Abstract
The nuclear receptor Peroxisome Proliferator-Activated Receptor γ (PPARγ) is essential for placental development. For insights into its functions in the placenta, we screened for PPARγ-regulated genes by integrating expression profiles of Pparg-null and Rxra-null placentas with those of WT and Pparg-null trophoblast stem cells differentiated in the presence or absence of a PPARγ agonist. Intersection of these paradigms identified high-probability PPARγ target genes. A few of these genes were previously reported as PPARγ targets in other tissues, but most are new in the context of either PPARγ or placental biology. Transcriptional profiling demonstrated a widespread role for the coactivator NCOA6/AIB3, but not MED1/PBP, in PPARγ-dependent placental gene expression. Spatial and temporal expression analyses revealed that PPARγ impacts genes in diverse trophoblast lineages and during different stages of differentiation. We further validated the Ldhb gene, which encodes the H isoform of lactate dehydrogenase, as a robust PPARγ target in trophoblasts, and propose a hypothetical model that integrates it with a network of PPARγ-regulated genes into a novel pathway of placental fuel metabolism. These findings offer insights not only into the placental functions of PPARγ, but also into unique, previously unsuspected biosynthetic functions of trophoblasts.
Keywords: Placenta, Trophoblast, PPARγ, NCOA6, LDHB
INTRODUCTION
The placenta is essential for the survival and development of all mammalian embryos, where it plays vital roles in transport of gases, nutrients and waste between the mother and the fetus, embryonic immune privilege and secretion of pregnancy hormones. Placental aberrations underlie a wide range of fetal and maternal pregnancy complications, including intrauterine growth restriction, preeclampsia and pregnancy loss. Gene targeting studies in mice revealed numerous genes and pathways that are indispensable for placental development and function (Rossant and Cross, 2001). Detailed molecular and cellular analyses of these gene networks are required for better understanding of placental physiology and pathology.
One of the critical regulators of placental development is the nuclear receptor Peroxisome proliferator-activated receptor γ (PPARγ) (Barak et al., 1999). Pparg-null placentas succumb to lethal pleiotropic defects that span arrested differentiation and lipid droplet depletion in labyrinthine trophoblasts, failed vascularization, maternal blood pool breakdown, and erythrophagocytosis by spongiotrophoblast (SpT) cells (Barak et al., 1999). In addition, these primary placental defects precipitate severe defects in embryonic heart development, which highlight the long-range effects of the placenta beyond its critical functions at the maternal-fetal barrier (Barak et al., 1999). Augmenting PPARγ activity in the placenta by treating pregnant mice with a PPARγ agonist interferes with placental development as well, causing substantial thinning of the SpT layer and dilation of the labyrinthine vasculature (Schaiff et al., 2007). These effects stress the importance of finely tuned PPARγ activity in this tissue. The robust induction of PPARγ during differentiation of trophoblast stem cells (TSC) further buttresses its tight association with placental development (Shalom-Barak et al., 2004).
Much has been learned about the functions of PPARγ in adipogenesis, energy metabolism and inflammation, but none of these functions provides clear mechanistic clues about its role in placental development. Considering that PPARγ is a transcription factor, the definitive answer lies in the identities of its transcriptional targets. Our earlier work identified Muc1 as a robust and highly specific PPARγ target gene in trophoblasts (Shalom-Barak, 2004). Placental MUC1 protein is confined to a single layer of labyrinthine trophoblasts around the maternal blood pools, which exhibit a partially penetrant dilated phenotype in Muc1-deficient placentas (Shalom-Barak et al., 2004). However, the normal survival and growth of Muc1-null embryos imply that other targets of PPARγ transduce its essential placental functions.
Here, a robust microarray-based screening strategy, which integrated non-redundant PPARγ dependence paradigms, identifies high-confidence PPARγ targets. Presumptive targets exhibit diverse expression patterns in the whole placenta and in TSC, suggestive of pleiotropic actions of PPARγ in different placental compartments. The vast majority of these genes are new in the contexts of both the placenta and PPARγ, and the few with known or deduced functions regulate intermediary metabolism and multiple endocrine signaling pathways. We further demonstrate that the transcription coactivator AIB3 (Ncoa6) affects the expression of the majority of placental PPARγ target genes, whereas the coactivator PBP (Med1) plays little to no role in their expression. Finally, we show that Ldhb, the gene encoding heart lactate dehydrogenase, is a tightly regulated PPARγ target in trophoblasts, which in the context of additional metabolic targets revealed here suggests a novel metabolic program in trophoblasts. These findings offer important new insights into the physiological functions and transcriptional mechanisms of PPARγ in the placenta.
MATERIALS AND METHODS
Mice
Pparg+/− mice (heterozygous for a lacZ knock-in allele; Barak et al., 1999) were maintained on two separate inbred backgrounds: 129SvImJ (129) and C57BL6/J (B6; N>14); placentas were procured from embryo progeny of 129-Pparg+/− sires with B6-Pparg+/− dams. Rxra+/− mice (Sucov et al., 1994) were backcrossed onto B6 (N>14); for placenta collection, B6-Rxra+/− females were mated with 129/SvImJ-WT males and placentas were procured from F2 embryo progeny of the resulting B6129F1-Rxra+/− mice. Med1+/− (Zhu et al., 2000) and Ncoa6+/− mice (Kuang et al., 2002) were introgressed onto B6 for six and two generations, respectively, prior to experiments. The Ppargyfp knock-in allele was generated using standard recombinant DNA technology and introduced into the Pparg locus by homologous recombination as described. Correct integration was validated by Southern blots with probes external to the homology arms of the vectors. Germ-line chimeras were established from one correctly targeted Ppargyfp/+ ES cell line, and heterozygotes subsequently introgressed onto a B6 background for over eight generations (N>8). To test in vivo effects of Rosiglitazone (Rosi) on placental gene expression WT pregnant mice were fed from E10.5-E18.5 either powdered chow alone or powdered chow containing 30mg/kg/d Rosi (Schaiff et al., 2007). All mouse studies at The Jackson Laboratory, Washington University and Magee-Womens Research Institute were approved by the respective Animal Care and Use Committees.
Placenta dissection and extraction
To collect WT and various mutant placentas pregnancies were timed by copulation plugs (noon of the day of plug detection = E0.5). Placentas were separated from embryos, carefully trimmed of excess decidual and yolk sac tissue, and kept individually refrigerated in RNALater (Sigma). Genotypes were determined via PCR of DNA from corresponding embryos, as described previously for each targeted allele. Placentas with similar genotypes were pooled in groups of three, and total RNA extracted using Tri-Reagent (Invitrogen).
Establishment and culture of trophoblast stem cells (TSC)
129-PparglacZ/+ males were mated with B6.129-Ppargyfp/+ females, and blastocysts collected at E3.5 and plated individually in 4-well dishes containing irradiated mouse embryo fibroblasts (MEF) in RPMI 1640 medium supplemented with 20% fetal bovine serum, 25ng/ml FGF4 and 1µg/ml heparin, as described (Tanaka et al., 1998). Blastocysts that attached to the substrate and developed cellular outgrowths were dissociated with trypsin and plated onto MEF in the same medium, which was changed every other day. Cultures were passaged to fresh plates every four days. Lines that exhibited morphological resemblance to TSC and a consistent expansion pattern were genotyped by PCR, and stocks were cryopreserved for further work.
For analysis of differentiated TSC, cells were passaged once in the absence of feeder cells in the aforementioned medium, supplemented with 70% embryonic fibroblast conditioned medium, and thereafter split for the various experiments. Cultures were maintained for another 24h in conditioned medium and FGF4, and differentiation was induced for the designated duration by withdrawing conditioned medium, FGF4 and heparin from the medium. Where indicated, cultures were supplemented with 1µM Rosi for the duration of differentiation. RNA was extracted with Tri-Reagent at the end of the indicated differentiation periods.
Affymetrix microarray analysis
Placental and TSC RNA samples were treated with RNase-free DNase and re-extracted. Biotin-labeled cRNA probes was generated according to manufacturer's instructions (Affymetrix, Santa Clara, CA) from: (i) Each of 3 pools of Pparg−/− placentas vs. 3 litter-matched pools of WT placentas; (ii) Each of 3 pools of litter-matched Rxra−/− vs. WT placentas; (iii) Each of 3 pools of litter-matched Med1−/− vs. litter-matched WT and Ncoa6−/− vs. litter-matched WT placentas; (iv) The three WT TSC lines described in this study were profiled after differentiation for two or four days in the absence or presence of 1µM Rosi (one probe per condition); Pparg-null TSC were profiled only in the presence of Rosi, to circumvent identification of genes that are induced by Rosi independent of PPARγ. Each cRNA probe was hybridized individually to a MOE430v2.0 GeneChip™ array in a Fluidics Station 450 (Affymetrix). Arrays were imaged with a GeneChip™ 3000 Laser Confocal Slide Scanner, quantified using GeneChip™ Operating Software v1.2 (Affymetrix), and expression values summarized using the robust multichip average (RMA) function. P values were assigned using analysis of variance (ANOVA) (Churchill, 2004), except for the three Rosi-treated vs. untreated WT TSC lines, which were compared pair-wise using LIMMA (Smyth, 2004). False discovery rates (FDR; q-values) were assigned to whole placenta data as described (Storey and Tibshirani, 2003), after first omitting all probe sets determined as ‘absent’ by the Affymetrix MAS5.0 algorithm in all test samples. Because of unsustainable variation, q-values for TSC data were determined only for genes that passed fold-change (FC) and q thresholds for WT vs. Pparg-null placentas. Data have been deposited in NCBI’s Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo), accession no. GSE39233. Abbreviated analysis files are available as Supplemental Tables S1 and S2.
Expression analyses
RT and quantitative PCR (qPCR) were performed with RT and SYBR Green PCR kits from Applied Biosystems according to manufacturer specifications using an ABI 9700 thermal cycler. Oligonucleotide pairs for individual genes (Table S3) were based on Primer Bank whenever available or designed using the Primer3 shareware. Only reactions that yielded appropriate dissociation curves are shown. qPCR reactions of placental RNA were normalized to 36B4; reactions of RNA from TSC were normalized to either the Dazap1 or Cwc15 genes, which were identified after extensive screens as the least changed genes throughout differentiation and Rosi treatment of all TSC lines. Relative expression values were determined by the ΔΔCt method (Livak and Schmittgen, 2001).
In situ hybridizations were performed as described (Barak et al., 1999). Pparg+/− (lacZ-KI) parent pairs were bred and concepti of each pregnancy were collected at E9.5, fixed in 4% paraformaldehyde and cryoembedded together in a single block of OCT. Individual concepti in each embryo cohort were genotyped by in situ hybridization of serial sections with antisense riboprobes for Pparg and lacZ, identifying the WT and Pparg-null allele, respectively. Additional serial sections were hybridized with antisense probes for the indicated genes.
Reporter assays
DOTAP-mediated transfection of CV1 cells was performed in a 48-well format as described (Shalom-Barak et al., 2004). In short, wells containing 50–70% confluent CV1 cells were lipofected in triplicates with combinations of nuclear receptors, reporters, and CMV-lacZ controls at 25, 62.5, and 125ng/well, respectively. Lipofection medium was replaced after 5–8 hours with DMEM containing 2% charcoal and resin-stripped fetal calf serum and the indicated ligand combinations. Cells were extracted 36–48 hours later, and assayed for luciferase and β-galactosidase activities. Data show mean and standard deviation of normalized luciferase/β-gal activity in triplicate wells from one representative experiment out of at least two.
Chromatin Immunoprecipitation (ChIP)
WT and Pparg-null TSC lines cultured were differentiated on 150 mm plates for four days in the presence of Rosi. Chromatin was cross-linked, extracted, fragmented and precipitated essentially as described (Hartman et al., 2002), except that the cells were sonicated by four, instead of three bouts of 15 seconds. Custom-made anti-PPARγ antibodies used in the assay were described previously (Kim et al., 2007). PCR fragments surrounding the three putative PPREs in the Ldhb promoter and first intron were each amplified from 10% of either the entire immunoprecipitate or 1% input in 30 cycles of 94°C – 15 seconds, 60°C – 30 seconds, 71.5°C – 55 seconds. Forward and reverse primer sequences were, respectively: RE1 – 5’-AGC TCG AGG CCT TTT GGA GTG ATG TAT CAG TAG C-3’ / 5’-TGA TTG GAA ACC CGA GAC GGG TCT ATC-3’; RE2 – 5’-GGT GTC ACG GAA CTG AAC AAG CTC C-3’ / 5’-CCC CAC ATT CCC ATC TCC TGT GG-3’; RE3 – 5’-GGG ACA AAA GAA AGA AAC GAG CTT GCT TCA G-3’ / 5’-GGA ACC ACA TCT CAC CGA ATG CAG G-3’.
RESULTS
Significant misexpression overlaps between Pparg-null and Rxra-null placentas
Genes whose placental expression is influenced by PPARγ were identified by Affymetrix microarray analysis of WT vs. Pparg-null placentas at E9.5. Statistical power and normalization of stochastic sample differences were achieved by comparing three pools of three Pparg-null placentas each to three litter-matched pools of three WT placentas. Loss of Pparg expression and a 4.5-fold reduction of Muc1 expression in microarrays of Pparg-null placentas provided technical and conceptual confidence in the dataset. To focus on biologically meaningful changes while minimizing false-positive calls, we defined significant differences as those that exceeded 1.3-fold with a false discovery rate (FDR; q-value) of ≤ 0.05. By these criteria, 481 genes were significantly down-regulated and 303 were significantly up-regulated in Pparg-null placentas.
Multiple phenotypic analogies between Rxra-null and Pparg-null placentas (Sapin et al., 1997; Barak et al., 1999), topped by the dramatic downregulation of Muc1 in Rxra-null placentas (Shalom-Barak et al., 2004), implicate RXRα as the primary heterodimeric partner of PPARγ in this tissue. Indeed, 203 of the genes that significantly decreased and 112 of those that increased in Pparg-null placentas, corresponding to 42% and 37% of the PPARγ-dependent transcriptome, were also differentially expressed in Rxra-null placentas (Fig. 1A). A significant reduction in Rxra mRNA levels and a threefold down-regulation of Muc1 in microarrays of Rxra-null placentas (p < 10−8) validated this arm of the analysis. These relative ratios between mono- and co-regulation by PPARγ and RXRα were rather consistent over a range of FC thresholds (Fig. 1A).
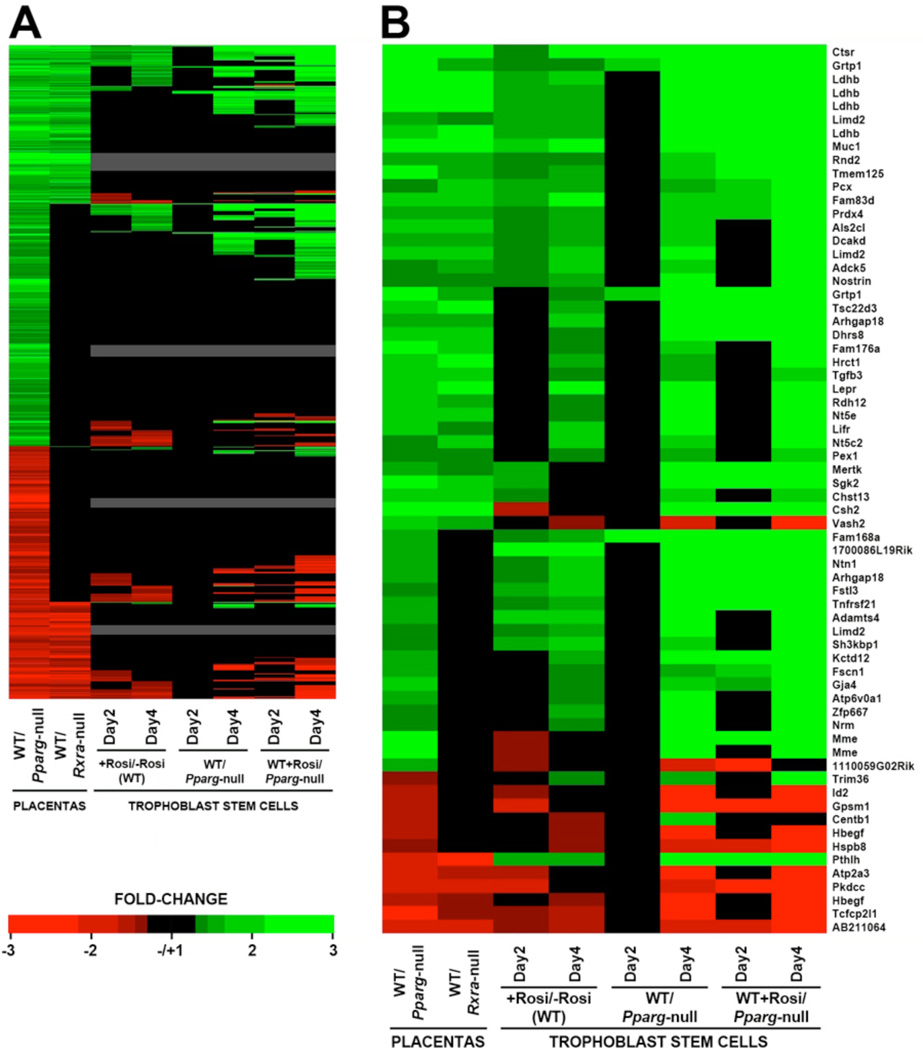
Fig. 1. Overlapping effects of PPARγ, RXRα and NCOA6 on placental gene expression.
(A) Proportioned Venn diagrams show the numbers of overlapping and mutant-specific probe sets down-regulated (left) or up-regulated (right) in Pparg-null and/or Rxra-null placentas at q<0.05 by over 1.3x (top numbers), 1.5x (middle) and 2.0x (bottom). (B) qPCR analysis of Pparg and Muc1 expression in WT, Pparg+/− and Pparg-null placentas. Each bar represents three-four pools of three placentas of the corresponding genotype. (C) Proportioned Venn diagrams of the relationships between PPARγ dosage sensitivity and RXRα dependence of representative genes down-regulated (left) or up-regulated (right) in Pparg-null placentas. PPARγ dosage sensitivity was defined as differential expression in Pparg+/− placentas that amounted to at least 25% of the differential expression in Pparg-null placentas, as determined by qPCR. RXRα data are based on microarray results. See Table S4 for the complete dataset. (D) Intersection of probe sets down-regulated (left) or up-regulated (right) in microarrays of Pparg-null, Rxra-null and Ncoa6-null placentas by more than 1.3x (top numbers), 1.5x (middle) and 2.0x (bottom), with q<0.05.
Effects of PPARγ dosage
The halving of Pparg and Muc1 levels in Pparg+/− placentas (Barak et al., 1999; Shalom-Barak et al., 2004; Fig. 1B) is consistent with the notion that at least some PPARγ-regulated genes respond to changes in PPARγ gene dosage. In contrast, genes that change only secondary to placental failure of Pparg-null embryos, and not due to bone fide changes in PPARγ signaling are not expected to be substantially influenced in the relatively healthy Pparg+/− placentas. Accordingly, qPCR analyses were performed to both validate microarray-inferred differential expression of genes in Pparg-null placentas and determine their relative levels in Pparg+/− placentas. Test samples included the six RNA pools used in the original microarrays, their litter-matched Pparg+/− placenta pools, and one additional pair of litter-matched WT and Pparg+/− placenta pools. Table S4 lists all tested genes and their expression values in Pparg-null and Pparg+/− relative to WT placentas. Our analyses confirmed the down-regulation of 99/104 and the up-regulation of 40/43 microarray-identified genes in Pparg-null placentas. For practical purposes, we defined dosage-sensitivity as a statistically significant (p < 0.05) expression change between WT and Pparg+/− placentas that amounted to at least 25% of the corresponding change in Pparg-null placentas. By this criterion, 44 of the 99 genes down-regulated in Ppargnull placentas (44%) displayed dosage sensitivity. These included 37 of the 75 genes that were also significantly down-regulated in Rxra-null placentas (49%), as opposed to 7 of the 24 genes (29%) that were unchanged in Rxra-null placentas (Fig. 1C). Among the 39 genes that were up-regulated in Pparg-null placentas, only 8 (21%) were significantly induced in Pparg+/− placentas. These were similarly divided between RXRα-dependent and independent genes (5/27 [19%] and 3/12 [25%], respectively) (Fig. 1C). Thus, dosage-sensitivity is a more frequent attribute of genes that are down-regulated in both Pparg-null and Rxra-null placentas, and less so of genes that are either refractory to RXRα status or induced upon PPARγ or RXRα deficiency.
AIB3 is a ubiquitous coactivator of placental PPARγ-dependent genes
Gene regulation by transcription factors depends on transcription cofactors, which cement cooperative interactions within transcriptional complexes, mediate interactions with the basal transcription machinery and modify chromatin structure of target promoters. Of the many cofactors whose interactions with PPARγ have been documented extensively, two coactivators – PBP (Med1) and AIB3 (Ncoa6) – stand out in terms of the placental defects and early lethality caused by their deficiency (Zhu et al., 2000; Kuang et al., 2002). To assess the contributions of MED1 and NCOA6 to placental PPARγ-dependent gene expression, we profiled gene expression of Med1-null and Ncoa6-null placentas following the same technical and analytical principles established with Pparg-null and Rxra-null placentas. Significantly reduced levels of Med1 or Ncoa6 in the microarrays of the respective null placenta pools provided technical confidence in both datasets. Against expectations, MED1 deficiency had a minor effect on placental gene expression, with only 19 genes down-regulated and 20 up-regulated by at least 1.3-fold, even with a more relaxed FDR threshold of up to 0.1. None of the down-regulated genes and only one of those that were up-regulated in Med1-null placentas were concordantly affected in both Pparg-null and Rxra-null placentas (see Table S1). In contrast, NCOA6 influenced the expression of over 2,500 placental genes by >1.3-fold and q < 0.05, confirming its major regulatory role in the tissue. Most importantly, 43% (210/481) of the genes significantly down-regulated in Pparg-null placentas were significantly suppressed in Ncoa6-null placentas. The Odds Ratio (OR) for such overlap by Fisher’s Exact Test was 15.12 with a P value of << 2.2E-16. Moreover, 127 of the 203 transcripts that were down-regulated in both Pparg-null and Rxra-null placentas (63%) were also down-regulated in Ncoa6-null placentas (Fig. 1D; OR=27.99; P << 2.2E-16). A smaller proportion of the genes that were up-regulated in Pparg-null placentas were significantly induced in Ncoa6-null placentas (48/303; 16%; OR=6.15; P < 2.2E-16), and only 33% of those that were co-up-regulated in Pparg-null and Rxra-null placentas recapitulating this trait in Ncoa6-null placentas (OR=14.25; P << 2.2E-16). These data strongly suggest that while MED1 is a relatively minor contributor to placental gene expression in general and to placental PPARγ-dependent transcription in particular, NCOA6 is a ubiquitous coactivator of PPARγ in the placenta.
Integration of in vivo and in vitro PPARγ-dependence paradigms
The essential placental functions of PPARγ are restricted to trophoblasts (Barak et al., 1999). However, the complex cellular composition of the placenta convolutes the interpretation of expression profiles in the whole tissue. Therefore, we sought to align the in vivo expression data with the effects of PPARγ manipulation in cultures of pure trophoblasts. TSC provide an ideal platform for this type of analysis, as they are able to differentiate into most trophoblast lineages. Moreover, PPARγ is potently induced and Muc1 is substantially up-regulated by the PPARγ agonist Rosi during TSC differentiation, demonstrating preservation of key placental PPARγ expression and activity patterns in these cells (Shalom-Barak et al., 2004).
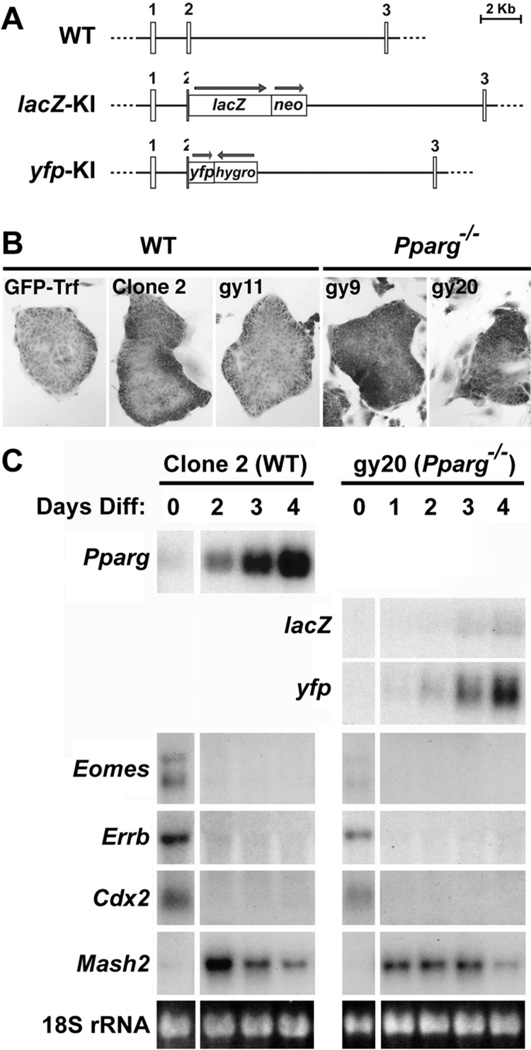
To this end, we established two new Pparg-null TSC lines, gy9 and gy20, derived from blastocysts carrying one Pparg::lacZ knock-in (KI) allele (Barak et al., 1999) and a second, Pparg::yellow fluorescent protein (yfp) KI allele (Fig. 2A). We also established a control WT TSC line, gy11, from sibling blastocysts. We studied these cell lines alongside two previously established WT TSC lines – GFP-Trf (Tanaka et al., 1998) and Clone 2 (Dr. T. Gridley, The Jackson Laboratory, unpublished data). All five cell-lines maintained a typical TSC morphology in the undifferentiated state (Fig. 2B). Moreover, expression of the trophoblast stemness markers Eomes, Cdx2, and Errb in the undifferentiated state and their suppression within 1–2 days of differentiation alongside induction of the Ascl2 (Mash2) gene authenticated the trophoblast identity of the new lines (Fig. 2C, and data not shown). Importantly, Pparg or the knocked-in lacZ and yfp were induced during differentiation of all WT or Pparg-null cell-lines, respectively (Fig. 2C), confirming the corresponding genotypes and indicating that the Pparg locus is induced during TSC differentiation independent of a functional PPARγ protein. As with Ascl2, the Pparg-null TSC did not exhibit consistent aberrations in the expression of other prototypic markers of the primary trophoblast lineages (see Table S2). Thus, WT and Pparg-null TSC provide a robust and malleable system, which retains key developmental features of trophoblasts without being muddled by other placental cell populations.
Fig. 2. Establishment of Pparg-null TSC.
(A) Schematic structures of WT Pparg and its two knock-in alleles used to derive TSC lines in this study. Numbers designate respective exons and arrows show the orientations of knocked in markers and drug selection cassettes. Both lacZ and yfp are in-frame with the interrupted Pparg sequence. (B) Representative undifferentiated colonies of the five studied TSC lines cultured in FGF4, heparin and MEF condition medium. (C) Time-course Northern blot analyses of Pparg and its lacZ and yfp surrogates, the stemness markers Eomes, Errb, and Cdx2, and the differentiated trophoblast marker Mash2 (Ascl2) during differentiation of WT TSC Clone 2 and Pparg-null clone gy20, respectively. 18S ribosomal RNA (rRNA) confirms normalization. Similar results were obtained with WT clones GFP-Trf and gy11 and Pparg-null gy9 cells, validating the TSC identity of the new lines.
Because expression of canonical PPARγ targets is expected to respond to both genetic and pharmacological manipulations of PPARγ, we screened for reproducible effects of both Pparg deficiency and PPARγ agonists among all five TSC lines. Accordingly, we profiled gene expression in five lines two and four days after induction of differentiation by withdrawal of FGF4, heparin and conditioned media, both in the presence and absence of Rosi. Confidence in the dataset was provided by: (i) high Pparg expression in all WT TSC and practically no expression in both Pparg-null TSC lines; (ii) down-regulation of Pparg by Rosi in each of the three WT lines, as observed previously (Shalom-Barak et al., 2004); (iii) induction of Muc1 and its further stimulation by Rosi during differentiation of all three WT, but none of the Pparg-null TSC lines (not shown).
Heterogeneous basal and induced expression of numerous genes among the five TSC lines yielded prohibitive false discovery rates when all transcripts were analyzed. Considering the limited physiological relevance of genes whose expression does not change in vivo, we therefore filtered the test data to the 784 probe sets that passed the differential expression threshold in Pparg-null placentas. These were subject to three primary analyses, with FC and q cutoffs of >1.3x and <0.05, respectively: (i) Responses to stimulation of PPARγ by Rosi, determined by pair-wise comparisons within each of the three WT TSC lines, to circumvent the variegated basal expression of individual genes among the lines. (ii) Effects of PPARγ deficiency, determined by variance analysis of the three untreated WT TSC lines vs. the two Rosi-treated Pparg-null ones. (iii) Compound effects of the two variables, determined by variance analysis of Rosi-treated WT vs. Pparg-null TSC. The power of the latter analysis is evident in the discovery of 99 additional differential transcripts that fell below the significance thresholds of the singular PPARγ stimulation or deficiency criteria.
The heat map in Fig. 3A breaks down the entire 784 probe set field. It demonstrates that transcripts induced by Rosi in WT TSC and/or down-regulated in Pparg-null TSC adhered with high consistency to three reliability criteria: (i) Concordant direction with in vivo regulation, ie down-regulation in Pparg-null placentas. (ii) Temporal continuity, ie maintenance of the expression differential from day 2 to day 4. Importantly, only eight transcripts were significantly different between WT and Pparg-null TSC on day 2, likely due to slow cumulative effect of PPARγ that was further compounded by variation between the cell lines. However, four days of differentiation unveiled 146 significant changes between WT and Pparg-null TSC lines, including all eight discovered on day 2. (iii) Recapitulation of the responses to Rosi or PPARγ deficiency alone in the compound difference between Rosi-treated WT and Pparg-null TSC. In contrast, transcripts that were significantly suppressed by Rosi were the least consistent with these criteria: 36–41% were down-regulated in Pparg-null placentas in vivo (discordant regulation), 57% lost their suppressed status from day 2 to day 4 (temporal discontinuity), and 41% fell short of the compound differential cutoff. These incongruities reduce the technical confidence in such readouts, and in conjunction with the prototypic transactivation properties of Rosi-stimulated PPARγ, in the reliability of purported Rosi suppression events.
Fig. 3. Integration of in vivo and in vitro PPARγ-dependence paradigms.
(A) Heat map of all probe sets that changed in Pparg-null placentas by greater than 1.3-fold with q<0.05 (left column), simulating their relative expression in Rxra-null placentas, Rosi-treated vs. untreated WT TSC, untreated WT vs. Rosi-treated Pparg-null TSC, and Rosi-treated WT vs. Pparg-null TSC, each after two and four days of differentiation. Black – individual differentials of less than 1.3-fold and/or q>0.05. Grey – probe sets below the Affymetrix MAS 5.0 expression threshold in all TSC samples. (B) An annotated distillation of the probe sets that abided by each of the PPARγ-dependence criteria. Abidance by each in vitro criterion at either day 2 or day 4 sufficed for inclusion in this heat map.
Fig. 3B hones in on all probe sets that passed the differential expression thresholds of both PPARγ deficiency and pharmacological stimulation in TSC. This cross-section of genes reveals enrichment for RXRα-dependent genes (64% vs. 40% in the starting gene pool) and for PPARγ-induced genes (53 vs. 12, compared to 481 vs. 303 in the starting dataset), reiterating the features of prototypic PPARγ targets.
Table 1 distills these data to the 35 genes that also changed significantly in Rxra-null placentas, further listing their responses to placental Pparg haploinsufficiency and Ncoa6 deficiency. At least 27 of the 30 positively regulated genes (90%) were significantly down-regulated in Ncoa6-null placentas, up from 43% and 63% of the genes showing down-regulation in Pparg-null placentas alone or both Pparg-null and Rxra-null placentas, respectively (see Fig. 1D). In addition, three of the five negatively regulated genes were also up-regulated in Ncoa6-null placentas. This progressive enrichment links the likelihood that a gene abided by all canonical PPARγ-dependence criteria to its dependence on NCOA6. In contrast, genes with PPARγ dosage sensitivity were not enriched compared to the broader pool of genes that changed in Pparg-null placentas irrespective of other criteria (Table S4), questioning the reliability of dosage sensitivity as a consistent staple of canonical PPARγ targets.
Table 1. Genes abiding by all PPARγ-dependence paradigms.
A compilation of all genes whose expression was significantly altered in each of Pparg-null placentas, Rxra-null placentas, Rosi-treated TSC and Pparg-null TSC. FC and statistical significance values are listed for all genes in each of these systems, as well as in Pparg+/− and Ncoa6-null placentas. For Pparg+/− placentas, the ratio of FC to the FC in Pparg-null placentas is listed as well.
| Gene |
Pparg-null placentas qPCR(FC; P) |
Rxra-null Placentas Affy (FC; q) |
Rosi effect in WT TSC Affy (FC; q) |
Pparg-null TSC Affy (FC; q) |
Pparg+/− placentas qPCR (FC; P; % of FC-null) |
Ncos6-null placentas Affy (FC; q) |
|---|---|---|---|---|---|---|
| Muc1 | −14.36; 0.000 | −3.13; 0.000 | +2.20; 0.014 √ | −2.40; 0.020 | −1.71; 0.001; 45 | −1.79; 0.004 |
| Csh2 | −3.79; 0.001 | −2.32; 0.000 | −1.64; 0.0102,3√ | −23.48; 0.004 √ | −1.90; 0.001; 64 | −3.36; 0.001 |
| Tmem166 | −3.52; 0.000 | −2.01; 0.000 | +1.34; 0.037 | −2.11; 0.029 | −1.56; 0.005; 50 | −2.10; 0.000 |
| Grtp1 | −2.63; 0.001 | −1.76; 0.000 | +1.44; 0.016 | −2.33; 0.015 √ | −1.82; 0.041; 73 | −1.42; 0.018 |
| Lepr | −2.40; 0.000 | −2.14; 0.000 | +3.70; 0.006 √ | −2.64; 0.015 √ | −1.63; 0.007; 66 | −1.69; 0.012 |
| Ldhb | −2.37; 0.000 | −3.13; 0.000 | +1.70; 0.014 √ | −7.13; 0.004 √ | −1.29; 0.024; 38 | −2.71; 0.001 |
| Ctsr | −2.24; 0.024 | −2.45; 0.000 | +2.68; 0.010 √ | −6.95; 0.005 √ | −1.52; 0.020; 61 | −1.68; 0.032 |
| Adck5 | −2.18; 0.001 | −1.56; 0.000 | +1.86; 0.010 √ | −1.69; 0.035 | −1.45; 0.013; 58 | −1.48; 0.002 |
| Pcx | −2.17; 0.002 | −1.72; 0.000 | +1.52; 0.016 √ | −1.58; 0.040 √ | −1.51; 0.011; 63 | −1.98; 0.000 |
| Als2cl | −1.99; 0.004 | −1.59; 0.000 | +1.57; 0.014 √ | −2.12; 0.023 | −1.21; 0.015; 35 | −2.85; 0.000 |
| Tsc22d3 | −1.90; 0.006 | −2.25; 0.000 | +1.45; 0.020 √ | −6.74; 0.012 | −1.41; 0.047; 61 | −2.00; 0.011 |
| Tgfb3 | −1.86; 0.001 | −1.81; 0.000 | +1.32; 0.045 | −1.58; 0.045 | −1.32; 0.041; 52 | −3.02; 0.000 |
| Dcakd | −1.62; 0.000 | −1.68; 0.000 | +1.51; 0.014 √ | −1.96; 0.035√ | −1.30; 0.004; 60 | −1.66; 0.001 |
| Nostrin | −1.34; 0.019 | −1.34; 0.000 | +1.63; 0.026 √ | −3.47; 0.021 | −1 21; 0 010; 69 | −1.28; 0.011 |
| Hsd17b11 | −1.82; 0.022 | −1.86; 0.000 | +1.39; 0.022 | −3.74; 0.009 | −1.43; 0.106; 67 | −1.99; 0.000 |
| Limd2 | −1.76; 0.005 | −1.59; 0.000 | +1.67; 0.014 √ | −3.89; 0.005 √ | −1.17; 0.072; 33 | −1.61; 0.002 |
| Prdx4 | −1.58; 0.0001 | −1.53; 0.000 | +1.54; 0.014 | −1.84; 0.017 | ND1 | −1.31; 0.009 |
| Nt5c2 | −1.45; 0.010 | −1.74; 0.000 | +1.99; 0.010 √ | −1.75; 0.025 √ | −1.18; 0.087; 50 | −1.54; 0.015 |
| Sgk2 | −4.96; 0.000 | −2.12; 0.000 | +1.58; 0.0032 √ | −2.42; 0.034 | +1.13; 0.233; −16 | −1.33; 0.024 |
| Rdh12 | −2.93; 0.002 | −1.60; 0.000 | +1.42; 0.022 | −2.53; 0.034 √ | −1.18; 0.284; 23 | −1.52; 0.009 |
| Hrct1 | −2.27; 0.024 | −2.35; 0.000 | +1.46; 0.018 √ | −1.55; 0.009 √ | −1.01; 0.467; 2 | −3.17; 0.000 |
| Rnd2 | −2.04; 0.001 | −1.49; 0.000 | +1.41; 0.022 √ | −1.82; 0.037 | −1.17; 0.125; 29 | −1.81; 0.003 |
| Tmem125 | −2.00; 0.001 | −1.62; 0.000 | +1.54; 0.014 √ | − 2.11; 0.049 | −1.12; 0.143; 22 | −1.76; 0.001 |
| Lifr | −2.00; 0.000 | −1.55; 0 001 | +1.67; 0.015 √ | −4.70; 0.008 | −1.02; 0.470; 3 | −1.57; 0.006 |
| Mertk | −1.79; 0.000 | −1.52; 0.000 | +1.57; 0.0032 √ | −6.13; 0.010 | −1.11; 0.146; 23 | −1.41; 0.045 |
| Vash2 | −1.69; 0.001 | −1.55; 0.002 | −1.31; 0.0243 | +1.86; 0.0353 | −1.11; 0.084; 25 | −2.70; 0.000 |
| Nt5e | −1.61; 0.016 | −1.72; 0.001 | +1.31; 0.039 | −3.62; 0.020 | −1.00; 0.496; 1 | −2.59; 0.003 |
| Chst13 | −1.45; 0.009 | −2.02, 0.000 | +1.36; 0.0122 | −1.67; 0.035 | +1.09; 0.169; 28 | −2.04; 0.000 |
| Fam83d | −1.73; 0.008 | −1.98; 0.000 | +2.49; 0.006 √ | −1.95; 0.020 √ | −1.13; 0.238; 27 | −1.28; 0.004 |
| Arhgap18 | −2.23; 0.000 | −1.62; 0.000 | +1.71; 0.020√ | −4.73; 0.004 √ | −1.06; 0.281; 10 | −1.07; 0.374 |
| L1td1 | +2.21; 0.000 | +1.66; 0.000 | −1.49; 0.016 | +1.93; 0.006 | +1.80; 0.003; 67 | +1.53; 0.014 |
| Pthlh | +1.81; 0.000 | +2.37; 0.000 | +1.63; 0.0363 | −6.09; 0.0053 √ | +1.34; 0.000; 42 | +4.98; 0.000 |
| Hbegf | +1.52; 0.003 | +1.33; 0.002 | −1.36; 0.023 | +2.19; 0.005 √ | +1.12; 0.008; 24 | +10.1; 0.499 |
| Pkdcc | +1.72; 0.018 | +1.71; 0.001 | −2.05; 0.0022 | +2.06; 0.022 | +1.08; 0.147; 11 | −1.58; 0.0093 |
| Atp2a3 | +2.54; 0.000 | +1.46; 0.001 | −1.49; 0.0142 | +2.36; 0.027 | +1.05; 0.374; 3 | +1.41; 0.005 |
Values for Pparg-null and Pparg+/− placentas are based on qPCR, except the Prdx4 gene (1), for which no adequate qPCR conditions were identified; ND – No data. Values for the remaining paradigms are based on Affymetrix microarray data.
TSC values are from the 4th day of differentiation, except genes whose differential expression was not statistically significant on that day, but day 2 data attained significance (2).
marks TSC data whose statistical significance was recapitulated in qPCR analyses.
Values with a statistically significant FC, yet opposite regulatory direction compared to Pparg-null placentas (referred to in the text as discordant regulation).
In Pparg+/− and Ncoa6-null placentas, differentials that were either very close to the FC threshold and statistically significant or well above the FC threshold with p ≤0.1 were classified as questionably significant and shaded grey.
Black shading – values that were considerably off the marks for FC and/or statistical significance and therefore lack significance. Genes were partitioned according to down-regulation vs. up-regulation in Pparg-null placentas, further clustered within these partitions according to significance patterns in Pparg+/− and Ncoa6-null placentas, and sorted within each cluster according to their FC in Pparg-null placentas.
Diverse temporal and spatial patterns of PPARγ-dependent trophoblast genes
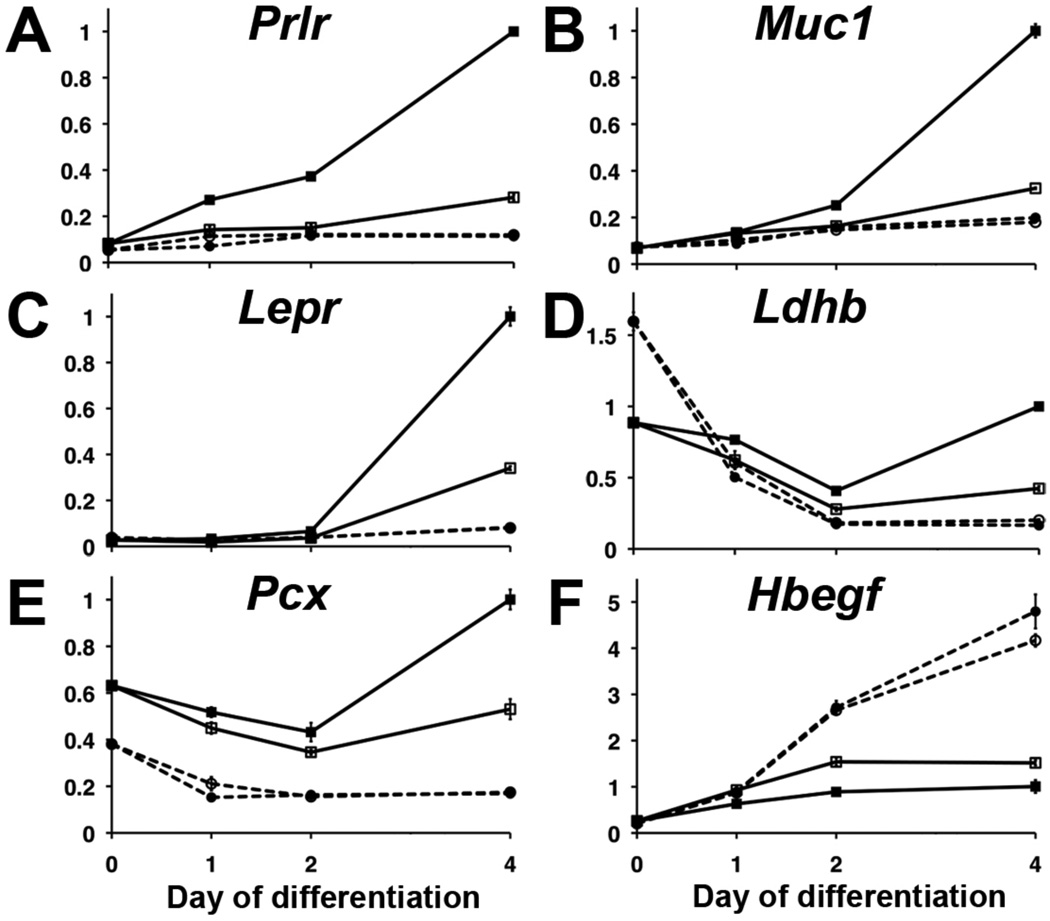
To analyze the timing and topology of PPARγ action during placental development, we interrogated the in vivo and in vitro expression patterns of representative genes that emerged from the analyses above. Supplemental Fig. S1 shows single qPCR measurements of 51 such genes throughout differentiation of the WT and Pparg-null TSC lines in the presence and absence of Rosi. Genes with representative expression prototypes were reconfirmed in one WT (GFP-Trf) and one Pparg-null TSC line by triplicate qPCR (Fig. 4). Several prototypic patterns emerged:
Canonical induction (Fig. S1,A–C): none-to-low expression in undifferentiated TSC, followed by incremental induction as early as one day or as late as four days from the onset of differentiation in WT but not Pparg-null TSC. Rosi typically enhanced such genes in WT TSC, and some genes could not attain significant expression in its absence. Prototypic examples shown are Prolactin Receptor (Prlr), Muc1, and Leptin Receptor (Lepr) (Fig 4,A–C).
A “bowl-shaped” response (Fig. S1,D): considerable, PPARγ-independent expression in undifferentiated TSC, followed by a significant drop during early differentiation, then resurgence and further stimulation by Rosi in WT, but not Pparg-null TSC. Variations of this pattern were exhibited by twelve of the tested genes, most of which encode metabolic functions. Shown are Lactate Dehydrogenase B (Ldhb) and Pyruvate Carboxylase (Pcx) (Fig. 4,D–E).
Biphasic induction (Fig. S1,E): PPARγ-dependent or independent up-regulation within one day of differentiation, down-regulation by the second day, and a subsequent PPARγ-dependent induction towards the fourth day of differentiation.
Variations of negative regulation by PPARγ and/or Rosi (Fig. S1,F), represented in Fig. 4F by Heparin-Binding EGF-like Growth Factor (Hbegf).
Fig. 4. Temporal patterns of PPARγ-regulated genes in TSC.
Expression of six prototypic PPARγ-dependent genes during differentiation of the WT TSC line GFP-Trf (Solid lines, squares) and the Pparg-null TSC line gy09 (dashed lines, circles) in the absence (empty squares/circles) or presence of Rosi (filled squares/circles). Each qPCR measurement was performed in triplicates, normalized to the Dazap1 gene and transformed to linear scale by the ΔΔCt method. Expression at day 4 in the presence of Rosi was assigned a relative value of 1.0.
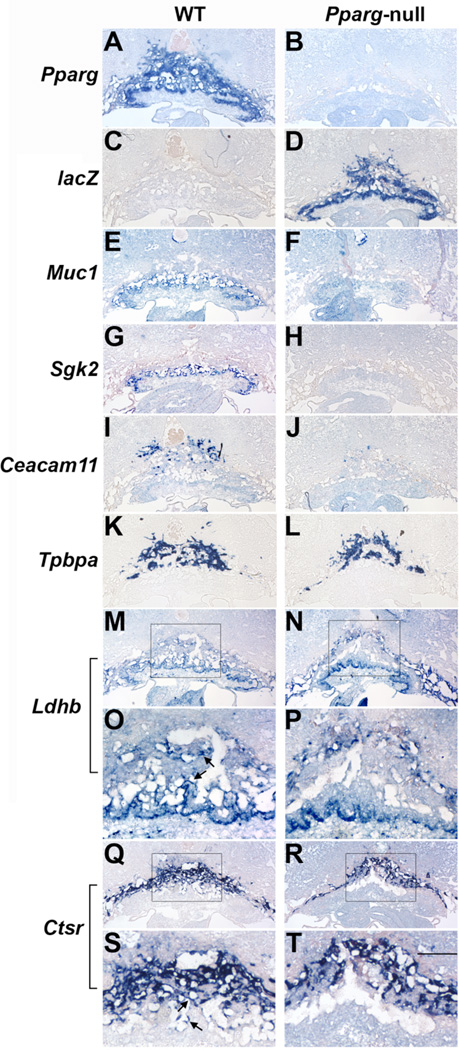
Placental distribution and PPARγ-dependence patterns exhibited marked variability as well; respective genotypes were determine by the expression of Pparg and/or lacZ (Fig. 5A–D):
Muc1 was expressed in a PPARγ-dependent fashion in the trophoblast layer flanking the labyrinthine maternal blood pools, as previously shown (Fig. 5E,F; Shalom-Barak et al., 2004).
Serum and Glucocorticoid-Induced Kinase 2 (Sgk2) was limited to a perichorionic trophoblast layer and absent from Pparg-null placentas (Fig. 5G,H).
Carcinoembryonic Antigen-Related Cell Adhesion Molecule 11 (Ceacam11), which registered the strongest decline of any gene in Pparg-null placentas (see Table S4), was expressed in a PPARγ-dependent fashion in scattered cells at the distal part of the SpT layer (Fig. 5I,J; compare spatial distribution to the canonical pan-SpT marker Tpbpa, Fig. 5K,L). Later in development Ceacam11-expressing cells populate parts of the SpT, and are distinct from glycogen trophoblasts (Kataoka et al., 2000 and data not shown).
Ldhb was expressed evenly throughout the chorion and trophoblast giant cells (TGC), as well as the labyrinth and SpT layers of WT placentas (Fig. 5M,O; arrows in 5O point to labyrinthine and SpT expression; see Muc1 in 5E for localization of the labyrinth and Tpbpa in 5K for the SpT). In Pparg-null placentas, its expression was enhanced in the chorion and TGC, but wiped out in the labyrinthine and SpT zones (Fig. 5N,P), conceivably driving the net reduction of 2.5x indicated by the underlying qPCR data.
In WT placentas cathepsin R (Ctsr) was robustly expressed in TGC, as well as in the SpT and around maternal blood pools in the labyrinth (Fig. 5Q,S; arrows in 5S pinpoint labyrinthine expression), as previously described (Simmons et al., 2007). Pparg-null placentas retained Ctsr expression in SpT and TGC, while completely losing it in the labyrinth (Fig. 5R,T), sufficing for a 2.2-fold lower expression overall, as indicated by qPCR.
Fig. 5. Spatial distribution of placental PPARγ-dependent genes.
Serial sections of E9.5 WT (left panels) and Pparg-null concepti (right) were hybridized with antisense RNA probes for Pparg (A–B), lacZ (C–D), Muc1 (E-F), Sgk2 (G–H), Ceacam11 (I–J), Tpbpa (K–L) Ldhb (M–P; arrows in O: expression in the labyrinth and SpT, absent in Pparg-null placentas [P]), and Ctsr (Q-T; arrows in S: expression around maternal blood pools in the labyrinth, absent in Pparg-null placentas [T]). Scale bar – 500 µm, except O,P,S,T – 200 µm.
Combined, these analyses demonstrate that PPARγ regulates diverse genes at distinct stages of differentiation and in various trophoblast lineages.
Ldhb is a target of PPARγ in trophoblasts
To gain insights into the placental activities of PPARγ, we sought functional themes among either the distilled targets in Table 1 or the larger set of all genes that changed in Pparg-null and Rxra-null placentas. Several patterns emerged: (i) PPARγ enhanced the expression of multiple rodent-specific, lineage-specific placental genes, including Ctsr, Prl2b1 (Prlpk), Prl3b1 (Csh2), Prl7b1 (Prlpn), and Ceacam11. (ii) PPARγ up-regulated multiple hormones or their receptors, particularly ones that regulate energy storage and metabolism, including Ghrh, Prlr, Lepr, Gcgr, Pthlh, and the mouse-specific prolactin family members mentioned above (Prl2b1, 3b1 & 7b1). (iii) PPARγ induced genes whose products are either known or logically relate to intermediary metabolism, including Ldhb, Pcx, Dgat1, Angptl4, Hsd17b4, Hsd17b11, and Dcakd.
Pcx, Dgat1 and Angptl4 have been previously implicated as direct PPARγ targets in adipocytes and/or macrophages (Jitrapakdee et al., 2005; Ruan et al., 2003; Koliwad et al., 2010; Yoon et al., 2000), and our findings extend these regulatory relationships to trophoblasts. Of the remaining genes, four reasons compelled us to focus on Ldhb. First, its strong abidance by each of the PPARγ dependence criteria and its dependence on NCOA6 were highly consistent with a key PPARγ-dependent function in trophoblasts. Second, unlike Pcx, Ldhb is poorly expressed in adipocytes in vivo and in vitro, and is refractory to either PPARγ stimulation or deficiency in differentiated 3T3-L1 cells (Suyeon Kim & YB, unpublished data), indicating that its dependence on PPARγ is tissue-specific. Third, follow-up qPCR analyses singled out Ldhb as the most up-regulated out of eleven PPARγ target candidates in placentas from Rosi-treated pregnant WT mice (Fig. 6A). Last, age-old studies that inferred the preference of LDHB for conversion of lactate to pyruvate (Cahn et al., 1962; Dawson et al., 1964) imply tandem synergy with the co-regulated PCX, which catalyzes carboxylation of pyruvate into oxaloacetate.
Fig. 6. Regulation of Ldhb by PPARγ.
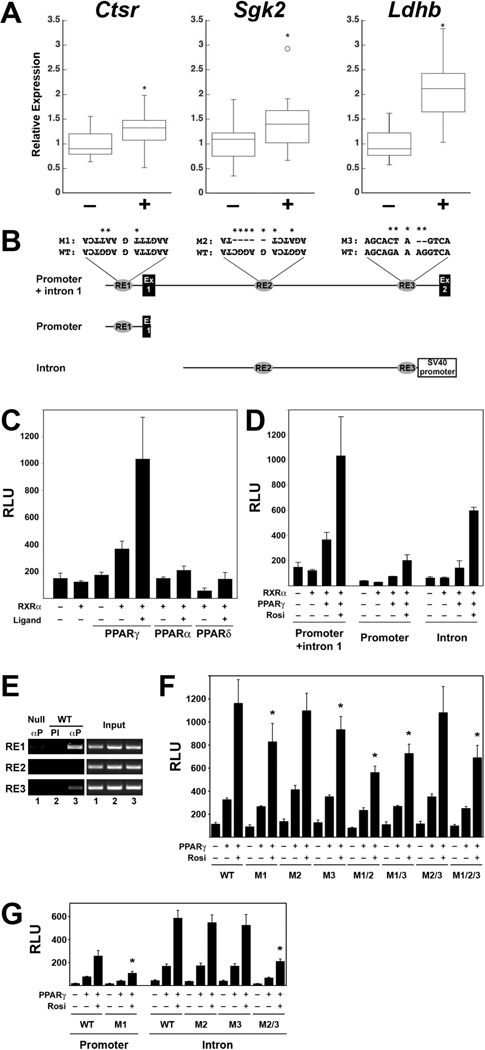
(A) RT-qPCR of RNA from WT E18.5 embryos from pregnant mice untreated (–) or treated with Rosi (+) from E10.5 to E18.5. Data are expressed as box plots showing median value, upper and lower quartiles, range, and outliers. Ctrl: n=23; Rosi: n=24. Asterisks – statistically significant differences according to Mann-Whitney test. (B) Structures of the full-length genomic Ldhb fragment (promoter + intron 1, top) and its two sub-fragments. Shown are relative positions of Exons 1 and 2 and the three PPRE-like elements (RE1, RE2 and RE3), including their native sequences and the sequences of inactivating mutations introduced into each (M1, M2, M3). Inverted lettering of RE1 and RE2 symbolizes their opposite orientation relative to Ldhb transcription. The proximal promoter-containing fragments were constructed into a promoterless pGL3-Basic vector, whereas the intronic fragment was placed upstream of an SV40 promoter-driven luciferase of the pGL3-Promoter vector. (C) CV1 cells were transfected with a luciferase reporter driven by the full-length Ldhb fragment, RXRα and the indicated combinations of PPARs, in the presence or absence of the cognate ligands, as indicated (1µM Rosi, 5µM Wy14643, and 1µM L165041 for PPARγ, PPARα, and PPARδ, respectively). (D) The indicated combinations of PPARγ and RXRα were cotransfected with the three different Ldhb promoter fragment-driven luciferase constructs, in the absence or presence of 1µM Rosi, as labeled. (E) ChIP analysis of PPARγ binding to each of the three PPREs in TSC differentiated for 4 days in the presence of Rosi. PCR-amplified Immunoprecipitated (left panel) or 1% of the input DNA (right) are shown for: Pparg-null TSC precipitated with anti-PPARγ antibody (αP, lane 1) and WT TSC with preimmune serum (PI, lane 2) or αP (lane 3). (F,G) Cells were transfected with full-length 2.7Kb Ldhb promoter fragments (F) or the disparate promoter and intronic fragments (G) harboring the designated native (WT) or mutant PPRE combinations, RXRα (all wells), PPARγ as indicated, and 1µM Rosi where labeled. Asterisks mark statistically significant differences of PPARγ+Rosi-stimulated reporter activity compared to the WT fragment. All assays in (C), (D), (F) and (G) were performed in triplicates. Luciferase activities was measured 48 hours post-transfection and normalized to β-galactosidase activity of co-transfected CMV-lacZ. (RLU – Relative luminescence).
A 2.7Kb genomic fragment encompassing the proximal promoter and first intron of Ldhb contains three PPRE-like elements (RE1, RE2, RE3; Fig. 6B). This fragment drove a strong, RXRα-dependent response of a luciferase reporter to PPARγ and Rosi, and much weaker responses to PPARα or PPARδ and their cognate agonists (Fig. 6C). The observed transcriptional activity was split between the proximal promoter, containing RE1, and the first intron, containing RE2 and RE3 (Fig. 6D). ChIP analysis indicated that RE1 and RE3 interact closely with PPARγ in Rosi-treated differentiated TSC; RE2 exhibited no detectable binding (Fig. 6E).
A complex pattern of unique and redundant contributions to the PPARγ response of Ldhb was revealed by mutating the putative PPREs, alone and in combination, in the contexts of the full 2.7Kb and the disparate promoter and intronic sub-fragments (see Fig. 6B for the mutant sequences). Mutating all three PPREs reduced the PPARγ response of the full promoter fragment by ~40% (Fig. 6F, M1/2/3). This indicated the importance of at least some of the PPREs, but also the contribution of additional promoter elements, either non-PPREs or non-obvious ones, to the PPARγ response. Such cooperation between PPRE-like and non-PPRE elements recapitulates our previous findings on the regulation of the Muc1 promoter by PPARγ (Shalom-Barak et al., 2004). It suggests that PPARγ utilizes alternative, non DNA-binding mechanisms to engage with and activate its native targets even when direct DNA binding is prohibited. Similar evidence of cooperation between PPREs and non-PPREs came to light when RE1 was mutated in the context of the proximal promoter fragment (Fig. 6G, M1), as well as when both RE2 and RE3 were mutated within the intronic fragment (Fig. 6G, M2/3). Consistent with the ChIP data, mutations of individual PPREs revealed significant, albeit modest contributions of RE1 and RE3, but complete dispensability of RE2 for activation of the full fragment when the other two REs remained intact (Fig. 6F, M1, M2 & M3). However, RE2 seems to function as a backup PPRE. Thus, within the 2.7Kb fragment both RE2 and RE3 were dispensable when RE1 was intact (Fig. 6F, M2/3), but loss of either alongside RE1 was functionally equivalent to the loss of all three PPREs (Fig. 6F, M1/2 & M1/3 vs. M1/2/3). In contrast, although elimination of both RE2 and RE3 blunted the response of the intronic fragment (Fig. 6G, M2/3), either was dispensable for PPARγ responsiveness in the presence of the other (Fig. 6G, M2 & M3).
These combinatorial regulatory contributions of the three PPREs alongside non-PPRE sequences strongly indicate that the regulation of Ldhb by PPARγ is remote from promiscuous. Added to our microarray, qPCR and in situ hybridization results, these analyses implicate Ldhb as an important part of a PPARγ-controlled metabolic program in differentiated trophoblasts.
DISCUSSION
Conceptual strengths and methodological limitations
Placental PPARγ function cannot be readily deduced from the complex phenotype of Pparg-null placentas or the extensive knowledge of PPARγ function in other tissues. Our rigorous screens aimed at overcoming this hurdle by pinpointing high-confidence downstream targets of PPARγ in trophoblasts. We integrated the four non-redundant PPARγ-dependence paradigms of altered expression in Pparg-null placentas, Rxra-null placentas, Rosi-treated WT TSC, and Pparg-null TSC, surmising that any genuine PPARγ target should abide by each of these criteria. We further buttressed robustness by careful biological replications within each of these categories. Specifically, in whole placentas we reined in random discoveries by comparing three pools of three WT to three pools of three Pparg-null or Rxra-null placentas, which normalized both placenta-to-placenta variation and uneven sample contamination with peri-placental tissues. In TSC, we aimed for robustness by aligning two WT lines from other laboratories with one WT and two Pparg-null lines that we generated for this study. While each of these lines exhibited basic cellular and molecular characteristics of TSC, their global gene expression patterns, levels and kinetics were markedly heterogeneous even within same-genotype groups. Because no single TSC line could be deemed more representative than others, we circumvented this limitation by centering on genes that were differentially expressed in Pparg-null placentas. This retained the robustness offered by the multiple TSC lines, while focusing on genes with immediate in vivo relevance. In additional post-hoc qPCR analyses that tested effects of PPARγ dosage in Pparg+/− placentas, less than 50% of all genes tested passed a liberal dosage sensitivity benchmark, and this ratio was not improved among genes that abided by all paradigms. While the primary reason is conceivably the statistically fickle nature of the rather minute differential anticipated of most of these genes, PPARγ dosage sensitivity could not be reasonably applied as an exclusion criterion, and had solely confirmatory value.
A critical component in paradigm integration was the choice of FC and q-value thresholds. To contain false negative calls while ensuring that only genuinely expressed genes impacted FDR without compromising exclusion of false positive data, we narrowed the viable data fields by omitting the all probe sets that scored ‘Absent’ in all samples by MAS5 (see Fig. S2 and legend for a detailed rationale). We then chose the cutoffs of 1.3x change and q<0.05. Our subsequent qPCR analyses confirmed that false discovery was indeed contained at <<5% (see Table S4); however, we suspect that quite a few true PPARγ targets may have still been excluded, including: (i) Genes that failed just a single criterion. For instance, potential targets may have exhibited prohibitive FDR in biological replications in cases of naturally variegated expression. (ii) Genes with no to sub-detectable expression in vitro. For example, Ceacam11, which exhibited the strongest differential in vivo, was excluded due to its complete absence from differentiated TSC. Another example is the Gcgr gene, which we confirmed as a PPARγ target during a parallel research pursuit (Shalom-Barak et al., in preparation). Despite a ~20-fold lower expression in Pparg-null placentas (q<0.05), Gcgr missed our cut because its expression in TSC was too close to background in Affymetrix microarrays; targeted qPCR analyses later confirmed its PPARγ response in TSC. (iii) Genes with transient PPARγ dependence outside the analyzed time points, e.g., earlier than E9.5 or prior to the second day of TSC differentiation.
While at present we have no foolproof statistical method for overcoming the listed limitations on a systems scale, a potential future approach could be the development and meta-implementation of a reliable algorithm for FDR calculation in intersecting datasets. For now, to avoid muddying our initial insights with false positive identifications, our analysis centered on genes that abided stringently by each of the aforementioned criteria.
Transcriptional mechanics of PPARγ
A considerable 40% of the gene expression changes in Pparg-null placentas recur in Rxra-null placentas, consistent with previous studies that implicated RXRα as the major heterodimeric partner of PPARγ in the tissue (Sapin et al., 1997; Barak et al., 1999). The higher percentage of RXRα dependence (64%) among transcripts that responded to PPARγ in TSC buttresses this assertion. Considering genome-wide interaction studies that demonstrated RXRα occupancy of nearly all PPARγ genomic binding sites in adipocytes and macrophages (Nielsen et al., 2008; Lefterova et al., 2008; Lefterova et al., 2010), the remaining non-overlap is probably not due to RXR-independent functions of PPARγ. A more likely explanation is the unique phenotypic aspects of Pparg-null placentas, such as their terminal failure, which are averted in Rxra-null placentas via functional redundancy with basal levels of RXRβ (Wendling et al., 1999). Redundancy could be indiscriminate across PPARγ target promoters, such that residual RXRβ sustains sufficient expression of essential targets (Sapin et al., 1997). Alternatively, some PPARγ targets could be RXR isoform-specific, with RXRα-specific targets severely affected and essential RXRβ-specific targets largely unchanged in Rxra-null placentas. Future assessment of these scenarios would require either direct expression data from Rxrb-null and Rxra/b compound null placentas or genome-wide placental DNA-binding screens of PPARγ in WT and Rxra-null placentas. The hundreds of genes whose expression is affected in Rxra-null, but not Pparg-null placentas likely represent direct and indirect targets of other RXR-dependent nuclear receptors, such as PPARδ, LXRs and RARs.
Although PPARγ has been strongly implicated in corepressor interaction and gene silencing akin to the related RAR and TR, mechanistic details and specific targets of PPARγ-mediated repression are rather ill-defined (Cohen, 2006). Our data offer a systems perspective on the issue. In whole placentas, 303 transcripts were up-regulated subsequent to PPARγ deficiency, compared to 481 down-regulated ones. However, in TSC, where one would expect a proportionate number of positive and negative direct targets, this ratio declined 3.6-fold (9/303 vs. 52/481). Moreover, the canonical paradigm for direct repression by nuclear receptors involves silencing in the absence of ligand and activation in its presence (Glass and Rosenfeld, 2000). By the same token, direct repression targets of PPARγ should conceivably be up-regulated in Rosi-treated WT TSC. Yet, all nine genes up-regulated in Pparg-null TSC were down-regulated by Rosi, inconsistent with repression by DNA-bound PPARγ. Instead, these genes are probably repressed either secondary to PPARγ action or through a previously proposed mechanism of ligand-dependent binding and stabilization of corepressor complexes anchored to DNA through other transcription factors (Pascual et al., 2005).
This study provided critical assessment of the purported contributions of the transcriptional coactivators PBP (MED1) and AIB3 (NCOA6) to placental gene regulation by PPARγ (Zhu et al., 2000; Kuang et al., 2002). MED1 affected the expression of surprisingly few placental genes at E9.5, only one of which, exhibiting a marginal differential response, was weakly PPARγ- and RXRα-dependent. These findings warrant thorough reevaluation of the Med1-null placental phenotype and its relationship to PPARγ function in this tissue. In contrast, NCOA6 deficiency affected over 2,500 transcripts in E9.5 placentas, including 63% of those that were down-regulated in both Pparg-null and Rxra-null placentas and 90% of the genes that abided by all primary criteria of positive regulation by PPARγ (see Table 1). These observations, and the remarkable phenotypic similarity between Pparg-null and Ncoa6-null placentas, suggest that the target(s) accounting for the essential placental functions of PPARγ are part of the NCOA6-coactivated contingent.
PPARγ function in the placenta
With a few notable exceptions, the majority of placental PPARγ-dependent genes have not been described as PPARγ targets outside this tissue. Moreover, within the placenta itself PPARγ-dependent genes are spatially and temporally variegated, with distinct expression patterns in different subsets of trophoblast lineages and different induction kinetics during TSC differentiation. This diversity suggests that the unique subset of trophoblast PPARγ target genes is further partitioned into distinct spatiotemporal target combinations within the placenta.
Some of these expression patterns in their own right offer potential leads into placental PPARγ function. For example: (i) The narrow perichorionic expression of Sgk2 and its PPARγ-dependent induction within one day of TSC differentiation suggest that PPARγ acts very early during trophoblast differentiation. (ii) The rodent-specific Ceacam11, which is expressed in a novel subset of SpT cells, is the single most down-regulated gene in Pparg-null, Rxra-null and Ncoa6-null placentas, as well as in the phenotypically similar αV-intergin (Itgav)-null placentas (Bader et al., 1998; data not shown). No other PPARγ-dependent gene is affected in Itgav-null placentas, suggesting that either Ceacam11 or, more likely, its host trophoblast lineage is at the crux of placental PPARγ and αV-integrin signaling in rodents. (iii) Similarly, PPARγ-dependent expression of the labyrinthine sinusoidal trophoblasts-specific Prl3b1 and Ctsr (Simmons et al., 2007) could be either a product of direct regulation or a reflection of key roles of PPARγ in differentiation or survival of the sinusoidal lineage. Such effects on placental lineage decisions may explain the pleiotropic outcomes of PPARγ deficiency. Notably, the insignificant expression changes of classical markers of the labyrinth, SpT and TGC in Pparg-null placentas suggest that if PPARγ affects any placental lineage decisions, these are limited to specialized lineage subtypes.
The cohort of metabolic PPARγ target genes, represented by Ldhb and Pcx, provides other exciting insights. First, these genes exhibit substantial PPARγ-independent expression in undifferentiated TSC, suppression during early differentiation, and resurgence in WT, but not Pparg-null cells. This “bowl-shaped” pattern suggests that both stem and differentiated trophoblasts share a core metabolic program, but regulate it through mutually exclusive mechanisms, switching from dependence on stem-specific factors in TSC to obligatory PPARγ dependence upon their differentiation. PPARγ-independent expression of Ldhb in the stem cell-rich chorion and its PPARγ-dependence in the differentiated labyrinth and SpT is conceivably the spatial analog of this pattern. There is an apparent contradiction between the PPARγ-independent expression of Ldhb in placental TGC and its PPARγ-dependence in differentiated TSC cultures, which are dominated by TGC-like cells. This discrepancy can be settled by reconsidering the possibility that these TGC-like cells in culture are more akin to labyrinthine sinusoidal cells, which would align with the aforementioned idea that PPARγ regulates gene expression in this lineage.
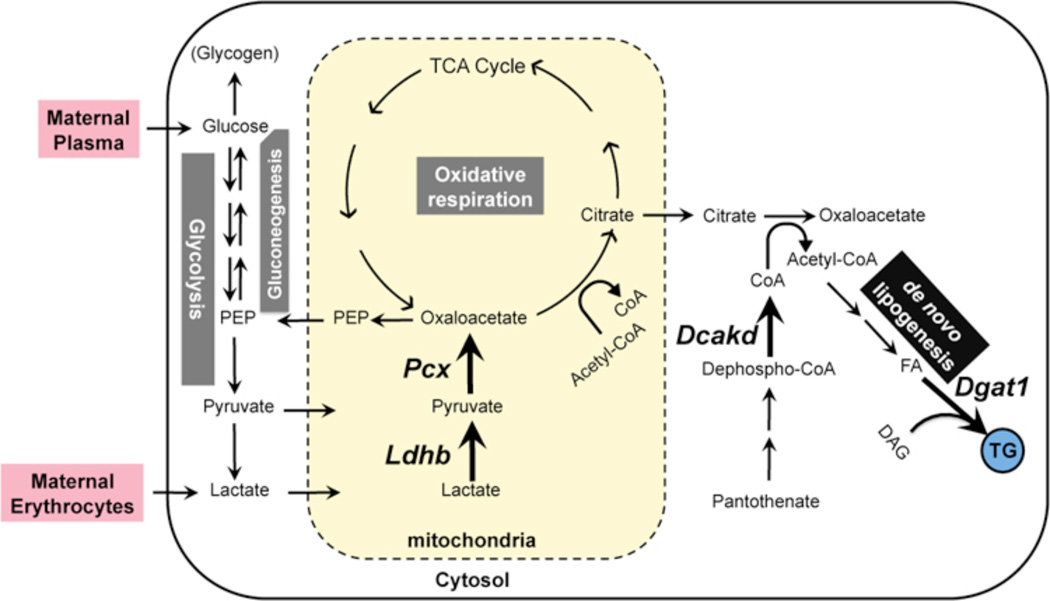
Second, the enzymatic functions of targets that fall into this regulatory category lead us to conjecture a unique placental metabolic cascade, illustrated in Fig. 7. The pivot point in this hypothetical program is pyruvate carboxylase, the product of Pcx, which converts pyruvate to oxaloacetate (OAA). OAA is a common precursor for multiple key metabolic pathways, including gluconeogenesis, anaplerotic replenishment of the tricarboxylic acid (TCA) cycle of oxidative respiration, and lipogenesis (Jitrapakdee et al., 2008). Of these, the most plausible conjecture is that PPARγ-regulated PCX catalyzes OAA formation in trophoblasts for the same primary purpose as in adipocytes, i.e. to prime de novo fatty acid synthesis (Jitrapakdee et al., 2005; Ballard and Hanson, 1967; Kajimoto et al., 2005). The similar regulatory pattern of Dgat1, which is the last and rate-limiting step in triglyceride synthesis (Yen et al., 2008), should maintain a forward equilibrium by utilizing the resulting fatty acids, implicating PPARγ in integrated control of placental lipogenesis. Therefore, and considering the glaring lack of evidence for PPARγ-regulated lipid uptake or binding genes in our screens, even with relaxed discovery criteria, impaired fatty acid synthesis is a likely explanation for the extreme lipid depletion of PPARγ and RXRα-null placentas (Barak et al., 1999; Sapin et al., 1997). The novel dephospho-CoA Kinase homolog Dcakd may keep this putative cascade rolling by maintaining a steady supply of coenzyme A, which keeps being consumed as a skeleton for the resultant fatty acyl chains. Finally, we hypothesize that unlike in adipocytes, the trophoblast PPARγ-driven lipogenic program initiates with lactate dehydrogenase B (LDHB). LDHB differs from the related LDHA in its kinetic and thermodynamic preferences for lactate to pyruvate conversion (Cahn et al., 1962; Dawson et al., 1964), invoking a PPARγ-coregulated enzymatic tandem of LDHB and PCX at the head of a pathway that recycles lactate into triglycerides. Integration of lactate into this cascade is in line with emerging evidence that other cells are able to efficiently utilize it as an alternate fuel source, as is the case with muscle during endurance exercise or neurons during hypoglycemia (Gladden, 2000; Wyss et al., 2011). Conceivably, the placenta may have adopted a similar program as a thrift mechanism that taps into the copious lactate waste hauled in by incoming maternal erythrocytes, where it is the dead-end product of housekeeping glycolysis (Bartlett, 1959). Notably, the absence of clinical symptoms in LDHB-deficient patients (Takatani et al., 2001) and the strictly postnatal symptoms of PC-deficient infants (Marin-Valencia et al., 2010) indicate that individual components of this program may be superfluous for placental development per se. Irrespective of whether this reflects dispensability, redundancy, or alternative metabolic pathways, the remarkable co-regulation of this gene network is highly suggestive of its primary significance in the placenta on an evolutionary scale.
Fig. 7. A model for PPARγ-regulated placental metabolism.
Multiple PPARγ-regulated nodes link the presumptive lactate substrate and the ultimate triglyceride product in an integrated, putative placental metabolic pathway. PPARγ-regulated genes and their corresponding directional arrows are in bold font. Abbreviations: CoA – Coenzyme A; DAG – Diacylglycerol; FA – Fatty scids; LPL – Lipoprotein Lipase; PEP – Phosphoenolpyruvate Carboxykinase.
On a final note, the absence of significant changes in canonical regulators of differentiation or development is surprising given the striking developmental phenotype of Pparg-null placentas. While one cannot formally exclude roles of novel genes from this screen in such tasks, we are increasingly compelled to consider the possibility that trophoblast differentiation is tied to successful execution of their unique metabolism. Conceivably, such link could be in the form of either metabolic fuel requirements or intermediate metabolites with dedicated signaling functions.
Supplementary Material
Highlights.
Placental PPARγ targets identified by integrating multiple regulatory paradigms.
Placental PPARγ-regulated genes are spatially and temporally diverse.
NCOA6, but not MED1, co-regulates a substantial cohort of placental PPARγ targets.
Ldhb is a direct target and part of a metabolic pathway regulated by placental PPARγ.
ACKNOWLEDGMENTS
We thank The Jackson Laboratory’s Gene Expression Service for Affymetrix microarray hybridization and scanning; Dr. Tom Gridley for the TSC line Clone 2; Jackson’s Computational Biology Resource for preliminary statistical analysis of microarray data; Dr. Suyeon Kim for help with in situ hybridizations. Ms. Patricia Cherry and Ms. Lee Rager for administrative assistance at Jackson and MWRI, respectively. Supported by NIH Grants HD044103 (to Y.B.), ES011597 (to Y.S.) and Pennsylvania Department of Health Research Formula Funds (to T.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was initiated at The Jackson Laboratory and finalized at Magee-Womens Research Institute
REFERENCES
- Bader BL, Rayburn H, Crowley D, Hynes RO. Extensive vasculogenesis, angiogenesis, and organogenesis precede lethality in mice lacking all alpha v integrins. Cell. 1998;95:507–519. doi: 10.1016/s0092-8674(00)81618-9. [DOI] [PubMed] [Google Scholar]
- Ballard FJ, Hanson RW. The citrate cleavage pathway and lipogenesis in rat adipose tissue: replenishment of oxaloacetate. J. Lipid Res. 1967;8:73–79. [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Bartlett GR. Human red cell glycolytic intermediates. J. Biol. Chem. 1959;234:449–458. [PubMed] [Google Scholar]
- Cahn RD, Kaplan NO, Levine L, Zwilling E. Nature and development of lactic dehydrogenases. Science. 1962;136:962–969. doi: 10.1126/science.136.3520.962. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Using ANOVA to analyze microarray data. Biotechniques. 2004;37:173–177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- Cohen RN. Nuclear receptor corepressors and PPARγ. Nucl. Recept. Signal.M. 2006;4:e003. doi: 10.1621/nrs.04003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DM, Goodfriend TL, Kaplan NO. Lactic dehydrogenases: functions of the two types. Science. 1964;143:929–933. doi: 10.1126/science.143.3609.929. [DOI] [PubMed] [Google Scholar]
- Gladden LB. Muscle as a consumer of lactate. Med. Sci. Sports Exerc. 2000;32:764–771. doi: 10.1097/00005768-200004000-00008. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Hartman HB, Hu X, Tyler KX, Dalal CK, Lazar MA. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 2002;277:19754–19761. doi: 10.1074/jbc.M201451200. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S, Slawik M, Medina-Gomez G, Campbell M, Wallace JC, Sethi JK, O'rahilly S, Vidal-Puig AJ. The peroxisome proliferator-activated receptor-gamma regulates murine pyruvate carboxylase gene expression in vivo and in vitro. J. Biol. Chem. 2005;280:27466–27476. doi: 10.1074/jbc.M503836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S, St-Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto K, Terada H, Baba Y, Shinohara Y. Essential role of citrate export from mitochondria at early differentiation stage of 3T3-L1 cells for their effective differentiation into fat cells, as revealed by studies using specific inhibitors of mitochondrial di- and tricarboxylate carriers. Mol. Genet. Metab. 2005;85:46–53. doi: 10.1016/j.ymgme.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Takata Y, Nakajima A, Saito S, Huh N. A carcinoembryonic antigen family cDNA from mouse placenta encoding a protein with a rare domain composition. Placenta. 2000;21:610–614. doi: 10.1053/plac.2000.0546. [DOI] [PubMed] [Google Scholar]
- Kim S, Huang L-W, Snow KJ, Ablamunits V, Hasham MG, Young TH, Paulk AC, Richardson JE, Affourtit J, Shalom-Barak T, Bult CJ, Barak Y. A mouse model of conditional lipodystrophy. Proc. Natl. Acad. Sci. USA. 2007;104:16627–16632. doi: 10.1073/pnas.0707797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliwad SK, Streeper RS, Monetti M, Cornelissen I, Chan L, Terayama K, Naylor S, Rao M, Hubbard B, Farese RV., Jr DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. J Clin. Invest. 2010;120:756–767. doi: 10.1172/JCI36066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Pereira FA, Yuan Y, DeMayo FJ, Ko L, Xu J. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J. Biol. Chem. 2002;277:45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferatoractivated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marin-Valencia I, Roe CR, Pascual JM. Pyruvate carboxylase deficiency: mechanisms, mimics and anaplerosis. Mol. Genet. Metab. 2010;101:9–17. doi: 10.1016/j.ymgme.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Børgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPARγ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Ruan H, Pownall HJ, Lodish HF. Troglitazone antagonizes tumor necrosis factor-alpha- induced reprogramming of adipocyte gene expression by inhibiting the transcriptional regulatory functions of NF-kappa. B. J. Biol. Chem. 2003;278:28181–28192. doi: 10.1074/jbc.M303141200. [DOI] [PubMed] [Google Scholar]
- Sapin V, Dolle P, Hindelang C, Kastner P, Chambon P. Defects of the chorioallantoic placenta in mouse RXRα null fetuses. Dev. Biol. 1997;191:29–41. doi: 10.1006/dbio.1997.8687. [DOI] [PubMed] [Google Scholar]
- Schaiff TW, Knapp FF, Jr, Barak Y, Biron-Shental T, Nelson MD, Sadovsky Y. Ligand-activated PPARγ alters placental morphology and placental fatty acid uptake in mice. Endocrinology. 2007;148:3625–3634. doi: 10.1210/en.2007-0211. [DOI] [PubMed] [Google Scholar]
- Shalom-Barak T, Nicholas JM, Wang Y, Zhang X, Ong ES, Young TH, Gendler SJ, Evans RM, Barak Y. PPARγ controls Muc1 transcription in trophoblasts. Mol. Cell. Biol. 2004;24:10661–10669. doi: 10.1128/MCB.24.24.10661-10669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 2007;304:567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXRα mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- Takatani T, Takaoka N, Tatsumi M, Kawamoto H, Okuno Y, Morita K, Masutani T, Murakawa K, Okamoto Y. A novel missense mutation in human lactate dehydrogenase B-subunit gene. Mol. Genet. Metab. 2001;73:344–348. doi: 10.1006/mgme.2001.3203. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Wendling O, Chambon P, Mark M. Retinoid X receptors are essential for early mouse development and placentogenesis. Proc. Natl. Acad. Sci. USA. 1999;96:547–551. doi: 10.1073/pnas.96.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss MT, Jolivet R, Buck A, Magistretti PJ, Weber B. In vivo evidence for lactate as a neuronal energy source. J. Neurosci. 2011;31:7477–7485. doi: 10.1523/JNEUROSCI.0415-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Chickering TW, Rosen ED, Dussault B, Qin Y, Soukas A, Friedman JM, Holmes WE, Spiegelman BM. Peroxisome proliferator-activated receptor γ target gene encoding a novel angiopoietin-related Protein associated with adipose differentiation. Mol. Cell. Biol. 2000;20:5343–5349. doi: 10.1128/mcb.20.14.5343-5349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.