Abstract
Genome-wide association studies (GWASs) have been used to search for susceptibility genes for type 2 diabetes and obesity in the Pima Indians, a population with high a prevalence of both diseases. In these studies, a variant (rs2025804) in the LEPR gene was nominally associated with BMI in 1082 subjects (P=0.03 adjusted for age, sex, birth year, and family membership). Therefore the LEPR and leptin overlapping transcript (LEPROT) genes were selected for further sequencing and genotyping in larger population-based samples for association analyses with obesity-related phenotypes. Selected variants (n=80) spanning these genes were genotyped in a sample of full-heritage Pima Indians (n=2842) and several common variants including rs2025804 were nominally associated with BMI (P=0.05-0.003 adjusted for age, sex, birth year, and family membership). Four common tag variants associated with BMI in the full-heritage Pima Indian sample were genotyped in a second sample of mixed-heritage Native Americans (n=2969) and 3 of the variants showed nominal replication (P=0.03-0.006 adjusted as above and additionally for Indian heritage). Combining both samples provided the strongest evidence for association (adjusted P=0.0003-0.0001). A subset of these individuals (n=403) had been metabolically characterized for predictors of obesity and the BMI risk alleles for the variants tagged by rs2025804 were also associated with lower 24 hour energy expenditure as assessed in a human respiratory chamber (P=0.0007 adjusted for age, sex, fat mass, fat free mass, activity, and family membership). We conclude that common non-coding variation in the LEPR gene is associated with higher BMI and lower energy expenditure in Native Americans.
INTRODUCTION
We have previously conducted genome-wide association studies (GWASs) in Pima Indians using the Affymetrix 100K and 1M genotyping arrays to identify genes that may increase the risk for type 2 diabetes or obesity in this population (1,2). In these studies an intronic G/A variant, rs2025804, located in the long isoform of the leptin receptor gene (LEPR) was identified that was nominally associated with body mass index (BMI) in 1082 GWAS subjects (P = 0.03 adjusted for age, sex, birth year, and family membership). Although the association with BMI was not among the strongest GWAS signals, LEPR is an excellent biological candidate gene. The leptin receptor (long isoform) is part of the leptin-melanocortin signaling pathway involved in food intake and energy expenditure, and mutations in several genes along this pathway, including LEPR, are known to result in monogenic forms of obesity (3,4,5). Other potential obesity candidate genes at this locus include the short isoforms of the LEPR and the leptin receptor overlapping transcript (LEPROT) which shares the same promoter and first 2 exons of the LEPR long isoform. Two of the short LEPR isoforms (Ra and Re) are involved in transporting leptin across the blood brain barrier and regulating leptin bioavailablity, respectively (6,7); and, it has been shown that knockdown of LEPROT in mice results in increased leptin signaling preventing the development of diet induced obesity (8). Therefore, the nominal evidence for association with BMI from our GWAS combined with the biological importance of the LEPRs and LEPROT in obesity-related pathways led us to further examine this region in Pima Indians.
METHODS AND PROCEDURES
Subjects and phenotypes for association analyses
Subjects included in the full-heritage Pima Indian genotyping sample (n = 2842) and the mixed-heritage Native American replication sample (n = 2969) are part of a longitudinal health study of the Gila River Indian Community in Central Arizona. (9). All longitudinally studied subjects that were selected had at least one BMI measurement available from a medical exam where the subject was >15 years of age and was determined to be non-diabetic by a 2 hour oral glucose tolerance test. If BMI measures were available from multiple exams that met these criteria, the maximum recorded BMI was used for analyses. The full-heritage Pima Indian population-based sample consisted of individuals whose heritage was reported as full Pima and/or Tohono O’odham (a closely related tribe). The mixed-heritage Native American sample consisted of all other individuals whose DNA had been obtained during the longitudinal study. Their reported heritage was on average ½ Pima and ¾ American Indian. A subset of the longitudinally studied subjects had also volunteered to take part in inpatient studies in our Clinical Research Center. These individuals were determined to be non-diabetic at the time of admission and participated in multiple metabolic tests to measure predictors of type 2 diabetes and obesity. Four hundred three subjects underwent studies of body composition and participated in a study in our human respiratory chamber to measure 24 hour energy expenditure (24hEE) by indirect calorimetry. Body composition was estimated by underwater weighing until January, 1996, and by dual energy X-ray absorptiometry (DPX-1, Lunar Radiation Corp, Madison, WI) thereafter. A conversion equation derived from comparative analyses was used to make estimates of body composition equivalent between the two methods (10). To measure 24hEE, volunteers entered the metabolic chamber after an overnight fast and remained there for 23 hours. Meals were provided at 7 am, 11 am, 4 pm, and 7 pm in equally divided caloric portions. Because of the confinement within the chamber, only 80% of the calories from the weight-maintaining diet provided while staying in our Clinical Research Center were provided in the respiratory chamber. Fresh air was drawn through the chamber and CO2 production and O2 consumption were measured and calculated every 15 min and extrapolated for a 24 hour interval. Energy expenditure over the 24 hours was calculated as previously described. Spontaneous physical activity was detected by radar sensors and expressed as percentage of time over the 24 hour period in which activity was detected. Sleeping metabolic rate (sleep energy expenditure) was defined as the average energy expenditure of all 15 minute periods during which spontaneous physical activity was <1.5%. Carbon dioxide production (VCO2) and oxygen consumption (VO2) were calculated for every 15 minute interval and extrapolated for the 24 hour interval (11,12). All subjects provided written informed consent prior to participation. This study was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Sequencing and genotyping
Sequencing of the LEPR and LEPROT genes in 96 obese subjects was done using Big Dye terminator (Applied Biosystems) on an automated DNA capillary sequencer (model 3730; Applied Biosystems). Subsequently, whole genome sequence (n = 10, sequence data generated by Complete Genomics Inc., Mountain View, CA) and whole exome sequence (n = 180, sequence data generated by ShanghaiBio US, North Brunswick, NJ) has become available for the Pima Indians. Genotyping of the variants in the two population-based samples was done by using either the SNPlex genotyping System 48-plex (Applied Biosystems) on an automated DNA capillary sequencer (model 3730; Applied Biosystems), the Illumina BeadXpress System, or by allelic discrimination PCR (Assays-on-Demand SNP Genotyping products; Applied Biosystems). All genotypic data included in the analyses met our quality control criteria which requires a successful call rate of >85%, a lack of deviation from Hardy-Weinberg equilibrium at P <0.001, and a discrepancy rate of <2.5% in masked duplicates. Genotyping error rates were determined using 330 and 100 blind duplicates for the population-based samples of full-heritage Pima Indians and mixed-heritage Native Americans respectively.
Statistical analyses
Statistical analyses were performed using the statistical analysis system of the SAS Institute (Cary, NC). For continuous variables, linear regression models were used to analyze the associations and P-values were adjusted for covariates including age, sex, and birth year. The logarithmic transformation of BMI was used in these analyses to reduce skewness. In the mixed-heritage Native American sample, the individual estimate of European admixture was also used as a covariate. These estimates were derived from 32 markers with large differences in allele frequency between populations (13) by using the method described in Hanis et al. (14). A combined test of association for the full-heritage Pima Indian and mixed-heritage Native American samples was conducted by the inverse variance method (15). Linkage disequilibrium (D′ and r2) was analyzed using the Haploview program (Haploview http:www.broad.mit.edu/mpg/haploview).
RESULTS AND DISCUSSION
To identify novel potentially functional variants, all of the exons, including the exon/intron boundaries, for LEPROT, LEPR long and short isoforms, 2 kb of the overlapping 5′ region for LEPROT/LEPR long isoforms, and 3.2 kb of the 5′ region for the short LEPR isoforms were sequenced in 96 obese Pima Indians with BMIs ranging from 50–80 kg/m2. None of these 96 subjects were first-degree relatives. One rare novel substitution in the long 3′-UTR of LEPROT was identified along with 5 common exon variants rs113700 (Arg109Lys), rs1137101 (Gln223Arg), rs1805134 (Ser343Ser), rs8179183 (Asn656Lys), and rs1805096 (Pro1019Pro). Analysis of recently obtained whole exome sequence data for 180 Pima Indians did not identify any additional rare coding variants (data not shown). Three LEPR variants previously described in the literature, Lys204Arg (3), Ser675Thr (16), and exon 17 splice site (17) were not detected in either the 96 obese subjects or the 180 (not selected for phenotype) exomes. Eighty variants, including several detected by sequencing as well as database tag SNPs (based on the Chinese HapMap, minor allele frequency ≥0.1 and a pair-wise r2 ≥0.8) which provided information for the non-sequenced regions, were genotyped in the population-based sample of 2842 full-heritage Pima Indians for association analysis with BMI (supplementary Table 1). The linkage disequilibrium pattern (D′ and r2) for the 80 variants as determined in the full-heritage Pima Indians is shown in supplementary figure 1. Several variants were nominally associated with BMI (P-values ranging from 0.05-0.003, adjusted for age, sex, and family heritage; supplementary Table 1). None of the 5 exon variants that were genotyped were associated with BMI. Most of the variants associated with BMI are located in the large intron 2 of the LEPR long isoforms which also partially overlaps with the 5′ region of the LEPR short isoforms (supplementary Fig. 1). Common polymorphisms associated with BMI could be tagged by 4 variants (Table 1). The GWAS polymorphism rs2025804 was among these tag variants associated with BMI in the full-heritage Pima Indian population sample; however, there was significant overlap between the GWAS sample and the population-based sample so this association was not an independent replication. To assess independent replication, the 4 tag variants were further genotyped in a second, non-overlapping population-based sample of mixed-heritage Native Americans (n = 2969). Three of the variants, rs2025804, rs12029311, and rs6662904, showed nominal replication (P = 0.03-0.006 adjusted as above and additionally for Indian heritage). Combining the full-heritage Pima Indian and mixed-heritage subjects (n = 5811) provided the strongest evidence for association with BMI (adjusted P = 0.0003-0.0001; Table 1). The rs2025804 variant has also been shown to be modestly associated with obesity in both a Caucasian population (18) and Chinese population (19). The obesity risk alleles for rs2025804 (G) and rs12029311 (A) are much more common in Pima Indians (frequencies of 0.73 and 0.25, respectively) as compared to individuals of Caucasian or African ancestry (Table 2). In particular, the risk allele for rs12029311 is extremely rare in Africans (ranging from 0.0-0.02) and has not been reported in Caucasians. In contrast, the frequencies of these two risk alleles in Asian populations are similar to that observed in Pima Indians. The risk allele for rs6662904 (G) is also more common in Pima Indians (0.70) than in Caucasians (0.54), but it is very common (0.91-0.97) in Asian and African populations.
Table 1. Association analyses with BMI, body weight, and height.
Association analyses for the 4 tag variants (D′ <0.99, r2 <0.85) with BMI, body weight, and height. Allele frequency is given for the designated risk allele. The risk alleles were consistent between the full-heritage and mixed-heritage samples; however, for some variants the risk allele was the major allele in the full-heritage sample but the minor allele in the mixed-heritage sample. For each variant, the mean BMI by genotype is shown in the 1st row, the mean body weight by genotype is shown in the 2nd row, and the mean height by genotype is shown in the 3rd row.
| Variant | Risk allele | Full-heritage Pima Indians (N = 2842) Means based on genotypes |
Mixed-heritage Native Americans (N = 2969) Mean based on genotypes |
Combined (N = 5811)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk AF | Risk/Risk | Risk/Non-risk | Non-risk/Non-risk | Pa | Risk AF | Risk/Risk | Risk/Non-risk | Non-risk/Non-risk | Pa | Pa | ||
| rs2025804 | G | 0.73 | 36.7 ± 8.2 | 35.6 ± 8.1 | 35.7 ± 8.9 | 0.004 | 0.62 | 34.9 ± 8.4 | 33.8 ± 8.5 | 33.0 ± 7.7 | 0.03 | 0.0003 |
| 100.3 ± 24.5 | 97.0 ± 23.9 | 97.0 ± 25.9 | 0.009 | 97.1 ± 25.9 | 93.9 ± 25.6 | 93.1 ± 23.7 | 0.10 | 0.002 | ||||
| 165.1 ± 8.2 | 164.8 ± 8.4 | 164.5 ± 8.6 | 0.30 | 166.4 ± 8.6 | 166.5 ± 8.4 | 167.9 ± 8.6 | 0.12 | 0.72 | ||||
| rs1171274 | C | 0.60 | 36.6 ± 8.2 | 36.3 ± 8.2 | 35.8 ± 8.7 | 0.05 | 0.49 | 34.8 ± 8.5 | 34.3 ± 8.7 | 33.3 ± 7.9 | 0.43 | 0.05 |
| 100.2 ± 24.6 | 98.8 ± 24.3 | 96.5 ± 25.0 | 0.05 | 96.5 ± 26.2 | 95.5 ± 26.5 | 93.4 ± 23.7 | 0.59 | 0.07 | ||||
| 165.3 ± 8.3 | 164.9 ± 8.1 | 164.0 ± 8.7 | 0.13 | 166.2 ± 8.7 | 166.6 ± 8.4 | 167.3 ± 8.5 | 0.15 | 0.99 | ||||
| rs12029311 | A | 0.25 | 38.6 ± 9.3 | 36.3 ± 8.0 | 35.9 ± 8.2 | 0.006 | 0.20 | 36.7 ± 9.7 | 34.8 ± 8.4 | 33.6 ± 8.4 | 0.006 | 0.0002 |
| 104.6 ± 26.4 | 99.3 ± 24.5 | 97.6 ± 24.1 | 0.03 | 101.6 ± 28.0 | 96.7 ± 26.1 | 94.1 ± 25.0 | 0.03 | 0.003 | ||||
| 164.7 ± 8.4 | 165.1 ± 8.1 | 164.8 ± 8.4 | 0.55 | 166.3 ± 8.7 | 166.4 ± 8.3 | 167.0 ± 8.6 | 0.67 | 0.45 | ||||
| rs6662904 | G | 0.70 | 36.7 ± 8.4 | 35.8 ± 7.8 | 36.1 ± 8.8 | 0.008 | 0.68 | 34.7 ± 8.5 | 33.5 ± 8.5 | 33.9 ± 7.7 | 0.007 | 0.0001 |
| 100.2 ± 24.9 | 97.5 ± 23.2 | 97.6 ± 26.1 | 0.02 | 96.8 ± 25.8 | 93.2 ± 25.9 | 94.8 ± 24.1 | 0.02 | 0.0007 | ||||
| 165.1 ± 8.4 | 164.9 ± 8.1 | 164.1 ± 8.4 | 0.38 | 166.8 ± 8.5 | 166.5 ± 8.5 | 167.1 ± 8.7 | 0.71 | 0.67 | ||||
P-values were adjusted for age, sex, birth year, and family membership, and in the mixed-heritage sample the P-values were additionally adjusted for Indian heritage
Table 2. Comparison of the risk allele frequencies for rs2025804, rs12029311, and rs6662904 in the Pima Indian and HapMap populations.
The allele frequencies for the HapMap populations were obtained from the HapMap SNPs track, UCSC Genome Browser, build 37.1.
| Population | rs2025804 | rs12029311 | rs6662904 |
|---|---|---|---|
|
| |||
| G | A | G | |
| Full-heritage Pima Indians, Arizona, USA | 0.73 | 0.25 | 0.70 |
| Utah residents with ancestry from northern and western Europe (CEU) | 0.40 | 0.00 | 0.54 |
| Toscani in Italia (TSI) | 0.27 | 0.006 | 0.51 |
| Yoruba in Ibadan, Nigeria (YRI) | 0.22 | 0.00 | 0.994 |
| Luhya in Webuye, Kenya (LWK) | 0.22 | 0.006 | - |
| Maasai in Kinyawa, Kenya (MKK) | 0.15 | - | 0.96 |
| African Ancestry in SW USA (ASW) | 0.19 | 0.02 | 0.91 |
| Han Chinese in Beijing, China (CHB) | 0.84 | 0.22 | 0.96 |
| Chinese in Metropolitan Denver, CO, USA (CHD) | 0.89 | 0.23 | 0.97 |
| Japanese in Tokyo, Japan (JPT) | 0.85 | 0.20 | 0.91 |
| Gujarati Indians in Houston, Texas, USA (GIH) | 0.21 | 0.02 | 0.65 |
| Mexican Ancestry in Los Angeles, CA, USA (MXL) | 0.34 | - | 0.69 |
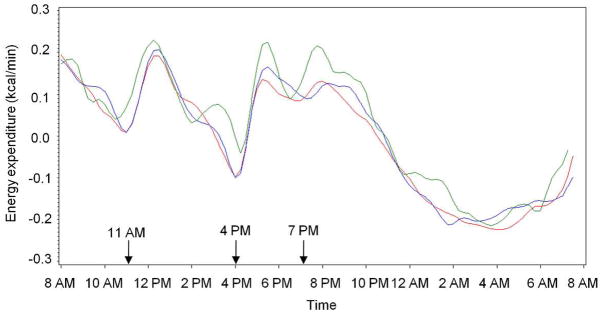
Data on mean 24 hour energy expenditure (24hEE) were also available for 403 subjects who were genotyped in this study. The rs2025804 variant is also significantly associated with 24hEE, where individuals carrying the risk (G) allele for obesity had a lower 24hEE (mean 24hEE based on genotypes for G/G = 2351 ± 401, G/A = 2353 ± 382, A/A = 2369 ± 446; P = 0.0007 adjusted for age, sex, fat mass, fat free mass, activity, and family membership). A plot of the energy expenditure data at 15 minute intervals based on genotypes for rs2025804 shows that the largest overall increases in mean energy expenditure occurred postprandially (Fig. 1). Individuals carrying no risk allele for rs2025804 (AA genotype; green line in Fig. 1) had a higher mean postprandial energy expenditure as compared to the individuals homozygous or heterozygous for the obesity risk allele (GG genotype red line and GA genotype blue line). DEXA measures of body composition were available on these same subjects. Individuals with the highest post-prandial energy expenditure (A/A genotype) had the lowest mean fat mass among the genotypic groups (32.7, 32.0, and 29.8 kg for G/G, G/A, and A/A, respectively). Individuals with the highest mean BMI (G/G genotype) had both the highest mean fat mass (means given above) and the highest mean fat-free mass (64.1, 62.0, and 62.8 for G/G, G/A, and A/A, respectively). However, these differences in fat and fat-free mass by genotype did not reach statistical significance in these metabolically characterized individuals.
Figure 1.
Energy expenditure over 24 hrs adjusted for age, sex, fat mass, fat free mass, and activity based on genotypes for rs2025804. Twenty four hour energy expenditure was measured by indirect calorimetry in a metabolic chamber. Oxygen consumption and carbon dioxide production were measured and energy expenditure calculated over 15 minute intervals. Arrows indicate when meals were given. Red line, homozygous G/G (n = 167); blue line, heterozygous G/A (n = 110); green line, homozygous A/A (n = 26).
The observed postprandial increase in energy expenditure (Fig. 1) is likely due to the thermic effect of food (TEF). Previous reports have shown that leptin levels increase following meals (20,21); and, studies using animal models have suggested that leptin may play a role in the TEF (via thermogenesis) in rodents (22) and sheep (23). An earlier study of overweight and obese women found that the LEPR Lys656Asn (rs8179183) variant was nominally associated with substrate carbohydrate oxidation in 38 women who had undergone measures of glucose-induced thermogenesis (GIT) (24). The association with substrate oxidation was observed following a 75 g oral glucose load but not in the fasted state, and they proposed that the LEPR may affect the thermic response to carbohydrate in glucose-induced thermogenesis (24). This small study in women is consistent with our hypothesis that variants in the LEPR affect energy expenditure by regulating the TEF in humans.
As part of our future genetic studies of obesity and type 2 diabetes, we are sequencing the entire genomes of several Pima Indians. Among the 10 genomes sequenced to date, 28 additional intronic variants were identified that are potentially in very high LD with rs2025804 (current data suggests perfect LD but sample size requires caution). One of these variants is novel (C to T substitution) and maps 371 bases centromeric to rs2025804 and was therefore genotyped in our sample of 2842 full-heritage Pima Indians. The novel C/T variant was confirmed to be in high LD with rs2025804 (D′ = 99 and r2 = 77) and was similarly associated with BMI (supplementary Table 1). These variants span 47 kb in the 5′ region of the short LEPR isoforms and this region contains a number of predicted DNA elements that may regulate transcription (supplementary Fig. 2). Some of the polymorphisms including the novel C/T variant and rs2025804 are located in predicted regulatory regions (supplementary Fig. 2); however, direct analysis between genotypes and in vivo LEPR expression levels is confounded by the multiple LEPR isoforms expressed in various human tissues. Future in vitro studies will include experiments to determine whether any of the variants located within a predicted regulatory element may affect LEPR expression.
In Pima Indians, the BMI association of the rs2025804 LEPR variant is comparable to the BMI association of rs9939609 in FTO (25). Genotypic data from the 2842 full-heritage Pima Indians analyzed in this study show similar levels of significance (P = 0.004 and 0.002) and effect sizes (beta estimate = 0.020 and 0.027) for rs2025804 and rs9939609. However, the association of the FTO variant with BMI “improves more” than the BMI association of the LEPR variant when the full-heritage Pima sample is combined with the mixed-heritage sample of Native Americans (P = 0.0003 and 0.00008 for LEPR and FTO, respectively) presumably because of the strong effect of FTO across all ethnic groups including Caucasians. The ability to detect an association between common LEPR variants and BMI may not be as robust as FTO across all ethnic groups, since some risk alleles are either much rarer or nonexistent (i.e. monomorphic for the non-risk allele) in individuals of African and Caucasian descent; however, our GWAS lead variant rs2025804 was also nominally associated with obesity in Caucasian (18) and Chinese (19) populations. Among Pima Indians, variation in LEPR may have a more important population effect than FTO because the risk allele for LEPR is very common (0.73) whereas the risk allele for FTO is much less common (0.14).
In summary, common variants in the LEPR gene are nominally but reproducibly associated with increased BMI in Native Americans, where the risk alleles for these variants are also associated with reduced 24 hour energy expenditure. Although it is known that the leptin receptor plays a role in food intake, we propose that it may have an additional affect on energy expenditure by regulating postprandial thermogenesis.
Supplementary Material
Acknowledgments
We thank the many persons who volunteered for these studies. This work was funded by the Intramural Research Program of NIDDK, NIH
Footnotes
DISCLOSURE
The authors declare that there is no conflict of interest associated with this manuscript.
References
- 1.Hanson RL, Bogardus C, Duggan D, et al. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra A, Kobes S, Knowler WC, Baier L, Bogardus C, Hanson RL. A genome-wide association study of body mass index in American Indians. Obesity. 2011 Jun 23; doi: 10.1038/oby.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement K, Vaisse C, Lahlou N, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 4.Farooqi IS. Monogenic human obesity. Front Horm Res. 2008;36:1–11. doi: 10.1159/000115333. [DOI] [PubMed] [Google Scholar]
- 5.Beckers S, Zegers D, Van Gaal LF, Van Hul W. The role of the leptin-melanocortin signalling pathway in the control of food intake. Crit Rev Eukaryot Gene Expr. 2009;19:267–287. doi: 10.1615/critreveukargeneexpr.v19.i4.20. [DOI] [PubMed] [Google Scholar]
- 6.Ziylan YZ, Baltaci AK, Mogulkoc R. Leptin transport in the central nervous system. Cell Biochem Funct. 2009;27:63–70. doi: 10.1002/cbf.1538. [DOI] [PubMed] [Google Scholar]
- 7.Osborn O, Sanchez-Alavez M, Brownell SE, et al. Metabolic characterization of a mouse deficient in all known leptin receptor isoforms. Cell Mol Neurobiol. 2010;30:23–33. doi: 10.1007/s10571-009-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couturier C, Sarkis C, Seron K, et al. Silencing of OB-RGRP in mouse hypothalamic arcuate nucleus increases leptin receptor signaling and prevents diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:19476–19481. doi: 10.1073/pnas.0706671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978;108:497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- 10.Norman RA, Thompson DB, Foroud T, et al. Genomewide search for genes influencing percent body fat in Pima Indians: suggestive linkage at chromosome 11q21-q22. Pima Diabetes Gene Group. Am J Hum Genet. 1997;60:166–173. [PMC free article] [PubMed] [Google Scholar]
- 11.Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C. Determinants of 24-hour energy expenditure in man: methods and results using a respiratory chamber. J Clin Invest. 1986;78:1568–1578. doi: 10.1172/JCI112749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schutz Y, Ravussin E, Diethelm R, Jequier E. Spontaneous physical activity measured by radar in obese and control subjects studied in a respiratory chamber. Int J Obes. 1982;6:23–28. [PubMed] [Google Scholar]
- 13.Tian C, Hinds DA, Shigeta R, et al. A genome-wide single nucleotide polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease in Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 15.Petitti DB. Statistical methods in meta-analysis. In: Petitti DB, editor. Meta-Analysis, Decision Analysis and Cost-Effectiveness Analysis: Methods for Quantitative Synthesis in Medicine. Oxford: Oxford University Press; 2000. pp. 94–118. [Google Scholar]
- 16.Echwald SM, Sorensen TD, Sorensen TI, et al. Amino acid variants in the human leptin receptor: lack of association to juvenile onset obesity. Biochem Biophys Res Commun. 1997;233:248–252. doi: 10.1006/bbrc.1997.6430. [DOI] [PubMed] [Google Scholar]
- 17.Roth H, Korn T, Rosenkranz K, et al. Transmission disequilibrium and sequence variants at the leptin receptor gene in extremely obese German children and adolescents. Hum Genet. 1998;103:540–546. doi: 10.1007/s004390050867. [DOI] [PubMed] [Google Scholar]
- 18.Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, Atwood LD. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8 (Suppl 1):S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HH, Lee WJ, Fann CS, Bouchard C, Pan WH. Severe obesity is associated with novel single nucleotide polymorphisms of the ESR1 and PPARg locus in Han Chinese. Am J Clin Nutr. 2009;90:255–262. doi: 10.3945/ajcn.2009.25914. [DOI] [PubMed] [Google Scholar]
- 20.Villanueva EC, Myers MG. Leptin receptor signaling and the regulation of mammalian physiology. Int J Obes (Lond) 2008;(Suppl 7):S8–12. doi: 10.1038/ijo.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris DL, Rui L. Recent advances in understanding leptin signaling and leptin resistance. Am J Physiol Endocrinol Metab. 2009;297:E1247–1259. doi: 10.1152/ajpendo.00274.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Matheny MK, Tümer N, Mitchell MK, Scarpace PJ. Leptin antagonist reveals that the normalization of caloric intake and the thermic effect of food after high-fat feeding are leptin dependent. Am J Physiol Regul Integr Comp Physiol. 2007;292:R868–874. doi: 10.1152/ajpregu.00213.2006. [DOI] [PubMed] [Google Scholar]
- 23.Henry BA, Dunshea FR, Gould M, Clarke IJ. Profiling postprandial thermogenesis in muscle and fat of sheep and the central effect of leptin administration. Endocrinology. 2008;149:2019–2026. doi: 10.1210/en.2007-1311. [DOI] [PubMed] [Google Scholar]
- 24.Wauters M, Considine RV, Chagnon M, et al. Leptin levels, leptin receptor gene polymorphisms, and energy metabolism in women. Obes Res. 2002;10:394–400. doi: 10.1038/oby.2002.54. [DOI] [PubMed] [Google Scholar]
- 25.Rong R, Hanson RL, Ortiz D, et al. Association analysis of FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761 and CDKN2B with type 2 diabetes and pre-diabetic traits in Pima Indians. Diabetes. 2009;58:478–488. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.