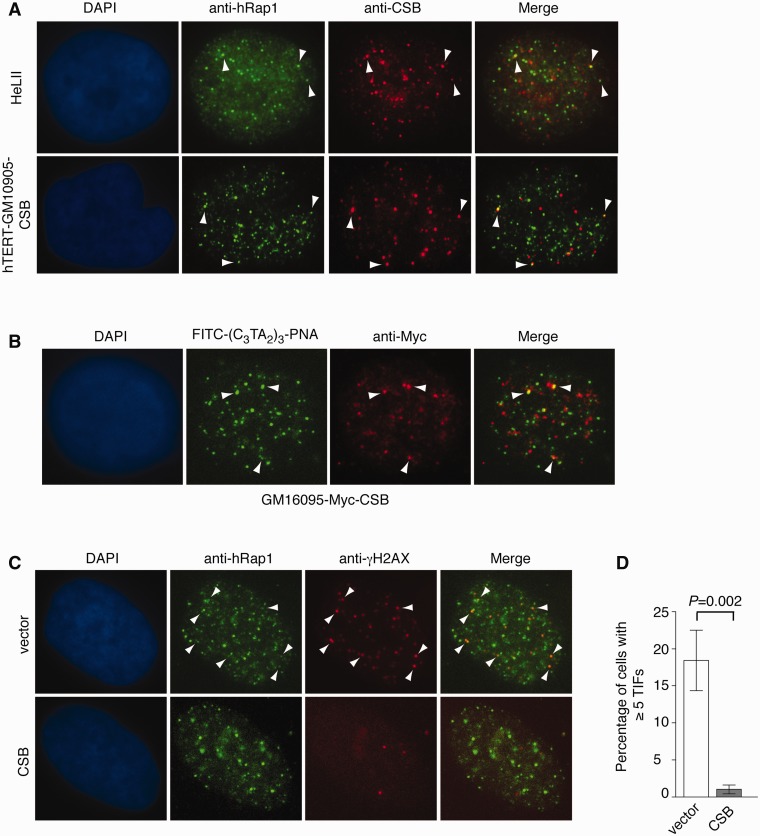
To investigate whether CSB may be associated with human telomeres, we performed dual indirect immunofluorescence with anti-CSB antibody in conjunction with anti-hRap1 antibody, a marker for interphase telomeres (41). We observed an overlap between several anti-hRap1 staining (green) and anti-CSB staining (red) foci in HeLa cells (Figure 2A). The co-localization of CSB with several hRap1 foci was also detected in CSB-complemented immortalized CS cells hTERT-GM10905 (Figure 2A). In addition, we also performed IF–FISH analysis with anti-Myc antibody in conjunction with a FITC-conjugated telomeric DNA-containing PNA probe in SV40-transformed CS cells GM16095 stably expressing Myc-tagged CSB. We again observed the presence of CSB (red) at several telomeres (green) (Figure 2B). Taken together, these results suggest that CSB may be associated with a small subset of human telomeres although we cannot rule out the possibility that observed costaining of CSB with telomeres may be coincidental.
Figure 2.
CSB localizes at a small subset of human telomeres and prevents the formation of TIFs in CS cells. (A) Analysis of indirect immunofluorescence (IF) on HeLaII and CSB-complemented hTERT-GM10905 cells. IF was perfromed with mouse anti-CSB (red) in conjunction with rabbit anti-hRap1 (green). Cells were extracted with detergent prior to fixation by paraformaldehyde to remove soluble proteins. Cell nuclei were stained with DAPI shown in blue. Arrowheads indicate the overlap between anti-CSB and anti-hRap1 staining. (B) Analysis of IF-FISH on GM16095 cells expressing Myc-CSB. IF–FISH analyses were performed with anti-Myc antibody (red) in conjunction with a FITC-conjugated (CCCTAA)3-containing PNA probe (green). Cell nuclei were stained with DAPI shown in blue. Arrowheads indicate the colocalization of CSB with telomeric DNA. (C) Indirect immunofluorescence using anti-hRap1 in conjunction with anti-γ-H2AX was performed with fixed hTERT-GM10905 cells expressing either the vector alone or wild-type CSB. Arrowheads indicate sites of colocalization of γH2AX and hRap1. (D) Quantification of percentage of cells with five or more TIFs. For each cell line, a total of 300 cells from three independent experiments were scored. Standard deviations from three independent experiments are indicated.
Dysfuntional telomeres are known to attract DNA damage response factors including γH2AX (7–10). To investigate whether CS cells may accumulate dysfunctional telomeres, dual indirect immunofluorescence was performed on hTERT-GM10905 expressing either CSB or the vector alone with anti-hRap1 antibody in conjunction with anti-γH2AX antibody. We observed an induction of TIFs in vector-expressing hTERT-GM10905 cells when compared to CSB-complemented hTERT-GM10905 cells (Figure 2C). While 18% of the vector-expressing hTERT-GM10905 cells exhibited five or more TIFs, such TIFs were detected in only 1% of CSB-complemented hTERT-GM10905 cells (Figure 2D). These results suggest that CSB is required for telomere protection.
Primary fibroblasts derived from CS patients carrying a CSB mutation show an accumulation of telomere doublets
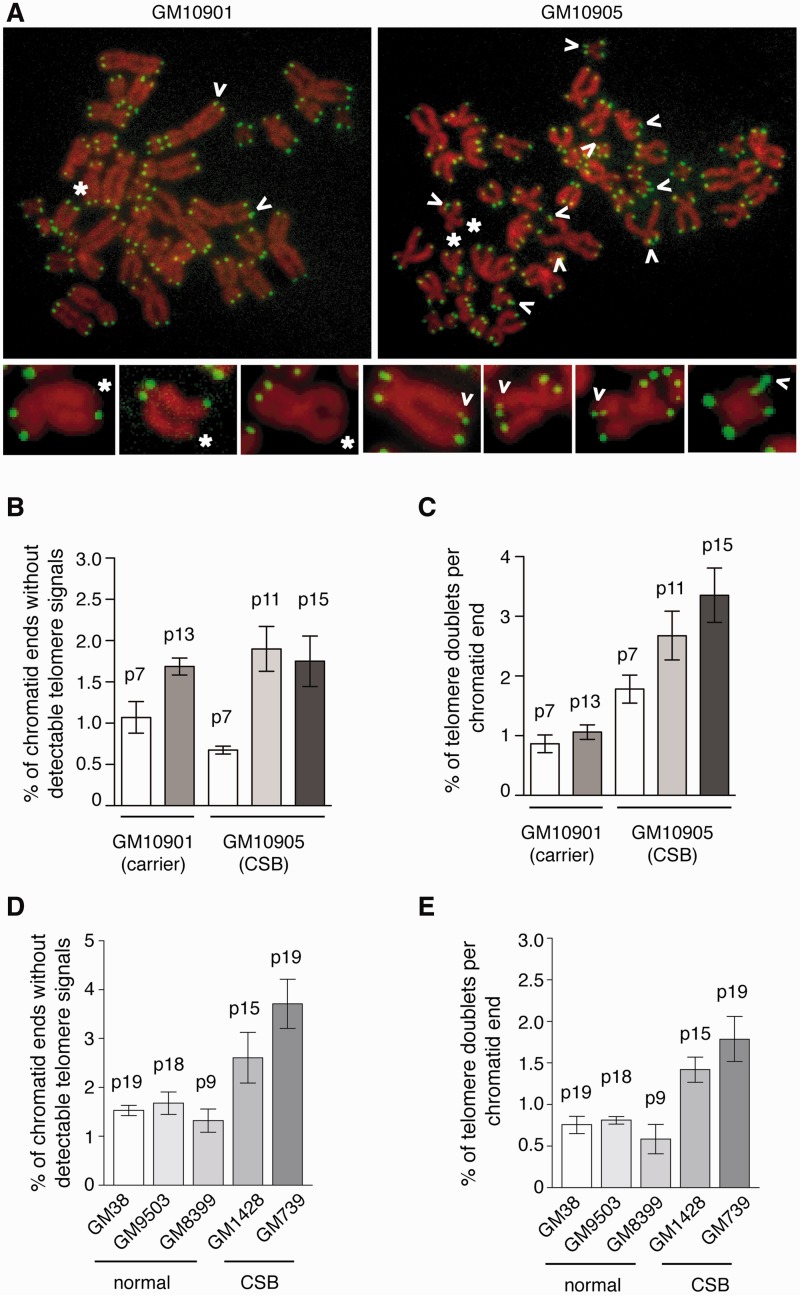
To investigate whether CSB may be required for maintaining telomere structure, we performed FISH analysis of metaphase spreads on two cell lines (GM10901 and GM10905) at various passages to inspect for the presence of any telomere abnormalities, including telomere loss (chromatid ends without a detectable telomeric signal), telomere fusions, telomere-containing double minute chromosomes (TDM) and telomere doublets/fragile telomeres (more than one telomeric signal at a single chromatid end). GM10901 and GM10905 are two respective primary fibroblast cell lines derived from a mother heterozygote for a CSB mutation and her CS offspring. We did not observe any significant accumulation of TDM and telomere fusions in either GM10901 or GM10905 (Figure 3A). While telomere loss was detected in both GM10901 and GM10905 (Figure 3B), no significant difference in the formation of telomere loss was found when GM10901 and GM10905 cells of various passages were compared (Figure 3B). In contrast, we found that various passages of GM10905 cells consistently exhibited an accumulation of telomere doublets when compared to the heterozygote GM10901 cells of similar passages (Figure 3C).
Figure 3.
CS primary fibroblasts carrying CSB mutations accumulate telomere doublets. (A) Analysis of metaphase chromosomes from GM10901 and GM10905. Chromosomes were stained with DAPI and false colored in red. Telomeric DNA was detected by FISH using a FITC-conjugated (CCCTAA)3-containing PNA probe (green). Open arrows represent telomere doublets whereas asterisks indicate telomere loss. Enlarged images of chromosomes with telomere doublets or telomere loss are shown at the bottom. (B–E) Quantification of telomere loss or telomere doublets from indicated cell lines. For each cell line, a total of 2410–2699 chromosomes from 60 metaphase cells were scored in a blind manner for the presence of telomere loss (B and D) as well as telomere doublets in (C and E). Standard deviations derived from three independent experiments are indicated. Passage numbers of cell lines used are indicated above the bars.
We also examined the presence of telomere loss and telomere doublets in two other CS cell lines GM1428 and GM739 in comparison to three fibroblast cell lines (GM38, GM9503, GM8399) derived from normal individuals. We found that when compared to the normal control cells, both GM1428 and GM739 displayed an increase in the formation of telomere loss and telomere doublets (Figure 3D and E), the latter consistent with our earlier finding. No full-length CSB was detected in any CS cells examined (Supplementary Figure S1). Taken together, these results suggest that CSB is required for maintaining the integrity of telomere structure.
Introduction of wild-type CSB into CS cells suppresses the formation of telomere doublets
Formally, it is possible that the increased formation of telomere doublets observed in CS primary fibroblasts might be due to the difference in the genetic background between CS cells and normal control cells. To address this question, we decided to examine telomere structures in several pairs of cell lines with isogenic background.
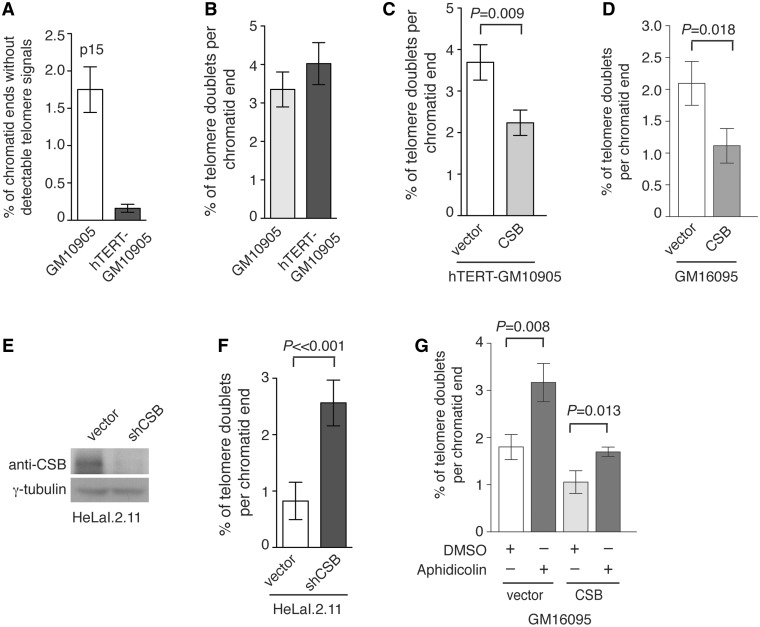
CS primary fibroblasts GM10905 was immortalized with exogenously expressed catalytic subunit of telomerase (hTERT) (Supplementary Figure S2A) to overcome poor growth and premature senescence associated with CS cells. Subsequently, retrovirus expressing either wild-type CSB or the vector alone was used to infect hTERT-GM10905 cells, generating two stable isogenic cell lines (hTERT-GM10905-vector and hTERT-GM10905-CSB). FISH analysis revealed that overexpression of hTERT drastically reduced telomere loss (Figure 4A and Supplementary Figure S2B); however, it had little effect on the accumulation of telomere doublets in GM10905 cells (Figure 4B and Supplementary Figure S2B). On the other hand, we found that introduction of wild-type CSB into hTERT-GM10905 cells led to a reduction in the formation of telomere doublets (Figure 4C and Supplementary Figure S3). We observed a 40% decrease (P = 0.009) in the formation of telomere doublets in CSB-complemented hTERT-GM10905 cells when compared to vector-expressing hTERT-GM10905 cells (Figure 4C).
Figure 4.
CSB is required to prevent the formation of telomere doublets. (A) Quantification of telomere loss from indicated cell lines. For each cell line, a total of at least 2649–2668 chromosomes from 60 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated. (B) Quantification of telomere doublets from indicated cell lines. For each cell line, a total of 2649–2668 chromosomes from 60 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated. (C) Quantification of telomere doublets from hTERT-GM10905 cells expressing indicated constructs. For each cell line, a total of 2707–2754 chromosomes from 60 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated. (D) Quantification of telomere doublets from GM16095 cells expressing indicated constructs. For each cell line, a total of 4774–4923 chromosomes from 60 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated. (E) Western analysis of CSB expression. CSB was stably knocked down in HeLaI.2.11 cells. Immunoblotting was performed with anti-CSB or anti-γ-tubulin antibody. The latter was used as a loading control. (F) Quantification of telomere doublets from HeLaI.2.11 cells expressing the vector alone or pRS-shCSB. For each cell line, a total of 2678–2961 chromosomes from at least 43 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated. (G) Quantification of telomere doublets from GM16095 cells expressing indicated constructs. Cells were treated with DMSO or aphidicolin (0.2 µM) for 16 h. For each cell line, a total of 3879–4321 chromosomes from 51 to 53 metaphase cells were scored in a blind manner. Standard deviations derived from three independent experiments are indicated.
We also examined the formation of telomere doublets in a second pair of isogenic CS cell lines (GM16095) complemented with either the vector alone or wild-type CSB. Introduction of wild-type CSB also resulted in a reduction in the formation of telomere doublets in GM16095 (Figure 4D). To further investigate the role of CSB in the formation of telomere doublets, we knocked down CSB in HeLaI.2.11 cells (Figure 4E) and found that depletion of CSB led to an induction of telomere doublets (Figure 4F and Supplementary Figure S4). Taken together, these results suggest that CSB prevents the formation of telomere doublets.
Aphidicolin, an inhibitor of DNA replication, has been shown to induce telomere doublets (9,51). We found that treatment with aphidicolin resulted in a further increase in the formation of telomere doublets in CS cells (GM16095) (Figure 4G), consistent with previous findings that the effect of aphidicolin was additive (9,46,48). We also observed an increase in the formation of telomere doublets in CSB-complemented GM16095 cells upon aphidicolin treatment, althought such increase was less than that observed in GM16095 cells expressing the vector alone (Figure 4G). These results suggest that telomere doublets observed in CS cells may have arisen from a defect associated with telomere replication.
Introduction of wild-type CSB into CS cells promotes telomerase-dependent telomere lengthening
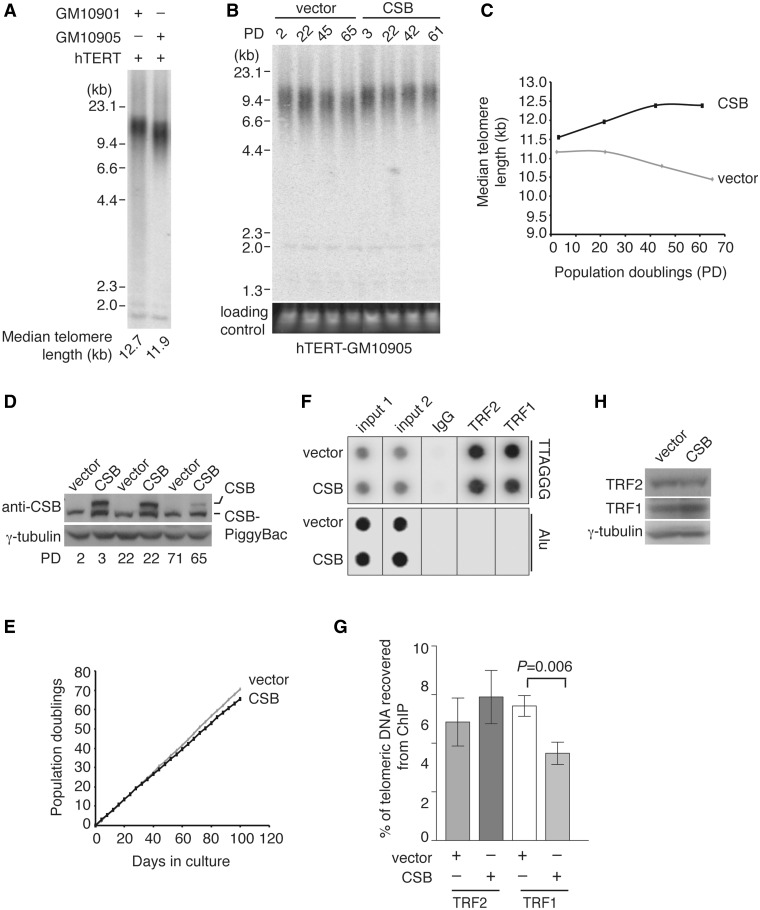
We observed that the median telomere length in hTERT-immortalized heterozygote mother GM10901 cells was longer than that in hTERT-immortalized CS offspring GM10905 cells (Figure 5A). Therefore, we decided to examine whether CSB might be involved in telomere length maintenance. To address this question, pools (not single cell clones) of hTERT-GM10905 cells stably expressing the vector alone or wild-type CSB were continously cultured for over 60 population doublings (PDs) and their telomere length dynamics was examined. Analysis of telomere restriction fragments revealed that the median telomere length in hTERT-GM10905 cells expressing the vector alone declined at a rate of ∼11.6 bp/PD, whereas the median telomere length increased at a rate of 21.5 bp/PD for the first 42 PDs and then plateaued in hTERT-GM10905 cells expressing wild-type CSB (Figure 5B and C). A decline in the level of CSB expression in hTERT-GM10905 CSB cells was noticed after PD60 (Figure 5D), suggesting that the loss of CSB expression may in part contribute to the plateau of the median telomere length seen between PD42 and PD61 in these cells. We did not observe any significant difference in the growth rate between hTERT-GM10905 vector and hTERT-GM10905 CSB cells (Figure 5E). Taken together, these results suggest that CSB is required for telomerase-dependent telomere elongation.
Figure 5.
CSB is required for telomere length maintenance. (A) Genomic blot of telomeric restriction fragments from hTERT-immortalized GM10901 and GM10905 cells. About 3 µg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. DNA molecular weight markers are shown on the left of the blot. Median telomere length of indicated cell lines are shown on the bottom of the blot. (B) Genomic blots of telomeric restriction fragments from hTERT-GM10905 cells expressing either the vector alone or wild-type CSB as indicated above the lanes. About 3 µg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. PDs are indicated above the lanes whereas DNA molecular weight markers are shown on the left of the blots. The bottom panel, taken from an ethidium bromide-stained agarose gel, is used as a loading control. (C) Median telomere length of indicated cell lines was plotted against PDs. (D) Western analysis of CSB expression in hTERT-GM10905 cells. Immunoblotting was performed with anti-CSB or anti-γ-tubulin antibody. The indicated CSB-PiggyBac fusion protein is a product of alternative splicing involving the first five exons of CSB and a conserved PGBD3 located within the intron 5 of the CSB gene (52). (E) Growth curve of hTERT-GM10905 cells expressing various constructs as indicated. The number of PDs was plotted against days in culture. (F) Dot blots of ChIPs with anti-TRF1 or anti-TRF2 antibody. ChIPs were performed with lysates from hTERT-GM10905 cells expressing either the vector alone or wild-type CSB. Anti-IgG ChIP was used as a control. (G) Quantification of ChIPs from (E). The signals from dot blots were quantified by ImageQuant (IQ) analysis. Standard deviations from three independent experiments are shown. (H) Western analysis of protein expression. Immunoblotting was carried out with anti-TRF1, anti-TRF2 or anti-γ-tubulin antibody.
We also performed ChIP analysis with an antibody against TRF1 or TRF2, both of which are mediators of telomere length maintenance (14–16,42). We found that introduction of wild-type CSB into hTERT-GM10905 cells had little effect on telomeric association of TRF2 (Figure 5F and G), but it led to a significant increase in TRF1 association with telomeric DNA (Figure 5F and G). When compared to CSB-complemented hTERT-GM10905 cells, we observed a 54% (P = 0.006) increase in the amount of telomere-bound TRF1 in hTERT-GM10905 cells expressing the vector alone (Figure 5G). The level of TRF1 in the vector-expressing hTERT-GM10905 cells was indistinguishable from that in the CSB-complemented hTERT-GM10905 cells (Figure 5H). These results suggest that association of TRF1 with telomeric DNA may be deregulated in CS cells carrying a CSB mutation.
CSB is required for maintaining the homeostatic level of TERRA
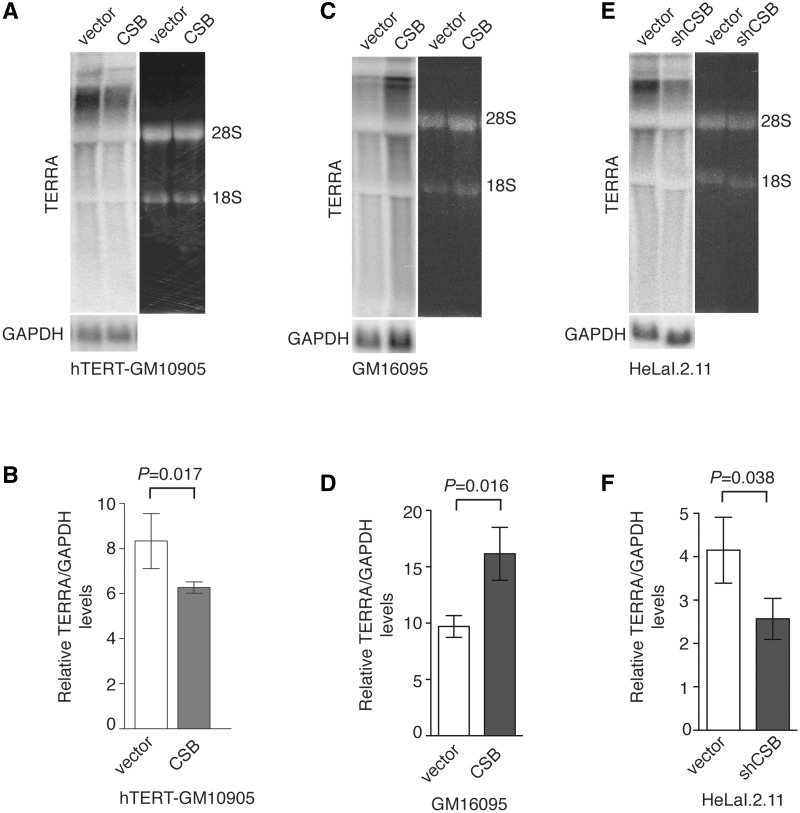
CSB has been implicated in transcription (32–35) and therefore we decided to examine whether CSB may be involved in regulating the expression of TERRA, a large non-coding telomere repeat-containing RNA (5). Northern analysis on three pairs of isogenic cell lines revealed a mis-regulation of TERRA associated with CS cells or CSB knockdown cells. We observed a 35% increase (P = 0.017) in the level of TERRA in hTERT-GM10905 expressing the vector alone when compared to hTERT-GM10905 cells complemented with wild-type CSB (Figure 6A and B). On the other hand, the level of TERRA in GM16095 cells expressing the vector alone was ∼45% (P = 0.016) less than that in GM16095 complemented with wild-type CSB (Figure 6C and D). Knockdown of CSB led to a 38% (P = 0.038) reduction in the level of TERRA in HeLaI.2.11 cells (Figure 6E and F). These results suggest that CSB is required for the homeostatic level of TERRA and that the level of TERRA may increase or decrease in CS cells depending upon the nature of CSB mutations.
Figure 6.
CSB is required for maintaining the homeostatic level of TERRA. (A) Analysis of TERRA expression from hTERT-GM10905 cells expressing the vector alone or CSB. Northern blotting was performed with a 32P-labeled telomeric DNA-containing probe shown on the left top panel. The northern blot of GAPDH shown on the left bottom panel was used as a loading control. The right panel was taken from the ethidium bromide-stained agarose gel. The position of 28S or 18S ribosomal RNA is indicated. (B) Quantification of relative TERRA levels from (A). The signals from northern blots were quantified by ImageQuant analysis. The TERRA signal from each lane was normalized to the GAPDH signal in the corresponding lane, giving rise to the relative level of TERRA to GAPDH. (C) Northern analysis of TERRA expression from GM16095 cells expressing the vector alone or wild-type CSB. The northern blot of GAPDH shown on the left bottom panel was used as a loading control. The right panel was taken from the ethidium bromide-stained agarose gel. The position of 28S or 18S ribosomal RNA is indicated. (D) Quantification of relative TERRA levels from (C). Quantification was performed as described in (B). (E) Northern analysis of TERRA expression from HeLaI.2.11 cells stably expressing the vector alone or pRS-shCSB. The northern blot of GAPDH shown on the left bottom panel was used as a loading control. The right panel was taken from the ethidium bromide-stained agarose gel. The position of 28S or 18S ribosomal RNA is indicated. (F) Quantification of relative TERRA levels from (E). Quantification was performed as described in (B).
DISCUSSION
CSB, a multifunctional protein, plays an important role in DNA repair, transcription and chromatin remodeling. In this report, we have uncovered a role for CSB in telomere maintenance and protection. We have shown that CSB interacts physically with TRF2, a key component of the shelterin complex essential for telomere maintenance. We have demonstrated that CS cells or CSB knockdown cells exhibit an accumulation of telomere doublets and an induction of TIF formation. We have shown that CS cells carrying a CSB mutation are defective in telomerase-dependent telomere elongation whereas introduction of CSB into CS cells results in telomerase-dependent telomere elongation, suggesting that CSB is required for telomere length maintenance. Furthermore, we have shown that the level of TERRA is misregulated in CS cells or CSB knockdown cells. Taken together, these results reveal an important role of CSB in the maintenance of telomere length and integrity. These results further imply that CS patients lacking functional CSB are defective in telomere maintenance, which is associated with cancer and aging.
Our co-immunoprecipitation studies suggest that a small percentage of endogenous TRF2 (estimated to be ∼1–5%) interacts with CSB and vice versa. This low level of interaction is similar to previously reported association between TRF2 and several other DNA repair proteins including XPF/ERCC1 and Mre11/Rad50/Nbs1 (40,43), indicating that CSB interaction with TRF2 may be dependent upon a specific functional requirement.
Analysis of domain mapping suggests that the TRFH domain of TRF2 is sufficient and required for its interaction with CSB. The TRFH domain of TRF2 has been shown to interact with proteins containing the Y/FxLxP motif (17,18). CSB contains one YxLxP motif corresponding to amino acids 402–406 but also seven degenerate Y/FxLxx motifs spread throughout the entire protein. Double mutations at positions L404 and P406 did not abrogate CSB interaction with TRF2 (T.R.H. Mitchell and X.D. Zhu, unpublished data). These results, in conjuction with our finding that multiple domains of CSB are engaged in its interaction with TRF2 raise the possibility that TRF2 might interact with degenerate Y/FxLxx motifs of CSB. Alternatively, TRF2 may interact with CSB through a mechanism independent of Y/FxLxP motifs. Future studies are required to investigate the mechanism underlying CSB interaction with TRF2.
The physical interaction between TRF2 and CSB raises the possibility that TRF2 may play a role in recruiting and/or modulating CSB function at telomeres. We have observed localization of CSB at a small subset of human telomeres. Several shelterin accessory proteins have been reported to localize at one or a few human telomeres, including HP1, BLM, PNUTS and MCPH1 (53–55). Perhaps, like these shelterin accessory factors, CSB might be needed by only a few telomeres at a given time although we cannot rule out the possibility that the colocalization of CSB with a few telomeres may be coincidental.
We have shown that overexpression of wild-type CSB has little effect on the telomere association of TRF2 but results in a reduction in the amount of telomere-bound TRF1, a negative mediator of telomerase-dependent telomere elongation. Perhaps, the reduction in the level of telomere-bound TRF1 may in part contribute to the telomerase-dependent telomere elongation observed in CSB-expressing hTERT-GM10905 cells. We have not been able to detect any interaction between CSB and endogenous TRF1 (T.R.H Mitchell and X.D. Zhu, unpublished data), suggesting that the effect of CSB on TRF1 binding to telomeric DNA may be indirect.
While we have observed a greater accumulation of telomere loss in CS primary fibroblast GM739 (p19) and GM1428 cells (p15) than in the control cells GM38 (p19) and GM9503 (p18), no significant difference in the formation of telomere loss has been detected between the heterozygote mother GM10901 and her CS offspring GM10905. It is possible that the lack of difference in telomere loss between the heterozygote mother and her CS offspring may be due to CSB haploinsufficiency. Alternatively, the level of accumulation of telomere loss observed in CS cells may vary depending upon their genetic background.
We have found that while knockdown of CSB leads to a reduction in the level of TERRA, overexpression of wild-type CSB can have an opposite effect on the level of TERRA in CS cells. Introduction of wild-type CSB into CS cells hTERT-GM10905 results in a decrease in the level of TERRA whereas introduction of wild-type CSB into CS cells GM16095 leads to an increase in the level of TERRA. Both CS cell lines carry a nonsense mutation (Supplementary Table S1), which converts R735 to a stop codon in GM10905 (22,56) and K337 to a stop codon in GM16095 (27). The level of overexpressed CSB in hTERT-GM10905 cells is comparable to that in GM16095 (N. Batenburg, T.R.H. Mitchell and X.D. Zhu, unpublished data), suggesting that it is unlikely that exogenously expressed CSB may account for its opposite effect on the level of TERRA in these two cell lines. Although both cell lines do not express full-length CSB, GM10905 cells express a CSB-PiggyBac fusion protein (Figure 5D) (52), which is not present in GM16095 (27). CSB-PiggyBac is a product of alternative splicing involving the first five exons of CSB and a conserved PiggyBac transposable element (PGBD3) located within the intron 5 of the CSB gene (52). How overexpression of CSB differentially affects the level of TERRA remains unknown. Our finding suggests that the nature of CSB mutations may play a role in influencing TERRA expression. Taken together, our data suggest that CSB is required for maintaining the homeostatic level of TERRA, excess expression or depletion of which has been shown to impair the maintenance of telomere length and integrity (5,6,57,58).
We have shown that CSB mutations or CSB depletion promotes the formation of telomere doublets, also known as fragile telomeres (9,51). It has been shown that fragile telomeres can arise from a defect in telomere replication (9,51). Consistent with this notion, we have observed that treatment with aphidicolin further induces the formation of telomere doublets in CS cells, suggesting that telomere replication is compromised in CS cells. It is likely that the compromised telomere replication in CS cells may be in part caused by misregulation of TERRA, an integral component of telomere heterochromatin. Perhaps misreglation of TERRA associated with CS cells could lead to an altered telomere heterochromatin, which could impede the progression of replication fork.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Cancer Research Society [16208 to X.-D.Z.]; Canadian Cancer Society [16066 to A.J.R.]; T.R.H.M is a holder of Ontario Graduate Scholarship. Funding for open access charge: Cancer Research Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for Titia de Lange (Rockefeller University) for providing various reagents, including TRF2 cDNA, retroviral vectors and antibodies to TRF1, TRF2 and hRap1. We thank John R. Walker for providing critical comments. N.L.B., T.R.H.M. and X.D.Z. designed experiments, interpreted the data and wrote the manuscript. N.L.B. and T.R.H.M. performed all experiments described. T.R.H.M. and D.M.L. conceptualized and performed initial experiments identifying the interaction between CSB and TRF2. D.M.L. and A.J.R. provided intellectual input.
REFERENCES
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects Mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 4.Walker JR, Zhu XD. Post-translational modification of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012;133:421–434. doi: 10.1016/j.mad.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 6.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 8.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell TR, Glenfield K, Jeyanthan K, Zhu XD. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 2009;29:4918–4934. doi: 10.1128/MCB.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 12.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 13.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 14.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1 [see comments] Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 15.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- 19.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 20.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 21.Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum. Mutat. 2009;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 23.van der Horst GT, van Steeg H, Berg RJ, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, et al. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 24.Leech RW, Brumback RA, Miller RH, Otsuka F, Tarone RE, Robbins JH. Cockayne syndrome: clinicopathologic and tissue culture studies of affected siblings. J. Neuropathol. Exp. Neurol. 1985;44:507–519. [PubMed] [Google Scholar]
- 25.Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 28.Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 30.Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J. Biol. Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 31.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 32.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 33.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl Acad.Sci. USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyng KJ, May A, Brosh RM, Jr, Cheng WH, Chen C, Becker KG, Bohr VA. The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene. 2003;22:1135–1149. doi: 10.1038/sj.onc.1206187. [DOI] [PubMed] [Google Scholar]
- 35.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl Acad. Sci. USA. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Zacal NJ, Rainbow AJ, Zhu XD. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair. 2007;6:157–166. doi: 10.1016/j.dnarep.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Yu ZJ, Sui JL, Bai B, Zhou PK. siRNA-mediated silencing of Cockayne Cyndrome group B gene potentiates radiation-induced apoptosis and antiproliferative effect in HeLa cells. Chin. Med. J. 2006;119:731–739. [PubMed] [Google Scholar]
- 40.Zhu XD, Kuster B, Mann M, Petrini JH, Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 42.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 43.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 44.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 45.Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 2007;14:832–840. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- 46.McKerlie M, Zhu XD. Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat. Commun. 2011;2:371. doi: 10.1038/ncomms1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 48.McKerlie M, Lin S, Zhu XD. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res. 2012;40:3975–3989. doi: 10.1093/nar/gks035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech. Ageing Dev. 2008;129:602–610. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell. 2003;14:5060–5068. doi: 10.1091/mbc.E03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4:e1000031. doi: 10.1371/journal.pgen.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S. A role for heterochromatin protein 1gamma at human telomeres. Genes Dev. 2011;25:1807–1819. doi: 10.1101/gad.17325211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- 56.Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-cacchione variant of xeroderma pigmentosum. Human Mol. Genet. 2000;9:1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- 57.Maicher A, Kastner L, Dees M, Luke B. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012;40:6649–6659. doi: 10.1093/nar/gks358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfeiffer V, Lingner J. TERRA Promotes Telomere Shortening through Exonuclease 1-Mediated Resection of Chromosome Ends. PLoS Genet. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.