Abstract
Epigenetic and chromatin modifications play particularly important roles in Embryonic and induced Pluripotent Stem cells (ES and iPS cells) allowing for the cells to both differentiate and dedifferentiate back to a pluripotent state. We analyzed how the loss of a key chromatin modifying enzyme, histone deacetylase 1(HDAC1), affects early and cardiovascular differentiation of both ES and iPS cells. We also investigated potential differences between these two cell types when differentiation is induced. Our data indicates an essential role for HDAC1 in deacetylating regulatory regions of key pluripotency-associated genes during early differentiation. Although HDAC1 functions primarily as a histone deacetylase, its loss also affects DNA methylation in ES and iPS cells both during pluripotency and differentiation. We show that HDAC1 plays a crucial, non-redundant role in cardiomyocyte differentiation and maturation. Our data also elucidates important differences between ES and iPS cells, when levels of this enzyme are reduced, that affect their ability to differentiate into functional cardiomyocytes. As varying levels of chromatin modifying enzymes are likely to exist in patient derived iPS cells, understanding the molecular circuitry of these enzymes in ES and iPS cells is critical for their potential use in cardiovascular therapeutic applications.
Keywords: Stem cells, induced Pluripotent Stem cells, Epigenetic regulation, Histone deacetylases, HDAC1, Cardiovascular differentiation
Introduction
The ability to isolate Human Embryonic Stem cells (ES cells) from unused In Vitro Fertilization (IVF) embryos opened a door of opportunities and hopes for their many potential uses in drug testing, use as models to help our understanding of various biological processes and most importantly their therapeutic potential in regenerative medicine. However, ethical, technical and regulatory issues as well as unavailability of autologous human ES cells for cell therapy applications limit the potential therapeutic utility of ES cells for cardiac repair in humans. Reprogramming of somatic cells into induced pluripotent stem cells (iPS cells) opened a new and exciting door of a cell type with the apparent plasticity of embryonic stem cell and the added advantage of patient specificity [1,2]. Since then the focus of iPS cell biology has shifted towards understanding the epigenetic regulation and cell signaling that underlie somatic cell dedifferentiation processes and the molecular mechanisms that are involved in the maintenance of the newly acquired pluripotent phenotype.
Induced Pluripotent Stem cells pose great potential for their use in clinical research and therapy. However, despite their promises and the new advancements made in the field, some challenges and unknowns still hinder the full realization of the clinical potential of these cells. Critical gaps in our knowledge of iPS cell biology include incomplete epigenetic and mechanistic characterization of their reprogramming and directed differentiation processes [3,4,5,6]. Additionally, while it is widely accepted that one of the first steps that allows these cells to differentiate is expression of pluripotency associated genes, such as Oct4, Nanog and Sox2, how these genes are turned off as differentiation is induced is poorly understood.
A cell's identity is defined by its epigenetic code, modifications of which directly influence gene expression or repression [8,9,10,11,12,13]. The epigenetic state of pluripotent cells is extremely complex as pluripotency needs to be tightly regulated and maintained during continuous proliferation yet developmental genes should be accessible enough for differentiation to occur rapidly once the differentiation machinery in the cell has started [11,12,13]. Histone Deacetylases (HDACs) have been identified as key players in both reprograming of somatic cells into iPS cells as well as important enzymes during differentiation of ES and iPS cells. However, most such studies rely on global inhibitors of HDACs, which in fact have different roles and direct differentiation into different lineages. The epigenetic similarity between iPS and ES cells and their pluripotent potential has also been recently questioned [11,14].
Methods
Cell types and Cell culture
iPS cells were generated from NIH3T3 cells using cell extract mediated reprogramming. The efficiency of our technique in generating these cells as well as their pluripotent nature has been previously analyzed and reported [15]. C57BL/6 murine ES cells were purchased from ATCC (Cat.# SCRC-1002) and iPS cells were cultured in 15% FBS, 50uM Beta-Mercaptoethanol, 1mM nonessential amino acids and 100 U/ml Pen/Strep supplemented Dulbeco's modified eagle medium (DMEM) in the presence of Leukemia Inhibitory factor (LIF; 10ng/mL).
Formation of Embryoid Bodies (EB)
Differentiation of iPS and ES cells through embryoid body formation was performed using standard hanging drop method. Briefly, a single-cell suspension of each cell line at a concentration of 2.5 × 105 cell/mL in 20 mL of differentiating media (Iscove's Modified Dulbecco's Medium (IDDM) supplemented with 15% FBS, 100 U/ml Pen/Strep, 200ug/ml transferrin, 0.5 mML-ascorbic acid and 4.5 × 10-4 M monothioglycerol) was deposited in 20ul hanging drops in 100 × 100 mm square petri dishes. After 2 days of being cultured in suspension, the cells were plated onto 0.1% gelatin coated dishes for continued differentiation.
Real-Time Arrays and mRNA expression
Expression analysis of epigenetic modifying enzymes and factors was performed using SABiosciences's RT2 Profiler™ PCR Array System according to manufacturer's instructions. Quantitative (Real-Time) RT-PCR was performed as described earlier [15] . Quantitative Real-Time PCR (Q-RTPCR or Q-PCR) was performed using gene specific labeled probes and primers. Array data was verified with independent Q-PCR.
HDAC1 Knock Down
The lentiviral-hdac1 shRNA vectors were purchased from Sigma-Aldrich® and transduction was performed according to manufacturer's instructions. Stably knocked-down clones were generated by puromycin selection for two weeks.
Immunofluorescence staining
Protein expression analysis through immunofluorescence staining was performed as described earlier [15].
Ca++ Studies
Calcium studies were performed mostly as previously described [16]. Isolated EBs were loaded with fluo-4AM (Invitrogen, 15 μmol/L, 20 minutes) and placed in an experimental chamber on the stage of a confocal microscope. Media was re-circulated for the remainder of the experiment. Laser scanning of cells in beating loci was accomplished with an LSM510 laser scanning confocal microscope (Zeiss Instruments) and allowed measurements of intracellular Ca2+ transients in individual myocytes during the experiment. Data were collected during spontaneous beating and external stimulation (350ms). Image J was used to visualize the beating profile.
Methylation and Pyrosequencing Verification
Methylation studies were performed as previously described [17]. PCR reactions were carried out using the Hotstart Taq polymerase kit (Qiagen) in 25 μL total volume and with 50 pm of forward primer and reverse primer. For each PCR reaction, 50 ng of the bisulfite converted DNA in 1 μL was used as a template. After 5 min of initial denaturation at 95°C, the cycling conditions of 44 cycles consisted of denaturation at 95°C for 15 s, annealing at 65°C for 30 s (NKX2.5, T and GATA4,) and 60°C for 30 s (TBX5) and elongation at 72°C for 45 s. The PCR products were stored at 4°C until ready for pyrosequencing. Pyrosequencing was performed using the PyroMark MD Pyrosequencing System (Biotage) as described previously. In brief, the PCR product was bound onto streptavidin-Sepharose HP beads (GE Healthcare). Beads containing the immobilized PCR product were denatured using a 0.2M NaOH solution and neutralized. Pyrosequencing primer at a concentration of 0.3 μM was annealed to the purified single-stranded PCR product at 28°C. Methylation quantification was performed using the manufacturer-provided software. The primers used in the PCR runs and pyrosequencing reactions are shown in supplementary Table 1.
Statistical analysis
Two-way ANOVA followed by a Bonferroni post-hoc test was used to analyze the data. P values of < 0.05 were used to assign significance.
Results
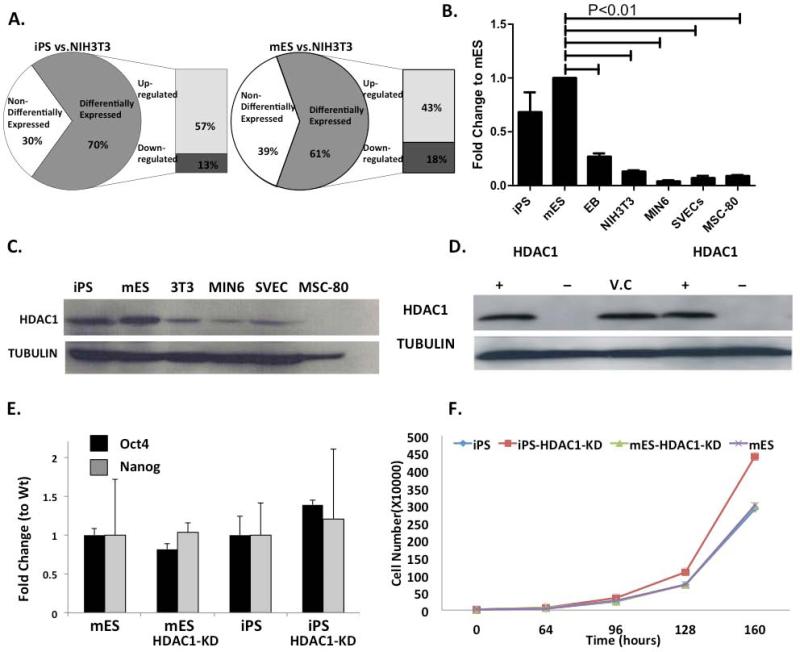
In order to identify key chromatin modifying factors and enzymes in mES and iPS cells, we performed quantitative Real Time PCR (Q-RT-PCR) array based analysis of the expression patterns of 172 chromatin modifying enzymes and factors in mES, iPS and NIH3T3 cells (Fig. 1A). ES and iPS cells had very similar expression profiles with less than 15% of genes showing significant difference between these two cell types. Additionally, the few genes that did show significant difference only showed a 2-3 fold difference. When comparing the two-pluripotent cell types to differentiated cells, the majority of epigenetic enzymes and factors, that were differentially expressed, were up-regulated in pluripotent cells indicating more dynamic chromatin modifications in these cells (Fig. 1A). We assessed the expression levels of several HDACs from all different HDAC classes. Only HDAC1 and to a lower extent HDAC2, were highly expressed in pluripotent cells. Independent real time PCR analysis verified this data (Fig.1B, Sup.Fig.1A-E). Based on this array data, we identified HDAC1 as one of the enzymes expressed at high levels in pluripotent cells. Independent quantitative RT-PCR and immunoblots confirmed that HDAC1 is expressed at high levels in pluripotent cells and the expression levels significantly go down in somatic cells representative of the three germ layers (Fig.1B, C).
Figure 1. HDAC1 is highly expressed in pluripotent cells and its expression decreases as cells differentiate.
A. Quantitative Real-Time PCR array-based expression pattern of 86 epigenetic modifying factors. B. Quantitative Real time PCR using Hdac1 primer/probe in iPS (induced pluripotent cells), mES (murine embryonic stem cells), SVEC (mesodermal lineage; endothelial cells), MIN6 (endodermal lineage; pancreatic beta cells), MSC-80 (ectodermal lineage; Schwann cells), NIH3T3 (fibroblasts) and Embryoid Body (day 4) cells (n=3). C.Western Blot for HDAC1 protein expression in iPS (induced pluripotent cells), mES (murine embryonic stem cells), SVEC (endothelial cells), MIN6 (pancreatic beta cells), MSC-80 (Schwann cells) and NIH3T3 (fibroblasts). D. Levels of HDAC1 protein in iPS wt (+), iPS HDAC1 knock down (-), Vector Control (V.C), mES wt (+) and mES HDAC1 KD (-) cells as measured by Western Blot. E. Quantitative RT-PCR of expression levels of Oct4 and Nanog mRNA in mES, iPS and respective HDAC1 knock down cells during their pluripotent state and normal self-renewing conditions. F. Cell proliferation rates for mES, iPS and their respective HDAC1 knock down cells.
This expression indicated a key role for HDAC1 in the pluripotency and differentiation plasticity of iPS cells. Additionally, it provided a model to study if slight differences at the chromatin levels in iPS cells would result in a biological deficiency of these cells to differentiate. To elucidate the role of HDAC1 in pluripotent cell differentiation, particularly into cardiomyocyte differentiation, we created shRNA-mediated stable HDAC1-knock down (HDAC1-KD) cell lines in both ES and iPS cells (Fig. 1D). To test whether there was a compensatory elevated expression of other HDACs when HDAC1 is knocked down, we analyzed RNA expression levels of HDAC2 (another class I HDAC) and HDAC5 (a class II HDAC). In both cell types, we did not observe any significant increase in the expression levels of these HDACs (Sup.Fig. 2A).
Interestingly, loss of HDAC1 in either iPS or ES cells did not alter the expression levels of two pluripotency associated genes (Oct4 and Nanog) under basal undifferentiated and self-renewal conditions (Fig. 1E). Additionally, cell proliferation (Fig. 1F) and cell division parameters did not significantly change between wild type and HDAC1-KD cells, under basal, undifferentiated conditions (Sup. Fig.2B-C). Even though iPS cells showed slightly different cell numbers as proliferation progresses, this difference was not statistically different (fig. 1F).
HDAC1 has been widely studied due to its implication in many disorders and has been shown to be important during development [18,19]. HDAC1 knockout mice are embryonic lethal at day 9 post fertilization. We next investigated a possible impact of HDAC1 deficiency in cardiovascular cell lineage differentiation of ES and iPS cells.
The embryonic lethality in HDAC1 KO mice and the observed high expression of this enzyme in iPS and ES cells indicated that HDAC1 potentially plays a role in the early stages of differentiation. HDAC1 is a histone deacetylase and as such is involved in silencing gene expression. Master regulators of pluripotency, such as OCT4, SOX2 and NANOG, maintain pluripotency by binding to activating regions of promoters of genes important in maintaining pluripotency, autologous regulation, and associate with complexes which keep developmental genes repressed [20,21,22]. Because lack of HDAC1 during early differentiation could result in persistent high levels of these pluripotency master regulators through lack of deacetylation at the regulatory regions of these genes, we hypothesized that HDAC1 could be involved in silencing pluripotency associated genes as the cells are induced to differentiate. In order to test this hypothesis, we induced differentiation through Embryoid Body (EB) formation in both sets of pluripotent cells. As differentiation progressed we analyzed changes in expression levels of pluripotency associated genes.
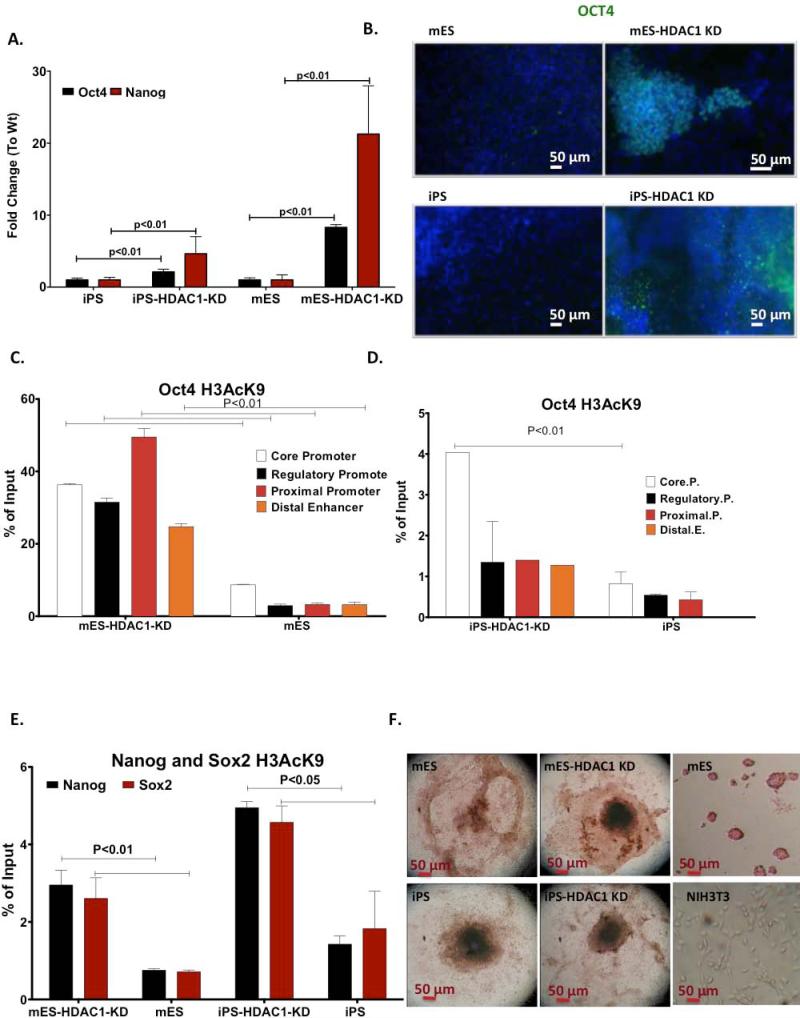
One of the first steps in the differentiation process is the silencing of pluripotency-associated genes. While levels of these genes vary slightly under self- renewal conditions (Fig.1E), we observed that upon the induction of differentiation expression levels of Oct4, Sox2 and Nanog dramatically decreased in wild type iPS and ES cells (Fig.2A-B). However, in HDAC1-KD cells, persistent high levels of these pluripotency-associated genes were observed (Fig. 2A-B). Since lack of HDAC1 could affect deacetylation of pluripotency associated genes, we checked acetylation levels at regulatory regions of these genes to test whether the persistent high levels of expression were due to failure of these regulatory regions of to get deacetylated by HDAC1. We analyzed the extent of acetylation of Histone H3 at lysine 9 (H3AcK9) in day 6 differentiating EBs of wt and HDAC1-KD cells by chromatin immune-precipitation (ChIP). Because expression levels of pluripotency-associated genes dramatically decrease in wt cells after differentiation has been induced, we chose day 6 of differentiation so as to be able to directly compare acetylation levels at these promoters between wt and HDAC1-KD cells. At later days of differentiation, acetylation levels of pluripotency associated genes in wt cells are undetectable. As expected, acetylation levels of all 4 regulatory regions analyzed for Oct4 were very high in HDAC1-KD cells (Fig. 2C,D). Based on expression data (Fig.1A) acetylation levels of these genes would be expected to stay high as differentiation progresses. Rather interestingly, acetylation of these regions in the iPS-HDAC1-KD cells was lower than in mES-HDAC1-KD cells (Fig.2D). Acetylation levels of Nanog and Sox2 promoter regions were also higher in mES and iPS HDAC1-KD cells compared to wt cells (Fig.2E). Differences in acetylation levels between wt iPS and ES cells were insignificant. Acetylation levels at promoter regions of Nanog and Sox2 follow the same pattern as with Oct4. However, iPS-HDAC1 KD cells show lower levels of acetylation at these promoters compared to ES-HDAC1 KD cells. This data suggests that HDAC1 plays a crucial role in deacetylating regulatory regions of pluripotency-associated genes during differentiation, resulting in their repression. We further assessed a direct physical association association of HDAC1 with OCT4 in both mES and iPS cells in their pluripotent, undifferentiated state (Sup. Fig.2F). HDAC1 is known to associate with two complexes, the NuRD and the NODE complex [23,24]. This change is likely to occur through association with the NuRD complex, rather than with the NODE complex since key members of the latter would not be present during differentiation [23,24]. The lack of HDAC1 leads to the deregulated suppression of pluripotency-associated genes thereby inhibiting mES and iPS cell differentiation.
Figure 2. HDAC1 is required for deacetylattion and turning off of pluripotency associated genes during differentiation.
A. Quantitative RT-PCR showing Oct4 and Nanog mRNA levels in Wt and HDAC1-KD mES and iPS cells at 6-8 days of differentiation. B. Immunofluorescence staining for OCT4 (green) in EBs derived from Wt mES, iPS and their respective HDAC1-KD cells. Image was taken close to the periphery of the EB at day 6 of differentiation. Nuclie were counterstained with dapi (blue). C. Chromatin immunoprecipation of H3AcK9 levels at different regulatory regions of Oct4 at day 6 of differentiation in Wt and HDAC1-KD mES cells. D. ChIP results of H3AcK9 levels at different regulatory regions of Oct4 at day 6 of differentiation in Wt and HDAC1-KD iPS cells. E. ChIP results of H3AcK9 levels at the promoter of Sox2 and Nanog at day 6 of differentiation in Wt and HDAC1-KD mES and iPS cells. F. Alkaline Phosphatase staining of Wt iPS and mES cells (controls) and HDAC1-KD iPS and mES cells at day 8 of differentiation. Staining of pluripotent mES cells and NIH3T3 cells serve as positive and negative staining controls, respectively. Images taken at 2.5X magnification.
Next we investigated whether this could lead to a higher differentiating potential of iPS cells even when HDAC1 had been knocked down. In the first stages of differentiation we observed EBs from cells in which HDAC1 had been knocked down failed to expand and grow compared to their respective Wt counterparts (Sup. Fig. 2D). In order to better visualize differentiation within the EB, we stained for Alkaline Phosphatase, an enzyme expressed in pluripotent cells. As an EB expands and differentiates cells in the periphery are more differentiated than cells in the core of the EB. As EBs derived from ES and iPS cells grew and differentiated they lost expression of Alkaline Phosphatase, an enzyme expressed in pluripotent cells. We observed higher expression, even in the periphery of ES cells when compared to their respective Wt cells (Fig.2F). However, iPS cells in which HDAC1 had been knocked down showed a pattern of Alkaline Phosphatase loss similar to their respective wt. This indicated a retention of limited differentiation ability in iPS cells in which HDAC1 had been knocked down compared to ES-HDAC1 KD cells.
Pluripotent ES and iPS cells are a promising source for potential therapeutic applications for regenerative medicine including cardiovascular repair and regeneration. In order to better understand the differentiation ability of iPS HDAC1-KD cells and how that compared to ES HDAC1-KD cells, we looked at iPS-HDAC1 KD cells potential to differentiate specifically into fully functional cardiomyocytes. Lack of HDAC1 also reduced the expression of early endodermal and to some extent ectodermal markers (Sup.Fig. 2E), however we were most interested in the effect of HDAC1 on cardiovascular differentiation, partly due to inconsistencies in the current knowledge about the role of HDAC1 in cardiovascular differentiation. HDAC1-KO mice are embryonic lethal with defects in heart formation but cardiac specific HDAC1 knock-out mice (under myosine heavy chain promoter :- a late marker of cardiac differentiation) do not present any overt cardiac phenotype although double HDAC1/HDAC2 cardiac specific KO mice did show arrhythmias, shortly after birth. Thus, we investigated the role of HDAC1 in the differentiation of iPS-HDAC1 KD cells into fully functional cardiomyocytes and how their differentiation compared to that of ES-HDAC1-KD cells.
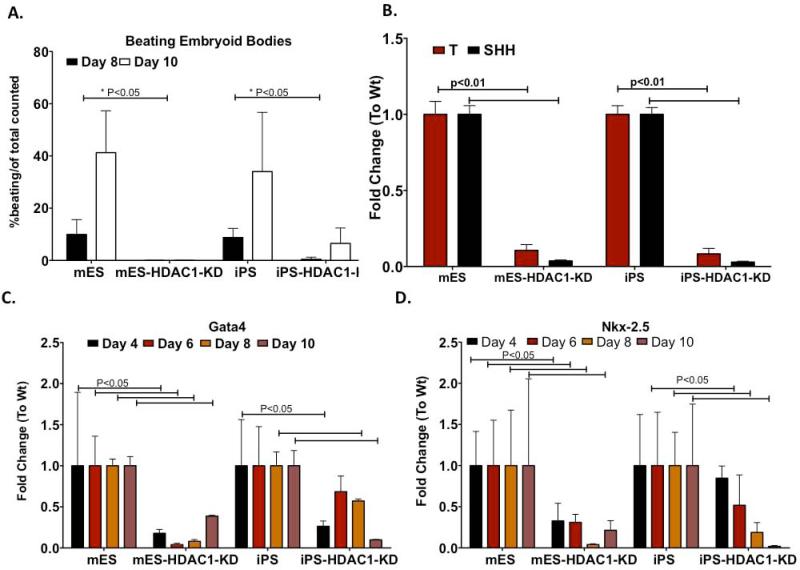
We determined the comparative effects of HDAC1-silencing on cardiomyocyte differentiation in both mES-HDAC1-KD and iPS-HDAC1-KD cells. As EBs differentiates, differentiated cardiomyocytes show spontaneous beating. Although wt mES and iPS cells differentiated similarly and showed similar kinetics for spontaneous beating, their HDAC1-KD counterparts displayed either complete loss or significantly reduced and delayed beating loci (Fig.3A). While about 30% of EBs generated from wt mES and iPS show loci of spontaneous beating, none of the EBs generated from ES-HDAC1-KD showed any spontaneous beating (Fig.3A, Sup. Videos1-2). However, some iPSHDAC1-KD cells did spontaneously beat, albeit the beating was delayed and significantly reduced when compared to wt iPS cells (Sup. Videos 1-2). This data shows that some iPS cells, even under very low/absent HDAC1 levels are able to differentiate into spontaneously beating cardimyocytes.
Figure 3. HDAC1-KD cells show reduced ability to differentiate into cardiovascular cells.
A. Percentage of beating EBs over total number of EBs counted, in mES, iPS, mES-HDAC1-KD and iPS-HDAC1-KD derived EBs. (n=3, total EB counted ~500). B. Quantitative RT-PCR showing expression levels of early mesodermal markers on day 6 of differentiation in Wt mES and iPS cells and the respective HDAC1-KD cells. Data shown reflects average expression levels of three independent differentiation experiments. C. Quantitative RT-PCR showing mRNA expression levels of cardiomyocyte- specific transcription factor, Gata4, in wt mES and iPS cells and the respective HDAC1-KD cells at various days during differentiation. Data shown is representative of three independent differentiation experiments. D. Quantitative RTPCR showing mRNA expression of cardiomyocyte-specific transcription factor Nkx2.5 at various times during differentiation. Data shown is representative of three independent differentiation experiments.
We wanted to investigate whether iPS-HDAC1 KD cells, unlike their mES counterpart, retained the ability to differentiate into fully functional cardiomyocytes and express cardiomyocyte specific markers. Expression of early mesodermal genes was significantly lower in both iPS and mES cells in which HDAC1 has been knocked down compared to the respective wild type (Fig. 3B). Expression levels of key cardiomyocyte markers was consistently lower in HDAC1-KD cells and the lack of expression of key markers was more pronounced in mES-HDAC1-KD cells (Fig 3C,D).
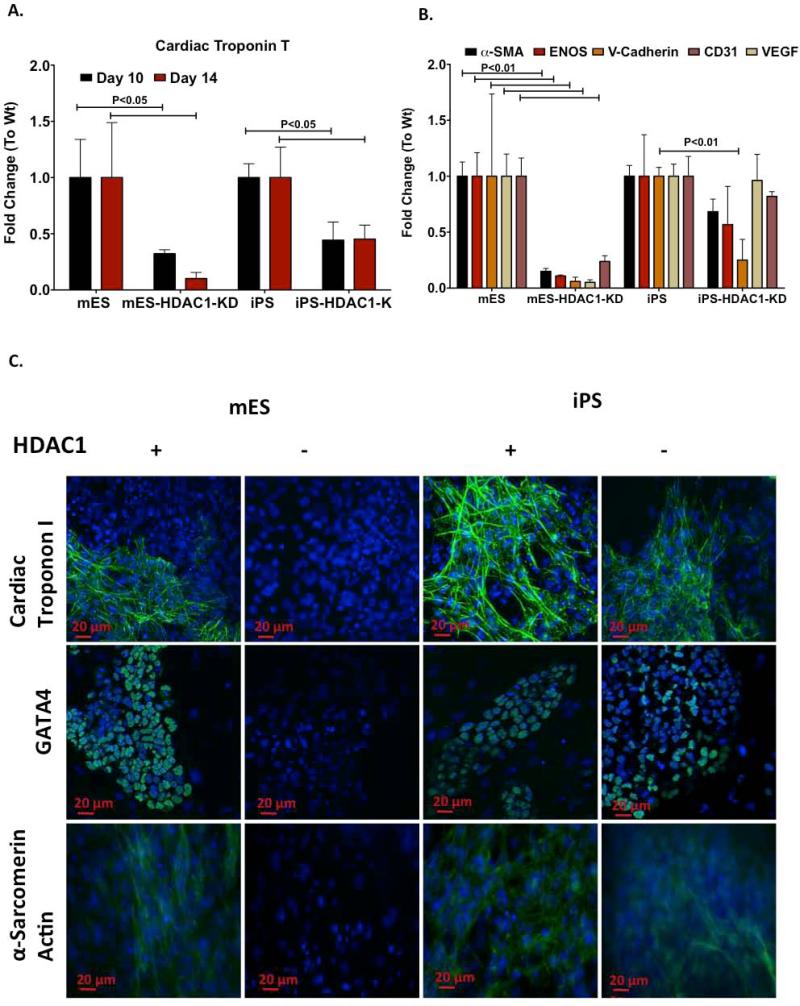
Expression of mature cardiomyocyte markers such as Cardiac Troponin T (CTT), and other cardiovascular specific proteins was also higher in iPS-HDAC1-KD cells than in mES-HDAC1-KD cells (Fig. 4A,B). Additionally, while mES-HDAC1-KD cells showed no expression of mature cardiomyocyte proteins essential for spontaneous contractility such as CTT and α-Sarcomeric Actinin, few of the iPS-HDAC1-KD cells derived cardiomyocytes did, (Fig.4C). In iPS-HDAC1-KD cells, fewer EBs developed spontaneously beating loci, thus global expression of cardiomyocyte specific proteins was lower than in iPS-HDAC1-KD derived EBs. However, expression of these proteins within beating loci in iPS-HDAC1-KD cell derived EBs is comparable to that of wt iPS cell derived EBs (Fig. 4C). This data indicates that while ES cells lose their ability to differentiate into cardiomyocytes or other cardiovascular lineages when HDAC1 is knocked down, iPS cells, to a certain extent, retain the ability to differentiate under the same conditions and cope better with the drastically reduced levels of HDAC1 during differentiation.
Figure 4. HDAC1-KD cells show reduced ability to express mature, cardiovascular specific markers.
A. Levels of Cardiac Troponin T mRNA as differentiation progresses in wt mES and iPS cells and their respective HDAC1-KD cells as assesed by Quantitative RT-PCR. B. Quantitative RT-PCR showing mRNA expression levels of cardiovascular lineage specific genes at day 12 of differentiation in wt- mES and iPS cells and their respective HDAC1-KD cells. C. Immunofluorescence photomicrograph depicting protein expression of mature cardiomyocyte markers-Cardiac Troponin I (CTI), GATA4 and alpha-Sarcomeric Actin at day 12 of differentiation in Wt mES and iPS cells and their respective HDAC1-KD cells.
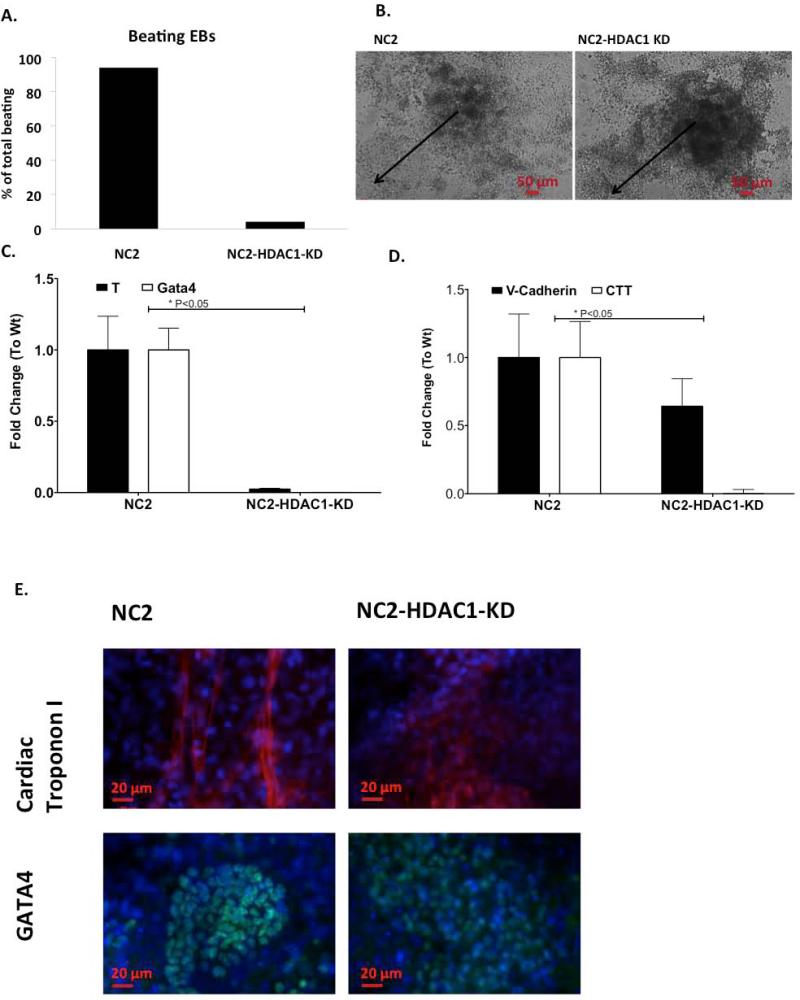
In order to investigate whether the observed effect and difference in behavior between iPS and ES cells under reduced levels of HDAC1 was specific to our iPS cell type, we repeated these experiments in an independently generated iPS cell line, NC2. We tested whether NC2 would also show restricted cardiovascular differentiation with the loss of HDAC1 compared to mES-HDAC1-KD cells, which consistently show very little differentiation. Similarly to the initial iPS cell line we analyzed, NC2 (iPS)-HDAC1-KD cells also showed delayed and reduced beating and repressed differentiation when compared to the Wt cells, and unlike mES-HDAC1-KD cells, NC2 (iPS)-HDAC1-KD cells show some beating and differentiation (Fig. 5A-D). This is more apparent at the protein level, where the few beating loci within EBs derived from NC2 (iPS)-HDAC1-KD cells show robust expression of key cardiomyocyte proteins (Fig. 5E) as opposed to mES-HDAC1-KD cells which show none at the protein level (Fig.4C).
Figure 5. Knockdown of HDAC1 has similar phenotype in an independently generated iPS cell line (NC2).
A. Spontaneous beating EBs/total EBs in NC2 and NC2 HDAC1 KD cells. B. Light microscopy images showing differentiation in EBs derived from NC2-iPS cells and NC2-iPS cells lacking HDAC1 at day 6 of differentiation. Black arrows indicate distance from the center of the EB to the periphery. C. Quantitative RTPCR showing expression levels of cardiovascular genes brachyury (T) and Gata4 and D. V-Cadherin and Cardiac Troponin T in NC2 and NC2- HDAC1-KD cells. Data shown reflects expression levels of three independent differentiation experiments. E. Immunofluorescence showing protein expression levels of Cardiac Troponin I (CTI) and Gata4 at day 12 of differentiation in Wt NC2-iPS cells and their respective HDAC1-KD cells.
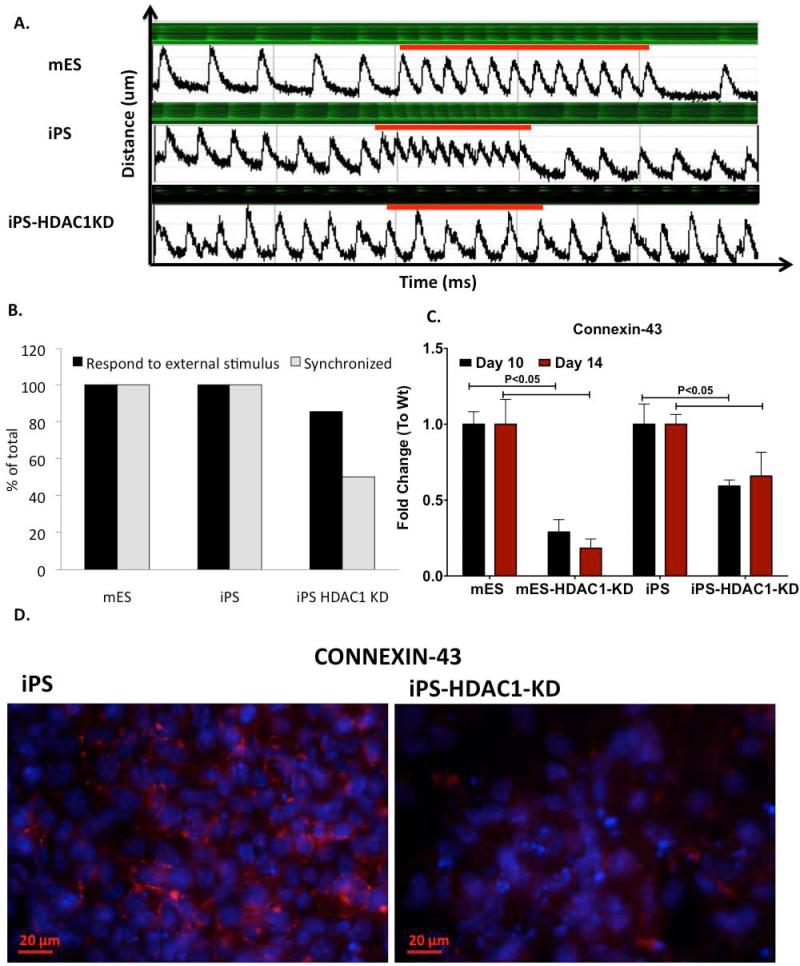
Since we observed delayed and reduced cardiomyocyte differentiation in iPSHDAC1-KD cells, we investigated whether the few iPS-HDAC1-KD cells that showed some beating were physiologically competent and had the ability to become fully functional cardiomyocytes. To test this we monitored calcium handling of the cells during beating in real time. Fully mature cardiomyocytes possess the ability to beat in synchrony with adjacent cells and beat at the rate determined by the pacemakers. Thus, the beating colonies were analyzed for these two crucial characteristics of mature cardiomyocytes: i) The ability to beat in synchrony and ii) the ability to respond to external stimuli. All wt mES and iPS cell derived beating cardiomyocytes analyzed had a synchronized intrinsic rate, responded very well to external stimuli (electric pulse at 350 ms) and recovered back to the initial intrinsic rate when the stimulus ceased (Fig.6A; Sup. Videos 3-4). Some iPS-HDAC1-KD derived beating EBs showed aberrant and non-synchronous calcium handling and did not respond to external stimuli (Fig.6A, B; S. videos 5, 6). While all analyzed beating loci derived from wt iPS cells showed 100% synchronization and response to the external stimulus, about 80% of beating loci derived from iPS-HDAC1-KD cells responded to the external stimulus and only 50% beat in synchrony (Fig.6B). We used electric stimulation to induce beating in the ESHDAC1-KD derived EBs, however, after several exposures these EBs did not show contraction. To investigate whether the reason for the poor synchronization and calcium handling in iPS-HDAC1-KD derived EBs is due to aberrant expression or localization of gap junction proteins, we analyzed the protein expression level and pattern of Connexin-43 (CX-43) in beating loci derived from these cells. In the wt cells, CX-43 is both highly expressed and adequately organized in the periphery of the cells (Fig.6C-D). In iPS cells in which HDAC1 had been knocked down expression of CX-43 was substantially reduced and disorganized (Fig.6C-D). This data suggests that while some iPS cells are able to overcome the need for HDAC1 in the first early stages of differentiation, HDAC1 is important for these cells to fully mature and maintain a cardiomyocyte phenotype.
Figure 6. HDAC1 is critical for the maintenance of a fully functional cardiomyocyte phenotype.
A. Calcium uptake and release experiments indicating synchrony and response to stimuli of beating EBs in mES, iPS and iPS-HDAC1-KD derived EBs B. Percentage of EBs that respond to external stimuli and are synchronized (example shown in A). C. Quantitative RT-PCR showing mRNA expression levels of Connexin-43 in mES-HDAC1-KD and iPS-HDAC1-KD EBs. Data shown reflects expression levels of three independent differentiation experiments. D. Organization of Connexin 43 in beating regions of EBs derived from Wt iPS and iPSHDAC1-KD cells.
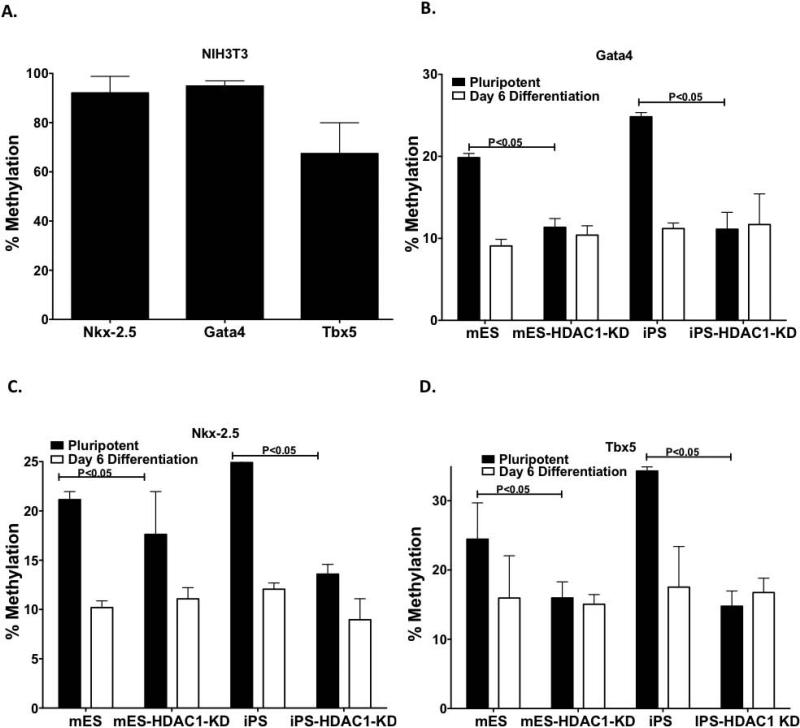
Clearly, iPS-HDAC1-KD cells coped better than mES-HDAC1-KD cells in terms of cardiomyocyte differentiation. In an attempt to explain this differential characteristic of iPS and mES cells, we analyzed if iPS cells had an epigenetic memory which allowed them to cope better with the lack of HDAC1 during differentiation. Because our iPS cells used in these experiments are derived from fibroblasts (of mesodermal origin), we tested promoter regions of 4 different mesodermal and cardiovascular genes to determine any differences in methylation patterns between mES, iPS and their respective HDAC1 KD counterparts, before and during differentiation. In NIH3T3 cells, as expected, these promoters were highly methylated (Fig.7A). However, the methylation pattern of these promoters in the four pluripotent cell types prior to and during differentiation was interesting. Unlike a recent report [14] we did not see any differences in the methylation pattern at any of these promoters when comparing wt iPS and mES cells (Fig.7B-D). Even during the undifferentiated state methylation levels of the promoters of all the genes analyzed were lower in HDAC1-KD cells compared to wt cells (Fig.7B-D). In the wt cells methylation levels of these genes went down as the cells differentiated, whereas HDAC1-KD cells maintained similar levels of methylation with very little change between pluripotent and differentiated states. While there was no expression of these genes in the pluripotent state of these cells, the low methylation levels indicate a cross talk between the histone acetylation and DNA methylation at these promoters during early differentiation.
Figure 7. HDAC1 affects DNA methylation levels of mesodermal/cardiac genes.
A. Methylation levels at the promoter regions of Nkx 2.5, Gata4 and Tbx5 in NIH3T3 cells as assessed by pyrosequencing (n=3, 50 repeats each time). Methylation levels of B. Gata4, C. NKX2.5, and D. Tbx5 in mES, iPS and the respective HDAC1-KD cells in their pluripotent state and at day 6 of differentiation.
Discussion
In summary, our data indicates that loss of HDAC1 in mES and iPS cells inhibits their ability to differentiate by suppressing the histone deacetylation of promoters of pluripotency associated genes, therefore resulting in their sustained expression and as a consequence repressed lineage specific differentiation. While mES cells show no differentiation, iPS cells show some ability to differentiate even when HDAC1 has been knocked down. Other reports have indicated a key role for HDACs in ES cell differentiation through global inhibition of HDACs using Trichostatin A (TSA)[22,25]. Although these reports have greatly extended the body of knowledge around histone deacetylases and differentiation, they were not designed to recognize and determine crucial differences between different members of the HDAC family. ES cells have very dynamic chromatin maintenance and modification machinery. Treatment with TSA, which inhibits all class I and II HDACs, results in more acetylated histones and thus a globally more active transcription state, which can result in both inhibition and promotion of differentiation of ES cells, depending on the time of the treatment. This may explain contradicting reports on the role of HDACs in inhibiting differentiation or in promoting differentiation, as both could be possible through different HDACs or different complexes they associate with [18,19,22,25,26]. Our data using HDAC1-KD pluripotent cells suggests that HDAC1 is specifically important in the early differentiation as it is required to deacetylate puripotency-associated genes when differentiation is induced. The prolonged expression of these pluripotent genes during differentiation results in delayed/absent expression of early differentiation genes.
Lack of HDAC1 during differentiation results in reduced cardiovascular differentiation and decreased or absent spontaneous contraction in differentiating ES and iPS cells. In accordance with recent reports, we saw a difference in differentiation ability between ES and iPS cells [14]. While mES-HDAC1-KD cells do not show any spontaneous beating during differentiation, iPS-HDAC1-KD cells do. Some of these cells show expression of cardiovascular markers and some differentiation into cardiomoycytes. However those cells that do differentiate into cardiomyocytes show partial absence of synchrony and do not always respond to external stimuli. This indicates a role for HDAC1 in the maturation to a fully functional cardiomyocyte phenotype by regulating expression of Gap junction protein Connexin-43 in iPS cells. Thus, even though these cells are able to differentiate when HDAC1 had been knocked down, they are unable to maintain a functional cardiomyocyte phenotype in its absence.
Recent reports have also shown differences in the methylation pattern of different gene regions between iPS cells and mES cells [14]. When we compared the methylation pattern of specific cardiovascular promoters (short regions in CpG islands close to promoters) we observed no difference between our iPS and mES cells. We did however see a difference in the methylation of cardiomyocyte genes in HDAC1-KD cells both before and during differentiation.
The process of repressing or expressing a gene involves histone modifications, DNA methylation and expression of various transcription factors and enhancers, which not only act in synchrony but also interact and cross talk to each other. HDAC1 affects histone acetylation and indirectly DNA methylation. Our observations indicate that analysis of epigenetic molecular mechanisms important in maintaining pluripotency is crucial to our understanding of what makes pluripotent cells pluripotent and what governs their differentiation. This body of knowledge, in the future, could lead to the development of a better translational strategy for the use of these powerful cells in regenerative medicine, including post-injury cardiovascular repair and regeneration.
Conclusion
ES and iPS cells carry great potential for therapeutic use. The epigenetic of these molecules however is not fully understood. We showed that HDAC1, a key chromatin modifying enzyme, is important in deacetylating pluripotency-associated genes during differentiation in both these cell types. We also show that this molecule plays non-redundant role during pluripotent cell-derived cardiomyocyte differentiation and maturation. Unlike ES-HDAC1-KD cells which do not show any cardiomyocyte differentiation ability, iPS-HDAC1 KD cells retain some ability to differentiate, albeit the derived cardiomyocytes are electro-physiologically incompetent. These data expand our knowledge of the chromatin modifications involved in the differentiation of ES and iPS cells as well as elucidate differences in differentiation plasticity that are observed when changes at the epigenetic level exist.
Supplementary Material
Supplementary Figure 1: Differential expression of Class 1-IV HDACs during pluripotency and differentiation. A. mRNA expression levels of class I HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. B. mRNA expression levels of class IIA HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. C. mRNA expression levels of class IIB HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. D. mRNA expression levels of class III HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. E. mRNA expression levels of class IV HDACs in pluripotent mES cells and differentiating cells at different days of differentiation.
Supplementary Figure 2: Loss of HDAC1 does not affect the expression levels of other HDACs but reduces the expression of endodermal and ectodermal lineage genes. A. Levels of HDAC2 and HDAC5 mRNA in iPS and mES cells and their respective HDAC1-KD cells. B. Number of divisions as time progresses for mES, iPS and respective HDAC1 knock down cells. C. Division time mES, iPS and respective HDAC1 knock down cells. D. Light microscopy images showing lack of differentiation in EBs derived from mES and iPS cells lacking HDAC1 at day 6 of differentiation. Black arrows indicates distance from the center of the EB to the periphery. E. Quantitative RTPCR showing mRNA expression levels of early endodermal and ectodermal genes in wt mES and iPS cells and respective HDAC1-KD cells. F. Western Blot for OCT4 following immunoprecipitation with HDAC1 showing physical association of OCT4 with HDAC1 in mES and iPS cells.
S.V1 Embryoid Bodies derived from iPS-HDAC1-KD cells show delayed/reduced beating. EBs derived from: SV1. iPS cells; SV2. iPS-HDAC1-KD cells.
S.V2. Embryoid Bodies derived from iPS-HDAC1-KD cells show delayed/reduced beating. EBs derived from: SV1. iPS cells; SV2. iPS-HDAC1-KD cells.
S.V3 Cells derived from iPS cells lacking HDAC1 are not synchronized and do not respond to external stimuli. EBs derived from: SV5. iPS cells and SV6. iPS-HDAC1-KD cells. Calcium uptake and release (flashes of red) were recorded during spontaneous contraction followed by stimulation at 350ms and then ceasing of external stimulus.
S.V4. Cells derived from iPS cells lacking HDAC1 are not synchronized and do not respond to external stimuli. EBs derived from: SV5. iPS cells and SV6. iPS-HDAC1-KD cells. Calcium uptake and release (flashes of red) were recorded during spontaneous contraction followed by stimulation at 350ms and then ceasing of external stimulus.
Supplementary Table 1. Sequences of primers used for pyrosequencing.
Acknowledgments
Sources of Funding
Work described in this manuscript was supported in part by National Institute of Health grants HL091983, HL105597, HL095874, HL053354 and HL108795 to R.K. and American Heart Association's pre-doctoral fellowship grant 11PRE7360065 to E.H.
Footnotes
Author Contribution:
Eneda Hoxha: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing.
Erin M. Lambers: manuscript writing
Veronica Ramirez: conception and design
Hehuang Xie: collection and/or assembly of data
Alexandre De Andrade: collection and/or assembly of data
Melissa A. Thal: conception and design
Suresh K. Verma: conception and design
Prasanna Krishnamurthy: conception and design
Marcelo B. Soares: conception and design
John A. Wasserstrom: collection and/or assembly of data
Raj Kishore: conception and design, final editing and approval of manuscript
Disclosures
The authors have nothing to disclose.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Rolletschek A, Wobus AM. Induced human pluripotent stem cells: promises and open questions. Biological Chemistry. 2009;390(9):845–849. doi: 10.1515/BC.2009.103. [DOI] [PubMed] [Google Scholar]
- 4.Stefanovic S, Abboud N, Desilets S, Nury D, Cowan C, Puceat M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. The Journal of Cell Biology. 2009;186(5):665–673. doi: 10.1083/jcb.200901040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 6.Xi S, Geiman TM, Briones V, Guang Tao Y, Xu H, Muegge K. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells. 2009;27(11):2691–2702. doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008 Jun 13;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(18):8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 11.Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28(11):1981–1991. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- 12.Karantzali E, Schulz H, Hummel O, Hubner N, Hatzopoulos A, Kretsovali A. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: transcriptomic and epigenetic analysis. Genome Biology. 2008;9(4):R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajasingh J, Lambers E, Hamada H, et al. Cell-free embryonic stem cell extract-mediated derivation of multipotent stem cells from NIH3T3 fibroblasts for functional and anatomical ischemic tissue repair. Circulation Research. 2008;102(11):e107–117. doi: 10.1161/CIRCRESAHA.108.176115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasserstrom JA, Sharma R, Kapur S, et al. Multiple defects in intracellular calcium cycling in whole failing rat heart. Circulation. 2009;2(3):223–232. doi: 10.1161/CIRCHEARTFAILURE.108.811539. [DOI] [PubMed] [Google Scholar]
- 17.Xie H, Wang M, Bonaldo Mde F, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Research. 2009;37(13):4331–4340. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Developmental Biology. 2008;319(1):110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & Development. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature Cell Biology. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nature reviews. Molecular Cell Biology. 2001;2(6):422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular cell. 1998 Dec;2(6):851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 24.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008 Jun;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 25.Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38(1):32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 26.Lagger S, Meunier D, Mikula M, et al. Crucial function of histone deacetylase 1 for differentiation of teratomas in mice and humans. The EMBO Journal. 2010;29(23):3992–4007. doi: 10.1038/emboj.2010.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Differential expression of Class 1-IV HDACs during pluripotency and differentiation. A. mRNA expression levels of class I HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. B. mRNA expression levels of class IIA HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. C. mRNA expression levels of class IIB HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. D. mRNA expression levels of class III HDACs in pluripotent mES cells and differentiating cells at various days of differentiation. E. mRNA expression levels of class IV HDACs in pluripotent mES cells and differentiating cells at different days of differentiation.
Supplementary Figure 2: Loss of HDAC1 does not affect the expression levels of other HDACs but reduces the expression of endodermal and ectodermal lineage genes. A. Levels of HDAC2 and HDAC5 mRNA in iPS and mES cells and their respective HDAC1-KD cells. B. Number of divisions as time progresses for mES, iPS and respective HDAC1 knock down cells. C. Division time mES, iPS and respective HDAC1 knock down cells. D. Light microscopy images showing lack of differentiation in EBs derived from mES and iPS cells lacking HDAC1 at day 6 of differentiation. Black arrows indicates distance from the center of the EB to the periphery. E. Quantitative RTPCR showing mRNA expression levels of early endodermal and ectodermal genes in wt mES and iPS cells and respective HDAC1-KD cells. F. Western Blot for OCT4 following immunoprecipitation with HDAC1 showing physical association of OCT4 with HDAC1 in mES and iPS cells.
S.V1 Embryoid Bodies derived from iPS-HDAC1-KD cells show delayed/reduced beating. EBs derived from: SV1. iPS cells; SV2. iPS-HDAC1-KD cells.
S.V2. Embryoid Bodies derived from iPS-HDAC1-KD cells show delayed/reduced beating. EBs derived from: SV1. iPS cells; SV2. iPS-HDAC1-KD cells.
S.V3 Cells derived from iPS cells lacking HDAC1 are not synchronized and do not respond to external stimuli. EBs derived from: SV5. iPS cells and SV6. iPS-HDAC1-KD cells. Calcium uptake and release (flashes of red) were recorded during spontaneous contraction followed by stimulation at 350ms and then ceasing of external stimulus.
S.V4. Cells derived from iPS cells lacking HDAC1 are not synchronized and do not respond to external stimuli. EBs derived from: SV5. iPS cells and SV6. iPS-HDAC1-KD cells. Calcium uptake and release (flashes of red) were recorded during spontaneous contraction followed by stimulation at 350ms and then ceasing of external stimulus.
Supplementary Table 1. Sequences of primers used for pyrosequencing.