Abstract
Changes in cell homeostasis, or cell ‘stress’, are thought to tax the ability of the Hsp90 chaperone to facilitate an array of processes critical for genome maintenance. Here, we review the current understanding of how Hsp90 chaperone machinery ensures the function of proteins important for DNA repair, recombination and chromosome segregation. We discuss the idea that cell ‘stress’ can overload Hsp90, resulting in genomic instability that may have important implications for stress adaptation and selection. The importance of Hsp90 in genome maintenance and its limited capacity to buffer the proteome may underlie the initiation or progression of diseases such as cancer.
Keywords: Hsp90, chaperone, cell stress, genome instability, adaptation
Chaperones and the cellular response to stress
Adaptation to stress is not only the driving force for species evolution but also a daily challenge that cells and organisms struggle against during their individual existence. An important mechanism for coping with stresses is the induction of molecular chaperones 1. Chaperone proteins, such as the Hsp70, Hsp90 and Hsp100 family members, assist in protein folding and complex assembly and act on mis-folded proteins induced by conditions such as heat or the presence of reactive oxygen species. Chaperone induction is a mode of adaption that does not involve genetic changes but rather reflects an evolved capacity to maintain protein homeostasis.
A second adaptive mechanism, which relies on genetic variation, was proposed for the Hsp90 family of molecular chaperones 2, 3. This theory evolved from the observation of phenotypic variation associated with Drosophila melanogaster bearing mutations in the Hsp90 gene hsp83 4. It proposes that the adaptive potential of a population resides in its pre-existing genetic variation and posits that Hsp90, as a result of its broad role in proteome fidelity and plasticity, allows the persistence of genetic variants in the population by buffering their potential deleterious effects. Thus, Hsp90 restrains genetic variation much as a capacitor restrains electrical charge; when the load (e.g., number of Hsp90 clients) exceeds Hsp90 capacity then the phenotypic effects of pre-existing genetic variation may be released (Figure 1A and 1B), providing the substrates from which the adaptive variants can be selected. This model assumes that the degree of genetic variation is not necessarily influenced by stress and that the chaperones do not directly contribute to adaption but instead gate the phenotypic expression of the potentially adaptive variants.
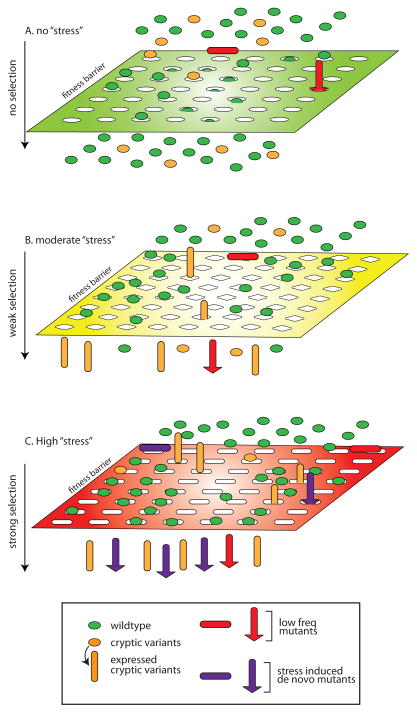
Figure 1.
A model for ‘stress’-induced hyper-mutation and adaptation. (A) Under normal growth conditions wild type cells (green ovals) and cells with Hsp90-buffered, cryptic mutations (orange ovals) are equally fit to pass through the fitness barrier. In contrast, pre-existing, unbuffered mutations (red shapes) in the population are unfit and cannot pass through the fitness barrier. (B) The imposition of moderate ‘stress’ changes the selection properties of the fitness barrier (note change in shape of grating), reducing the number of wild type cells that pass the barrier. The moderate stress increases Hsp90 ‘load’ and allows some cryptic mutations to express phenotypes (denoted by the conversion of orange ovals to cyclinders) that provide growth advantage. In rare cases, pre-existing unbuffered mutations may also have an advantage under selection. (C) High ‘stress’ imposes an even more selective fitness barrier and increases the frequency of de novo mutants (purple shapes) in the population. Under the increased stringency of selection wild type cells are not fit but a fraction of cryptic variants and de novo mutants that express advantageous phenotypes pass the fitness barrier.
In prokaryotes, it has been proposed that the rate of genetic variation is dramatically elevated in a stressed population to fuel evolutionary adaptation 5. Genetic variants are generated de novo as a result of activation of error-prone DNA synthesis and repair or increased frequency of transposon mobilization. Such stress-induced mutagenesis differs from the Hsp90-capacitor model in that the deleterious effect -- expected of most forms of mutations -- need not be buffered to maintain population fitness under conditions of low stress, and thus could potentially allow phenotypic leaps conferred by large-effect mutations under strong selective conditions (Figure 1C). Interestingly, recent studies have shown that stress conditions, especially those correlating with reduced Hsp90 function, also can induce diverse forms of mutations in eukaryotes, providing a new source of phenotypic diversity that potentially speeds up stress adaptation 6–9. Below we review the role of Hsp90 in genome stability and the diverse types of mutations that may be induced by Hsp90 stress. We speculate on the possible significance of these findings in cellular adaptation and disease and discuss outstanding questions for future research.
Hsp90 chaperone machinery in protein homeostasis
One emerging role for Hsp90 and its associated family of co-chaperone proteins (the Hsp90 chaperone machinery) is in orchestrating the spatial and temporal order of protein interactions 10. In the crowded, protein-rich environment of the cell, newly synthesized proteins must properly fold and successfully interact with their binding partners, and recycled proteins must appropriately disassociate from one partner and interact with another. To buffer mistakes and ensure the quality of protein assemblies, the Hsp90 chaperone machinery performs three main functions under normal cellular conditions.
First, it specifically interacts with a vast array of cellular substrates, termed clients. This is accomplished in part through adapter co-chaperones that bridge the interaction between Hsp90 and a particular subset of clients. Cdc37 and Sgt1 are examples of adapter co-chaperones that link Hsp90 to cellular kinases or to large multi-protein machines such as the kinetochore, respectively 11–14. Second, the Hsp90 chaperone machine stabilizes specific folding intermediates that allow the client to interact with binding partners, such as a receptor ligand or a protein subunit within a multi-protein complex. The mechanism of Hsp90 client folding is not fully understood but it is clear that many of the more than 20 co-chaperones modulate client interactions as well as the ATPase activity of Hsp90 15. Third, the chaperone machinery and clients are linked to ubiquitin-mediated proteasome degradation, which explains why many clients are turned over when mis-folded or mis-assembled in the absence of Hsp90 function. The ability of the Hsp90 chaperone machine to link protein assembly to degradation represents a quality control mechanism but in some cases may also provide plasticity for dynamic protein complexes - the ability of proteins to change functions from one complex to another.
Clients of Hsp90 are required for many crucial cellular functions. Of relevance to this review, studies to date have revealed many protein complexes involved in genome transmission and repair to be dependent on Hsp90 for function or stability, making Hsp90 an important player in the maintenance of genome stability.
Hsp90 and the coordination of DNA repair pathways
Recent evidence points to a critical role for Hsp90 chaperone machinery in DNA repair complexes required to cope with damage arising from normal replication stress or from mutagenic conditions. Below we will focus on cases where there is clear biochemical evidence for the role of Hsp90 and an obvious impact on DNA integrity when Hsp90 is inhibited.
Genomic DNA is constantly exposed to both exogenous (e.g., UV light) and endogenous (e.g., oxidative stress) insults that frequently give rise to lesions that cannot be repaired by high fidelity DNA polymerases. Translesion synthesis (TLS) is an essential process conserved from bacteria to eukaryotes that bypasses such blocks and prevent large DNA lesions (insertions and deletions) by switching between a family of specialized low fidelity polymerases (i.e., Y-polymerases or Y-pols) that can bypass the lesion and high-fidelity polymerases required for accurate DNA synthesis16,17. Careful regulation by Hsp90 and protein degradation may be important to ensure that Y-pols do not mis-target and induce global error-prone DNA synthesis18–20. The finding that Hsp90 co-purifies with one of the Y-pols, Polη, led to a series of experiments that showed that Hsp90 is required for Polη folding and its interaction with mono-ubiquitin-PCNA (ub-PCNA), as well as for targeting TLS to sites of UV-induced damage. Inhibition of Hsp90 decreased cell survival and increased Polη-dependent, UV-induced mutagenesis21. Another Y-pol, Rev1, has also been shown to require Hsp90 activity 22. Rev1 has a more specialized polymerase activity, limited to dCMP incorporation 23; however, Rev1 has been implicated in recruiting other TLS polymerases to UV-induced lesions 24–26. The dynamic interaction between several specialized DNA polymerases suggest that the mechanisms underlying TLS are complex and there remains much to be understood about how Hsp90 inhibition contributes to Y-pol-mediated mutagenesis.
The ability of PCNA, Polη and Rev1 to prevent large DNA lesions depends on the Fanconi anemia (FA) complex 27,28, which is also regulated by Hsp90. There are approximately 15 proteins associated with the FA complex and cells from patients with mutations in FA genes are susceptible to DNA damage from cross-linking reagents 29. The FA complex also plays a major role in resolving DNA cross-links that arise during normal replication. Indeed, normal human cells exhibit physically linked sister chromatids in anaphase that are sensitive to replication stress and are associated with FA protein complexes 30. The main role of the FA complex is mediated through its ubiquitin ligase activity, required to mono-ubiquitinate FANCD2, and PCNA as discussed above. Ub-FANCD2 in turn recruits BRCA2/FANCD1 to coordinate homologous recombination with TLS in the repair of double-strand breaks (DSBs) that arise from various forms of DNA cross-links (Figure 2C). Consistent with the general characteristics of Hsp90 clients, the FA complex is transiently activated during replication stress or DNA damage, its protein levels are tightly regulated through expression and ubiquitin-mediated proteasome degradation, and nuclear import/export is precisely controlled. For example, Hsp90 associates with FANCA (Figure 2B), a key component of the FA complex, and stabilizes the protein in the nucleus 31. Inhibition of Hsp90 results in the degradation of FANCA, a reduction in Ub-FANCD2 following DNA cross-linking and increased cytotoxicity following replication stress 31. Given the role of Hsp90 in both Pol n action and in FA-mediated recombination, it is tempting to speculate that high fidelity DNA repair requires the chaperone machinery to coordinate these activities (Figure 2).
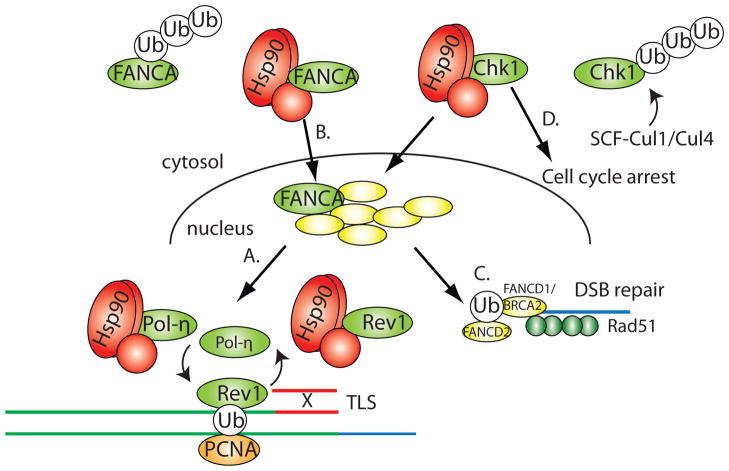
Figure 2.
Hsp90 coordinates DNA repair pathways. (A) Hsp90 interacts with the Y-polymerases, Polη and Rev1, that are critical for accurate translesion synthesis (TLS) repair. (B) Hsp90 interacts with FANCA to stabilize it in the nucleus and mediate its assembly with other Fanconi anemia (FA) proteins. The FA complex acts as a ubiquitin ligase and targets both PCNA and FANCD1/BRCA2 for mono-ubiquitination. (C) Hsp90 is required to stabilize FANCD1/BRCA2, which in turn recruits Rad51 to single-stranded DNA for homologous recombination-mediated repair of double-strand breaks (DSB). (D) Hsp90 is required for the activity of the Chk1 kinase, which is involved in regulating the stability of stalled replication forks, homologous recombination and cell cycle arrest. Hsp90 also coordinates the activity of Chk1 with its SCF-mediated turnover to allow recovery of cells after DNA damage.
The integrated nature of DNA repair pathways means that inhibition of Hsp90 can not only lead to an array of primary effects on clients but also to secondary effects on the associated protein network. For example, it is likely that the defects in FA following Hsp90 inhibition prevents Brca2 from accumulating at sites of damage, which in turn alters the ability of Rad51 to participate effectively in homologous recombination (Figure 2C) 32, 33. This may explain the observation that Hsp90 inhibition causes contraction of human CAG repeats in vivo 7. In addition, Hsp90 directly interacts with upstream signaling molecules, such as the Chk1 kinase, and balances its transient activation during DNA damage with its turnover via Cul1/Cul4 ubiquitin ligases (Figure 2D) 34–36. Chk1 is recruited to sites of DNA damage and serves to delay cell cycle progression, prevent origin firing, stabilize stalled replication forks and activate FA for DNA repair 37. Thus, it is almost certain that changes in Hsp90 activity, either through chemical inhibition or increased Hsp90 load due to stress, will impact how cells respond to genotoxic stress and the ability to accurately repair damaged DNA. We also note that, in considering the role of Hsp90 in genome maintenance, even chemical inhibition of Hsp90 only partially inhibits client function and thus modulation of Hsp90 activity in response to cell stress is unlikely to fully inactivate DNA repair complexes. Moreover, the variety of pathways affected by Hsp90 prevents simple predictions for how DNA integrity might be affected under stress conditions.
Hsp90 and transposon-mediated mutagenesis in the germline
Transposons and other mobile genetic elements have been widely recognized to accelerate evolutionary adaptation in both pro- and eukaryotic organisms 38. Transposon excision and insertion events can lead to gain or loss of gene function depending on the sites of transposition. A recent study in Drosophila uncovered a role for Hsp90 in preventing transposon-mediated mutagenesis and phenotypic variation 6. Mutations in hsp83 or Hsp90 inhibition with geldanamycin was found to cause elevated synthesis of a transcript encoded by the Stellate repeat elements in primary spermatocytes. The change in transcript levels was accompanied by increased mobility of several transposable elements in the F1 progeny of male parents homozygous for hsp83 mutations. About 1% of the Hsp90 defective flies exhibit phenotypic deviation from the wild-type and one of the phenotypic variants was shown to contain a disruption of the noc gene by the I transposable element, resulting in the noc-dependent Scutoid phenotype.
The expression of Stellate and other transposable elements are normally silenced by piRNA, a type of small non-coding RNAs expressed in germline cells named after the ability to interact with Piwi protein 39. Indeed, the transposon mobilization following Hsp90 inhibition and the subsequent phenotypic variation can be recapitulated with a mutation known to affect piRNA biogenesis 6. Morphological variation caused by Hsp90 inhibition had previously been reported in several studies and attributed to uncovering of cryptic genetic variation or alteration in epigenetic traits 4, 40. Transposon mobilization after inhibition of Hsp90 adds another possible mechanism for Hsp90 stress-induced phenotypic variation. A more recent study found that the Hsp90 co-chaperone Hop may link Hsp90 and Piwi together in a tripartite complex 41. Inhibition of Hsp90 by mutation or by geldanamycin does not affect Piwi protein stability but instead prevents Piwi phosphorylation, although the precise mechanism and consequence of Piwi phosphorylation remain to be elucidated. Interestingly, this study also examined morphological variation using an eye outgrowth assay and the data suggest that the observed variation in this particular trait in Hsp90 or piwi mutant background is likely to be due to cryptic mutations that alter the epigenetic regulation of developmental genes rather than to transposon-mediated mutagenesis. These studies highlight the complexity in the potential mechanisms by which Hsp90 stress brings out phenotypic variation even through a single downstream target (Piwi).
Hsp90 and the chromosome segregation machinery
Studies in both yeast and mammalian cells have shown that the Hsp90 chaperone machine is critical for chromosome transmission fidelity through its role in kinetochore assembly (though new evidence suggests that it may also be important for chromatin structure 42). Similar to DNA repair complexes, kinetochores are dynamic multi-protein complexes that must be properly assembled during the cell cycle and specifically targeted to form uniquely at centromeric (CEN) DNA. These qualities may speak to the more general requirement for quality control in multi-protein complex assembly.
Budding yeast cells assemble kinetochores on a small CEN DNA template (125bps) that contains one or two specialized nucleosomes (i.e., CenpA nucleosomes). In contrast, higher eukaryotes’ centromeres contain kilobases to megabases of heterochromatin with a large number of CenpA nucleosomes. Also unique to budding yeast is a multi-protein, sequence-specific CEN-DNA binding complex, CBF3 43, which acts as a bridge between inner CEN-chromatin and outer kinetochore complexes that attach to microtubules and move chromosomes 44. The assembly of CBF3 is balanced with its turnover through the coordinated action of Hsp90 and an SCF ubiquitin ligase complex that contains Skp1 45–47. An F-box motif in the core CBF3 subunit, Ctf13, interacts with Skp1 and in turn Skp1 interacts with the Hsp90 co-chaperone, Sgt1 48, 49. Although the precise assembly steps are not completely understood, the pre-assembly complex (Ctf13-Skp1-Sgt1-Hsp90) favors a Ctf13 conformation that allows it to interact with the other two CBF3 subunits, Cep3 and Ndc10 46. The final assembly step is accompanied by nuclear import and only the fully assembled complex can bind CEN-DNA. Mutations that block formation of the pre-assembly complex favor turnover of Ctf13 in an SCF ubiquitin ligase-dependent manner 45, 46. SCF ub-ligases contain both the Skp1 and Sgt1 subunits and their targeting subunits also contain F-box motifs 50, 51, raising the possibility that the activities of SCF and CBF3 are coordinated somehow by Hsp90. Although mutations that block CBF3 assembly eventually deplete CEN-bound CBF3 and compromise kinetochore function, the short-term effect is to compromise a non-centromere-associated pool of CBF3 complex 52 that interacts with chromosome passenger proteins and functions on the anaphase spindle and in cytokinesis 53, 54. These findings suggest that Hsp90 not only aids in assembly of multi-protein complexes but also contributes to the plasticity of proteins that function in more than one biochemical setting.
The same principle applies to human cells, where Hsp90-Sgt1 is involved in the assembly and turnover of the Mis12 complex. Even though the Mis12 complex has no obvious sequence homology with CBF3 and does not interact directly with CEN-DNA, it nonetheless serves to bridge inner kinetochore complexes to outer microtubule-interacting complexes (Figure 3) 55, 56. The Mis12 complex consists of four subunits that assemble on kinetochores in mitosis and Hsp90 inhibition reduces the levels of one subunit, Dsn1 57. Knockdown of Sgt1 causes a dramatic decrease in Dsn1 as well as the other subunits in the complex (Mis12, Nsl1 and Nnf1; Figure 3), arguing that Sgt1 protects Mis12 from the degradation pathway during Hsp90-mediated assembly. Co-inhibition of Sgt1 and SCF ubiquitin ligases by knockdown of Skp1 or Cul1 restores Mis12 levels, revealing that like CBF3, Mis12 assembly is balanced with SCF-mediated degradation. Sgt1 in mammalian cells also acts as an adapter linking Hsp90 to the Mis12 complex. The conservation of the biochemical pathway but not the kinetochore complex per se highlights the importance of quality control in assembling multi-protein complexes that bridge inner and outer kinetochore elements (Figure 3A). Indeed, balanced assembly and turnover of Mis12 is important for the efficiency and fidelity of kinetochore assembly. In the absence of Sgt1, Mis12 complex assembly is delayed so that kinetochore-microtubule attachments form slowly and cells arrest via the mitotic checkpoint. Skp1 co-depletion rescues Mis12 levels and restores the timely assembly of kinetochores but at a price: kinetochores assembled in the absence of both Sgt1 and Skp1 contain fewer microtubule-binding sites, which may result in chromosome segregation errors.
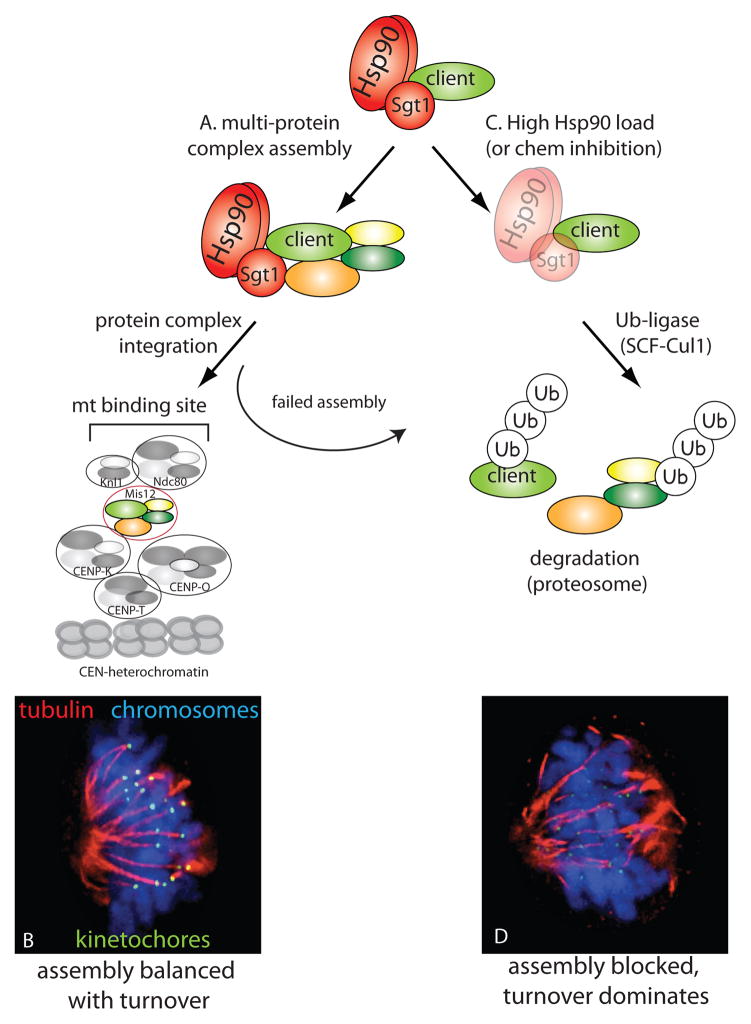
Figure 3.
Hsp90-Sgt1 balances kinetocore assembly and turnover. (A) Hsp90-Sgt1 is required for the assembly of the Mis12 kinetochore complex. Mis12 assembly precedes its integration into the kinetochore machine (consisting of multiple distinct protein complexes). (B) The Mis12 complex bridges inner and outer kinetochore complexes to ensure the efficient assembly of microtubule binding sites (green) that captures spindle microtubules (red) and allows chromosomes (blue) to properly align/segregate. (C) Inhibition of Hsp90, Sgt1 or increased Hsp90 ‘load’ prevents efficient Mis12 assembly, favoring SCF-Cul1-mediated turnover of Mis12 and other kinetochore proteins. (D) The resulting decrease in kinetochore assembly dramatically reduces microtubule binding and chromosomes are inefficiently aligned and segregated [staining as in (B)]. Images in (B) and (D) provided by A. Davies and K. Kaplan.
Hsp90-mediated assembly of kinetochores implies a potential linkage between cell stress and chromosome instability. In both yeast and mammals, it is unlikely that increased Hsp90 load would radically impair kinetochore function and lead to lethality. Rather, a shift in the balance between Hsp90 capacity and load more likely delays assembly of kinetochores or create kinetochores with non-optimal microtubule binding sites. One can imagine at least two ways these conditions could contribute to a rise in chromosome instability. First, delays in kinetochore assembly will engage the mitotic checkpoint and extended checkpoint activation is associated with mitotic slippage and gross chromosome mis-segregation 58. Second is the possibility that cell stress can lead to fewer kinetochore-microtubule binding sites, which may contribute to premature detachment of segregating sister chromosomes, an event that may escape normal checkpoint surveillance.
Hsp90 stress-induced chromosome instability and stress adaption
A direct outcome of erroneous mitosis as a result of faulty kinetochore function is the generation of aneuploid progeny, although recent studies found that mis-segregation could also lead to chromosome breaks and pulverization 59, 60. For multicellular organisms, germline aneuploidy is detrimental probably due to disruption of normal developmental programs or cellular differentiation. Indeed, aneuploidy is commonly known to be associated with birth defects and cancer 61, 62. On the cellular level, recent studies in unicellular organisms demonstrate that aneuploidy is a form of large-effect mutation capable of drastically altering cellular fitness under diverse growth conditions 63. The phenotypic effect of aneuploidy can be attributed to the altered stoichiometry of gene expression caused by gene copy number changes in aneuploids compared to parental euploids 61. The presence of a single aneuploid chromosome (gain or loss) affects the expression of tens to hundreds of genes located on that chromosome at a level directly proportional to the relative dosage of the chromosome 64–70. This primary effect can also lead to more dramatic changes in the expression of their downstream genes throughout the genome 71. As a result, a karyotypically heterogeneous aneuploid cell population can exhibit dramatic phenotypic variation 68,9, providing the substrate for evolutionary selection under stress.
In a recent study in budding yeast, chemical or mutational inhibition of Hsp90, or stress due to heat shock, potently induced aneuploidy probably through erroneous mitosis. The loss rate for an artificial chromosome was measured at hundreds of times above the control, even with an Hsp90 inhibitor (radicicol) concentration that only moderately retards growth 9. The adaptive value of the Hsp90 stress-induced aneuploidy was readily observed from the emergence of radicicol-resistant colonies when a diploid stain was put under the selection of nearly lethal drug concentration (higher than the levels needed to induce aneuploidy). All radicicol-resistant colonies examined were aneuploid, with different karyotypes but sharing a common feature: 1 or 2 extra copies of Chr XV, which carries the genes encoding Sti1, the yeast homolog of the Hsp90 co-chaperone Hop, Pdr5 (a pleiotropic efflux pump), and Sgt1. The extra copies of STI1 and PDR5 genes were necessary and partially sufficient for the observed radicicol resistance. Furthermore, cell populations with highly divergent karyotypes as a result of prior growth in a moderate concentration of radicicol were much more adaptive to other unrelated drugs, such as fluconazole, benomyl and tunicamycin, than a diploid population, and colonies resistant to the same drug exhibit common karyotypic patterns. For example, most of the fluconazole-resistant colonies gained Chr VIII, carrying ERG11, encoding an enzyme required for ergosterol biosynthesis and the target of the clinical antifungal drug fluconazole. Gain of the chromosome carrying ERG11 is in fact a frequent genetic change observed in fluconazole-resistant isolates of human pathogenic fungi Candida albicans and Crytoccocus Neoformans 72–75. A recent study in C. albicans reported that elevated temperature, mimicking fever in the host body, causes increased loss of heterozygosity due to chromosome mis-segregation 8. Heat also induced karyotype mosaicism in near tetraploid embryonic carcinoma cells, which may be subsequently selected to adapt to culture conditions 76. Thus, aneuploidy-mediated phenotypic variation may be a common consequence of Hsp90 stress in diverse eukaryotic organisms.
Concluding remarks
Above we have discussed several examples where the Hsp90 chaperone machinery acts as the key facilitator in cellular complexes that maintain genome stability. These findings predict that genome instability arises when the demand for Hsp90 exceeds its functional capacity, leading to genetic variation in the population that includes point mutations, transposon mobilization, contraction/expansion of repeat elements, chromosome rearrangements, and aneuploidy (Figure 4). It remains to be determined, however, whether the genetic variation observed following inhibition of Hsp90 could fuel stress adaption in a wide range of organisms. For example, the fitness gain or loss associated with aneuploidy in unicellular versus multi-cellular organisms may be quite different and thus simple generalization about the evolutionary advantage of such a mechanism is likely to be difficult.
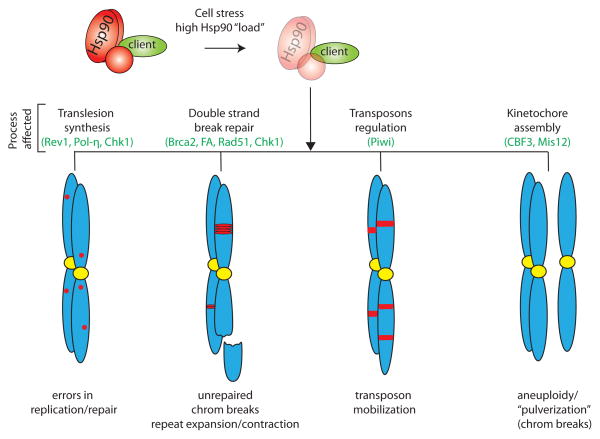
Figure 4.
Elevated Hsp90 ‘load’ can lead to genomic instability through diverse mechanisms. Under high cell stress, the ability of Hsp90 to chaperone its entire range of clients is compromised. This may inhibit pathways involved in trans-lesion synthesis (TLS), double-strand break repair, transposable element regulation and kinetochore assembly. Defects in these processes can contribute to increases in DNA replication errors, unrepaired chromosome breaks as well as the expansion/contraction of repetitive sequence elements, transposon mobilization, and aneuploidy.
A second question that remains is how Hsp90 function may become limited under physiological or pathological conditions. Cell stress - defined as a condition(s) that moves cells away from homeostasis established under laboratory or physiological conditions - has been suggested to increase the ‘load’ for Hsp90 and its co-chaperones. This notion assumes that Hsp90 has a limited capacity to interact with all or certain clients. Consistently, it has long been appreciated that a variety of stress conditions (e.g., heat, starvation, etc.) increase chaperone expression, and cancer cells, presumed to be under higher stress than normal cells, have more active Hsp90 chaperone complexes 77. In addition, studies suggest that Hsp90 and many co-chaperones undergo post-translational modifications upon cell stress78–80, and this may lead to biased assembly of Hsp90 chaperone complexes targeted to a subset of Hsp90 clients. The preferential chaperoning of some proteins over others could allow the Hsp90 machinery to restore homeostasis in a way tailored to a particular stress. A change in specificity also implies that certain clients may no longer be chaperoned, thus uncovering the phenotypic effect of cryptic genetic variation81 or causing genome instability.
Understanding Hsp90’s role in stress adaptation and genome stability has important implications for human disease. Although the susceptibility to disease may be genetically determined, a reasonable precept is that the actual transition to the disease state at the cellular level occurs when the assembly and plasticity of the encoded protein networks are compromised due to certain stress or age-related deterioration. In this view, the primary function of the Hsp90 chaperone machinery may be to buffer against such transitions and thus disease. The very assumptions that underlie the idea of chaperone load and limited capacity form the rationale for a broad array of clinical trials aimed at inhibiting Hsp90 in cancers 82. In light of the mechanisms reviewed here, it is reasonable to raise concerns about a therapeutic approach based solely on Hsp90 inhibition. For example, inhibition of Hsp90 impairs kinetochore assembly without a dramatic increase in cell death, which would lead to the emergence of drug resistance through aneuploidy. If so, inhibiting Hsp90 may initially slow tumor growth but ultimately favor adaptation to chemotherapy or even metastasis. A more sound approach may lie in the simultaneous inhibition of Hsp90 and proteasome-dependent protein degradation 83, which may push pathways to complete loss-of-function and fast cell death, or simultaneously inhibiting Hsp90 and mitosis to prevent the emergence of chromosomally abnormal cell progeny. However, such drastic treatments are likely to increase unintended side effects on patients. With a better understanding of Hsp90 client specificity and regulation under stress, more targeted approaches may become feasible to prevent cancer cell proliferation as well as the potential for adaptive mutations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sangster TA, et al. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 3.Jarosz DF, et al. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annual review of genetics. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 5.Galhardo RS, et al. Mutation as a stress response and the regulation of evolvability. Critical reviews in biochemistry and molecular biology. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Specchia V, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 7.Mittelman D, et al. Hsp90 modulates CAG repeat instability in human cells. Cell stress & chaperones. 2010;15:753–759. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forche A, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio. 2011;2 doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, et al. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochimica et biophysica acta. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Catlett M, Kaplan K. Sgt1p Is a Unique Co-chaperone That Acts as a Client Adaptor to Link Hsp90 to Skp1p. The Journal of biological chemistry. 2006;281:33739–33748. doi: 10.1074/jbc.M603847200. [DOI] [PubMed] [Google Scholar]
- 12.Kadota Y, Shirasu K. The HSP90 complex of plants. Biochimica et biophysica acta. 2012;1823:689–697. doi: 10.1016/j.bbamcr.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Hunter T, Poon RY. Cdc37: a protein kinase chaperone? TICB. 1997;7:157–161. doi: 10.1016/S0962-8924(97)01027-1. [DOI] [PubMed] [Google Scholar]
- 14.Kimura Y, et al. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes and Development. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 15.Echtenkamp FJ, Freeman BC. Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochimica et biophysica acta. 2012;1823:668–673. doi: 10.1016/j.bbamcr.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg EC, et al. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Molecular cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Hirota K, et al. Simultaneous disruption of two DNA polymerases, Poleta and Polzeta, in Avian DT40 cells unmasks the role of Poleta in cellular response to various DNA lesions. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntyre J, et al. The spectrum of spontaneous mutations caused by deficiency in proteasome maturase Ump1 in Saccharomyces cerevisiae. Curr Genet. 2007;52:221–228. doi: 10.1007/s00294-007-0156-8. [DOI] [PubMed] [Google Scholar]
- 19.McIntyre J, et al. Analysis of the spontaneous mutator phenotype associated with 20S proteasome deficiency in S. cerevisiae. Mutat Res. 2006;593:153–163. doi: 10.1016/j.mrfmmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Skoneczna A, et al. Polymerase eta is a short-lived, proteasomally degraded protein that is temporarily stabilized following UV irradiation in Saccharomyces cerevisiae. Journal of molecular biology. 2007;366:1074–1086. doi: 10.1016/j.jmb.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 21.Sekimoto T, et al. The molecular chaperone Hsp90 regulates accumulation of DNA polymerase eta at replication stalling sites in UV-irradiated cells. Molecular cell. 2010;37:79–89. doi: 10.1016/j.molcel.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Pozo FM, et al. Molecular chaperone Hsp90 regulates REV1-mediated mutagenesis. Molecular and cellular biology. 2011;31:3396–3409. doi: 10.1128/MCB.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 24.Guo C, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. The EMBO journal. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tissier A, et al. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA repair. 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Ohashi E, et al. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, et al. Regulation of Rev1 by the Fanconi anemia core complex. Nature structural & molecular biology. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 29.Valeri A, et al. Fanconi anaemia: from a monogenic disease to sporadic cancer. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2011;13:215–221. doi: 10.1007/s12094-011-0645-6. [DOI] [PubMed] [Google Scholar]
- 30.Chan K, et al. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature cell biology. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 31.Oda T, et al. Hsp90 regulates the Fanconi anemia DNA damage response pathway. Blood. 2007;109:5016–5026. doi: 10.1182/blood-2006-08-038638. [DOI] [PubMed] [Google Scholar]
- 32.Mittelman D, et al. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones. 2010;15:753–759. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noguchi M, et al. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 34.Arlander SJH, et al. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. The Journal of biological chemistry. 2003;278:52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- 35.Leung-Pineda V, et al. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–2637. doi: 10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YW, et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Molecular cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Enders GH. Expanded roles for Chk1 in genome maintenance. The Journal of biological chemistry. 2008;283:17749–17752. doi: 10.1074/jbc.R800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nature reviews Genetics. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 39.Juliano C, et al. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annual review of genetics. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nature genetics. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 41.Gangaraju VK, et al. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nature genetics. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kakihara Y, Houry WA. The R2TP complex: discovery and functions. Biochimica et biophysica acta. 2012;1823:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 44.McAinsh AD, et al. STRUCTURE, FUNCTION, AND REGULATION OF BUDDING YEAST KINETOCHORES. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. http://dx.doi.org/10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan KB, et al. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 46.Rodrigo-Brenni MC, et al. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Molecular Biology of the Cell. 2004;15:3366–3378. doi: 10.1091/mbc.E03-12-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stemmann O, et al. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8585–8590. doi: 10.1073/pnas.082223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitagawa K, et al. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Molecular cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 49.Lingelbach L, Kaplan K. The Interaction between Sgt1p and Skp1p Is Regulated by HSP90 Chaperones and Is Required for Proper CBF3 Assembly. Molecular and cellular biology. 2004;24:8938–8950. doi: 10.1128/MCB.24.20.8938-8950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 51.Feldman RM, et al. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 52.Thomas S, Kaplan KB. A Bir1p Sli15p kinetochore passenger complex regulates septin organization during anaphase. Molecular Biology of the Cell. 2007;18:3820–3834. doi: 10.1091/mbc.E07-03-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillis A, et al. A novel role for the CBF3 kinetochore-scaffold complex in regulating septin dynamics and cytokinesis. The Journal of Cell Biology. 2005;171:773–784. doi: 10.1083/jcb.200507017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozelle DK, et al. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. The Journal of Cell Biology. 2011;193:285–294. doi: 10.1083/jcb.201011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheeseman I, et al. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 57.Davies AE, Kaplan KB. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. The Journal of Cell Biology. 2010;189:261–274. doi: 10.1083/jcb.200910036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossio V, et al. Adapt or die: how eukaryotic cells respond to prolonged activation of the spindle assembly checkpoint. Biochemical Society transactions. 2010;38:1645–1649. doi: 10.1042/BST0381645. [DOI] [PubMed] [Google Scholar]
- 59.Janssen A, et al. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 60.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon DJ, et al. Causes and consequences of aneuploidy in cancer. Nature reviews Genetics. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 62.Iourov IY, et al. Somatic genome variations in health and disease. Current genomics. 2010;11:387–396. doi: 10.2174/138920210793176065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pavelka N, et al. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Current opinion in cell biology. 2010;22:809–815. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Upender MB, et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer research. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao C, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nature genetics. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 67.Birchler JA. Reflections on studies of gene expression in aneuploids. The Biochemical journal. 2010;426:119–123. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- 68.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Springer M, et al. A general lack of compensation for gene dosage in yeast. Molecular systems biology. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selmecki A, et al. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selmecki AM, et al. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS genetics. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sionov E, et al. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS pathogens. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Semighini CP, et al. Deletion of Cryptococcus neoformans AIF ortholog promotes chromosome aneuploidy and fluconazole-resistance in a metacaspase-independent manner. PLoS pathogens. 2011;7:e1002364. doi: 10.1371/journal.ppat.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta RK, Srinivas UK. Heat shock induces chromosomal instability in near-tetraploid embryonal carcinoma cells. Cancer biology & therapy. 2008;7:1471–1480. doi: 10.4161/cbt.7.9.6428. [DOI] [PubMed] [Google Scholar]
- 77.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 78.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochimica et biophysica acta. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyata Y, Nishida E. CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Molecular and cellular biology. 2004;24:4065–4074. doi: 10.1128/MCB.24.9.4065-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bansal PK, et al. Sgt1 dimerization is negatively regulated by protein kinase CK2-mediated phosphorylation at Ser361. The Journal of biological chemistry. 2009;284:18692–18698. doi: 10.1074/jbc.M109.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 82.Jhaveri K, et al. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochimica et biophysica acta. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frankland-Searby S, Bhaumik SR. The 26S proteasome complex: an attractive target for cancer therapy. Biochimica et biophysica acta. 2012;1825:64–76. doi: 10.1016/j.bbcan.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]