Abstract
The activating transcription factor 4 (ATF4) promotes transcriptional upregulation of specific target genes in response to cellular stress. ATF4 expression is regulated at the translational level by two short open reading frames (uORFs) in its 5′-untranslated region (5′-UTR). Here, we describe a mechanism regulating ATF4 expression in translation termination-deficient human cells. Using microarray analysis of total RNA and polysome-associated mRNAs, we show that depletion of the eucaryotic release factor 3a (eRF3a) induces upregulation of ATF4 and of ATF4 target genes. We show that eRF3a depletion modifies ATF4 translational control at regulatory uORFs increasing ATF4 ORF translation. Finally, we show that the increase of REDD1 expression, one of the upregulated targets of ATF4, is responsible for the mTOR pathway inhibition in eRF3a-depleted cells. Our results shed light on the molecular mechanisms connecting eRF3a depletion to mammalian target of rapamycin (mTOR) pathway inhibition and give an example of ATF4 activation that bypasses the signal transduction cascade leading to the phosphorylation of eIF2α. We propose that in mammals, in which the 5′-UTR regulatory elements of ATF4 mRNA are strictly conserved, variations in translation termination efficiency allow the modulation of the ATF4 response.
INTRODUCTION
The activating transcription factor 4 (ATF4 also known as CREB-2) is an ubiquitous basic/leucine zipper domain transcription factor that belongs to the cAMP-responsive element binding (CREB) protein family (1,2). ATF4 plays a central role in evolutionarily conserved mammalian stress response pathways. These stress response pathways, collectively referred to as the integrated stress response (3), involve phosphorylation of the α-subunit of eukaryotic initiation factor-2 (eIF2) by a family of eIF2 kinases that are each activated by different stresses. One of the well-characterized stress responses is the unfolded protein response (UPR), an endoplasmic reticulum (ER) stress induced by the accumulation of unfolded proteins resulting from conditions, such as glucose deprivation, altered calcium levels and hypoxia. UPR enhances the activity of the eIF2α kinase PERK (4,5). Phosphorylation of eIF2α is also enhanced by three other kinases, GCN2 in response to amino acid limitation, UV irradiation and proteasome inhibition (6–8), HRI, which is regulated by heme deficiency and oxidative stress (9,10) and PKR, a major component of the interferon antiviral response that is stimulated by dsRNA (11,12). Phosphorylation of eIF2α at Ser51 inhibits general protein synthesis by blocking the exchange of eIF2–GDP to eIF2–GTP, which is required for binding of initiator tRNA to the small ribosomal subunit. Concomitant, with the general inhibition of translation, phosphorylation of eIF2α selectively promotes translational upregulation of a specific subset of mRNAs that carry short open reading frames (uORFs) in the 5′-untranslated region (5′-UTR) preceding the functional coding sequence. ATF4 mRNA 5′-UTR contains two such uORFs that modulate ATF4 protein expression when eIF2α is phosphorylated. It has been clearly established that the organization of these 5′-UTR uORFs is essential for the ATF4 response to stress (13–15). When the levels of eIF2–GTP are high, in non-stressed cells, ribosomes translate the 5′-proximal uORF1. After completing translation termination, the small ribosomal subunit resumes scanning downstream uORF1, recharges the eIF2–GTP–Met-tRNAMet ternary complex and reinitiates translation at the next coding region, uORF2, which overlaps the ATF4 ORF. Thus, translation of uORF2 prevents translation of ATF4 ORF. When the levels of eIF2–GTP are low, i.e. upon phosphorylation of eIF2α in response to stress, small ribosomal subunits that resume scanning after completing translation of uORF1, are not recharged with eIF2–GTP–Met-tRNAMet when they reach the uORF2 AUG ∼90 bases downstream uORF1 and thus bypass uORF2. The additional 100 bases separating uORF2 AUG from the following AUG, that is ATF4 ORF start codon, are sufficient to allow the binding of the ternary complex to the small ribosomal subunit and the translation of ATF4 ORF (14,15). It has been observed that the organization of ATF4 5′-UTR uORFs and the distances separating uORF1, uORF2 and ATF4 ORF AUGs are strictly conserved among vertebrates (15). The increased translation of ATF4 during stress conditions induces the transcriptional activation of specific target genes that contain ATF4 responsive elements such as the amino acid response element (AARE) in their promoter (16). Microarray analyses in cultured mammalian cells showed that ATF4 controls many genes involved in amino acid metabolism, in transport and in redox chemistry (3). Recent reviews on the pathways activated in response to stresses point out the complexity of these adaptive responses serving to limit the detrimental effects of stress and to remedy the cellular disturbance (17,18). In mammalian cells, multiple stress sensors initiate complementary signaling pathways, all of which can regulate transcription. Moreover, it is now clear that these stress response pathways are interconnected into a complex signaling network that plays a central homeostatic role in normal vertebrate physiology (19–21).
Translation terminates when a stop codon enters the A site of the elongating ribosome. In eukaryotes, two release factors, eRF1 and eRF3, associated in a complex are required for this process: eRF1 recognizes stop codons and activates the ribosome peptidyl transferase center, which triggers the release of the polypeptide, whereas eRF3 facilitates eRF1 activity through its GTPase activity (22,23). In connection with its role in translation termination, eRF3 has also been implicated in mRNA decay through its interaction with the poly(A)-binding protein PABP (24), and in the nonsense-mediated mRNA decay pathway through its interaction with Upf1 (25) when termination takes place at premature stop codons. Depletion of release factors decreases translation termination efficiency and promotes translational readthrough of stop codons (23,26,27). In mammals, two distinct proteins, eRF3a and eRF3b, are able to function as release factors. However, it has been shown that eRF3a is the main factor acting in translation termination (26). The GSPT1 gene encoding eRF3a is expressed in a proliferation-dependent manner (28), and eRF3a is subjected to proteasomal degradation when not associated to eRF1 (29), suggesting that eRF3a plays a key role in the modulation of translation termination efficiency.
eRF3 was first identified in a screen for mutants affected in the G1-to-S-phase transition in Saccharomyces cerevisiae (30). Consistent with this report, experiments utilizing RNA interference to deplete eRF3a in human cells have shown that eRF3a depletion inhibited the mTOR signaling pathway, thereby causing cell cycle arrest at G1 phase (31). Although these data suggested that eRF3 may have non-translational functions, the fact that eRF3a GTP-binding activity was required to restore G1-to-S-phase progression in eRF3a-depleted cells indicated that eRF3a involvement in mTOR inhibition was a consequence of translation termination disruption. Here, we further characterize the consequences of human eRF3a depletion with the aim to understand the mechanism responsible for mTOR pathway inhibition. First, using microarray analysis of total RNA and polysome-associated mRNAs, we show that eRF3a depletion induces upregulation of the transcriptional activator ATF4 and of genes that are under the control of ATF4. Second, we show that eRF3a depletion modifies ATF4 translational control at upstream regulatory ORFs (uORFs) increasing ATF4 ORF translation. Finally, we show that the increased REDD1 expression, one of the upregulated targets of ATF4, is responsible for the mTOR pathway inhibition in eRF3a-depleted cells. Our results shed light on the molecular mechanisms connecting eRF3a depletion to mTOR pathway inhibition and give an example of ATF4 activation that bypasses the signal transduction cascade leading to the phosphorylation of eIF2α. We propose that in mammalian cells, in which the 5′-UTR regulatory elements of ATF4 mRNA are strictly conserved, variations in translation termination efficiency allow the modulation of the ATF4 response.
MATERIALS AND METHODS
Plasmids and short interfering RNA
Plasmid p5′-UTR-ATF4-FLUC was constructed by reverse transcription of HCT116 cell total RNA followed by PCR amplification of human ATF4 cDNA using ATF4 5′-UTR-fw and -rev oligonucleotides (Table 1) containing NheI and BamHI restriction sites for cloning in the NheI and BamHI sites of pTK-FLUC expression vector. Plasmid pTK-FLUC is a derivative of pRL-TK vector (Promega) that contains the firefly luciferase gene instead of Renilla luciferase gene. Plasmids expressing small interfering RNAs sh-3a1 and sh-3a9 (targeting eRF3a mRNA) have been previously described (26). Plasmids ptRNAam, ptRNAop and ptRNAoc expressing amber, opal and ochre suppressor tRNAs, respectively, have been described in (31). Plasmids pSUPERpuro-hUpf1/I and pSUPERpuro-hUpf1/II targeting two different sequences in human Upf1 (32) and pSUPERcontrol with the non-silencing shRNA sequence (5′-ATTCTCCGAACGTGTCACG-3′) used as negative control shRNA (sh-Ctrl) were a generous gift of O. Mühlemann.
Table 1.
Oligonucleotides used in this study
| Name | Sequence 5′→3′ |
|---|---|
| ATF4 probe | GCTTCCTATCTCCTTCAGTGATAT CCACTTCACTGCCCAG |
| B2M probe | TTCAAACCTCCATGATGCTGCTTACA TGTCTCGATCCCAC |
| FLUC probe | TCCTTAGAGAGGGGAGCGCCACCA GAAGCAATTTCGTGTA |
| ATF4 5′UTR-fw | TTCTAGCTAGCCCGCCCACAGATGTA GTTTTC |
| ATF4 5′UTR-rev | CGCGGATCCACAGCCAGCCATTCG GAGGA |
| ATF4 qPCR fw | CCAACAACAGCAAGGAGGAT |
| ATF4 qPCR rev | GGGGCAAAGAGATCACAAGT |
| REDD1 qPCR fw | GAGGAAGACACGGCTTACCT |
| REDD1 qPCR rev | CCACCTGGCTTACCAACTG |
| CHOP qPCR fw | CAGAACCAGCAGAGGTCACA |
| CHOP qPCR rev | AGCTGTGCCACTTTCCTTTC |
| TRIB3 qPCR fw | TGGTACCCAGCTCCTCTACG |
| TRIB3 qPCR rev | GACAAAGCGACACAGCTTGA |
| ASNS qPCR fw | ATCACTGTCGGGATGTACCC |
| ASNS qPCR rev | CTTCAACAGAGTGGCAGCAA |
| B2m qPCR fw | TATCCAGCGTACTCCAAAGA |
| B2m qPCR rev | GACAAGTCTGAATGCTCCAC |
| GSPT1 qPCR fw | GAGGAGGAAGAGGAAATCCC |
| GSPT1 qPCR rev | TCCTTTTGTCAACCATTCCA |
| ACTB qPCR fw | TCCCTGGAGAAGAGCTACGA |
| ACTB qPCR rev | AGCACTGTGTTGGCGTACAG |
| Hs_DDIT4_6 (R6) | CAGTACTGTAGCATGAAACAAa |
| Hs_DDIT4_9 (R9) | AAGAAGCTGTACAGCTCGGAAa |
aTarget sequence in human REDD1 gene.
Predesigned short interfering RNAs (siRNAs) directed against human DDIT4/REDD1: Hs_DDIT4_6 siRNA (si-R6) and Hs_DDIT4_9 siRNA (si-R9) and AllStars Negative Control siRNA were obtained from Qiagen (see target sequences in Table 1).
Cell culture, electroporation and transfection
The HCT116 cell line (ATCC number : CCL-247) was maintained in McCoy medium (Invitrogen) supplemented with 10% fetal calf serum, 100 µg/ml streptomycin and 100 U/ml penicillin at 37°C under 5% CO2 atmosphere. The 2XAARE-TRB3-TK-LUC construct used for the stable HeLa cell line was previously described (33). This construct was cloned into a lentiviral expression vector (34) and HeLa cells were then infected with these lentiviral particles. These cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 100 µg/ml streptomycin and 100 U/ml penicillin at 37°C under 5% CO2 atmosphere.
Electroporation of cells was performed as described (26) with a Gene pulser II electroporation system (Bio-Rad) using 4.8 × 106 cells and 10 µg of plasmid DNA. eRF3a depletion by RNAi was performed with sh-3a1 and sh-3a9 as described (26). Electroporated cells were collected 24–72 h after electroporation as indicated. Knockdown of hUpf1 by RNAi was induced by co-transfection of an equimolar mix of pSUPERpuro-hUpf1/I and pSUPERpuro-hUpf1/II plasmids (32). When electroporation was followed by transfection, electroporated cells were seeded in 6-well plates. Transfection was carried out using Lipofectamine 2000 (Invitrogen) and either 0.1–0.2 µg/well of plasmid DNA (Luciferase experiments) or 100 pmol/well of predesigned siRNA (REDD1 silencing experiments) according to the manufacturer's instructions. Typically, cells were treated for further analyses 24 h after transfection.
Polysome fractionation, RNA isolation, mRNA stability and northern blot analysis
Isolation of polysomes and polysome analysis were performed according to (31). RNA isolation from polysomal fractions was performed as described in (35). After ethanol precipitation, the RNA pellets corresponding to polysome fractions, typically fractions 1–9; see Figure 8 in (32) were resuspended in water and pooled.
For mRNA stability analysis, 48 h after electroporation, the cells were incubated for various times in fresh culture medium containing actinomycin D (5 µg/ml). RNA isolation and northern blotting were performed as described (35). The membranes were hybridized with oligonucleotide probes (Table 1) 32P-labeled with T4 polynucleotide kinase (Biolabs) and purified using QIAquick Nucleotide Removal kit (Qiagen). A mix of two oligonucleotides (Table 1), including β2-microglobulin (B2M) probe that served as a loading control, was used for hybridization. The membranes were washed under stringent conditions, exposed to phosphor screen and signals were analyzed using the Storm 860 Imaging system (Molecular Dynamics) and the ImageQuant software.
Microarray hybridization and data analysis
For gene expression profiling, we processed two independent replicates for total RNA and three independent replicates for polysome-associated RNA from control (sh-Ctrl) and eRF3a-depleted (sh-3a1) cell preparations. cRNA was synthesized, amplified and purified using the Illumina TotalPrep RNA Amplification Kit (Ambion) following manufacturer’s recommendations. Briefly, 200 ng of RNA was reverse transcribed. After second strand synthesis, the cDNA was transcribed in vitro and cRNA labeled with biotin-16-UTP. Labeled probes were hybridized in triplicate to HumanHT12-v3 Expression BeadChips (Illumina) using Illumina's protocol. The Beadchips were scanned on the Illumina IScan using Illumina IScan image data acquisition software. Illumina GenomeStudio software was used for preliminary data analysis. Raw microarray intensity data were background subtracted and normalized using the ‘normalize quantiles’ function in the GenomeStudio Software. We used the detection P-vals as flags, flag = 0, if P > 0.05 and flag = 1, if P ≤ 0.05. Each tested probe list have been created filtering probes flagged ‘1’ for at least half of the chips involved in the comparison. The group comparisons were done using Student’s t-test. To estimate the false discovery rate (FDR), we filtered the resulting P-values at 5% and used the Benjamini and Hochberg FDR correction (36). Microarray data have been deposited at MIAME and can be accessed at http://www.ebi.ac.uk/arrayexpress/. Experiment name: OJeanJean_eRF3a_ATF4, ArrayExpress accession: E-MEXP-3512.
Real-time quantitative PCR analysis
Total RNA was extracted 72 h after electroporation using NucleoSpin RNAII Kit (Macherey-Nagel) and treated with DNase I, Amp Grade (Invitrogen) prior to cDNA synthesis. Total RNA (1 µg) was reverse transcribed with 200 U of SuperScript® II Reverse Transcriptase (Invitrogen) using 100 µM of random hexamer primers according to manufacturer’s instructions. Real-time PCRs were performed on a LightCycler®480 System (Roche Applied Science) in 96-well plates. A master mix containing 0.5 µM of forward primer, 0.5 µM of reverse primer and 5 µl LightCycler SYBR Green I Master (Roche Diagnostics) was prepared for each target or reference gene mRNA. Primer pairs are listed in Table 1. Reactions were carried out in 10 µl with 4 µl of cDNA from a 100× dilution of the RT reaction as PCR template and 6 µl of master mix. Serial dilutions of a mixture of all cDNAs were used to generate a PCR standard curve. The LightCycler run protocol was as follows: denaturation and hot-start enzyme activation program (95°C for 5 min), amplification and quantification program repeated 50 times (95°C for 10 s, 60°C for 15 s and 72°C for 15 s with a single fluorescence measurement), melting curve program (65–95°C with a heating rate of 0.1°C/s and a continuous fluorescence measurement), and finally a cooling step to 40°C. All reactions were performed in triplicate in each 96-well plate. A negative control without cDNA template was run with every assay to assess the overall specificity. Absence of genomic DNA contamination was assessed using enzyme lacking reverse transcriptase reactions as template (no-RT). Data were analyzed using the Roche LightCycler®480 software and CP was calculated by the Second Derivative Maximum Method. The relative expression ratio of a target gene was calculated as described (37), based on real-time PCR efficiencies. The amount of the target mRNA was examined and normalized to the Actb (β-actin) and B2m reference gene mRNAs. Results were expressed as the ratio of mRNA of eRF3a-depleted cells relative to that of control cells.
Luciferase assays
For dual luciferase assay, transfection of electroporated cells was carried out in 6-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A mix of 100 ng of pRL-TK and 200 ng of p5′-UTR-ATF4-FLUC was used for transfection. Twenty-four hours after transfection, cells were resuspended in passive lysis buffer (Promega) and the firefly and Renilla luciferase activities were measured in triplicate using the Dual-luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions with a Wallac 1420 VICTOR2 luminometer (PerkinElmer). The average values and the ratios of firefly to Renilla luciferase activity were calculated.
For the 2XAARE-TRIB3-TKLUC cell line, 48 h after electroporation with sh-3a1 directed against eRF3a mRNA or control shRNA, cells were resuspended in passive lysis buffer (Promega) and the firefly luciferase activities were measured in triplicate using the Dual luciferase Reporter Assay kit (Promega) as described above. Protein concentrations in extracts were determined using Bradford Reagent (Sigma), bovine serum albumin being used as standard. Luciferase activity was calculated as the ratio of light units to microgram of protein and expressed as relative light units (RLUs).
Western blot analysis
Cell pellets were resuspended in 25–100 µl of a lysis buffer (50 mM Tris–HCl, pH 8, 120 mM NaCl, 1% NP40, 1 mM EDTA) containing 2× Complete EDTA-free cocktail of protease inhibitors and 1× phosphatase inhibitor cocktail (Roche) and 1 µg/ml pepstatine. Cells were lysed by sonication on ice, centrifuged for 20 min at 16 000g and supernatant retained as cell extracts. Protein concentrations of extracts were determined using Bradford Reagent (Sigma), bovine serum albumin being used as standard. For each sample, 30 µg of total protein were loaded on polyacrylamide gel and subjected to electrophoresis. Western blotting was then performed as described (31). Antibodies directed against human eRF3a were previously described (26). Rabbit polyclonal antibodies against human REDD1 (10 638-1-AP) were purchased from Proteintech. Antibodies against Asparagine synthetase (ASNS, C-14), ATF4 (sc-200) and β-actin (sc-7210) were purchased from Santa Cruz Biotechnology. Antibodies directed against eIF2α, phospho-eIF2α (Ser51), 4E-BP1, phospho-4E-BP1 (Thr37/46), S6, phospho-S6(Ser240/244), S6K1, phospho-S6K1 (Thr389) were purchased from Cell Signaling Technology. The anti-α-tubulin antibody (DM1A) was from GE Healthcare. Proteins were detected by chemiluminescence and exposure to X-ray film. Signal intensities from each western blot were quantified using the ImageJ 1.44 p software (NIH) after densitometric scanning of the films.
RESULTS
Microarray analysis of eRF3a depletion in human cells
We previously showed that eRF3a depletion in the human colon carcinoma cell line HCT116 p53+/+ induces mTOR signaling pathway inhibition (31), suggesting that eRF3a belongs to a regulatory process that modulates mTOR activity. However, the molecular mechanism and the precise role of eRF3a remained to be determined. Here, to elucidate the mechanism dictating mTOR signaling pathway inhibition, we aimed to define the transcript expression and polysome profiles of HCT116 cells after siRNA-mediated depletion of eRF3a. Three days after HCT116 cell electroporation with either sh-3a1, which was previously shown to effectively silence eRF3a (26), or non-silencing control shRNA (sh-Ctrl), total and polysome-associated RNAs were extracted and we conducted a microarray analysis of differentially expressed genes using the Illumina HumanHT-12 Expression BeadChips. Informative genes were selected as having a ‘present’ call in at least half of the samples in each condition (sh-3a1 and sh-Ctrl). The resulting gene lists were filtered such that only probes with a detection P < 0.05 and an average differential expression score >1.5 were included [see Supplementary files S1-RNA and S2-Polysome, and ‘Materials and Methods’ section for the details of the statistical analyses]. The P-value of the moderated t-test was adjusted with a Benjamini and Hochberg multiple test correction procedure (36) to identify statistically significant differentially expressed genes. We then selected all genes that had an FDR < 0.05 and fold change larger than 2. Using this metric, 36 genes were identified in the total RNA fraction as upregulated in cells with sh-3a1 targeting eRF3a compared to cells with the control sh-RNA (Table 2). This list included multiple genes involved in amino acid metabolism and transport and genes known to contain an AARE and to respond to ATF4 binding, e.g. ASNS, DDIT4 (also known as REDD1), TRIB3, DDIT3 also known as CHOP (16). In this analysis, ATF4 was also found to be upregulated but at a lower level (Table 2, the knockdown of GSPT1/eRF3a which is targeted by si-3a1 is also shown). Surprisingly, ATF3 that also contains an AARE and responds to ATF4 binding (38), and GADD34 that seems to be not directly regulated by ATF4 but is a downstream target of the eIF2α kinase/ATF4-dependent stress response pathways, were not significantly upregulated, ∼1.17- and ∼1.28-fold, respectively. The analysis of polysome-associated RNA confirmed the results obtained with total RNA. Using the same requirements to identify significant differentially expressed genes as described above for total RNA, we obtained a list of 194 genes that were upregulated more than 2-fold in the polysomal fraction of eRF3a-depleted cells (see Table 3 for the top 50 of these genes and Supplementary File S2-polysome for the entire list). As shown in Table 3, a number of these genes are also involved in amino acid metabolism, transport and respond to ATF4 binding, including ASNS, DDIT4/REDD1, TRIB3, DDIT3/CHOP. These results indicated that eRF3a depletion induced the ATF4 response.
Table 2.
Genes differentiality expressed following eRF3a depletion, total RNA
| Gene | Probea | Fold change | BHb | Function |
|---|---|---|---|---|
| UPP1 | ILMN_1742764 | 4.46 | 0.00001 | Uridine phosphorylase 1 |
| PCK2 | ILMN_1802699 | 4.01 | 0.00155 | Phosphoenolpyruvate carboxykinase 2 |
| IL20RB | ILMN_1765668 | 4.01 | 0.00473 | Interleukin 20 receptor beta |
| SERPINA3 | ILMN_1788874 | 3.90 | 0.04803 | Serpin peptidase inhibitor |
| GCNT3 | ILMN_1712082 | 3.64 | 0.02760 | Glucosaminyl (N-acetyl) transferase 3 |
| FAM129A | ILMN_1667966 | 3.54 | 0.00166 | Family with sequence similarity 129 |
| DDIT4, REDD1 | ILMN_1661599 | 3.36 | 0.00144 | DNA damage-inducible transcript 4 |
| CDKN2B | ILMN_1670111 | 3.17 | 0.02120 | Cyclin-dependent kinase inhibitor 2B |
| SLC7A11 | ILMN_1655229 | 3.08 | 0.00436 | Solute carrier family 7 |
| TSC22D3 | ILMN_2376403 | 3.01 | 0.01653 | TSC22 domain family, member 3 |
| OR2A9P | ILMN_2203544 | 2.99 | 0.00049 | Olfactory receptor, family 2, pseudogene |
| TRIB3 | ILMN_1787815 | 2.92 | 0.03081 | Tribbles homolog 3 (Drosophila) |
| LAMB3 | ILMN_1715684 | 2.84 | 0.00902 | Laminin, beta 3 |
| ASNS | ILMN_1796417 | 2.84 | 0.00113 | ASNS |
| ARRDC4 | ILMN_2184064 | 2.70 | 0.01060 | Arrestin domain containing 4 |
| PSAT1 | ILMN_1692938 | 2.60 | 0.02332 | Phosphoserine aminotransferase 1 |
| C6ORF48 | ILMN_2285708 | 2.57 | 0.01694 | Chromosome 6 ORF 48 |
| LCN2 | ILMN_1692223 | 2.55 | 0.04327 | Lipocalin 2 |
| DDIT3, CHOP | ILMN_1676984 | 2.54 | 0.02806 | DNA damage-inducible transcript 3 |
| CLIC4 | ILMN_1671250 | 2.46 | 0.00570 | Chloride intracellular channel 4 |
| SLC6A9 | ILMN_1714445 | 2.35 | 0.00242 | Solute carrier family 6 |
| GPT2 | ILMN_1684158 | 2.33 | 0.00232 | Glutamic pyruvate transaminase |
| PHGDH | ILMN_1704537 | 2.22 | 0.000001 | Phosphoglycerate dehydrogenase |
| LAMC2 | ILMN_1701424 | 2.22 | 0.00242 | laminin, gamma 2 |
| WFDC3 | ILMN_1740415 | 2.20 | 0.03031 | WAP four-disulfide core domain 3 |
| COX19 | ILMN_1656656 | 2.19 | 0.01029 | Cytochrome c oxidase assembly homolog |
| ZNF473 | ILMN_1782730 | 2.15 | 0.01393 | Zinc finger protein 473 |
| XPOT | ILMN_2188374 | 2.11 | 0.00049 | Exportin |
| KCNG1 | ILMN_1673769 | 2.10 | 0.01655 | Potassium voltage-gated channel |
| OR2A20P | ILMN_2081269 | 2.09 | 0.01344 | Olfactory receptor, family 2, pseudogene |
| TP53BP2 | ILMN_2270299 | 2.06 | 0.03046 | Tumor protein p53 binding protein |
| ARRDC3 | ILMN_2198516 | 2.05 | 0.00220 | Arrestin domain containing 3 |
| BTC | ILMN_2081070 | 2.05 | 0.00529 | β-Cellulin |
| CDCP1 | ILMN_1688670 | 2.03 | 0.02511 | CUB domain containing protein 1 |
| CARS | ILMN_2367469 | 2.01 | 0.00049 | Cysteinyl-tRNA synthetase |
| HSPA13 | ILMN_2231985 | 2.01 | 0.03081 | Heat shock protein 70 kDa family |
| ATF4 | ILMN_2358457 | 1.81 | 0.01043 | ATF4 |
| GSPT1 (eRF3a) | ILMN_1750130 | 0.36 | 0.00220 | Eukaryotic release factor 3 a |
aWhen more than one probe was identified for a single gene, only the probe giving the highest fold change is shown.
bBenjamini and Hochberg multiple test correction, FDR < 0.05.
Table 3.
Genes differentiality expressed following eRF3a depletion, polysomes, top50
| Gene | Probea | Fold change | BHb | Function |
|---|---|---|---|---|
| DDIT4, REDD1 | ILMN_1661599 | 11.12 | 0.00017 | DNA damage-inducible transcript 4 |
| C9ORF150 | ILMN_1701361 | 9.05 | 0.00034 | Chromosome 9 ORF 150 |
| SESN2 | ILMN_1751598 | 8.15 | 0.00000 | Sestrin 2 |
| PCK2 | ILMN_1802699 | 7.96 | 0.00110 | Phosphoenolpyruvate carboxykinase 2 |
| TSC22D3 | ILMN_2276952 | 7.08 | 0.00009 | TSC22 domain family, member 3 |
| ASNS | ILMN_2398107 | 6.47 | 0.00007 | ASNS |
| IL8 | ILMN_2184373 | 5.76 | 0.00000 | Interleukin 8 |
| CCL20 | ILMN_1657234 | 4.70 | 0.00011 | Chemokine (C–C motif) ligand 20 |
| TRIB3 | ILMN_1787815 | 4.61 | 0.00000 | Tribbles homolog 3 (Drosophila) |
| PRDM13 | ILMN_1698753 | 4.52 | 0.00198 | PR domain containing 13 |
| PHGDH | ILMN_1704537 | 4.50 | 0.00083 | Phosphoglycerate dehydrogenase |
| UPP1 | ILMN_1742764 | 4.10 | 0.00000 | Uridine phosphorylase 1 |
| DDIT3, CHOP | ILMN_1676984 | 3.94 | 0.00000 | DNA damage-inducible transcript 3 |
| NR4A2 | ILMN_2339955 | 3.88 | 0.00001 | Nuclear receptor subfamily 4 |
| GFPT1 | ILMN_2220184 | 3.60 | 0.00009 | Glutamine-fructose-6-phosphate transaminase 1 |
| EIF3C | ILMN_2330410 | 3.38 | 0.02239 | Eukaryotic translation initiation factor 3, subunit C |
| GCNT3 | ILMN_1712082 | 3.36 | 0.00008 | Glucosaminyl (N-acetyl) transferase 3, mucin type |
| STC2 | ILMN_1691884 | 3.29 | 0.00013 | Stanniocalcin 2 |
| TARS | ILMN_1685480 | 3.23 | 0.00093 | Threonyl-tRNA synthetase |
| OR2A1 | ILMN_2074432 | 3.20 | 0.00557 | Olfactory receptor, family 2, subfamily A, member 1 |
| PREPL | ILMN_1666615 | 3.19 | 0.00362 | Prolyl endopeptidase like |
| SARS | ILMN_1786972 | 3.18 | 0.00076 | Seryl-tRNA synthetase |
| ALDH2 | ILMN_1793859 | 3.11 | 0.00269 | Aldehyde dehydrogenase 2 family |
| MVP | ILMN_2344373 | 3.08 | 0.03346 | Major vault protein |
| ARHGEF1 | ILMN_1772370 | 3.03 | 0.00013 | Rho guanine nucleotide exchange factor 1 |
| CARS | ILMN_2367469 | 3.03 | 0.00081 | Cysteinyl-tRNA synthetase |
| MARS | ILMN_1799819 | 2.93 | 0.00167 | Methionyl-tRNA synthetase |
| IL20RB | ILMN_1765668 | 2.92 | 0.00012 | Interleukin 20 receptor beta |
| GSN | ILMN_1801043 | 2.92 | 0.00005 | Gelsolin (amyloidosis, Finnish type) |
| NFIL3 | ILMN_1707312 | 2.89 | 0.00034 | Nuclear factor, interleukin 3 regulated |
| SLC7A11 | ILMN_1655229 | 2.86 | 0.01229 | Solute carrier family 7 |
| LOC728689 | ILMN_1725984 | 2.84 | 0.02796 | Similar to eukaryotic translation initiation factor3, subunit8 |
| OR2A9P | ILMN_2203544 | 2.78 | 0.00069 | Olfactory receptor, family 2, pseudogene |
| ACOX1 | ILMN_1765767 | 2.71 | 0.00995 | Acyl-Coenzyme A oxidase 1, palmitoyl |
| TNFAIP3 | ILMN_1702691 | 2.69 | 0.00046 | Tumor necrosis factor, alpha-induced protein 3 |
| ULBP1 | ILMN_1738407 | 2.67 | 0.00001 | UL16 binding protein 1 |
| EPSTI1 | ILMN_2388547 | 2.65 | 0.00516 | Epithelial stromal interaction 1 (breast) |
| KIFAP3 | ILMN_2119224 | 2.62 | 0.00140 | Kinesin-associated protein 3 |
| AARS | ILMN_1662364 | 2.60 | 0.03870 | Alanyl-tRNA synthetase |
| FLYWCH1 | ILMN_1690035 | 2.59 | 0.00000 | FLYWCH-type zinc finger 1 |
| HEY1 | ILMN_1788203 | 2.58 | 0.00008 | Hairy/enhancer-of-split related with YRPW motif 1 |
| PAPSS1 | ILMN_2224103 | 2.57 | 0.03062 | 3-phosphoadenosine 5-phosphosulfate synthase 1 |
| TTC27 | ILMN_1757730 | 2.57 | 0.01883 | Tetratricopeptide repeat domain 27 |
| FAM129A | ILMN_1810725 | 2.53 | 0.00009 | Family with sequence similarity 129, member A |
| CTNNA1 | ILMN_2230902 | 2.53 | 0.02117 | Catenin (cadherin-associated protein), alpha 1 |
| VPS8 | ILMN_1678268 | 2.51 | 0.00180 | Vacuolar protein sorting 8 homolog |
| SP100 | ILMN_2390586 | 2.50 | 0.02301 | SP100 nuclear antigen |
| PSAT1 | ILMN_1692938 | 2.49 | 0.00079 | Phosphoserine aminotransferase 1 |
| ARHGEF2 | ILMN_1703477 | 2.49 | 0.00006 | Rho/rac guanine nucleotide exchange factor |
| ATF4 | ILMN_2358457 | 1.69 | 0.00334 | ATF4 |
| GSPT1, eRF3a | ILMN_1750130 | 0.57 | 0.00225 | Eukaryotic release factor 3 a |
aWhen more than one probe was identified for a single gene, only the probe giving the highest fold change is shown.
bBenjamini and Hochberg multiple test correction, FDR < 0.05.
Validation of the microarray results
qRT-PCR assays were used to verify the results of the gene expression changes measured using microarrays, with particular focus on ATF4 and classic ATF4 target genes, including ASNS, REDD1, TRIB3 and CHOP. The mRNA for B2M, which was found to be stable in the microarray analysis (fold change = 1.03), was used as the endogenous reference mRNA in the qRT-PCR assays. GSPT1 gene encoding eRF3a was used as a control to assess efficient eRF3a depletion. As shown in Figure 1A, based on three independent electroporation experiments, the ATF4, ASNS, REDD1, TRIB3 and CHOP genes were significantly upregulated in HCT116 cells electroporated with sh-3a1 compared to control cells electroporated with sh-Ctrl. Equivalent results were obtained using another shRNA, sh-3a9 (Supplementary Figure S1), which was previously shown to efficiently target eRF3a mRNA (26). qRT-PCR data correlated well with the microarray results although the expression changes were higher than those observed in microarray analysis. This was likely due to differences in method sensitivity.
Figure 1.
eRF3a depletion activates the ATF4 response. (A) qRT-PCR performed on total RNA of HCT116 cells 3 days after electroporation with shRNA targeting eRF3a mRNA (sh-3a1) or control shRNA (sh-Ctrl). The ratio of mRNA levels in sh-3a1 versus sh-Ctrl electroporated cells was calculated. Bars and error bars correspond to averages and SD from at least three independent experiments. mRNA levels of ATF4, selected targets of ATF4 (ASNS, REDD1, TRIB3 and CHOP) and GSPT1-encoding eRF3a are shown. (B) Western blot analysis of the time course of expression of eRF3a, total S6K1, S6K1 phosphorylated form (P-S6K1), ATF4, ASNS, REDD1 and the phosphorylated form of eIF2α (P-eIF2α) at 24, 48 and 72 h, respectively, after electroporation of cells with sh-3a1 or sh-Ctrl as indicated; α-TUB and β-ACT served as loading controls. Quantification of the signals (see ‘Materials and Methods’ section) are shown below each panel. Each signal was divided by the signal elicited by the reference protein, either α-TUB or β-ACT. For each time point, the control experiment was set to 1 (ND, not detected). (C) The 2XAARE-TRIB3-TKLUC cell line that expresses a Luc reporter driven by two AAREs upstream of the TK minimal promoter was electroporated with sh-3a1 or sh-Ctrl; (NT) corresponds to non-treated cells. Luciferase activity was calculated as the ratio of light units to microgram of protein and expressed as RLU. Bars and error bars correspond to averages and SD from three independent experiments. P-values (two-tailed Student’s t-test) are indicated for the relevant samples.
For selected genes, microarray results were also confirmed at the protein level by western blot analysis. The time course of ATF4, ASNS and REDD1 protein levels was examined in eRF3a-depleted cells and control cells (Figure 1B, sh-3a1 and sh-Ctrl, respectively; an additional experiment is presented in Supplementary Figure S2) at 24, 48 and 72 h after electroporation. We have previously shown that p70 S6 kinase 1 (S6K1) and the eucaryotic initiation factor 4E-binding protein 1 (4E-BP1), two direct targets of mTOR kinase activity, were hypophosphorylated upon eRF3a depletion (31). Thus, we examined S6K1 and 4E-BP1 phosphorylation status to monitor the inhibitory effect of eRF3a depletion on mTOR activity. For each western blot experiment, either α-tubulin (α-TUB) or β-actin (β-ACT) levels served as the loading control. Consistent with the results of microarrays and qRT-PCR analyses, the levels of ATF4, ASNS and REDD1 were significantly increased at 48 and 72 h of eRF3a depletion. Concomitantly, the phosphorylation of S6K1 (Figure 1B and Supplementary Figure S2) and 4E-BP1 (supplementary Figure S2) was reduced. Since the phosphorylation of eIF2α is an established marker in the cellular response to a variety of stresses promoting ATF4 activity (18), the eIF2α phosphorylation status was also examined. Interestingly, the level of eIF2α phosphorylation (P-eIF2α) was not increased in eRF3a-depleted cells when compared to control cells (Figure 1B and Supplementary Figure S2), suggesting that ATF4 upregulation was not induced through the activation of one of the four kinases which converge to P-eIF2α/ATF4 axis.
To assess the increase of ATF4 transcriptional activity in eRF3a-depleted cells, we used a stable cell line (2XAARE-TRIB3-TKLUC cells) expressing a luciferase gene under the control of two copies of TRIB3 AARE core sequence inserted upstream of the minimal herpes simplex virus promoter for thymidine kinase (TK). It has been demonstrated in vitro (39) and in vivo (33) that ATF4 binds the TRIB3 AARE in response to ER stress and to amino acids starvation. The 2XAARE-TRIB3-TKLUC cell line was electroporated with either sh-3a1 directed against eRF3a mRNA or control shRNA (sh-Ctrl) and 48 h later luciferase activity was monitored. As shown in Figure 1C, eRF3a depletion induced a ∼5-fold increase in luciferase activity suggesting that ATF4 binding to TRIB3 AARE was enhanced. Together with the results of microarrays, qRT-PCR and western blot analyses, this observation indicated (i) that ATF4 expression was upregulated in eRF3a-depleted cells and (ii) that this upregulation did not involve eIF2α phosphorylation.
Remembering that ATF4 mRNA 5′-UTR contains uORFs that modulate ATF4 ORF translation (14,15), and that the presence of these uORFs also promote ATF4 mRNA degradation by the nonsense-mediated decay (NMD) pathway (40,41), we set out to test whether eRF3a depletion modifies ATF4 mRNA translation and stability.
Modulation of ATF4 mRNA stability in eRF3a-depleted cells
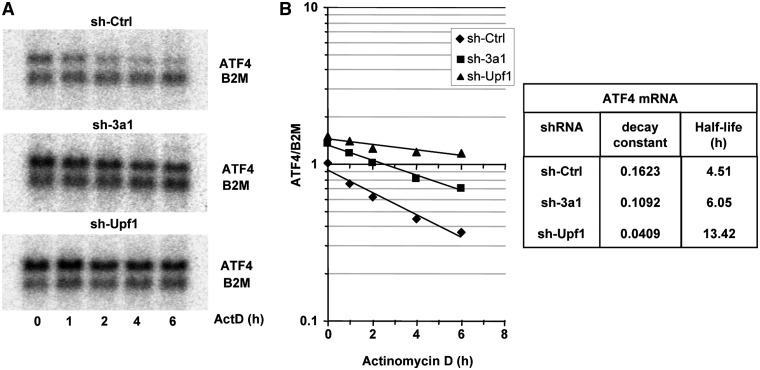
To assess whether eRF3a depletion influences ATF4 mRNA stability, we performed stability assays with mRNA from cells electroporated with sh-Ctrl and sh-3a1 and treated for various times (0–6 h) with Actinomycin D. We then examined ATF4 mRNA decay by northern blotting using B2M mRNA as a control. In a parallel experiment, we used shRNAs directed against Upf1 mRNA (32) to measure ATF4 mRNA decay when NMD is inhibited. As shown in Figure 2, depletion of eRF3a moderately increased (∼1.5 fold) the steady-state abundance and half-life of ATF4 mRNA. By comparison, Upf1 depletion induced a 3-fold extension of ATF4 mRNA half-life. This suggested that the prolonged ATF4 mRNA half-life in eRF3a-depleted cells could be due to a partial inhibition of ATF4 mRNA decay driven by the NMD pathway. Indeed, it is well documented that, due to the presence of uORFs in its 5′-UTR, ATF4 mRNA is sensitive to NMD (40). However, by decreasing the translation termination efficiency and promoting translational readthrough (26,27), eRF3a depletion could also affect the regulation of ATF4 translation that is dependent on uORFs translation.
Figure 2.
ATF4 stability in HCT116 cells after electroporation with control shRNA (sh-Ctrl) or shRNAs targeting either eRF3a mRNA (sh-3a1) or Upf1 mRNA (sh-Upf1). Two days after electroporation, cells were treated with Actinomycin D (5 µg/ml) for various time as indicated and total RNA was extracted. (A) After RNA separation on denaturing agarose gel, northern blot analysis was performed using ATF4 and B2M probes (B2M probe served as a loading control). (B) The signals were quantified using the Storm 860 Imaging system (Molecular Dynamics) and the ImageQuant software, and plotted as the ratio of ATF4 to B2M signals. Half-lives and decay constants were calculated from the regression curves.
eRF3a depletion induces translational readthrough at ATF4 mRNA uORF1 and increases ATF4 ORF translation
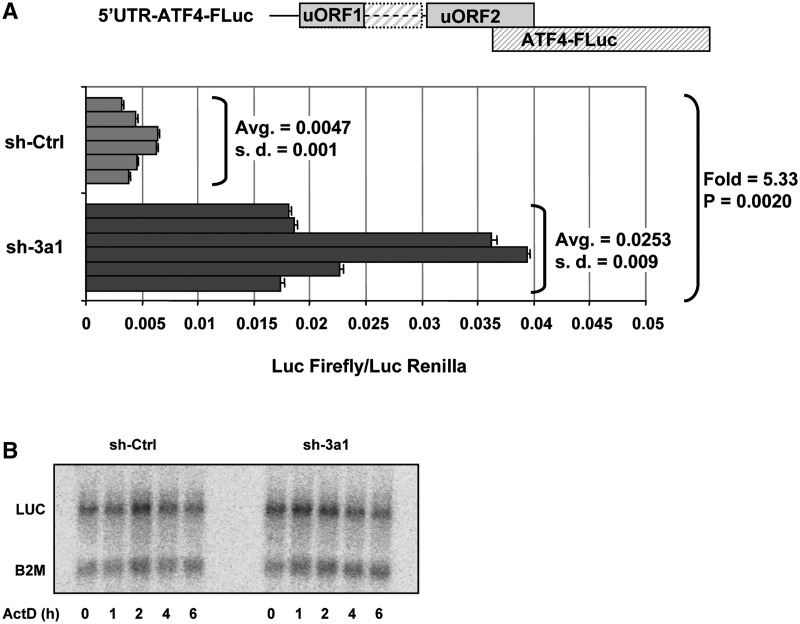
In ATF4 5′-UTR, the two uORFs, uORF1 and uORF2, are separated by ∼90 bases and an additional 100 bases separate uORF2 and ATF4 ORF initiation codons. To determine the effect of shRNA-mediated depletion of eRF3a on ATF4 ORF translation, we constructed a luciferase expressing vector containing the human ATF4 5′-UTR and the 5′-end of ATF4 ORF fused to firefly luciferase (FLuc) gene (Figure 3A). HCT116 cells were electroporated with either sh-3a1 or sh-Ctrl and 48 h later, cells were cotransfected with the 5′-UTR-ATF4-FLuc construct and the control plasmid expressing Renilla Luc gene using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, Renilla and Firefly luciferase activities were measured in cell extracts. The depletion of eRF3a mediated by sh-3a1 led to a significant ∼5-fold increase in Firefly luciferase relative activity (Figure 3A). As steady-state level and stability of the 5′-UTR-ATF4-FLuc mRNA were not affected in cells electroporated with sh-3a1 when compared to cells electroporated with sh-Ctrl (Figure 3B), our results suggested that eRF3a depletion increased translation of ATF4 ORF by inducing readthrough at uORF1 stop codon. The readthrough at uORF1 UAG stop codon extended translation to the next in frame stop codon, a UAA codon located only three bases upstream of uORF2 initiator AUG. This very short distance is not sufficient to allow the binding of the eIF2–GTP–Met-tRNAMet ternary complex to the small ribosomal subunit and translation reinitaition at uORF2 AUG. Conversely, the 100 bases separating the UAA stop codon of the elongated uORF1 from ATF4 ORF start codon are sufficient to allow the small ribosomal subunits that have resumed scanning, to be recharged with the ternary complex and to reinitiate translation at ATF4 AUG. This mechanism could explain how eRF3a depletion promotes translational upregulation of ATF4.
Figure 3.
eRF3a depletion increases ATF4 ORF translation. Two days after electroporation with shRNA targeting eRF3a mRNA (sh-3a1) or control shRNA (sh-Ctrl), cells were cotransfected with plasmids expressing firefly (p5′-UTR-ATF4-FLuc) or Renilla (pRL-TK) luciferases. (A) The situation of ATF4 uORF1, uORF2 and ATF4-FLuc fusion ORF in plasmid p5′-UTR-ATF4-FLuc is represented above the graph; the hatched box correspond to the elongated uORF1. Dual luciferase assays were performed in triplicate at 24 h after transfection. The graph shows six independent transfection experiments. For each experiment, bars and error bars correspond to averages and SD of the luciferase assay triplicates. The average values (avg) and SD of the six experiments are indicated on the graph with the P-value (two-tailed Student’s t-test) and fold change of luciferase activity for sh-3a1 versus sh-Ctrl electroporated cells. (B) Stability of FLuc reporter mRNA in HCT116 cells after electroporation with shRNA targeting eRF3a mRNA (sh-3a1) or control shRNA (sh-Ctrl). Three days after electroporation, cells were treated with Actinomycin D (5 µg/ml) for various time as indicated. Total RNA was extracted and separated on a denaturing agarose gel prior northern blot analysis. The membrane was probed with LUC probe and B2M probe that served as a loading control.
It has been previously reported that the two uORFs present in the 5′-non-coding portion of the ATF4 mRNA are conserved among vertebrates including the length of the uORFs and the distances separating their initiation codons (15). Re-examining ATF4 5′-UTR of various species, we found that uORF1 UAG stop codon and the following in frame UAA stop codon preceding uORF2 were also remarkably conserved in mammals (see the alignment in Supplementary Figure S3). This conservation strongly suggested that, in addition to the distance separating uORF1 and uORF2, both stop codons (uORF1 UAG and in frame UAA) and their distance to ATF4 AUG also played a crucial role in ATF4 translational control.
The increase in REDD1 expression is responsible for mTOR inhibition in eRF3a-depleted cells
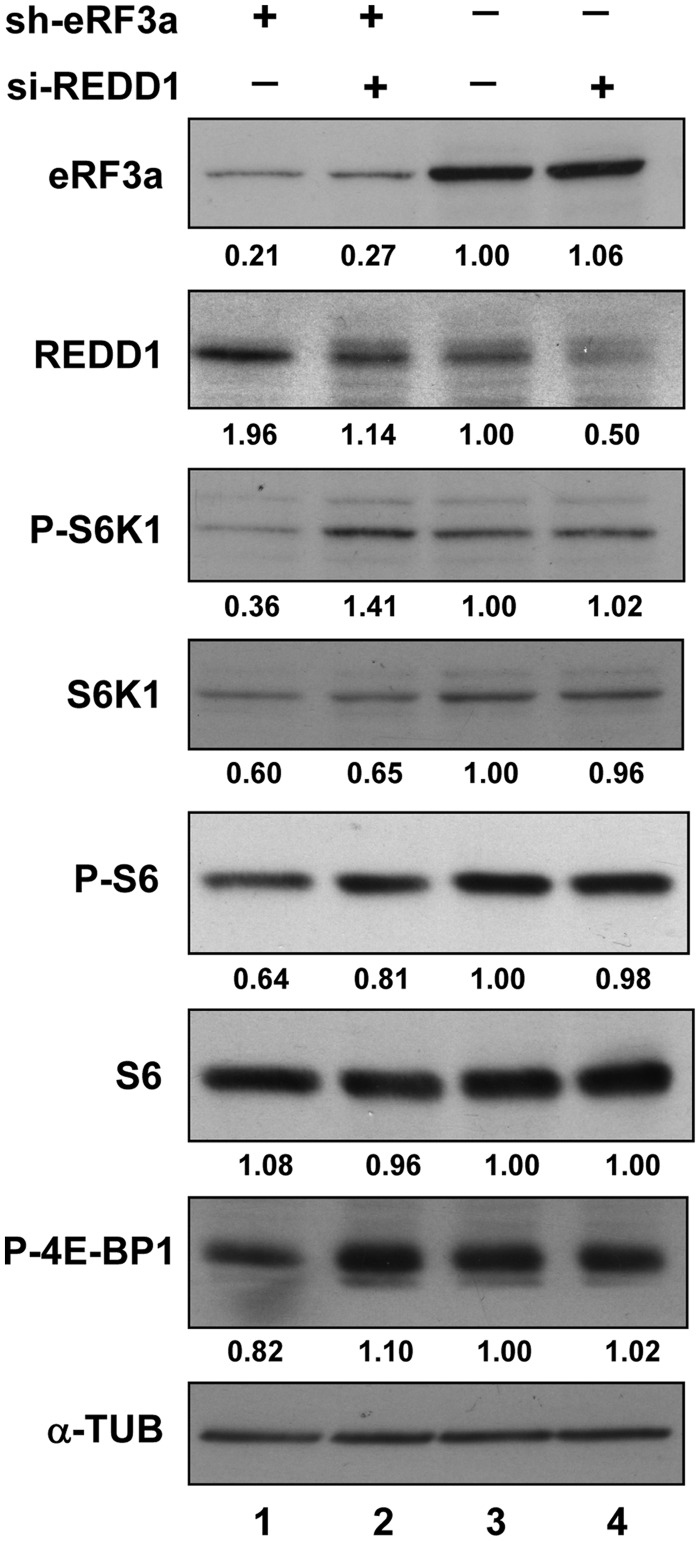
REDD1 was originally identified during a screen for genes whose expression was altered in response to hypoxia in cultured cells (42) and in subsequent studies, REDD1 expression was shown to be upregulated in response to a wide variety of cell stresses including ER stress (43). It has also been shown that increased ATF4 expression is both necessary and sufficient for upregulation of REDD1 expression in response to ER stress (43). Besides, other recent studies have shown that REDD1 is an upstream regulator of TSC1/TSC2 complex activity, which acts to repress signaling through the mammalian target of rapamycin complex 1 (mTORC1) in mammalian cells (44–46). In view of our earlier finding that eRF3a depletion inhibits mTOR activity (31), and of the results presented above showing that eRF3a depletion induced ATF4 response, we further investigated whether REDD1 overexpression triggered by ATF4 activation was responsible for mTOR inhibition in eRF3a-depleted cells. We have tested whether REDD1 knockdown affected the phosphorylation status of three well-known markers of mTORC1 activity, 4E-BP1 (Thr37/46), S6K1 (Thr389) and ribosomal protein S6 (Ser240 and Ser244), in eRF3a-depleted cells. At 24 h after electroporation with sh-3a1 or sh-Ctrl, HCT116 cells were transfected with either predesigned siRNAs directed against REDD1 mRNA or control siRNA using Lipofectamine 2000 (Invitrogen). Western blot analyses performed 24 h later showed that the knockdown of REDD1 in eRF3a-depleted cells restored S6K1, S6 and 4E-BP1 phosphorylation at levels comparable to those of control cells (Figure 4 and Supplementary Figure S4). These results strongly suggested that the inhibition of mTORC1 activity induced by eRF3a depletion was due to the increase in REDD1 expression.
Figure 4.
REDD1 knockdown alleviated mTOR inhibition in eRF3a-depleted cells. At 24 h after electroporation with sh-3a1 (lanes 1 and 2) or sh-Ctrl (lanes 3 and 4), HCT116 cells were transfected with either predesigned si-R9 siRNA directed against REDD1 (lanes 2 and 4) or control siRNA (lanes 1 and 3) using Lipofectamine 2000 (Invitrogen). The predisigned si-R9 siRNA allows an efficient knockdown of REDD1 (see Supplementary Figure S4). Western blot analysis was performed 24 h later to measure the levels of REDD1, phosphorylated S6K1 (P-S6K1), total S6K1, phosphorylated ribosomal protein S6 (P-S6), total ribosomal protein S6 (S6), phosphorylated 4E-PB1 (P-4E-BP1) and α-TUB that served as loading control. Quantification of the signals (see ‘Materials and Methods’ section) are shown below each panel. Each signal was divided by the signal elicited by the α-TUB reference protein. The control experiment (lane 3) was set to 1.
DISCUSSION
Completing our previously published data showing the inhibition of mTOR pathway in human eRF3a-depleted cells (31), we elucidate here the mechanism leading to this inhibition. Using large-scale analyses of gene expression, we first show that eRF3a depletion induces the expression of a number of genes known to be upregulated during the ER stress and amino acid deprivation, and to be under the control of the transcriptional activator ATF4. However, some genes that were also described to be activated by ATF4, including ATF3, GADD34, SNAT2 (SLC38A2) and CAT-1 (SLC7A1), are not present in the list of the transcripts upregulated by a factor 2 upon eRF3a depletion. Nevertheless, these transcripts are upregulated by a factor of at least 1.3 in both total RNA and polysome-associated RNA of eRF3a-depleted cells (data not shown). It has been reported that elevated ATF4 expression alone is sufficient to trigger the amino acid-responsive transcriptional control program (47). However, these results also suggest that other factors may serve to enhance ATF4 function. Recently, Dey et al. (48) addressed the question of the regulatory mechanisms governing the variable ATF4 expression in response to eIF2α phosphorylation and different stress conditions. Their major conclusion is that the versatility of gene expression during stress responses is likely due to a combined effect of transcriptional and translational control of ATF4. They also suggest that, in addition to ATF4, other target genes activated by eIF2α phosphorylation may function in conjunction with ATF4 to modulate the cellular response to stress. Indeed, genes containing 5′-uORF in their transcript are good candidates to be upregulated in response to eIF2α phosphorylation (48). The answer to the question of what is responsible for the upregulation of genes during stress is not obvious. Genes, such as ASNS and CHOP, contain both 5′-uORFs in their transcript (41) and ATF4-responsive elements in their promoter (16). Thus, it is difficult to evaluate the contribution of each of these two elements in the upregulation of their expression. In our experiments, we also found an upregulation of well-known ATF4-activated genes as well as other genes that are not considered to be under ATF4 control. As an example, the transcript of IFRD1, which is a transcriptional co-repressor that controls the growth and differentiation of various cell types (49), is upregulated in total RNA as well as in polysome-associated mRNA of eRF3a-depleted cells (1.81- and 2.21-fold, respectively; see Supplementary Files S1-RNA and S2-polysome). It was recently shown that a 5′-uORF regulates IFRD1 mRNA translation during cellular stress (50). Therefore, we suspected that the effect of eRF3a depletion has something in common with the response to stress.
However, the experimental conditions we used here differ from those used to induce the stress response. The differential expression of genes was monitored after a 72 h long depletion of eRF3a, whereas most of the cellular stresses, such as tunicamycin treatment or amino acid deprivation, are applied for shorter periods of time, currently, 4–12 h (51,52). When compared to the ATF4 response to stress, the differences we observed in the ATF4 response to eRF3a depletion could be due to these differences in timing and duration that may allow complex regulatory mechanisms, including feedback control loops, to take place. In order to have an efficient knockdown of eRF3a expression, our analysis of the effects of eRF3a depletion was performed 72 h after electroporation of shRNA targeting eRF3a. As shown in Supplementary Figure S1B for mRNA levels and Figure 1B for ASNS and REDD1 protein levels, the time course of the increase in the expression of the ATF4 target genes follows that of the decrease in eRF3a.
Our results showing that the phosphorylation status of eIF2α is not modified in eRF3a-depleted cells indicate that the ATF4 response to eRF3a depletion is not due to the activation of stress response pathways. Alternatively, our results demonstrate that the ATF4 response is caused by translation termination deficiency. The defect in eRF3a leads to a decrease in translation termination efficiency and induces translational readthrough of stop codons (26,27). In the case of ATF4 mRNA, this increase in readthrough modifies its translational regulation that relies on the presence of two 5′-uORFs (14,15). It has been shown previously that the change of ATF4 uORF1 stop codon into sense codon increased translation of the downstream ATF4 ORF, by-passing the inhibitory uORF2 (14). The situation is identical in the case of readthrough at uORF1 stop codon. This means that the efficiency of readthrough and thereby the efficiency of translation termination modulate ATF4 expression at the translational level. It is interesting to note that the distance separating the in-frame UAA stop codon of the elongated uORF1 from ATF4 ORF is optimal for reinitiation at ATF4 initiation codon in the absence of stress, i.e. when eIF2α phosphorylation is low. Reinforcing the previous observation of Vattem and Wek (15), that the regulatory elements of ATF4 5′-UTR are conserved among vertebrates, we observed that, not only uORFs and the distance in between, but also, the number of stop codons, the nature of these stop codons and their nucleotide context, which greatly influence the efficiency of readthrough (53), are well conserved at least in mammals (see Supplementary Figure S3). The conservation of the regulatory elements in ATF4 5′-UTR further substantiates the notion that translation termination efficiency modulates ATF4 expression in mammals.
It has been reported that ATF4 mRNA was subject to degradation by the NMD pathway (40,41). In this study, we observe an increase in ATF4 mRNA half-life upon eRF3a depletion (Figure 2), and we hypothesize that this could be due to a partial inhibition of the ATF4 mRNA decay driven by the NMD pathway. ATF4 mRNA is likely targeted by NMD due to the presence of uORF1 stop codon, which plays the role of a premature stop codon. The second element required for NMD to take place in mammalian cells is the presence of an exon junction complex (EJC) downstream of the premature stop codon. The EJC is assembled on a newly formed mRNA at a region 24-nt upstream of splice junctions and premature stop codons have been shown to elicit NMD when positioned >50-nt upstream of an exon–exon boundary (54). Examining ATF4 gene from various mammalian species, we note that a conserved first intron is located within the 5′-UTR sequence, at ∼90 nt of uORF1 stop codon, immediately downstream of uORF2 initiation codon. Thus, during the first round of translation, the ribosome that has translated uORF1 can trigger NMD before reinitiating at uORF2. In the case of readthrough at uORF1 stop codon, the ribosome pursues translation in the region separating uORF1 from uORF2 and removes the EJC, thus preventing ATF4 mRNA degradation by NMD. However, this happens only when the readthrough occurs at the pioneer round of translation, explaining the moderate increase in ATF4 mRNA half-life when compared to that in Upf1-depleted cells (Figure 2). Nevertheless, in addition to enhance ATF4 ORF translation, the readthrough of stop codon induced by eRF3a depletion may also stabilize ATF4 mRNA, which could contribute to the increase in ATF4 expression. Together, the ∼2-fold increase in ATF4 mRNA steady-state level observed by microarray and qPCR analyses and the ∼5-fold increase in ATF4 ORF translation (Figure 3A) are in good agreement with the ∼9-fold increase in ATF4 protein observed by western blot in eRF3a-depleted cells (Figures 1B and Supplementary Figure S2).
We have previously shown that, in contrast to eRF3a depletion, suppressor tRNA overexpression had no effect on cell cycle progression in HCT116 cells (31). Since eRF3a depletion and suppressor tRNA overexpression can both induce readthrough of stop codons, we anticipated that the modifications of cell cycle observed in eRF3a-depleted cells were not the consequence of the effect of stop codon readthrough on proteins essential for cell cycle progression. This conclusion was re-evaluated in the light of the finding presented here of an effect of eRF3a depletion on ATF4 expression. Consistent with the results obtained on cell cycle, overexpressed amber, ochre or opal suppressor tRNAs neither increased ATF4 and REDD1 protein expression nor induced ATF4 transcriptional activity in 2XAARE-TRIB3-TKLUC cells (Supplementary Figure S5A and S5B, respectively). Nevertheless, ATF4 ORF translation was increased ∼1.7-fold by amber suppressor tRNA overexpression, which can suppress ATF4 uORF1 amber UAG stop codon, when compared to opale suppressor tRNA overexpression (Supplementary Figure S5C). This result is rather surprising when compared to the 5-fold increase in ATF4 ORF translation obtained with eRF3a depletion. Of course, the context surrounding ATF4 uORF1 stop codon could be more favorable for the readthrough induced by eRF3a depletion than for the readthrough induced by amber suppressor tRNA overexpression. However, the mechanisms of stop codon readthrough induced by either suppressor tRNAs or eRF3a depletion are profoundly different. The overexpressed artificial amber suppressor tRNA recognizes the UAG stop codon as does a cognate tRNA for a sense codon and translation elongation continues until the next non-UAG stop codon is encountered, then translation termination takes place with normal efficiency. In the case of eRF3a depletion, normal cellular tRNAs that are primarily needed for reading their cognate sense codons, recognizes UAG stop codons as near cognate tRNAs, slowing down the elongation process. At the next stop codon, either the same mechanism takes place or alternatively, translation terminates with a low efficiency due to the defect of translation termination complexes. These differences can profoundly modify the mechanism of reinitiation at a downstream initiation codon and could explain the results obtained for ATF4 ORF translation, which relies on a readthrough-reinitiation process in both cases, suppressor tRNA overexpression and eRF3a depletion. Although the overexpressed amber suppressor tRNA induced a slight increase in ATF4 ORF translation, it was surprising that it did not induce the ATF4 response. Examining ATF4 mRNA level, we repeatedly found that it slightly decreased in suppressor tRNAs overexpressing cells (Supplementary Figure S5D), suggesting that ATF4 was transcriptionally repressed in response to suppressor tRNA overexpression. These results remind those obtained in response to UV irradiation, for which, despite an increased ATF4 mRNA translation, the lowered availability of the ATF4 transcript significantly reduces the amount of ATF4 synthesized, thus blocking ATF4 induction (48).
One of the consequences of the increase in ATF4 synthesis in response to eRF3a depletion is an elevated expression of REDD1. Recent studies have now confirmed the role of REDD1 as an inhibitor of mTOR pathway, showing that overexpression of REDD1 is sufficient to downregulate mTOR substrate phosphorylation, whereas REDD1 disruption in mice prevents mTOR substrate dephosphorylation under stress condition (44–46). Confirming this role of REDD1 on mTOR, our results demonstrate that the elevated level of REDD1 in eRF3a-depleted cells is the cause of mTOR inhibition (Figure 4 and Supplementary Figure S4). Since eRF3a depletion promotes the ATF4-induced overexpression of REDD1 that leads to mTOR inhibition, the efficiency of translation termination can be added to the growing list of mechanisms controlling the mTOR pathway.
However, are there physiological circumstances for which this so far unappreciated regulatory mechanism could take place? Such situations require the depletion of eRF3a. The mTOR kinase plays a key role in cell growth and cell cycle progression and, from yeast to mammals, genes encoding eRF3 have long been considered as cell cycle regulatory genes (28,30). In addition, eRF3a abundance that is controlled by the ubiquitin–proteasome degradation pathway, regulates the translation termination complex formation and thus is a major determinant of translation termination efficiency (29). We have also reported that eRF3a depletion causes cell cycle arrest at G1 phase via mTOR inhibition (31). There is at least one reported physiological situation for which eRF3a depletion is connected with an arrest of cell division. Studying human articular chondrocytes differentiation, Tallheden et al. (55) have shown that the transition from proliferation to differentiation, a process which takes place without change in cell number, was associated with a 5- to 10-fold decrease in eRF3a gene (GSPT1) expression. Whether the decrease of eRF3a in these cells is accompanied by a decrease in translation termination efficiency and by an increase in the ATF4 response remain to be elucidated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5, Supplementary Data sets 1 and 2.
FUNDING
Association pour la Recherche sur le Cancer [4891]; Ministère de l’Enseignement Supérieur et de la Recherche (fellowship to H.A.G.). Funding for open access charge: the Université Pierre et Marie Curie, and the Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Oliver Mühlemann for the generous gift of the plasmids-encoding Upf1-targeting shRNA, Valérie Carraro for technical assistance, and the IFR 83 at UPMC for real-time PCR facilities.
REFERENCES
- 1.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 2.Lee KA, Hai TY, SivaRaman L, Thimmappaya B, Hurst HC, Jones NC, Green MR. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc. Natl Acad. Sci. USA. 1987;84:8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman RJ. Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem. Sci. 2004;29:152–158. doi: 10.1016/j.tibs.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 8.Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem. J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale M, Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 14.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaveroux C, Lambert-Langlais S, Cherasse Y, Averous J, Parry L, Carraro V, Jousse C, Maurin AC, Bruhat A, Fafournoux P. Molecular mechanisms involved in the adaptation to amino acid limitation in mammals. Biochimie. 2010;92:736–745. doi: 10.1016/j.biochi.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malabanan KP, Khachigian LM. Activation transcription factor-4 and the acute vascular response to injury. J. Mol. Med. 2010;88:545–552. doi: 10.1007/s00109-010-0615-4. [DOI] [PubMed] [Google Scholar]
- 19.Pereira ER, Liao N, Neale GA, Hendershot LM. Transcriptional and post-transcriptional regulation of proangiogenic factors by the unfolded protein response. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 2008;283:10848–10857. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inge-Vechtomov S, Zhouravleva G, Philippe M. Eukaryotic release factors (eRFs) history. Biol. Cell. 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 23.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell. 2002;94:205–216. doi: 10.1016/s0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D, Jean-Jean O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 2005;25:5801–5811. doi: 10.1128/MCB.25.14.5801-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janzen DM, Geballe AP. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 2004;32:4491–4502. doi: 10.1093/nar/gkh791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoshino S, Miyazawa H, Enomoto T, Hanaoka F, Kikuchi Y, Kikuchi A, Ui M. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 1989;8:3807–3814. doi: 10.1002/j.1460-2075.1989.tb08558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chauvin C, Jean-Jean O. Proteasomal degradation of human release factor eRF3a regulates translation termination complex formation. RNA. 2008;14:240–245. doi: 10.1261/rna.728608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi Y, Shimatake H, Kikuchi A. A yeast gene required for the G1-to-S transition encodes a protein containing an A-kinase target site and GTPase domain. EMBO J. 1988;7:1175–1182. doi: 10.1002/j.1460-2075.1988.tb02928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauvin C, Salhi S, Jean-Jean O. Human eukaryotic release factor 3a depletion causes cell cycle arrest at G1 phase through inhibition of the mTOR pathway. Mol. Cell. Biol. 2007;27:5619–5629. doi: 10.1128/MCB.00035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carraro V, Maurin AC, Lambert-Langlais S, Averous J, Chaveroux C, Parry L, Jousse C, Ord D, Ord T, Fafournoux P, et al. Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2alpha/ATF4 pathway. PLoS One. 2010;5:e15716. doi: 10.1371/journal.pone.0015716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schambach A, Galla M, Maetzig T, Loew R, Baum C. Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol. Ther. 2007;15:1167–1173. doi: 10.1038/sj.mt.6300152. [DOI] [PubMed] [Google Scholar]
- 35.Verrier SB, Jean-Jean O. Complementarity between the mRNA 5' untranslated region and 18S ribosomal RNA can inhibit translation. RNA. 2000;6:584–597. doi: 10.1017/s1355838200992239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ord D, Ord T. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochem. Biophys. Res. Commun. 2005;330:210–218. doi: 10.1016/j.bbrc.2005.02.149. [DOI] [PubMed] [Google Scholar]
- 40.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 42.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res. Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 46.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan J, Ord D, Ord T, Kilberg MS. Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J. Biol. Chem. 2009;284:21241–21248. doi: 10.1074/jbc.M109.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vietor I, Huber LA. Role of TIS7 family of transcriptional regulators in differentiation and regeneration. Differentiation. 2007;75:891–897. doi: 10.1111/j.1432-0436.2007.00205.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhao C, Datta S, Mandal P, Xu S, Hamilton T. Stress-sensitive regulation of IFRD1 mRNA decay is mediated by an upstream open reading frame. J. Biol. Chem. 2010;285:8552–8562. doi: 10.1074/jbc.M109.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaveroux C, Lambert-Langlais S, Parry L, Carraro V, Jousse C, Maurin AC, Bruhat A, Marceau G, Sapin V, Averous J, et al. Identification of GCN2 as new redox regulator for oxidative stress prevention in vivo. Biochem. Biophys. Res. Commun. 2011;415:120–124. doi: 10.1016/j.bbrc.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 52.Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Tallheden T, Karlsson C, Brunner A, Van Der Lee J, Hagg R, Tommasini R, Lindahl A. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthr. Cartilage. 2004;12:525–535. doi: 10.1016/j.joca.2004.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.