Abstract
The Exon Junction Complex (EJC) plays a critical role in multiple posttranscriptional events, including RNA subcellular localization, nonsense-mediated decay (NMD), and translation. We previously reported that knockdown of the EJC core component Eukaryotic initiation factor 4a3 (Eif4a3) results in full-body paralysis of embryos of the frog, Xenopus laevis. Here, we explore the cellular and molecular mechanisms underlying this phenotype. We find that cultured muscle cells derived from Eif4a3 morphants do not contract, and fail to undergo calcium-dependent calcium release in response to electrical stimulation or treatment with caffeine. We show that ryr transcripts are incorrectly spliced in Eif4a3 morphants, and demonstrate that inhibition of Xenopus Ryanodine receptor (Ryr) function similarly results in embryonic paralysis. These results suggest that the EJC mediates muscle cell function via regulation of pre-mRNA splicing during early vertebrate embryogenesis.
Keywords: Eif4a3, EJC, Xenopus laevis, ryanodine receptor, RNA processing, splicing
Introduction
RNA processing plays a fundamental role in the regulation of gene expression (Moore and Proudfoot, 2009). Recent studies have implicated the multi-protein Exon Junction Complex (EJC), which binds upstream of exon splice junctions in a sequence-independent fashion, in a number of RNA processing events, including localization, splicing, translation, and degradation (Le Hir et al., 2001; Le Hir et al., 2000; Tange et al., 2004). The EJC consists of several core proteins and numerous accessory factors (Bono and Gehring, 2011; Moore and Proudfoot, 2009); Eif4a3 is the EJC core component thought to directly contact RNA (Ballut et al., 2005; Shibuya et al., 2004). Eif4a3 was first identified in animal cells in a screen for transcripts upregulated in the ventral ectoderm of gastrula stage Xenopus embryos (Weinstein et al., 1997). Eif4a3 expression is quite dynamic during subsequent stages of development, suggesting diverse, tissuerestricted activities for this factor (Weinstein et al., 1997). Eif4a3 misexpression is sufficient to drive epidermal induction in cells otherwise destined to adopt a neural fate (Weinstein et al., 1997). Loss-of-function studies suggest that the requirement for Eif4a3 during development is more nuanced: morpholino-mediated knockdown of Eif4a3 leads to defects in heart looping and melanophore development and, strikingly, complete embryonic paralysis (Haremaki et al., 2010).
Although it is clear that Eif4a3 is essential for normal embryogenesis, the mechanisms underlying this requirement are not well understood. Knockdown of the EJC core components Y14 and Magoh give rise to defects similar to those seen in Eif4a3 morphants, suggesting that the embryonic requirement for Eif4a3 is mediated through its role in the EJC (Haremaki et al., 2010; Kenwrick et al., 2004); however, the pleiotropic effects seen following Eif4a3 loss-of-function may reflect the disruption of one or several distinct RNA processing functions for this protein, acting on one or more target transcripts.
In order to address the specific requirements for Eif4a3 during development, we have focused on one aspect of the Eif4a3 morphant phenotype: we describe here the cellular and molecular mechanisms underlying the paralysis that results from Eif4a3 knockdown. We find that muscle cell cultures derived from Eif4a3 morphant embryos fail to contract in response to electrical or chemical stimulation; this appears to result from a defect in calcium-dependent calcium release. We demonstrate that the Ryanodine receptor 1 (Ryr1), a key mediator of intracellular calcium release, is required for embryonic movement in Xenopus; furthermore, we show both that Ryr1 is dramatically downregulated following Eif4a3 knockdown, and that ryr1 transcripts are improperly spliced in Eif4a3 morphants. Our results thus implicate Eif4a3 in muscle cell function, via regulation of ryr1 pre-mRNA splicing.
Materials and methods
Eif4a3 constructs and morpholinos
For Myc-Eif4a3, the coding sequence of X. laevis Eif4a3 was amplified by PCR using the following primers: Xeif4a3-5mycU: AGGCCTGCGGCCGCAGCTGTTGCAG; Xeif4a3-5mycD: AGGCCTCAAATAAGATCAGCAACGTTC and subcloned downstream of six Myc (EQKLISEEDLNEM) epitopes in the StuI site of CS2+MT. X. laevis Eif4a3 morpholino (Eif4a3MO) and the five-mismatch control morpholino (MM) were described previously (Haremaki et al., 2010). A construct containing only the coding sequence of X. laevis Eif4a3 was described previously (Weinstein et al., 1997); both this and the Myc-Eif4a3 construct lack the Eif4a3MO-binding site.
Muscle cell culture
X. laevis muscle cell cultures were prepared as described (Nacira Tabti, 1998). For electrical charge assays, cells were stimulated directly by silver electrodes at 10V, 20V, or 200V 6Hz using an Isolated Pulse Stimulator (model# 2100, A-M Systems). Immunocytochemistry for Xenopus muscle cell culture was performed as described (Campbell et al., 2006). Anti-myosin antibody (MF20, Developmental Studies Hybridoma Bank) was used at 1:60 dilution; FITC-conjugated secondary antibody (Jackson ImmunoResearch) was used at 1:50 dilution.
Calcium imaging
Stock solution (1mM) of the membrane-permeant form of fluo-4 (Invitrogen) was prepared in DMSO, then added to culture media to give a final concentration of 3 υM. The nonionic detergent Pluronic F-127 (0.03% final) was added to increase solubility (Prada et al., 2005). Two days after culture preparation, cells were incubated with fluo-4 for 30 minutes at room temperature, then washed 3 times in culture media. 10V 6Hz 2 second electrical stimulation was applied to the cells; simultaneously, images of fluo-4 treated cells were taken by 2 second exposure using a Leica DMIL inverted microscope with filter module I3 and a DFC420c camera, and analyzed by ImageJ.
Caffeine and Ionomycin treatment
For caffeine treatment, stock solutions (300mM) were diluted into 62.5 mM in 1.2 mL of culture medium removed from the cultured cells; this was then added back to the remaining 1.8 mL medium. For Ionomycin treatment, stock solutions (2mM) were diluted into 200uM with PBS then added to culture medium at a concentration of 1:100.
Ryanodine receptor genomic sequence and splice-blocking morpholinos
X. laevis partial ryr1 cDNA sequence (Contig043167) was obtained from XDB3.2 (http://xenopus.nibb.ac.jp/).To acquire ryr1 genomic sequence, X. laevis genomic DNA was amplified by PCR using the following primers: XlRYRprobeU: TCTGTCCATTCTGGGACACT, XlRYRprobeD: GACCAGTGTGTTCCGTTTCA. The amplified 3.2kb fragment was cloned into the T-vector (Promega) and fully sequenced. Exon-intron structures were predicted by comparison with Xenopus tropicalis ryr1 (JGI v4.1, ID: 469327). A splice-blocking morpholino (RyrMO)(AGATAATGTTCTCTGACCTGTTTGC) was designed at the exon 99-intron 99 boundary. The effects of this morpholino on ryr1 transcripts were confirmed by RT-PCR using the following primers: XlRYRprobeU: TCTGTCCATTCTGGGACACT, RYRnewD: CAGCTCTCCAAATGCATCAA. The Ryr5MM morpholino (AGAaAATcTTgTCTcACCTcTTTGC ) introduced 5 base pair mismatches to the RyrMO sequence.
RNA co-immunoprecipitation (RIP)
1ng Myc-eif4a3 RNA was injected with and without 21ng Eif4a3MO into 2-cell stage embryos, which were subsequently harvested at stage 27. RNA co-immunoprecipitation assays were performed as described (Vishnu et al., 2011). Precipitated RNA was purified by RNA-Bee (Tel-Test, Inc.). After DNase treatment, cDNA was synthesized with random hexamers by MMLV Reverse Transcriptase (Promega). mRNA levels were assayed by PCR using the following primers:
Xlryr5'F1: GAGGAGATCCAGTTCCTCAG,
Xlryr5'R4: GAATAGCATGGCCATAGAGC,
RYRnewU: CTGGCCGTGGTTGTTTATCT,
RYRnewD: CAGCTCTCCAAATGCATCAA,
ProbeD: GACCAGTGTGTTCCGTTTCA,
RYRE101F: TGCTATCTCTTCCACATGTA (exon 101 primer),
RYRI101F: GAACCCGAAAATACCCCATC (intron 101 primer),
RYRE102R: CTCCATGTCCTCCTTTACTTG (exon 102 primer),
FoxD3U: TGTGGAGCGTAACTGGAATG,
FoxD3D: GTTCTTGGGCTTGTTCTGGA,
GSK3BP_U: TTCTTGCGTGAGGGGTAGAA,
GSK3BP_D: CATTGCACGGTTGTCTCAGT,
CYP1C_U: GCCCCATCTCACCTTTTGTA,
CYP1C_D: GAAGTCAAGCGCAGGAAAAC.
RT-PCR and Western blot analysis
Western blot analysis was performed as described (Hama et al., 2002). Antibodies that detect both Ryanodine receptor-1 and -2 isoforms (34C, Developmental Studies Hybridoma Bank) were used at 1:50 dilution. Antibodies against β-tubulin (T8660, Sigma) were used at 1:200 dilution. Secondary antibodies (donkey anti-mouse IgG coupled to horseradish peroxidase) (Jackson ImmunoResearch Laboratories) were used at 1:1,000 dilution.
RT-PCR was performed as described (Wilson and Hemmati-Brivanlou, 1995). Primers used in this study are as follows:
RYRnewU: CTGGCCGTGGTTGTTTATCT,
RYRnewD: CAGCTCTCCAAATGCATCAA,
ODC-F: AATGGATTTCAGAGACCA,
ODC-R: CCAAGGCTAAAGTTGCAG
Results
Eif4a3 is required for muscle cell contraction
We previously reported that knockdown of the EJC component Eukaryotic initiation factor 4a3 (Eif4a3) results in full-body paralysis in embryos of the frog Xenopus laevis (Haremaki et al., 2010). Paralysis can arise from a variety of causes, including defects in skeletal muscle and/or neuronal development. Eif4a3 morphants have sensory neuron defects that may underlie the lack of touch response in these embryos; however, this is not a likely cause of paralysis (Haremaki et al., 2010). While we observed no marked differences in somite structure between Eif4a3 morphants and control embryos, we have not yet addressed directly the effects of Eif4a3 knockdown on muscle cell activity.
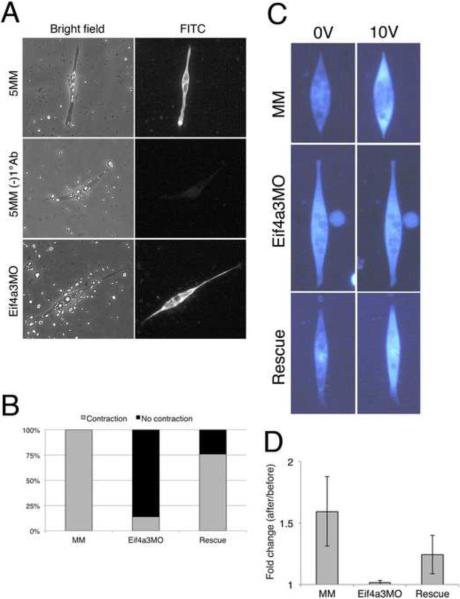
To investigate the potential requirement for Eif4a3 in muscle development and function, muscle cell cultures were prepared from stage 23 dorsal explants of uninjected embryos, Eif4a3 morpholino (Eif4a3MO)-injected embryos, or embryos injected with a control morpholino that differs at 5 base pairs and does not affect Eif4a3 translation (Eif4a3MM) (Haremaki et al., 2010; Nacira Tabti, 1998). Spindle-shaped cells obtained from control and morphant embryos were morphologically indistinguishable; staining with the anti-myosin specific antibody MF20 confirmed that these were indeed muscle cells (Fig. 1A)(Bader et al., 1982). To assess muscle cell contractility, we subjected cultures to electrical stimulation (Xie et al., 1997). All Eif4a3MM-injected spindle-shaped muscle cultures tested contract in response to electrical stimulation of 20V (100%; n=34); muscle cultures derived from Eif4a3MO-injected embryos, however, largely failed to contract following 20V stimulation (14%; n=25) (Fig. 1B; also see video files). These latter cultures also failed to contract in response to 200V stimulation (0%; n=12; data not shown). The morpholino effect was rescued by co-expression of 1ng morpholino-insensitive eif4a3 RNA (76%; n=25) (Haremaki et al., 2010) (Fig. 1B). These results suggest that a defect in muscle fiber contraction contributes to the Eif4a3 morphant paralysis phenotype.
Fig. 1.
Impaired contraction of Eif4a3 morphant muscle cell cultures. (A) Labeling of spindle-shaped cultured cells by the myosin-specific MF20 antibody. Cell shown in middle panel was processed without primary antibody. Cells were derived from Eif4a3MO and Eif4a3MM (5MM) morphant embryos. (B) Eif4a3 knockdown inhibits contraction of muscle cells in response to electrical stimulation. Muscle cell cultures were derived from stage 23 embryos injected with Eif4a3 morpholino (Eif4a3MO), five base pair mismatch control morpholino (MM), or both Eif4a3MO and eif4a3 RNA (Rescue) (n=25, 34, 25, respectively). Number of cells observed to contract, as a percentage of the total, are shown. (C) Eif4a3 knockdown inhibits increase of intracellular calcium in muscle cells following electrical stimulation. Spindle shaped muscle cells were obtained from stage 23 embryos injected with EIf4a3 morpholino (Eif4a3MO), five base pair mismatch control morpholino (MM), or both Eif4a3MO and eif4a3 RNA (Rescue) and incubated with the fluorescent calcium indicator, Fluo-4. (D) Quantification of studies shown in (C). Fluorescence intensity was compared before and after 10V electrical stimulation. Each of five cultures was measured and averages of fold change (after/before) were plotted. Error bars indicate S.D.
Eif4a3 is required for calcium release from the sarcoplasmic reticulum
To investigate the basis for the lack of response to electrical stimulation in Eif4a3-depleted cells, we used a membrane-permeant form of Fluo-4, a calcium-sensitive fluorescent dye, to compare intracellular calcium levels in control and Eif4a3 morphant cultures. Muscle cells from embryos injected with Eif4a3MM, Eif4a3MO, or Eif4a3MO and eif4a3 RNA (“Rescue”) were treated with Fluo-4, and calcium levels were measured following electrical stimulation. A significant increase in fluorescence was observed in Eif4a3MM-derived muscle cells following application of 10V stimulation; however, increases were not detected in Eif4a3MO-derived muscle cells following stimulation (Figs. 1C, D). eif4a3 RNA overexpression rescued this effect (Figs. C, D). Taken together, these studies suggest that Eif4a3 is required for normal intracellular calcium release. We note that, while post-stimulation fluorescence in the Eif4A3MO, EIF4a3MM, and Rescue cells are indistinguishable, pre-stimulation (baseline) fluorescence is higher in the Eif4a3MO (74.6+/−11.0) than in the Eif4a3MM (49.7+/−13.4) or Rescue cultures (51.9+/−5.5)(data not shown); these data suggest that the diminished response to electrical stimulation following Eif4a3 knockdown may result, at least in part, from a leak in intracellular Ca2+ stores.
Eif4a3 is required for ryanodine receptor expression
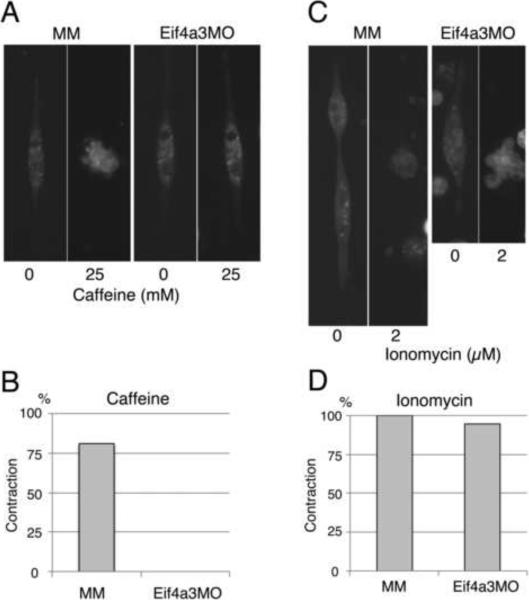
Ca2+-induced Ca2+ release from the sarcoplasmic reticulum (SR) is essential for muscle cell contraction; the Ryanodine receptor 1(Ryr1) is a critical regulator of this process in skeletal muscle (Hamilton, 2005). To examine the potential involvement of Ryr1 in Eif4a3-mediated muscle cell contraction, we treated muscle cells with the Ryr activator caffeine (Stephenson, 2008). Cell contractions were observed in 81% of Eif4a3MM cultures (n=51) following treatment with 25 mM caffeine; in Eif4a3MO muscle cultures, however, caffeine treatment did not induce cell contraction (n=32)(Fig. 2A, B). Addition of the calcium ionophore Ionomycin, however, induced muscle cell contractions in 100% of Eif43MM cultures (n=27) and 95% of Eif4a3MO cultures (n=20) (Fig. 2C, D). Taken together, these results suggest that loss of Eif4a3 impairs contractility through inhibition of calcium release, and may act via inhibition of Ryr function.
Fig. 2.
Eif4a3 morphant muscle cells are insensitive to caffeine, and contract in response to the calcium ionophore Ionomycin. (A) Eif4a3 knockdown inhibits caffeine-mediated contraction in muscle cell cultures. Muscle cells were prepared from Eif4a3MM (MM)- or Eif4a3MO- injected stage 23 embryos and treated with 25mM caffeine. (B) Graph summarizing caffeine experiment results. (C) Ionomycin induces contractility in both wild-type and Eif4A3 morphant cell cultures. Muscle cells were prepared from Eif4a3MM (MM)- or Eif4a3MO- injected stage 23 embryos and treated with 2uM Ionomycin. (D) Graph summarizing Ionomycin experiment results.
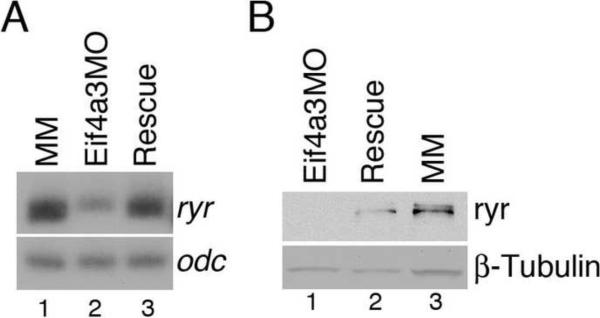
To address this possibility, we examined ryr mRNA and protein in stage 27 Eif4a3 morphants and control embryos. ryr1 mRNA levels were reduced in Eif4a3-depleted embryos relative to that seen in Eif4a3MM-injected controls; this effect was rescued by co-expression of eIf4a3 RNA (Fig. 3A). Ryr protein was clearly detected in uninjected embryos, and in embryos injected with Eif4a3MM; Ryr was reduced to undetectable levels in Eif4a3 morphants (Fig. 3B and data not shown). This effect was rescued by co-expression of eif4a3 RNA (Fig. 3B); Eif4a3 activity thus appears to be required for expression of ryr1 mRNA and protein.
Fig. 3.
Eif4a3 knockdown leads to a reduction in ryanodine receptor (ryr) RNA and protein levels. (A) Semi-quantitative RT-PCR analysis of ryr (25 cycles); odc was used as a loading control. RNA was purified from stage 27 embryos injected with mismatch control morpholinos (MM), Eif4a3 morpholinos (Eif4a3MO), or co-injected with Eif4a3MO and eif4a3 RNA (Rescue). (B) Western blot analysis with anti-Ryr antibody (34C, Developmental Studies Hybridoma Bank). β-Tubulin, visualized with an anti-β-Tubulin antibody, was used as an internal control.
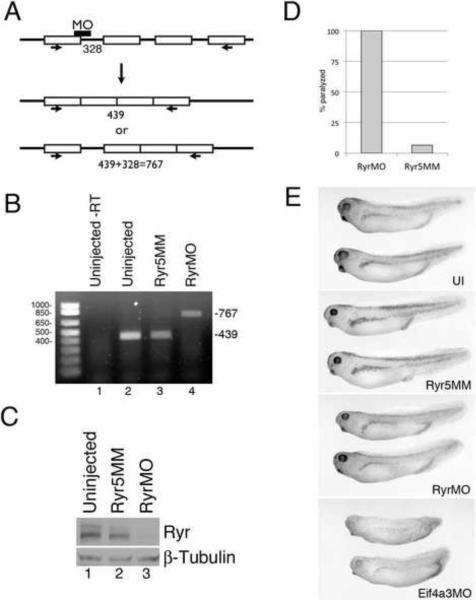
The Ryanodine receptor is required for embryonic movement
To directly investigate the role of the Ryanodine receptor in muscle contraction in Xenopus laevis, we utilized a splice-blocking Ryr morpholino oligonucleotide (RyrMO), designed to bind the putative exon-intron boundary 99 of X. laevis ryr1 (Fig. 4A). As expected, PCR and sequence analysis demonstrated that the target intron is retained in RyrMO-injected, but not in uninjected embryos or embryos injected with a control morpholino that differs from RyrMO at 5 base pairs (Ryr5MM)(Fig. 4B, and data not shown). Western blot analysis with anti-Ryr antibody showed a strong decrease in Ryr protein following injection of RyrMO, but not Ryr5MM, in stage 27 embryos (Fig. 4C). Injection of RyrMO, but not Ryr5MM, results in complete paralysis in stage 27 embryos (n=23)(Fig. 4D). These data further support our hypothesis that Ryanodine receptor inhibition underlies the paralysis observed in Eif4a3 morphants.
Fig. 4.
Ryr is required for embryonic movement in X. laevis. (A) Schematic showing the binding site for the Ryr splice-blocking morpholino (RyrMO); numbers indicate expected PCR product size with indicated primers (arrows). (B) RT-PCR analysis of stage 27 embryos injected with either RyrMO or a 5 base pair mismatch Ryr morpholino (Ryr5MM). (C) Western blot analysis using an anti-Ryr antibody. β-tubulin was used as a loading control. (D) Ryr knockdown leads to complete paralysis in Xenopus embryos. Graph depicting percentage of touch-unresponsive stage 27 embryos injected with 20ng (RyrMO/embryo (n=23); these animals showed no spontaneous movement at later stages (data not shown). Control embryos were injected with 20ng Ryr5MM/embryo, and were largely responsive to touch (n=17). (E) Uninjected (UI), Ryr5MM, RyrMO and Eif4a3MO-injected embryos at stage 32.
The vertebrate ryr receptor genes code for proteins of approximately 5000 amino acids (Zalk et al., 2007); no full-length ryr cDNAs have been isolated from X. laevis or X. tropicalis; transcript size in the latter organism is predicted to exceed 15Kb (JGI v.4.1 ID: 469327). We have not successfully generated full-length ryr RNA in vitro from any species; thus, we have not been able to test directly whether Ryr is epistatic to Eif4a3 in the context of embryonic movement. We previously reported that Eif4a3 is also required for embryonic pigmentation and cardiac looping (Haremaki et al., 2010); RyrMO-injected embryos, in contrast, do not display pigmentation or cardiac defects, suggesting that the developmental requirement for Eif4a3 is not mediated solely through ryr (Fig. 4E and data not shown).
Eif4a3 is required for accurate splicing of the ryanodine receptor pre-mRNA
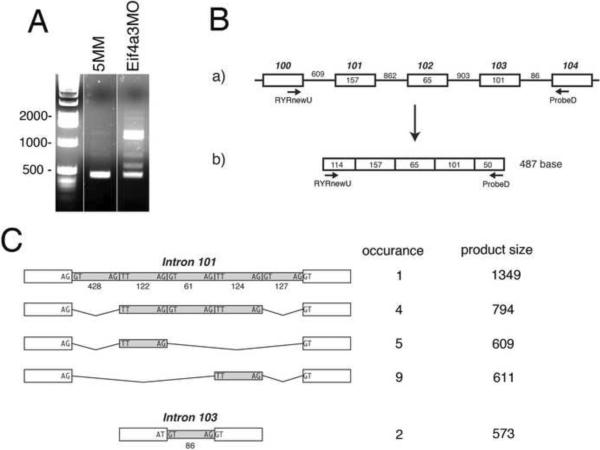
RT-PCR analysis of the ryanodine receptor 1 transcript revealed several larger-than-expected PCR products following Eif4a3 knockdown (Figs. 5A, B). Interestingly, sequence analysis revealed that larger products contain ryanodine receptor intronic sequences: PCR products were identified with either complete or partial retention of intron 101, or with complete retention of intron 103. Notably, a mutation in the region of mammalian ryr corresponding to Xenopus exon 101 leads to a severe form of central core disease in humans, and inhibition of Ca2+ release in the mouse (Zvaritch et al., 2007). Retention of introns 100 or 102 were not observed (Figure 5C and data not shown). All partially retained introns include the sequences “TT” “AG” at their 5' and 3' termini, respectively. For these studies, only the C-terminal-encoding sequence of Xenopus ryr1 was available for analysis; therefore, there may be Eif4a3 knockdown-dependent splicing defects in other parts of the transcript that were not identified here. Nevertheless, our data demonstrate that Eif4a3 is required for correct splicing of the ryr1 transcript.
Fig. 5.
Eif4a3 is required for correct splicing of ryanodine receptor (ryr) pre-mRNA. (A) RT-PCR analysis of ryr RNA from Eif4a3MM (5MM) and Eif4a3MO-injected stage 27 embryos; primer pairs are indicated in (B). 29 cycles of PCR was used for this analysis, and samples were not normalized. (B) a) Intron-exon configuration of the X. laevis ryanodine receptor gene from exons 100 to 104. Exons and introns are represented by boxes and lines, respectively; exon size is given in boxes and intron size is given above lines. Exon number is indicated above the boxes, and is based on the X. tropicalis ryr sequence. b) Expected RT-PCR product after complete splicing. (C) Observed retention patterns of introns 101 and 103. PCR products from Eif4a3MO-injected embryos that were larger than 487bp were cloned and sequenced. The schematic shows the four identified retention patterns of intron 101. For intron 103, only complete retention was observed. The size of each intron subdomain is indicated. Sequences of exon-intron and intron subdomain boundaries are indicated.
To determine the fraction of ryr mRNA that is aberrantly spliced following Eif4a3 knockdown, we analyzed wild-type and Eif4a3 morphant embryos derived from five different frogs, comparing the relative levels of exon 101-exon 102 and intron 101-exon 102 products generated under each condition, using semi-quantitative RT-PCR and primers specific to each region. In wild-type embryos, the intron-exon (“incorrectly spliced”) product was virtually non-existent; in Eif4a3 morphants, however, the intron-exon product averaged 41.4 +/–16.4% of the ryr total (exon101-exon102 & intron101-exon102), on a molar basis (data not shown). This analysis takes into account only intron 101 retention; the total incorrectly spliced ryr fraction following Eif4a3 knockdown may thus be significantly higher than reported here.
Eif4a3 binds to ryanodine receptor mRNA
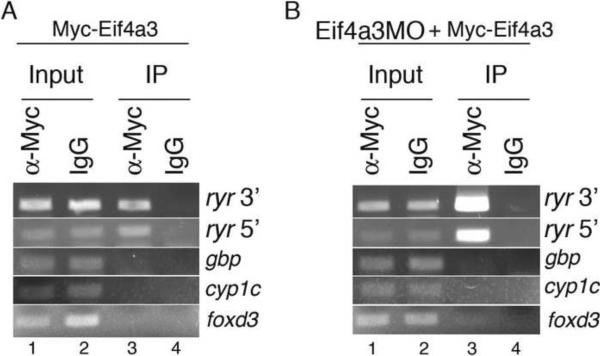
Our studies implicate Eif4a3 in ryr splicing fidelity, suggesting the potential for direct association between Eif4a3 and the ryr transcript (Ballut et al., 2005; Shibuya et al., 2004). To address this possibility, we injected early cleavage stage embryos with RNA encoding Myc epitope-tagged eif4a3, and performed immunoprecipitations with an anti-Myc antibody at stage 27. Reverse transcription, followed by PCR using primers against 5' and 3' exonic regions of ryr, were used to amplify co-precipitated ryr RNA. We find that ryr is present in anti-Mycimmunoprecipitated samples, but not in control samples in which IgG was substituted for anti-Myc antibody (Fig. 6A). This trend was even more apparent when native Eif4a3 was depleted by co-injection of Eif4a3MO (Fig. 6B). The EJC has been shown to bind RNA upstream of exonexon boundaries (Le Hir et al., 2001; Le Hir et al., 2000); we therefore used the transcripts of intronless genes as negative controls in this study; as expected, PCR products of cDNA derived from the intronless genes gbp, cyp1c and foxd3 were not detected in immunoprecipitated samples (Figs. 6A, B). These results indicate that Eif4a3 can bind ryr mRNA, and supports a model in which Eif4a3 regulates embryonic movement via proper splicing of ryr pre-mRNA.
Fig. 6.
Eif4a3 associates with ryr mRNA. Cell extracts from stage 27 embryos injected with Myc-Eif4a3 (A) or Myc-Eif4a3 with Eif4a3MO (B) were incubated overnight with anti-Myc antibody or mouse IgG. Following immunoprecipitation, Myc-Eif4a3-associated RNA was purified, reverse transcribed, and amplified using gene-specific primers. “Input RNA”: non-immunoprecipitated RNA.
Discussion
Eif4a3 is essential for normal embryonic development, and has been implicated, as a component of the multi-protein EJC, in multiple RNA processing events (Haremaki et al., 2010; Tange et al., 2004). We find that Eif4a3 is required for the contraction of cultured muscle cells and mediates the proper splicing of ryr1, knockdown of which leads to complete embryonic paralysis, a phenotype we previously observed following knockdown of the EJC components Eif4a3, Y14, or Magoh (Haremaki et al., 2010). Taken together, our studies suggest that Eif4a3, via the actions of the EJC, is required for appropriate splicing of ryr transcripts and, as a result, for Ryr function, muscle cell contraction, and embryonic movement.
While we predict that aberrant splicing of ryr contributes to a loss of embryonic mobility via effects on muscle cell contractility, mammalian ryr1 has been shown to be required for calcium release in neuronal populations, as well (De Crescenzo et al., 2012). Ryr dysregulation in neuronal tissue may thus also contribute to the Eif4a3 morphant phenotype. Efforts are underway to determine whether neuronal defects observed in Eif4a3 morphants are mediated by Ryr loss of function (Haremaki et al., 2010).
During the course of these studies, two papers were published demonstrating a requirement for Eif4a3 in the splicing of the Drosophila ERK MAP kinase transcript (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). In these studies, whole-genome analyses were used to demonstrate that only a small subset of transcripts are affected by Eif4a3 loss-of-function; those with large introns are particularly sensitive to splicing defects (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). The apparently normal early development of Xenopus Eif43 morphants suggests that many splicing events are unaffected by loss of Eif43 during vertebrate embryogenesis, as well (Haremaki et al., 2010). It will be interesting to determine whether intron size or number correlate with Eif4a3-dependent splicing in Xenopus. We have not observed alterations in ERK splicing in Eif4a3 morphants, despite the fact that ERK, based on the predicted Xenopus tropicalis genomic structure, includes a 19 Kb intron (JGI v.4.1, ID 155435) (data not shown). We have yet to examine all eight predicted introns in Xenopus ERK, however, and our studies do suggest a differential sensitivity among Xenopus ryr introns to Eif43 knockdown (Fig. 5). Establishment of the distinguishing features of Eif4a3-regulated transcripts and introns in Xenopus will be an important step in defining the mechanisms by which Eif4a3 and the EJC regulate pre-mRNA splicing in the vertebrate embryo.
Eif4a3 knockdown leads to defects in ryr splicing, and a reduction of Ryr, in vivo; Eif4a3 knockdown may also lead, via alternative splicing, to the production of distinct Ryr isoforms that contribute to embryonic paralysis or other phenotypes seen in the Eif4a3 morphants. Alternative splicing has been shown to play important roles during embryogenesis including, for example, in Drosophila sex determination (Salz, 2011). Recent studies in human embryonic stem (ES) and induced pluripotent stem (iPS) cells have demonstrated, remarkably, that alternative splicing of FOXP1 produces two proteins with distinct DNA binding specificities that together regulate the transcriptional network governing stem cell pluripotency and differentiation (Gabut et al., 2011). Loss of EJC components, including Eif4a3, has recently been shown to promote the production of proapoptotic splice variants of several Bcl family members in cultured cell lines, demonstrating that Eif4a3 can regulate alternative splicing in vitro (Michelle et al., 2012). Studies are in progress to address whether Eif4a3 knockdown also generates alternative splice variants of ryr that possess biological activity.
Our studies implicate splicing dysregulation as a primary mechanism underlying paralysis in Eif4a3 morphants; they do not, however, preclude distinct requirements for Eif4a3 in other regulatory mechanisms during embryogenesis, nor do they eliminate the possibility that ryr splicing defects are an indirect consequence of Eif4a3 loss-of-function. Future studies will address whether Eif4a3 knockdown-mediated defects in melanophore and cardiac development are a result of errors in NMD, splicing, and/or other RNA processing events.
Supplementary Material
Acknowledgments
We thank H Hirata, A Kulozik, and H Takeshima for gifts of reagents, F Ono for advice on experimental design, and M Birne for care of the Xenopus colony. This work is supported by PHS grant R01-GM61671 (DCW) and funds from Queens College of the City University of New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrancois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, Therrien M. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring NH. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA biology. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell NR, Podugu SP, Ferrari MB. Spatiotemporal characterization of short versus long duration calcium transients in embryonic muscle and their role in myofibrillogenesis. Dev Biol. 2006;292:253–264. doi: 10.1016/j.ydbio.2005.11.040. [DOI] [PubMed] [Google Scholar]
- De Crescenzo V, Fogarty KE, Lefkowitz JJ, Bellve KD, Zvaritch E, MacLennan DH, Walsh JV., Jr. Type 1 ryanodine receptor knock-in mutation causing central core disease of skeletal muscle also displays a neuronal phenotype. Proc Natl Acad Sci U S A. 2012;109:610–615. doi: 10.1073/pnas.1115111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Hama J, Suri C, Haremaki T, Weinstein DC. The molecular basis of Src kinase specificity during vertebrate mesoderm formation. J Biol Chem. 2002;277:19806–19810. doi: 10.1074/jbc.M110637200. [DOI] [PubMed] [Google Scholar]
- Hamilton SL. Ryanodine receptors. Cell Calcium. 2005;38:253–260. doi: 10.1016/j.ceca.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Haremaki T, Sridharan J, Dvora S, Weinstein DC. Regulation of vertebrate embryogenesis by the exon junction complex core component Eif4a3. Dev Dyn. 2010;239:1977–1987. doi: 10.1002/dvdy.22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev Dyn. 2004;229:289–299. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. Embo J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, Le Hir H, Klinck R, Elela SA, Prinos P, Chabot B. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol. 2012;32:954–967. doi: 10.1128/MCB.06130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nacira Tabti JA, Mu-ming Poo. Culturing spinal neurons and muscle cells from Xenopus embryos. In: Gary Banker KG, editor. Culturing Nerve Cells. 2nd edition 1998. pp. 237–259. [Google Scholar]
- Prada C, Udin SB, Wiechmann AF, Zhdanova IV. Stimulation of melatonin receptors decreases calcium levels in xenopus tectal cells by activating GABA(C) receptors. Journal of neurophysiology. 2005;94:968–978. doi: 10.1152/jn.01286.2004. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK. Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21:395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Stephenson DG. Caffeine - a valuable tool in excitation-contraction coupling research. J Physiol. 2008;586:695–696. doi: 10.1113/jphysiol.2007.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Vishnu MR, Sumaroka M, Klein PS, Liebhaber SA. The poly(rC)-binding protein alphaCP2 is a noncanonical factor in X. laevis cytoplasmic polyadenylation. RNA. 2011;17:944–956. doi: 10.1261/rna.2587411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein DC, Honore E, Hemmati-Brivanlou A. Epidermal induction and inhibition of neural fate by translation initiation factor 4AIII. Development. 1997;124:4235–4242. doi: 10.1242/dev.124.21.4235. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- Xie K, Wang T, Olafsson P, Mizuno K, Lu B. Activity-dependent expression of NT-3 in muscle cells in culture: implications in the development of neuromuscular junctions. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:2947–2958. doi: 10.1523/JNEUROSCI.17-09-02947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annu Rev Biochem. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- Zvaritch E, Depreux F, Kraeva N, Loy RE, Goonasekera SA, Boncompagni S, Kraev A, Gramolini AO, Dirksen RT, Franzini-Armstrong C, Seidman CE, Seidman JG, Maclennan DH. An Ryr1I4895T mutation abolishes Ca2+ release channel function and delays development in homozygous offspring of a mutant mouse line. Proc Natl Acad Sci U S A. 2007;104:18537–18542. doi: 10.1073/pnas.0709312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.