Abstract
The peptidoglycan cell wall maintains turgor pressure and cell shape of most bacteria. Cell wall hydrolases are essential, along with synthases, for growth and daughter cell separation. Recent work in diverse organisms has uncovered new cell wall hydrolases that act autonomously or on neighboring cells to modulate invasion of prey cells, cell shape, innate immune detection, intercellular communication, and competitor lysis. The hydrolases involved in these processes catalyze the cleavage of bonds throughout the sugar and peptide moities of peptidoglycan. Phenotypes associated with these diverse hydrolases reveal new functions of the bacterial cell wall beyond growth and division.
Keywords: cell wall hydrolase, peptidoglycan, cell shape, innate immune sensing, bacterial communication, bacterial pathogenesis, bacterial predation
Uncovering new peptidoglycan hydrolases and functions
Bacterial populations experience a range of osmolarities as environmental conditions shift. To prevent lysis, most bacteria synthesize a sturdy but flexible peptidoglycan (PG) cell wall that can withstand high turgor pressure. One unit of PG consists of an N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) disaccharide with a pentapeptide chain linked to MurNAc; the pentapeptide is typically L-Ala-D-Glu-2,6-diaminopimelic acid (m-DAP)-D-Ala-D-Ala, but variations exist, such as replacement of m-DAP with Lys [1, 2]. The disaccharide-pentapeptide PG subunits are polymerized by the transglycosylase activity of PG synthesis complexes to form glycan strands. Transpeptidase activity of PG synthesis complexes links peptide chains from neighboring strands to form a net-like continuous molecule surrounding the cell called the PG sacculus. This highly conserved essential bacterial structure is a common antibiotic target [3] and an important innate immune activator in animals via multiple PG recognition proteins (reviewed in [4, 5]).
Bacteria must modify their cell wall to grow and divide. To insert new strands or recycle old PG, hydrolases cleave covalent bonds that hold the PG together. Two main classes of PG hydrolases have been described. Glycosidases cleave the glycan backbone in either the middle (endolytic) or end of a strand (exolytic). Amidases cleave the bond between the glycan strand and peptide chain or bonds within the peptide chains and crosslinks [6] (Figure 1). PG amidases can be further subdivided into carboxypeptidases, which remove the C-terminal residue of peptides, or endopeptidases, which cleave internal linkages [2]. Methods to study PG hydrolases include global analysis of the relative abundance of different disaccharide-peptide species (muropeptides) within the PG sacculus and pulse-chase experiments that reveal sites of new PG incorporation on a subcellular level, but few tools exist to detect subcellular structural modification of existing PG (Box 1). Furthermore, a single hydrolase can have multiple activities, making hydrolase research challenging.
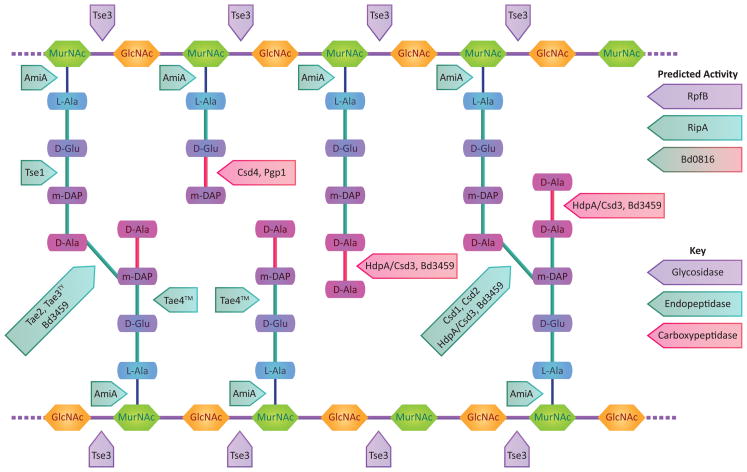
Figure 1.
Cleavage specificities of PG hydrolases described in this review. PG hydrolase class is color coded as indicated in the key. With the exception of AmiA, cleavage specificity is supported by biochemical data as cited in the main text.
Box 1. Methods for analyzing PG modification.
A major challenge in the study of the bacterial PG sacculus and the enzymes that modify it involves the diversity of modifications present (which are still being discovered). Here we describe assays currently used to detect and exploit these PG modifications.
Composition analysis by HPLC and mass spectrometry [54]
Whole sacculi can be isolated from cells by detergent extraction and enzymatic digestions of protein and nucleic acid. Extensively washed sacculi are then digested with glycosidases producing a pool of uncrosslinked disaccharide monomers with their attached peptides as well as dimers, trimers, etc. crosslinked by peptide crossbridges of varying types. Different oligomeric states, peptides, and modifications to the sugar and peptide moieties allow separation by HPLC and the individual peaks can be identified by tandem mass spectrometry. Once the PG profile of a bacterial strain is elucidated, HPLC can be used to compare the global abundance of different species in mutant strains, giving clues to activity of the gene mutated. Definitive elucidation of enzymatic specificity requires incubation of purified enzymes with cell wall material and analysis of the reaction products.
Detection of PG fragment release by hydrolases [26]
After labeling with Remazol Brilliant Blue, a dye which binds carbohydrates, highly oligomeric cell wall can be separated from soluble hydrolysis products by centrifugation. Thus purified enzymes can be incubated with labeled cell wall material and release of dye labeled soluble material can be monitored by spectrophotometry. However, the chemical nature of the released fragments is not resolved with this technique.
Localization of new synthesis [55]
Bacterial cells can incorporate D-Cys in the sacculus by a periplamic amino acid exchange reaction with terminal D-Ala. Treatment with –SH specific biotinylation reagents allow antibody mediated visualization by epifluorescence or electron microscopy. New PG incorporation can be visualized by growth for several doublings in the presence of D-Cys followed by a short chase period without D-Cys. In this case the sacculi are uniformly labeled except in areas of new growth.
Localization of pentapeptides (and new synthesis) [56]
This method takes advantage of the binding specificity of the antibiotic vancomycin for the terminal D-Ala-D-Ala of PG pentapetides. In many bacteria, pentapeptides are rapidly eliminated during formation of crosslinks or through hydrolysis by carboxypeptidases. Thus for these organisms, sites of fluorescein coupled vancomycin (Van-FL) binding reveal sites of new PG incorporation.
The cell wall of Gram-positive organisms is exposed on the cell surface and therefore accessible to both antibiotics and hydrolase toxins including self-targeting autolysins (reviewed in [7]) and secreted toxins such as lysostaphin, an enzyme secreted by Staphylococcus simulans that targets Staphylococcus aureus PG [8, 9]. The outer membrane of Gram-negative bacteria protects the PG sacculus from hydrolase toxins and facilitates efficient recycling of PG turnover products, presumably minimizing PG fragment release from the cell. Indeed disruption of PG recycling attenuates Shigella flexineri immune evasion during pulmonary and systemic infection in mouse models [10]. Conversely, several Gram-negative organisms actively release disaccharide-tetrapeptide monomers, including tracheal cytotoxin (TCT) of Bordetella pertussis, [11, 12], through the action of multiple lytic transglycosylases [13, 14]. This PG monomer targets select eukaryotic cells, promoting Nesseria gonorrhoeae and B. pertussis pathogenesis in humans and Vibrio fischeri symbiosis with its squid host (reviewed in [15]). Thus in addition to their role in cell growth and division, PG hydrolases can act as toxins directly by attacking the cell wall of neighboring bacteria or indirectly by liberating biologically active muropeptides from the PG sacculus.
In this review we highlight recent work in diverse organisms that has expanded the number of recognized PG hydrolases and our understanding of their range of biological functions. We focus on enzymes that do not play a major role in cell division, but rather modulate other bacterial behaviors. The predatory bacterium Bdellovibrio bacteriovorus expresses amidases required to alter the shape of its prey, facilitating entry into prey cells [16]. Several newly characterized amidases in Helicobacter pylori [17–20] and Campylobacter jejuni [21] modulate cell shape and promote host colonization through effects on motility and innate immune activation. Proteobacteria in several clades have now been shown to utilize type VI secretion systems to deliver multiple classes of PG hydrolases to the periplasm of neighboring bacteria to promote their own expansion within mixed bacterial communities [22, 23], extending the paradigm of interspecies PG hydrolytic toxins beyond Gram-positive organisms. Mycobacterium tuberculosis appears to require a combination of hydrolases to ‘resuscitate’ from latency [24–26]. The use of multiple classes of hydrolases simultaneously may arise from the need for cleavage of distinct linkages to alter the PG in specific ways. Additionally, liberation of specific PG fragments may be a signaling mechanism among bacteria that the host exploits through innate immune receptors.
Perturbation of shape
Predatory bacteria such as B. bacteriovorus establish a replicative niche in the periplasm of Gram-negative prey cells. Invasion appears to be driven by retraction of type IV pili pulling against the rigid PG of the prey cell [27]. During invasion, a shape transition occurs; a normally rod shaped prey, such as Escherichia coli, is converted to a round structure called the bdelloplast within 15 minutes of invasion. The predator then replicates in the remodeled periplasm, and four to nine progeny are released several hours after initial invasion [28].
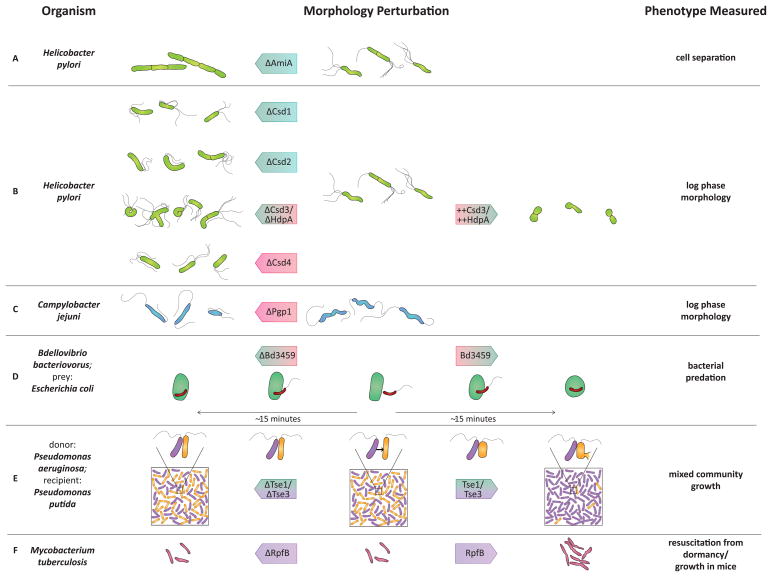
B. bacteriovorus has three genes predicted to encode proteins related to E. coli penicillin-binding protein 4 (PBP4 or DacB), a DD-endo/carboxypeptidase. One is constitutively transcribed, but transcription of the other two (bd0816 and bd3459) is induced during invasion, suggesting their gene products may play a role in the formation of a round bdelloplast [16]. Incubation of purified Bd3459 with pentapeptide-rich cell wall material drives conversion to monomeric tetrapeptides suggesting both DD-carboxypeptidase activity on pentapeptide monomers and DD-endopeptidase on tetra–pentapeptide and tetra–tetrapeptide crosslinks (Figure 1). Periplasmic expression of Bd3459 in E. coli leads to cell rounding independent of B. bacteriovorus invasion, and B. bacteriovorus strains missing either or both PG hydrolases produce bdelloplasts that are less round than those produced by wild-type. The Δbd0816Δbd3459 double mutant also shows delayed invasion of some prey and an increased number of simultaneous infections, or ‘tailgating’, which are rarely observed during wild-type infections [16]. Atomic force microscopy studies showed decreased stiffness of the bdelloplast relative to uninfected E. coli cells [29]. Thus, decreased PG hydrolysis (and thus increased crosslinking) in the double mutant may cause increased torsional stress from a more rigid sacculus, which may in turn cause the slower invasion phenotype as well as enhanced tailgating that normally is prevented when the sacculus becomes flaccid upon bdelloplast formation.
Since periplasmic overexpression of Bd3459 in the E. coli periplasm eventually causes cell lysis [16], PG hydrolysis by Bd0816 and Bd3459 during B. bacteriovorus invasion must be tightly controlled such that the sacculus softens enough to allow invasion and deter additional predators from entering, but remains sufficiently intact to prevent cell lysis prior to predator replication. The observed tight temporal transcriptional regulation [16] could be sufficient to limit hydrolytic activity. Both peptidases have predicted signal sequences but the mechanism by which they traverse both predator and prey outer membranes is not yet clear. It is also unclear if the round structure of the wild-type bdelloplast has a biological function or is merely a consequence of the crosslinking relaxation-mediated softening of the PG. While bdelloplast specific peptidases clearly contribute to predation, their sufficiency for bdelloplast formation and the associated reduction in cell stiffness remains to be determined.
While B. bacteriovorus hydrolases alter prey shape, H. pylori and C. jejuni, both helical rod-shaped bacteria that colonize the gastrointestinal tract, express PG hydrolases that intrinsically alter shape cell. Two H. pylori enzymes, amidase A (AmiA) and Helicobacter D,D-peptidase A (HdpA), were identified by searching the genome for candidate PG hydrolases [17, 18]. HdpA was independently identified through a genetic screen for shape mutants [19] and named cell shape determinant 3 (Csd 3). This screen identified three additional PG peptidases: Csd1, Csd2, and Csd4 [19, 20]. Screens of C. jejuni libraries for mutants with altered binding to calcofluor white identified several virulence factors, including a PG carboxypeptidase, peptidoglycan peptidase 1 (Pgp1), which is homologous to H. pylori Csd4 [21].
Genetic and biochemical evidence suggest both distinct and overlapping activities for these enzymes (Figure 1). Csd1, Csd2, and Csd3/HdpA belong to the M23/LytM family of metallopeptidases. Δcsd1, Δcsd2, and Δcsd3/hdpA strains accumulate tetra–pentapeptide dimers, suggesting these enzymes may act as endopeptidases [17, 19]. Indeed, purified HdpA/Csd3 catalyzes the cleavage of tetra–pentapeptide dimers to tetra- and pentapeptide monomers [17]. HdpA/Csd3 also has carboxypeptidase activity on pentapeptide monomers in vitro [17]. Δcsd4 and Δpgp1 strains accumulate tripeptide monomers, and purified Csd4 and Pgp1, which contain M14 peptidase domains, catalyze the carboxypeptidase cleavage of tripeptides to dipeptides [20, 21]. During the stationary phase transition to coccoid morphology, wild-type H. pylori accumulate dipeptide monomers while tripeptide levels decrease [30]. ΔamiA H. pylori accumulates substantially less dipeptide and tripeptide levels are stable during this transition, suggesting that AmiA, a homologue of enzymes that cleave the N-acetylmuramoyl-L-alanine bond, is directly or indirectly involved in this PG remodeling process [18]. The variety of PG species found in the H. pylori and C. jejuni sacculi suggest the presence of additional peptidases unidentified at this time.
Many of the PG hydrolase deletion strains have strikingly altered shapes (Figure 2) in spite of normal or only slightly attenuated growth rates [17, 19–21], leading to the hypothesis that these PG hydrolases collectively remodel the sacculus to generate helical shape. The deletion strain morphologies range from straight rods for Δcsd4 and Δpgp1 [20, 21], to curved rods for Δcsd1 and Δcsd2 [19], to irregular c-shaped or branched cells for Δcsd3/hdpA, depending on the strain background [17, 19]. ΔamiA H. pylori is still helical rod-shaped, but is defective for cell separation and unable to modulate its shape during stationary phase [18]. Overexpression of HdpA/Csd3 causes cell rounding [17], reminiscent of the rounding of prey cells by B. bacteriovorus PBP4 homologues that show similar PG cleavage specificity [16].
Figure 2.
Observed functions of representative PG hydrolases described in this review. Rows (a)–(c) depict a steady-state shape phenotype during logarithmic growth in deletion mutants (left), wild-type (middle), and overexpression strains (right). Rows (d)–(f) depict dynamic processes resulting from hydrolase activity. The middle shows the starting state. Cells to the right demonstrate wild-type behavior, while cells to the left depict the same behavior for the indicated hydrolase deletion mutant. Color coding of hydrolases is the same as in Figure 1.
Deletion of csd genes from H. pylori or pgp1 from C. jejuni leads to colonization defects in animal infection models [17, 19–21]. Cell wall integrity is intact in many of the deletion strains, as assessed by tolerance to antimicrobials and extreme pH and osmolarity, suggesting a role for PG hydrolases in colonization that is independent of cell protection. For H. pylori, helical shape is hypothesized to allow efficient motility through the gastric mucosa [31, 32]. While motility deficits are subtle, Sycuro et al. [20] demonstrated a correlation between severity of shape phenotype and motility defect; the straight and c-shaped mutants have more pronounced motility defects than the curved mutants. C. jejuni straight mutants also have a slight motility defect in soft agar [21]. Thus colonization defects seen for H. pylori and C. jejuni PG hydrolase mutants may arise from shape-dependent reduced swimming efficiency through mucus that diminishes access to preferred niches on or in gastrointestinal epithelial cells.
Modulation of immune activation
For bacteria residing in mammalian hosts, loss of PG hydrolase function may produce or eliminate PG agonists of host innate immune receptors, provoking or dampening immune responses. PG subunits containing m-DAP (i.e. tripeptide or longer) are agonists of the cytosolic innate immune receptor Nod1, while dipeptide monomer is an agonist of Nod2 [4, 5]. Regulated turnover of the sacculus in the periplasm by lytic transglycosylases may provide a source of various PG agonists, depending on the composition of the sacculus.
Inflammation appears critical for H. pylori induced pathology [33]. Viala et al. [34] showed that H. pylori induces proinflammatory cytokine secretion in epithelial cells by activating Nod1 and that pulse-labeled cell wall material enters the epithelial cell cytosol via the cag type IV secretion system (T4SS). Transfection of Nod reporter cell lines with digested PG from stationary cells gave a lower Nod1 and higher Nod2 response than seen for PG from log phase cells, consistent with the shift to lower tripeptide abundance and higher dipeptide abundance during the coccoid shape transition [18]. Additionally, cag T4SS-dependent signaling was partially attenuated in a mutant of the soluble lytic transglycosylase (slt) [34–36], which shows reduced exolytic cleavage of the glycan backbone [37]. However, interleukin-8 (IL-8) secretion by gastric epithelial cells (AGS) in response to wild-type and Δcsd4 H. pylori is similar, despite the accumulation of tripeptide in the Δcsd4 sacculus [20]. Thus the precise structure and source of PG fragments delivered by the cag T4SS is unclear and may require multiple hydrolases in the bacterium, and perhaps the host cell, to generate the Nod1 agonist.
On the flip side, organisms may evade the host innate immune system by regulating the nature and overall amount of PG released. Unlike H. pylori, which primarily remains extracellular, C. jejuni efficiently invades epithelial cells and intracellular bacterial killing may directly deliver PG fragments to cytosolic Nod receptors. Indeed, Δpgp1 C. jejuni, which similar to Δcsd4 H. pylori accumulates tripeptide at the expense of dipeptide in the sacculus, produces a significantly higher IL-8 response in human epithelial cells (INT407) than wild-type C. jejuni [21]. As Nod2 is not highly expressed in epithelial cells, the trimming of tripeptides to dipeptides by Pgp1 and the AmiA-mediated decrease of tripeptide during stationary phase may help C. jejuni and H. pylori minimize immune detection.
PG release may also be controlled by regulating PG hydrolase activity. Liu et al. recently determined that enhanced entry C (EnhC), a Legionella pneumophila protein, binds and inhibits a soluble lytic transglycosylase (SltL) [38]. EnhC is expressed during stationary phase and is required for growth in macrophages [39]. Strains that overexpress SltL and the ΔenhC strain are restricted for growth in macrophages, suggesting that higher than wild-type SltL activity may increase PG release, leading to recognition and clearance of the pathogen. Consistent with this hypothesis, SltL-overexpressing strains elicit a higher signaling response than wild-type L. pneumophila when assayed by challenge of a Nod1 reporter cell line [38]. EnhC is also required for cellular integrity during growth in amoeba, the main L. pneumophila host. Thus, in L. pneumophila it appears that although SltL activity is required for efficient growth [38], EnhC decreases SltL activity and thus PG turnover during stationary phase. This results in a stronger cell wall and less innate immune signaling, which in turn allows for infection. The decrease in agonist release during stationary phase demonstrated here for L. pneumophila mirrors that of H. pylori, albeit by a different mechanism.
Manipulation of multispecies bacterial communities
The outer membrane of Gram-negative bacteria has long been thought to protect the PG from secreted hydrolase toxins. However recent work in Gram-negative bacteria showed that type VI secretion systems (T6SS) deliver effectors directly from the donor cell cytoplasm across the donor inner and outer membranes as well as the outer membrane of a neighboring recipient bacterium and into its periplasm [22, 23, 40]. Furthermore, effector delivery to recipient cells providesa competitive growth advantage in mixed bacterial communities [41, 42]. Significant insight into both T6SS secretion mechanisms and the mechanisms of effector mediated growth advantage came from the study of two PG hydrolase effectors, T6 secretion exported 1 (Tse1) and Tse3, which are translocated by Pseudomonas aeruginosa haemolysin co-regulated protein secretion island I (HSI-I)-encoded T6SS (Figure 2) [22].
Comprehensive motif searches of known substrates of the HSI-I T6SS revealed that Tse1 has homology to NlpC/p60 family proteins, some of which trim PG peptides, and Tse3 resembles glycosidases [22]. Studies with purified enzymes showed that Tse1 cleaves between D-isoglutamic acid (D-iGlu) and m-DAP of tetra–tetrapeptide crosslinks and is specific for the donor stem, while Tse3 cleaves between MurNAc and GlcNAc in a hydrolytic fashion similar to lysozyme (Figure 1). Though some secretion systems encode PG hydrolases that promote assembly of the secretion machine, which traverses the PG layer, neither Tse1 nor Tse3 are required for T6SS function. The hydrolytic activities of Tse1 and Tse3 can be inhibited by periplasmically localized immunity proteins (Tsi1 and Tsi3, respectively) encoded adjacent to each effector [22]. Competition experiments demonstrate that T6SS immunity proteins Tsi1 and Tsi3 are required when cells are in contact and thus able to self-intoxicate by T6SS-mediated translocation of Tse1 and Tse3 into the periplasm of neighboring cells. No growth defect is seen for immunity protein mutants grown in liquid culture, which are not in contact and therefore secrete effectors into the media, but cannot translocate them into other cells. Thus a cell’s own effectors do not enter its periplasm during secretion [22]. Indeed, cryo-electron tomography studies of Vibrio cholerae T6SS reveal a cytoplasmic structure similar to a contractile phage tail connected via a flared bell-shaped base to a conical shape density that traverses the periplasm and the outer membrane that may provide a protected conduit for effectors during secretion, as predicted by the above competition experiments [43].
Targeting Tse1 or Tse3 to the periplasm at expression levels above those of the enzymes expressed from the native loci resulted in cell lysis, while periplasmic localization at native expression levels did not [22]. Similar to B. bacteriovorus (Bd3459) and H. pylori (Csd3/HdpA) endopeptidases, Tse1 leads to cell rounding prior to lysis. Expression of both Tse1 and Tse3 contributes to fitness in a partially-redundant fashion during co-culture of P. aeruginosa and Pseudomonas putida. Lysis may not be necessary for the observed competitive growth advantage; release of specific PG fragments that have a signaling function on the donor cell could represent an alternate mechanism. Many bacteria have signaling proteins containing PG binding domains [44]. Indeed P. aeruginosa induces production of the antimicrobial pyocyanin in response to exogenous GlcNAc or high molecular weight PG [45]. Combined action of Tse1 and Tse3 would be expected to liberate monomeric disaccharide-dipeptide and disaccharide-hexapeptide. While dipeptides are known to activate mammalian Nod2 receptors, no bacterial receptors for these fragments have been described.
Additional studies show the use of PG hydrolases as effectors may be widespread; the presence of putative hydrolase effector–immunity pairs associated with candidate T6SS was detected in many species in Beta-, Delta-, and Gamma-proteobacteria lineages. Proteomic analysis of a T6SS secretome and computational prediction identified 51 candidate PG peptidase effectors that fall into four families based on primary sequence homology [23]. At least one effector from each family has been biochemically characterized. T6 amidase effector 2 (Tae2) from Bukholderia thailandensis, a family 2 member, breaks a different bond (DD-bond between m-DAP and D-Ala) compared to the family 1 effector Tse1 described above. A family 3 member, Tae3TY from Salmonella enterica serovar Typhi, showed the same enzymatic specificity as Tae2. A family 4 effector, Tae4™ from Salmonella enterica serovar Typhimurium, breaks peptide crosslinks at the γ-D-glutamyl-m-DAP DL-bond, similar to Tse1, but targets the acceptor, not the donor, stem and can additionally cleave monomeric tetrapeptide stems at the same position. Thus the newly identified PG hydrolase effector families show overlapping as well as distinct cleavage specificities, with all characterized hydrolase effectors breaking interstrand crosslinks and Tae4™ additionally modifying uncrosslinked peptides (Figure 1).
Functional analysis of the immunity proteins revealed T6 amidase immunity 3TY (Tai3TY) can protect against both Tae3TY and Tae2BT, two effectors with the same enzymatic specificity, but Tai2BT only protects against Tae2BT [23]. Tai3TY did not confer protection against Tae2TY and thus is not active against all DD-endopeptidases. While all identified hydrolase effectors have cognate immunity proteins, many orphan immunity proteins were also identified. Collectively, these observations indicate a complex co-evolution of effector and immunity proteins that may reflect a continual arms race among bacterial species and strains. In addition to selective pressure from immunity proteins, the different families of hydrolases that target different bonds within peptide crosslinks may imply other mechanisms of effector resistance, such as substrate modification, that have not yet been identified. Russell et al. note that organisms harboring hydrolase effectors appear enriched for those that inhabit complex communities such as the gastrointestinal tract, perhaps suggesting a role in kin discrimination or establishing bacterial community structure [23]. Further studies observing differentially marked strains could shed light on how these effectors influence mixed communities.
Regulation of latency
A particularly vexing problem for the treatment of chronic bacterial infections is the ability of many bacteria to enter a quiescent state of low metabolic activity with little or no replication. Mycobacterium tuberculosis, which causes acute and latent lung infections, encodes several putative PG hydrolases homologous to resuscitation promoting factor (Rpf) from Micrococcus luteus (Figure 2). Rpf was first described as a bacterial cytokine that could promote growth of dormant, nongrowing cells of M. luteus and Mycobacterium smegmatis [46]. Structural and biochemical studies revealed that M. tuberculosis RpfB has shared features with c-type lysozymes and soluble lytic transglycosylases and thus would be expected to cleave the PG sugar backbone between MurNAc and GlcNAc residues. While the purified enzyme did not induce cell lysis or release labeled cell wall material, mutation of the proposed catalytic glutamate abolished resuscitation activity of purified protein [47]. M. luteus rpf is essential, but none of the five M. tuberculosis rpf homologues are required for growth in broth culture and a quintuple mutant is viable [48]. Mutants lacking rpfB or rpfE showed attenuation in a mouse infection model in spite of normal growth in peripheral blood mononuclear cells suggesting an important role during lung infection [48].
RpfB forms a complex with an essential protein, resuscitation promoting factor interacting protein A (RipA) [26], which has homology to NlpC/p60 family proteins (similar to some T6SS hydrolase effectors described above). Purified RipA releases labeled cell wall material and addition of RpfB further stimulates cell wall solubilization [24]. RipA localizes to sites of cell growth and division where it can form a complex with the PG synthase PBP1a [25]. PBP1a seems to compete with RpfB for RipA binding and inhibits RpfB stimulation of cell wall hydrolysis. These results suggest RipA participates in two distinct complexes, a synthesis complex with PBP1A and a cell wall fragment release complex with RpfB (and/or RpfE, which also interacts with RipA). As ripA is essential while the quintuple rpf mutant is viable, it seems likely that the synthesis complex is required for viability and the Rfp complex participates in cell signaling.
As described for P. aeruginosa above, there is increasing evidence that bacteria can sense and respond to PG. Perception of the Rpf signal in M. tuberculosis is mediated by the transmembrane serine/threonine protein kinase B (PknB), which likely recognizes PG fragments via extracellular penicillin and Ser/Thr kinase associated (PASTA) domains [49–51]. PknB phosphorylates a pseudokinase domain of the putative peptidoglycan lipid II flippase MviN/MurJ [52] promoting its binding to a forkhead domain protein (FhaA) that negatively regulates PG synthesis [53]. MviN depletion inhibits cell growth and alters cell morphology while FhaA depletion results in increased PG incorporation at poles and the cells become shorter and fatter. Thus proper Rpf signaling may regulate both growth and cell shape.
In spite of considerable progress defining the Rpf signaling cascade, the actual PG fragment(s) recognized remains elusive. MurNAc-linked tripeptides containing m-DAP and D-Gln in the second position (a modification seen in M. tuberculosis PG) show modest binding to PknB PASTA domain in vitro, but these same peptides had very modest resuscitation activity [49]. While the precise specificity and cleavage products have yet to be determined for either RpfB or RipA, their predicted combined activities would be expected to release disaccahride–dipeptide, similar to the expected combined activity of Tse1 and Tse3 from P. aeruginosa. Perhaps the lack of disaccharide in the substrates tested in the Mir et al. study underlies the poor activity observed by the peptides tested [49]. Alternatively, PknB may not recognize a soluble PG species, but a structural deformation of the adjacent sacculus produced by the action of these PG hydrolases.
Concluding remarks
The studies reviewed here reveal new roles for PG hydrolases that cause perturbation of the cell wall in regulation of diverse bacterial behaviors including predation, motility, immune evasion, bacterial community structure, and latency. Many questions remain regarding the precise mechanisms through which these hydrolases exert effects on these biological processes (Box 2). One intriguing possibility is that structural perturbations of the sacculus have underappreciated regulatory consequences. As noted above, mammalian innate immune receptors such as Nod1 and Nod2 recognize very specific monomeric PG disaccharide-peptide species. Innate immune receptors are thought to target immutable pathogen components essential for survival in the host environment. While the PG sacculus is clearly an essential structure continually turned over during growth, the PG species sensed are not in high abundance in the polymerized sacculus and cell growth does not require their release. A common theme of hydrolases described here is coincident enzymes whose combined activity could produce or eliminate Nod protein agonists. For example, H. pylori appears to minimize Nod1 agonist expression in some growth states while promoting inflammation through Nod1 in others. We speculate that Nod2 may sense a factor released by T6SS-expressing Proteobacteria during growth (and competition) in complex host communities and produced by M. tuberculosis to signal favorable growth conditions and exit from latency. Thus mammalian hosts may be tuned to sense not merely the presence of growing bacteria, but those engaged in specific behaviors.
Box 2. Outstanding questions.
What are the identities of the currently predicted, yet unidentified, PG hydrolases used by cells to generate the diversity of PG structures seen in cell sacculi and released from cells? How are these PG hydrolases distributed and regulated among the diversity of bacteria?
Many of the processes described here require the activity of several PG hydrolases. How do the various PG hydrolases work together to make complex changes to the PG sacculus and release specific signaling molecules? Which of the many phenotypes associated with their activities are most critical for survival in different environments?
What regulatory networks exist to balance PG hydrolase activity and synthase activity during growth, repair, and modulation of the PG sacculus?
Which secretion mechanisms are used to deliver hydrolases or the molecules derived from hydrolytic cleavage of PG to allow bacteria to influence the co-inhabitants of their environment?
Acknowledgments
We thank members of the Salama lab and Peter Wyckoff for helpful discussions and funding from NIH grant R01AI094839 and the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program (JAT). The contents are solely the responsibility of the authors and do not necessarily represent the official views of these funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vollmer W, et al. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Vollmer W, et al. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 3.Bugg TD, et al. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 5.Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol Rev. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 6.Firczuk M, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev. 2007;31:676–691. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- 7.Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 8.Kumar JK. Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol. 2008;80:555–561. doi: 10.1007/s00253-008-1579-y. [DOI] [PubMed] [Google Scholar]
- 9.Schindler CA, Schuhardt VT. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nigro G, et al. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell Microbiol. 2008;10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 11.Cookson BT, et al. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun. 1989;57:2223–2229. doi: 10.1128/iai.57.7.2223-2229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cookson BT, et al. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry. 1989;28:1744–1749. doi: 10.1021/bi00430a048. [DOI] [PubMed] [Google Scholar]
- 13.Adin DM, et al. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2009;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloud-Hansen KA, et al. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol. 2008;190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloud-Hansen KA, et al. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 16.Lerner TR, et al. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 2012;8:e1002524. doi: 10.1371/journal.ppat.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonis M, et al. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol. 2010;78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 18.Chaput C, et al. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sycuro LK, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sycuro LK, et al. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frirdich E, et al. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog. 2012;8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell AB, et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell AB, et al. A widespread type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11:583–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hett EC, et al. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hett EC, et al. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog. 2010;6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hett EC, et al. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 27.Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 28.Fenton AK, et al. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol. 2010;192:6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volle CB, et al. Quantitative changes in the elasticity and adhesive properties of Escherichia coli ZK1056 prey cells during predation by Bdellovibrio bacteriovorus 109. J Langmuir. 2008;24:8102–8110. doi: 10.1021/la8009354. [DOI] [PubMed] [Google Scholar]
- 30.Costa K, et al. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 1999;181:3710–3715. doi: 10.1128/jb.181.12.3710-3715.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg HC, Turner L. Movement of microorganisms in viscous environments. Nature. 1979;278:349–351. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- 32.Hazell SL, et al. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 33.Sayi A, et al. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J Immunol. 2009;182:7085–7101. doi: 10.4049/jimmunol.0803293. [DOI] [PubMed] [Google Scholar]
- 34.Viala J, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 35.Nagy TA, et al. Helicobacter pylori regulates cellular migration and apoptosis by activation of phosphatidylinositol 3-kinase signaling. J Infect Dis. 2009;199:641–651. doi: 10.1086/596660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy TA, et al. beta-Catenin and p120 mediate PPARdelta-dependent proliferation induced by Helicobacter pylori in human and rodent epithelia. Gastroenterology. 2011;141:553–564. doi: 10.1053/j.gastro.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaput C, et al. Characterization of Helicobacter pylori lytic transglycosylases Slt and MltD. J Bacteriol. 2007;189:422–429. doi: 10.1128/JB.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M, et al. The Legionella pneumophila EnhC protein interferes with immunostimulatory muramyl peptide production to evade innate immunity. Cell Host Microbe. 2012;12:166–176. doi: 10.1016/j.chom.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, et al. Legionella pneumophila EnhC is required for efficient replication in tumour necrosis factor alpha-stimulated macrophages. Cell Microbiol. 2008;10:1906–1923. doi: 10.1111/j.1462-5822.2008.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman JM, et al. Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol. 2012 doi: 10.1146/annurev-micro-121809–151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz S, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basler M, et al. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeats C, et al. The PASTA domain: a beta-lactam-binding domain. Trends Biochem Sci. 2002;27:438. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 45.Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193:909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukamolova GV, et al. A bacterial cytokine. Proc Natl Acad Sci U S A. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen-Gonsaud M, et al. The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat Struct Mol Biol. 2005;12:270–273. doi: 10.1038/nsmb905. [DOI] [PubMed] [Google Scholar]
- 48.Kana BD, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mir M, et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010;75:1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 51.Shah IM, et al. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gee CL, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glauner B, et al. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 55.de Pedro MA, et al. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]