RNA-binding proteins that target mRNA coding regions are emerging as regulators of post-transcriptional processes in eukaryotes. Here the authors describe a newly identified RNA-binding protein, RBP42, which targets the coding region of mRNAs in the insect form of the African trypanosome, Trypanosoma brucei. RBP42 is an essential protein and associates with polysome-bound mRNAs in the cytoplasm. By use of HITS-CLIP, target RNA sequences were identified and quantified using high-throughput RNA sequencing. Analysis revealed that RBP42 bound mainly within the coding region of mRNAs that encode proteins involved in cellular energy metabolism. It is therefore possible that RBP42 plays a critical role in modulating T. brucei energy metabolism.
Keywords: trypanosomes, mRNA translation, NTF-2 containing proteins, HITS-CLIP analysis
Abstract
RNA-binding proteins that target mRNA coding regions are emerging as regulators of post-transcriptional processes in eukaryotes. Here we describe a newly identified RNA-binding protein, RBP42, which targets the coding region of mRNAs in the insect form of the African trypanosome, Trypanosoma brucei. RBP42 is an essential protein and associates with polysome-bound mRNAs in the cytoplasm. A global survey of RBP42-bound mRNAs was performed by applying HITS-CLIP technology, which captures protein–RNA interactions in vivo using UV light. Specific RBP42–mRNA interactions, as well as mRNA interactions with a known RNA-binding protein, were purified using specific antibodies. Target RNA sequences were identified and quantified using high-throughput RNA sequencing. Analysis revealed that RBP42 bound mainly within the coding region of mRNAs that encode proteins involved in cellular energy metabolism. Although the mechanism of RBP42's function is unclear at present, we speculate that RBP42 plays a critical role in modulating T. brucei energy metabolism.
INTRODUCTION
African trypanosomes undergo a complex life cycle requiring morphological and metabolic changes controlled by their gene expression programs. In the absence of regulated transcription, trypanosome gene expression is regulated mainly at the post-transcriptional level, where environmental signals are likely integrated with mRNA maturation, localization, translation, and decay (Fernandez-Moya and Estevez 2010; Kramer and Carrington 2011). In trypanosomes, post-transcriptional reactions are likely extensive and intricately networked to account for the large number of transcripts and proteins that fluctuate in abundance during different stages of the parasite life-cycle (for review, see Siegel et al. 2011). It is unclear how post-transcriptional reactions drive differential gene expression and enable the parasite to respond to environmental changes in a cell with little, if any, differential transcription.
RNA-binding proteins interact with their target mRNAs, via cis-acting elements, during many post-transcriptional reactions that lead to regulated mRNA stability and translation. In trypanosomes, cis-acting elements have been identified mainly in 3′ UTRs of stage-specific or cell cycle–specific mRNAs (Fernandez-Moya and Estevez 2010; Kramer and Carrington 2011). Some of the most extensively studied stage-specific mRNAs in trypanosomes are the procyclin mRNAs (Acosta-Serrano et al. 2001). GPEET, EP1, and EP2 procyclins are the major surface glycoproteins encoded by the procyclic mRNAs; they cover the Trypanosoma brucei surface when the parasite resides in the midgut of its tsetse vector. The 3′ UTRs of EP1 and GPEET contain three stem–loop structures encoding several cis-acting elements that regulate mRNA translation and stability. Specifically, EP1 translation is increased when the RNA-binding protein TbZPF3 binds to two of the cis-acting elements in the 3′ UTR of EP1 (Walrad et al. 2009). Similarly, GFP reporter gene translation is increased when RNA-binding proteins ALB3 and ALB4 bind to a cis-acting 3′ UTR element in a chimeric mRNA containing the GFP open reading frame (ORF) flanked by the GPEET 3′ UTR (Mani et al. 2011). Intriguingly, in both cases the effect of the RNA-binding protein–3′ UTR interaction is modulated by coding region sequences within the target mRNAs. Additional RNA-binding proteins probably contribute to translational efficiency of procyclin. Furthermore, extremely little is known about the translational regulation of mRNAs that encode nonmembrane proteins (Kramer et al. 2010).
In addition to playing a role in translational regulation, RNA-binding proteins contribute to mRNA stability in trypanosomes. The best-characterized RNA-binding protein involved in mRNA stability regulation is T. brucei DRBD3, also called PTB1, which stabilizes a cohort of mRNAs by binding within their 3′ UTRs (Estevez 2008; Stern et al. 2009). In RNA decay experiments, depletion of DRBD3/PTB1 resulted in a reduced half-life of a subset of this RNA cohort. These alterations in mRNA stability may explain why DRBD3/PTB1 is essential for parasite viability. Interestingly, one study indicated that many of the DRBD3-modulated mRNAs encode cellular membrane proteins (Estevez 2008). This finding is consistent with a growing literature verifying post-transcriptional coregulation of related genes in yeasts, Drosophila, and mammalian cells (Keene 2007).
Here, we investigated an uncharacterized RNA-binding protein, named RBP42 (Gene identification Tb927.6.4440), to determine its role in trypanosome RNA metabolism. (RBP means RNA-binding protein, and 42 clarifies that RBP42 is the 42nd identified RNA-binding protein in trypanosomatids.) The RBP42 protein sequence suggests that RBP42 is a distant homolog of the metazoan RAS-GTPase activating protein SH3-binding protein, G3BP (Parker et al. 1996; Irvine et al. 2004). G3BP is a broadly functioning mRNA-binding protein known to integrate extracellular signals with intracellular metabolic responses. The intriguing possibility that RBP42 plays an important role in trypanosome mRNA metabolism inspired us to characterize its RNA targets. To accomplish this, we utilized a highly powerful technique, named HITS-CLIP, which traps protein–RNA interactions in vivo by UV crosslinking (Ule et al. 2003; Licatalosi et al. 2008). A global picture of RBP42–RNA interactions demonstrates that RBP42 binds within mRNA coding sequences. These interactions occur within a cohort of mRNAs that function in cellular energy metabolism.
RESULTS
RBP42 is similar to mammalian RNA-binding protein, G3BP
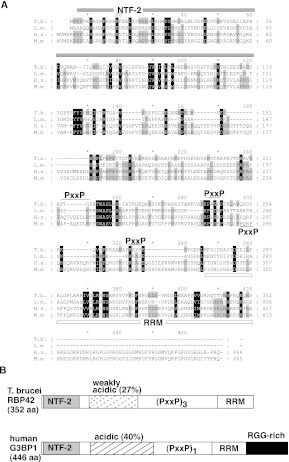
RBP42 was originally identified as a copurifying protein with RNA polymerase II subunit RPB4 in T. brucei procyclic parasites (Das et al. 2006). Protein sequence analysis revealed that RBP42 harbors two regions similar to known protein domains. The N-terminal 110 amino acids display homology with the NTF2-like protein domain (PROSITE domain PS50177), and the C-terminal 90 amino acids display homology with the RNA recognition motif (RRM) protein domain (PROSITE domain PS50102) (Fig. 1A,B; Bandziulis et al. 1989). NTF2-like domains are found in proteins with diverse activities but are usually involved in protein–protein interactions. RRM domains are the most common RNA-binding domains found in eukaryotic RNA-binding proteins.
FIGURE 1.
Alignment of T. brucei RBP42 and human G3BP1. (A) Multiple sequence alignments of two trypanosomatid RBP42s and two metazoa G3BP1s. Gene annotations are T. brucei (Tb927.6.4440), Leishmania major (LmjF.30.3090), Homo sapiens (G3BP1,Q13283, Gene identification 10146), and Mus musculus (P97855). The NTF-2 domain is overlined by a gray bar; the RNA recognition motif (RRM) domain is underlined by a white bar; and the three PxxP motifs in T. brucei are overlined with a black line. The single PxxP motif in the human and mouse genes is underlined. Numbering at the ends of lines identifies amino acid location for each organism. (B) Schematic comparison of T. brucei RBP42 and human G3BP1. RBP42 is 79% of the size of G3BP1 (drawing is approximately to scale). The NTF-2 domain (light gray), weakly acidic and acidic regions (dashed-stripe and striped), triple or single PxxP motifs (PxxP), RRM domain (white), and RRG-rich region (black) are shown. RBP42 and G3BP1 are similar but not identical; therefore, the trypanosome protein is named in accordance with the published trypanosomatid RNA-binding protein designation scheme (De Gaudenzi et al. 2005). Within the RRM, the RNP1 signature sequence motif (K/R) G (F/Y)(G/A)FVX(F/Y) is present in T. brucei RBP42 as KGYVFFDF (amino acids 311–319).
A search for proteins with a similar domain architecture revealed that RBP42 closely resembles the mammalian G3BP proteins and their fly homolog, Rasputin (Parker et al. 1996; Irvine et al. 2004). G3BP interacts with RAS GTPase-activating protein p120, a protein involved in the mammalian RAS signaling pathway. G3BP is essential for mouse development, and its expression is increased in many human cancer cell types (Barnes et al. 2002; Kim et al. 2007). Interestingly, G3BP is reported to have multiple activities related to cellular RNA metabolism, including RNA helicase activity, phosphorylation-dependent endoribonuclease activity, and cytoplasmic stress granule facilitator activity (Atlas et al. 2007; Solomon et al. 2007; Ortega et al. 2010).
In spite of the similar domain architecture between RBP42 and G3BP, noticeable differences exist. First, only 25% of the primary amino acid sequence is conserved between T. brucei RBP42 and mammalian G3BPs. Second, the RNA-binding domain, RRM, contains two conserved helical regions in G3BPs but only one conserved helix in RBP42. Finally, the arginine-glycine-rich (RGG) domain, present at the C terminus of G3BPs, is absent from RBP42. The PxxP motifs, which often modulate protein–protein interactions, vary in number among different mammalian G3BP family members, as well as among different trypanosomatid RBP42 orthologs.
RBP42 is essential for normal cell growth
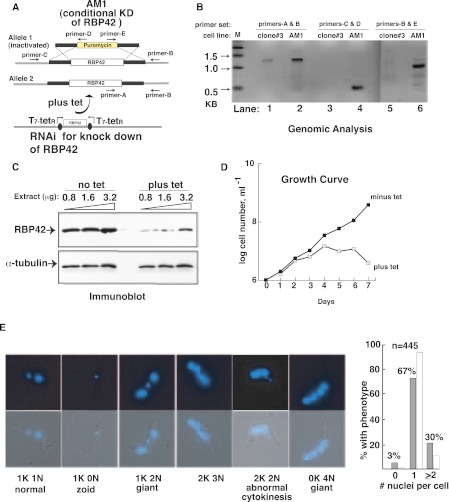
To determine whether RBP42 is essential for parasite viability, we used conditional double-stranded RNAi-mediated knockdown (RNAi) of RBP42 in a procyclic cell line with only one RBP42 allele; the second allele was insertionally inactivated by a puromycin-resistant cassette (Fig. 2A,B). (A cell line lacking both RBP42 alleles could not be established despite several trials.) Three clonal cell lines were selected, each of which exhibited a slow growth phenotype and altered morphology after 2 d of RNAi induction. These data indicate that RBP42 is an essential protein for procyclic T. brucei survival. Immunoblot analysis of one typical clone, called AM1, showed the expected reductions in the RBP42 protein (Fig. 2C). Depletion of RBP42 caused AM1 cells to stop dividing, as reflected in the growth curve (Fig. 2D). Microscopic observation of RBP42-depleted AM1 cells on day 4 revealed noticeable phenotypic alterations, with cells exhibiting an abnormal shape (Ploubidou et al. 1999; Hammarton 2007). Cells exhibited apparent cytokinesis defects, as inferred from the large number of nuclei in many parasites (Fig. 2E). DAPI-stained nuclei and kinetoplast were photographed and counted on a per cell basis; ∼30% of the RBP42 depleted cells were aberrantly shaped, containing two or more nuclei (including 1K2N, 2K3N, 2K2N, and 0K4N). Approximately 3% of the cells were zoids, containing no nucleus (1K0N). In comparison, noninduced cells, with intact RBP42, contained <1% of aberrant cells with multiple nuclei and <1% zoids. Interestingly, a recent RNAi-based screen designed to identify essential T. brucei genes (Alsford et al. 2011) indicated that RNAi knockdown of RPB42 resulted in a defect in cellular fitness of bloodstream and procyclic form parasites, as well as a defect in life cycle differentiation. Taken together, these data suggest that decreased levels of RBP42 negatively impact parasite morphology and growth at multiple stages of their life cycle.
FIGURE 2.
Effect of RBP42 depletion on cell viability. (A) RNAi-dependent ablation of RBP42 mRNA in T. brucei strain AM1. This strain contains a single intact RBP42 allele, as well as the RNAi construct. (B) Analysis of genomic DNA to assess the AM1 genotype. PCR reactions utilized primers-A, -B, -C, and -D located as shown in panel A. The 0.6-kb DNA in lane 4 and the 0.9-kb DNA in lane 6 confirm that AM1 contains a puromycin cassette in place of one of the two endogenous RBP42 alleles. The 1.3-kb band in lane 2 shows that the remaining RBP42 allele is intact. (Lanes 1,3,5) Controls showing the presence of intact RBP42 alleles in the parental clone 3 cell line. (C) RBP42 is decreased after ablation of mRNA by tetracycline-dependent RNAi induction, as demonstrated by immunoblot analysis. α-Tubulin levels are shown as a loading control. (D) T. brucei requires RBP42 for cell viability. Tetracycline, which activates the RNAi-dependent ablation of RBP42, was added at day 0. Parasites were directly observed and counted daily using a hemocytometer. Three clones were studied and AM1 chosen as a representative example of the observed phenotype. (E) Cellular morphology and cytokinesis are altered as a result of RBP42 depletion. Examples of the predominant phenotypes are shown as fluorescent micrographs containing DAPI-stained nuclei and kinetoplasts (both in blue) of parasites that represent each phenotype observed on day 4 after RBP42 depletion. Quantification of DAPI-stained nuclei per cell is shown for n = 445 cells in the histogram to the right of micrographs. Open bars, normal phenotypic distribution.
RBP42 is a cytoplasmic protein and associates with polysomes
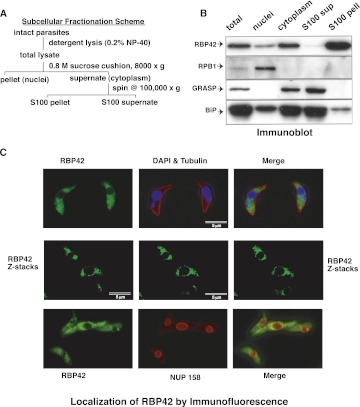
To assess the cellular distribution of RBP42, we fractionated cellular extracts by differential centrifugation and assayed each fraction by immunoblot analysis using specific antibodies. Known nuclear and cytoplasmic proteins fractionated as expected; RPB1, the largest subunit of RNA polymerase II, served as a nuclear marker and Golgi Reassembly Stacking Protein (GRASP) and the ER chaperone BiP served as cytoplasmic markers. Our fractionation analysis demonstrated that RBP42 is predominantly present in parasite cytoplasm (Fig. 3A,B). Further separation, by high-speed centrifugation, of cytoplasmic extracts into a soluble fraction and particulate fraction, which predominantly consists of ribosomes and polysomes, resulted in RBP42. Immunofluorescence microscopy of fixed parasites indicated that RBP42 localization was consistent with the observation above; immunostained RBP42 was visible throughout the cytoplasm and had a dotted granular appearance (Fig. 3C). RBP42 was not observed within the nucleus. Some of the dotted stains appeared to outline the nuclear periphery. However, RBP42 did not colocalize with the nuclear pore complex protein, NUP158.
FIGURE 3.
RBP42 is within the S100-precipitated fraction in the cellular cytoplasm. (A) Subcellular fractionation scheme is shown. (B) Immunoblotting of fractionated lysates identifies RBP42 as cytoplasmic and associated with the ribosome-containing subcellular fraction (S100 pellet). Protein detection was performed using the four antibodies listed on the left side of the panel. RPB1, the largest subunit of RNA polymerase II, is a nuclear marker; GRASP and BiP are cytoplasmic markers that fractionate with the S100 supernatant due to the early detergent step in the fractionation (Ho et al. 2006). (C) Immunofluorescent detection of fixed parasites shows cytoplasmic localization of RBP42. Four of 15 Z-stacks are shown. Z-stacks were collected to refine resolution.
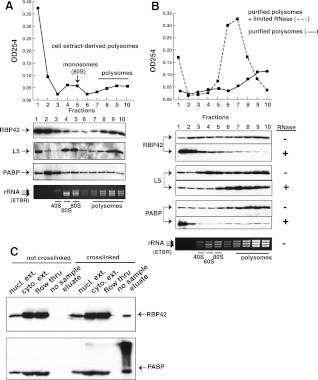
To determine whether RBP42 is associated with polysomes, we fractionated polysomes from monosomes by sucrose-density gradient centrifugation of cytoplasmic extracts and tested for the presence of RBP42 in polysomal fractions. Distribution of rRNA and the large ribosomal protein L5 showed that the polysomes were enriched in fractions 7–10 and the 40S and 60S ribosomal subunits, and the 80S ribosome monomers were enriched in fractions 3–5 (Fig. 4A). Immunoblot assays of polysomal fractions revealed that a significant portion of RBP42 is associated with polysomes. Poly(A)-binding protein (PABP), which binds to mRNA in polysomes, serves as a control for the presence of mRNA in the polysomal fractions. To test whether RBP42's interaction with polysomes was mRNA-dependent, purified polysomes were treated with limiting amounts of RNase prior to fractionation. RNase treatment clipped the mRNAs within the polysomes and released RBP42 and PABP into lower-molecular-weight complexes (Fig. 4B). When purified polysomes were fractionated without prior RNase treatment, RBP42 and PABP remained tightly associated with polysomes in control experiments. The release of RBP42 from polysomes by limiting RNase treatment indicated that RBP42 association with polysomes is mediated by protein–mRNA interactions.
FIGURE 4.
RBP42 fractionates predominantly with polysomes. (A) Polysome fractionation profile is shown. Immunoblotting of fractionated material was accomplished using the RBP42, L5, and PABP antibodies listed on the left side of the panel. L5, a component of the 60S ribosomal subunit, is a monosome (80S) and polysome marker; PABP, a cytoplasmic poly(A)-binding protein, is an mRNA marker. Locations of the small subunit (40S), large subunit (60S), intact monosomes (80S), and polysomes in the gradients was accomplished using an ethidium bromide (ETBR)–stained Agarose gel. T. brucei 28S is naturally present as two large fragments, which migrate below 18S rRNA. (B) Purified polysomes obtained from fractions 7–10 in A were further fractionated after mock (solid line) or limited RNase A treatment (dotted line). Immunoblotting was done as shown in A. (C) Immunoblotting of fractionated lysates before or after in vivo protein–RNA crosslinking by UV light. An equal amount of each fraction was present in all lanes to show the relative abundance of RBP42 and PABP in each fraction. Fractions, defined at the top of each lane, were obtained after oligo-d(T) cellulose chromatography.
To confirm that RBP42 interacts with mRNA, parasite proteins were crosslinked to substrate RNAs in vivo using UV light. Purified parasite protein crosslinked to poly(A)+ RNA were then isolated using oligo-d(T) cellulose chromatography. As shown by immunoblot analysis, RBP42 copurified with poly(A)+ RNA, indicating a direct interaction with cellular mRNA in vivo (Fig. 4C). PABP, which directly binds to mRNA, served as a control and copurified with the poly(A)+ fraction (Fig. 4C). In a control experiment, a similar purification of poly(A)+ mRNA from noncrosslinked parasites did not copurify either RBP42 or PABP. Taken together, these data indicate that RBP42 directly associates with translating mRNAs within the parasite.
To assess whether RBP42 affects mRNA translation in T. brucei, we measured the overall translation rate in normal or RBP42-depleted cells. Protein synthesis rates were measured by the incorporation of 35S-methionine/cysteine into cells (Supplemental Fig. 1). RBP42-depleted cells showed a 34% diminished rate of protein synthesis compared with normal cells. (Protein synthesis rate was linear for the duration of the assay and 35S-incorporation was similar at two different cell densities.) Although reduced protein synthesis may reflect the slow-growth rate of RBP42-depleted cells, it may be a direct effect of RBP42 on mRNA translation (Barquilla et al. 2008; Freire et al. 2011).
Crosslinking immunoprecipitation of RBP42 RNA targets
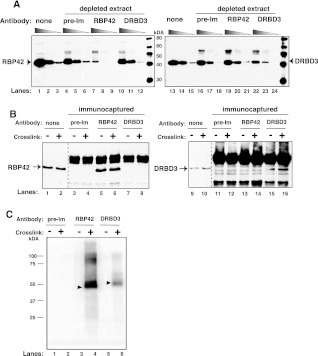
Having established that RBP42 directly binds to mRNA, we sought to identify its cellular substrates using crosslinking immunoprecipitation (CLIP) methodology. We reasoned that identification of RBP42's mRNA substrates would help reveal its function in trypanosome gene expression. To set up CLIP, we established optimum conditions for specific and efficient immunoprecipitation of RBP42 protein (Fig. 5). Anti-RBP42 antibody specifically and efficiently removes RBP42 from cellular extracts, as indicated by immunoblot analysis (Fig. 5A, cf. lanes 4–6 and 7–9). In addition, we tested a control antibody that recognizes a known RNA-binding protein, DRBD3 (Estevez 2008). The anti-DRBD3 antibody was equally specific and efficient in removing DRBD3 from cellular extract (Fig. 5A, cf. lanes 16–18 and 22–24). The specificity of the anti-RBP42 and anti-DRBD3 antibodies was further verified by immunoblot analyses of the immunocaptured proteins: Anti-RBP42 antibody only captured RBP42, and no DRBD3 was detected (Fig. 5B, cf. lanes 5,6 and 13,14). Similarly, the anti-DRBD3 antibody only captured DRBD3, and no RBP42 was detected (Fig. 5B, cf. lanes 15,16 and 7,8).
FIGURE 5.
RBP42 antibody recognizes RBP42 in cell extracts. (A) Cell extracts were depleted using either preimmune serum or RBP42 or DRBD3 antibodies. Immunoblots were probed with the indicated antibody (anti-RBP42 was used in lanes 1–12, and anti-DRBD3 was used in lanes 13–24). None indicates no-antibody-treated samples. (Lanes 1,4,7,10,13,16,19,22) Undiluted samples; (lanes 2,5,8,11,14,17,20,23) 1:5 diluted samples; and (lanes 3,6,9,12,15,18,21,24) 1:25 diluted samples from either starting or immunodepleted extracts. (B) Proteins were captured from crosslinked (+) or not crosslinked (−) parasites using the antibodies indicated above the lanes. Immunocaptured proteins were detected with anti-RBP42 (lanes 1–8) or anti-DRBD3 (lanes 9–16) antibodies. None indicates no-antibody-treated samples. (C) Immunoprecipitated protein–RNA complexes were 5′-end radiolabeled on their RNA, separated by denaturating-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. A PhosphorImager scan of protein– 32P-radiolabeled RNA complexes is shown. RBP42 and DRBD3 migrate similarly as the two proteins are similar in molecular mass, and the RNA fragments associated with the proteins are 30–50 nt long.
RNA targets of RBP42 or DRBD3 were purified by immunoprecipitating protein–RNA complexes using anti-RBP42 or anti-DRBD3 antibodies. RNAs were detected after radiolabeling their 5′ ends (Fig. 5C). RNA was obtained in protein–RNA complexes after crosslinking (lanes 4,6). No RNAs were observed in the absence of crosslinking (lanes 3,5). In control experiments, preimmune serum did not immunoprecipitate any protein–RNA complexes either before or after crosslinking (lanes 1,2). Protein-bound RNAs were subsequently extracted, reverse transcribed into cDNAs, and subjected to high-throughput DNA sequencing (HITS-CLIP).
RBP42 predominantly binds mRNAs within their coding regions
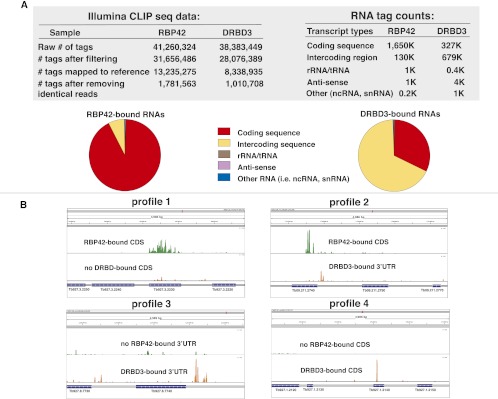
HITS-CLIP of RBP42 and DRBD3 cDNA libraries was performed on the Illumina platform. A total of 41 M and 38 M reads were obtained for RBP42 and DRBD3, respectively. Unique reads were mapped to the T. brucei genome for RBP42 and DRBD3 (1.8 M and 1.1 M, respectively). The distribution of mapped reads to different chromosomal features revealed that a surprisingly large number (>94%) of the RBP42 reads mapped within mRNA coding sequences. In contrast, only ∼30% of the DRBD3 reads mapped within mRNA coding regions (Fig. 6A). Neither RBP42 nor DRBD3 targeted noncoding RNAs (including rRNAs, tRNAs, and snRNAs). The distribution of RBP42 reads within mRNA sequences indicates that RBP42 predominantly binds within mRNA coding regions in trypanosomes.
FIGURE 6.
HITS-CLIP data, RNA read assignments, and quantification. (A) Illumina CLIP sequencing data and RNA tag count data. Pie charts depict the relative abundance of the RNA targets, which are derived from the RNA tag count data. Coding sequences are defined as the annotated open reading frames of mRNAs in version 4 of the T. brucei brucei 927 genome at TriTrypDB and GeneDB (tritrypdb.org/tritrypdb and www.genedb.org). Intercoding sequences contain 5′ and 3′ UTRs, as well as sequences that may contain primary transcripts. rRNA/tRNA, anti-sense, and other RNAs are as defined in the database referenced above. Reads with poor quality and sequence ambiguity, as well as reads generated by PCR amplification, were eliminated prior to mapping. (B) RBP42–mRNA interactions and DRBD3–mRNA interactions (control protein–RNA interactions) within specific genes in the T. brucei genome. (Profile 1) RBP42-bound sequences within the coding region of succinyl-CoA synthetase (α-subunit shown here; Tb927.3.2230). Two upstream genes (Tb927.3.2250 and Tb927.2240) and one downstream gene (Tb927.2220) that are not bound by RBP42 reveal binding specificity, as does the absence of DRBD3 binding to all four genes shown. (Profile 2) RBP42-bound sequences within the CDS and DRBD3-bound sequences within the 3′ UTR of Gim5B (Tb09.211.2740). The absence of the reciprocal binding, as well as the absence of binding to the flanking genes, indicates the binding specificities. (Profile 3) DRBD3-bound sequences within the (predicted) 3′ UTR region of MFS mRNA (major facilitator superfamily protein; Tb927.8.7740). DRBD3-targeting of the MFS mRNA 3′ UTR was initially discovered by Estevez (2008). (Binding specificity is revealed by the low binding of DRBD3 or RBP42 to the CDS of this gene, as well as the low binding of RBP42 to the 3′ UTR of this gene. The small amount of signal indicating DRBD3 interactions with the 3′ UTR of the upstream gene, Tb927.8.7730, was not investigated.) (Profile 4) DRBD3-bound sequences within the coding region of a hypothetical protein mRNA (Tb927.1.2140). Binding specificity is revealed by the absence of RBP42 binding to this gene, as well as the absence of DRBD3 and RBP42 binding to two upstream flanking genes (Tb927.1.2120 and Tb927.1.2130) and the downstream flanking gene (Tb927.1.2150). Scales, 0–1500 (Profile 1), 0–300 (Profile 2), 0–100 (Profile 3), and 0–600 (Profile 4). Green denotes RBP42-bound sequences, and orange denotes DRBD3-bound mRNA sequences.
Visualization of RBP42 and DRBD3 reads in the IGV browser showed four types of profiles (Fig. 6B). Profile 1, an example of the most frequently observed profile, shows RBP42 binding within the coding region of a gene and no DRBD3 binding. Profile 2, another frequently found profile, shows RBP42 bound within the coding region and DRBD3 bound to the 3′ UTR of the same gene. Profile 3 shows DRBD3 bound to the 3′ UTR of a gene and no RBP42 binding. Profile 4 shows an example of DRBD3 bound to the coding region of a gene.
To verify that nonspecific associations did not cause the binding of RBP42 to mRNA coding sequences during the CLIP procedures, we determined whether the number of RBP42 reads correlated with mRNA length or with mRNA cellular abundance. The comparison of RBP42 read number with mRNA transcript lengths indicated no correlation between RBP42 binding and mRNA length (R2 = 0.07) (Supplemental Fig. 2). Similarly, the comparison of RBP42 read number with mRNA levels, obtained from a transcriptome study of procyclic parasites (Siegel et al. 2010), indicated that the RBP42 read number was independent of mRNA abundance. Whereas a large number of reads were obtained for many low abundant mRNAs, very few reads were detected for several highly abundant mRNAs (R2 = 0.4) (Supplemental Fig. 2).
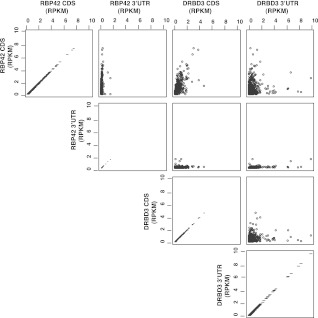
RBP42 binding to coding regions was further verified by analyzing HITS-CLIP data using an 835-gene set for which the 3′ UTRs could be determined with high confidence (Siegel et al. 2010, 2011). The total unique reads for RBP42 were 253,315; of which 237,261 (∼94%) were within CDS regions and 16,054 (∼6%) were within 3′ UTRs. Total unique reads for DRBD3 were 87,811, of which 38,603 (∼43%) were within CDS regions and 49,208 (∼57%) were within 3′ UTRs. The number of RBP42 and DRBD3 reads within CDS and 3′ UTR was plotted as pairwise comparisons on a scatterplot matrix (Fig. 7). These analyses confirmed that RBP42 strongly prefers to bind within mRNA coding sequences.
FIGURE 7.
A comparison of RBP42 and DRBD3 unique reads within the CDS and 3′ UTRs of 835 genes with defined 3′ UTRs. RBP42 and DRBD3 unique reads within the CDS and 3′ UTR of 835 genes were normalized over gene length and read counts to obtain RPKM (reads per kilobase per million mapped reads) values. Gene length for each gene was determined by adding CDS length and 3′ UTR length. The highest RPKM value obtained for RBP42 CDS regions was 7.4; for RBP42 3′ UTR region, 1.4; for DRBD3 CDS, 4.6, and for DRBD3 3′ UTR, 9.9. Pairwise comparisons of RPKM values for each protein over CDs and 3′ UTR regions are shown as a scatterplot matrix. The axes show the RPKM values for the reads mapped to the CDs and 3′ UTR in the RBP42 and DRBD3 experiments.
Genes involved in cellular energy metabolism are robust RBP42 targets
To identify the mRNA substrates of RBP42, we analyzed our HITS-CLIP data using the F-Seq algorithm, which uses kernel density measurements to define peak regions of binding (Boyle et al. 2008). F-Seq analysis detected peaks of RBP42 reads for over 2000 genes with peak-enrichment values ranging from 1.7–169 over background. To identify the most reliable RBP42 targets, we compared the F-seq analysis with the RBP42 read profiles in the IGV browser. On the basis of this inspection, we considered that F-Seq values ≥25 represent the most reliable binding, with easily detectable peak binding regions. These criteria resulted in the identification of 188 mRNAs with F-Seq enrichment values ≥25 and corresponding to the top ∼3% of RBP42 targets. These 188 genes were selected for further analysis.
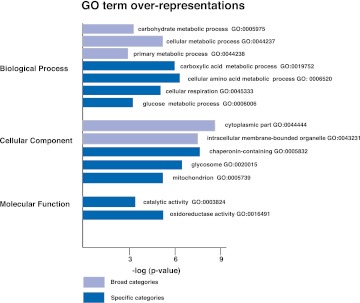
Among the 188 gene targets, 137 (73%) genes have a known function, annotated in TriTrypDB; http://tritrypdb.org/tritrypdb/. The rest are unknown trypanosome-specific genes annotated as hypothetical. GO terminology was used to categorize the 137 genes according to their biological process, cellular component, and molecular function (Fig. 8; Supplemental Fig. 3). Significantly enriched GO terms (P < 0.01) include several broad categories, for example, carbohydrate metabolic process (7 × 10−3), cellular metabolic process (2 × 10−5), primary metabolic process (1 × 10−3), cytoplasmic part (3 × 10−8), and intracellular membrane–bounded organelle (4 × 10−7). Interestingly, the 137-gene set is overrepresented by genes involved in glucose and proline metabolism. GO terms describing carboxylic acid metabolic process (1 × 10−6), cellular amino acid metabolic process (4 × 10−6), cellular respiration (1 × 10−5), glucose metabolic process (3 × 10−3), glycosome (8 × 10−6), mitochondrion (3 × 10−5), catalytic activity (8 × 10−3), and oxidoreductase activity (5 × 10−5)—processes integral to glucose and proline metabolism—are significantly overrepresented. This overrepresentation of genes involved in cellular energy generation pathway indicates that RBP42 may play a key role in the regulation of these mRNAs and may determine cellular metabolic responsiveness.
FIGURE 8.
Gene Ontology (GO) assignments for RBP42-bound mRNAs. Highly significant (P < 0.01) associations of RBP42 targets with GO terms are shown. GO terms are derived from a GO Slim set designed to incorporate trypanosome-specific processes and cellular compartments (Alsford et al. 2011). GO terms that describe broad categories are indicated by light blue bars; GO terms that describe specific categories are indicated by dark blue bars. Statistical analysis of GO term associations was carried out using 137 annotated genes. Since annotated genes lack a comprehensive set of appropriate GO terms, each significant association was verified.
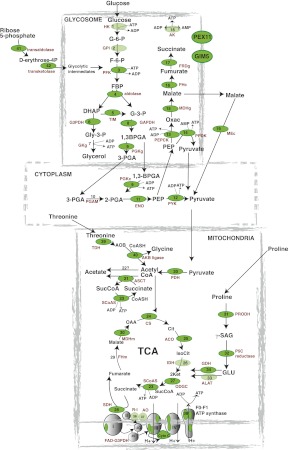
The main energy sources for procyclic parasites are D-glucose and L-proline. These nutrients are metabolized by a combination of glycolytic enzymes in the glycosomes, TCA cycle enzymes, and the respiratory chain in the mitochondrion, and as intermediate steps in the cytoplasm (Besteiro et al. 2005). Recent RNAi and gene knockout studies have contributed to a current model of procyclic energy metabolic pathways (Bringaud et al. 2006; Zikova et al. 2009). Mapping of the RBP42 targets to this procyclic-specific metabolic pathway is shown in Figure 9.
FIGURE 9.
A combination of visual and computer-based analysis reveals that the most significant RBP42-bound mRNAs encode proteins that function in energy production. Metabolism-associated genes with the highest enrichment values (top 10%) for RBP-42–bound RNA tags are depicted as dark green ovals; genes with above-background values are depicted as light green ovals. Enzymes are (1) hexokinase (HK); (2) glucose-6-phosphate isomerase (GPI); (3) phosphofructokinase (PFK); (4) aldolase; (5) triose-phosphate isomerase (TIM); (6) glycerol-3-phosphate dehydrogenase (G3PDH); (7) glycerol kinase-glycosomal (GKg); (8) glyceraldehyde-3-phosphate dehydrogenase (GAPDH); (9) phosphoglycerate kinase (cytosolic, PGKc, and glycosomal [PGKg]); (10) phosphoglycerate mutase (PGAM); (11) enolase (ENO); (12) pyruvate kinase (PYK); (13) phosphoenolpyruvate carboxykinase (PEPCK); (14) pyruvate phosphate dikinase (PPDK); (15) malate dehydrogenase-glycosomal (MDHg); (16) fumarase-cytosolic (FHc); (17) NADH-dependent fumarate reductase (FRDg); (18) adenylate kinase-glycosomal (AK), GIM5-glycosomal membrane protein (GIM5), and PEX11-glycosomal membrane protein (PEX11); (19) malic enzyme-cytoplasmic (MEc); (20) pyruvate dehydrogenase complex (PDH); (21) acetate:succinate CoA transferase (ASCT); (22) unknown enzyme; (23) succinyl CoA synthetase (SCoAS); (24) citrate synthase (CS); (25) aconitase (ACO); (26) isocitrate dehydrogenase (IDH); (27) 2-ketoglutarate dehydrogenase (ODGC); (28) succinate dehydrogenase (SDH); (29) fumarase-mitochondrial (FHm); (30) malate dehydrogenase-mitochondrial (MDHm); (31) proline dehydrogenase (PRODH); (32) pyrroline-5-carboxylate reductase (P5C reductase); (33) L-alanine aminotransferase (ALAT); (34) glutamate dehydrogenase (GDH); (35) glycerol-3-phosphate oxidase (FAD-G3PDH); (36) rotenone-insensitive NADH dehydrogenase (R–I); (37) alternative oxidase (AO); and (38) F0/F1-ATP synthase (β-, γ-subunits and subunit 9) in complexes I–IV of the respiratory chain (in complex I, subunit 20-kDa subunit is indicated; in complex II, no subunits are indicated; in complex III, the Rieske iron-sulfur protein is indicated; in complex IV, the cytochrome oxidase subunits IV, V, and VII–IX are indicated; cytochrome C [Cyto C]); (39) threonine-3-dehydrogenase (TDH); (40) 2-amino-3-ketobutyrate CoA ligase (ABK ligase); (41) transaldolase; and (42) transketolase.
In the glycosome, which is a trypanosome-specific peroxisome-like organelle, there are 14 enzymes involved in glucose metabolism (enzymes 1–9, 13–18) (Parsons 2004; Besteiro et al. 2005; Coustou et al. 2008). Seven were among the top 3% of RBP42 targets (enzymes 3–6, 13–15). Four additional enzymes were among the top 10% of RBP42 targets (enzymes 8, 9, 16, 17). In addition, the glycosomal membrane proteins, PEX11 and GIM5, which maintain glycosomal integrity, were RBP42 targets. Among the five cytosolic glycolytic enzymes involved in pyruvate production (enzymes 9–12, 19), four enzymes were within the top 5% of RBP42 targets (enzymes 9, 11, 12, 19). In the mitochondrion, pyruvate is metabolized to acetyl CoA, which enters two main pathways. Energy is generated during acetate production by six enzymes (enzymes 20–23, 39, 40) directly or indirectly involved in substrate-level ATP production. Five were in the top 3% of RBP42 targets. The sixth enzyme, enzyme 22, is unidentified in trypanosomes (Riviere et al. 2004). The TCA cycle contains eight enzymes (enzymes 23–30), of which six enzymes were within the top 5% of RBP42 targets. Of the additional two enzymes not within the top 5%, one enzyme (enzyme 26) was enriched above background, and the other (enzyme 29) was not a RBP42 target. In the case of multi-subunit TCA-cycle enzymes, many of the enzyme subunits were RBP42 targets (including enzymes 20, 23, 27). Among the four enzymes that metabolize proline to 2-ketoglutarate (enzymes 31–34), all four were within the top 5% of RBP42 targets. Finally, multiple respiratory chain components were among the top 5% of RBP42 targets as well. These included several of the cytochrome oxidase subunits of complex IV, at least four of the F-type ATPase subunits (Zikova et al. 2009), cytochrome C, 20-kDa subunit of complex I, and the Rieske iron–sulfur protein in complex III. Taken together, these findings show that RBP42 targets a large portion of cellular mRNAs that encode energy generating enzymes and function to maintain cell viability.
DISCUSSION
RNA-binding proteins play a pivotal role in trypanosome gene expression. Although several important trypanosome RNA-binding proteins have been studied to date, their comprehensive set of target mRNAs remain unknown. In this report we characterized a new RNA-binding protein, RBP42, and identified its cellular targets using a global unbiased approach. We found that RBP42 binds to the coding region of many mRNAs involved in cellular energy metabolism. RBP42 is essential for procyclic (Fig. 2) and bloodstream-form viability (Alsford et al. 2011). We propose that this newly characterized protein plays important roles in mRNA metabolism during the parasite's life cycle.
The identification of RBP42 targets revealed that RBP42 binds to a large number of mRNAs that encode enzymes involved in energy metabolism. Modulation of cellular energy metabolism in response to changing nutritional sources requires adjustments of metabolic pathways. In procyclic parasites, both glucose and amino acids are utilized as primary nutritional sources. When glucose is the major nutrient source, glycolytic enzymes in the glycosome and mitochondrion are involved in energy production. When amino acids are the main nutritional sources, additional mitochondrial enzymes are utilized. As trypanosomes lack differential transcription, post-transcriptional reactions must play a defining role in the adjustment of these metabolic pathways. We predict that RBP42 helps regulate the cellular response to nutritional sources by binding to mRNAs that encode these metabolic enzymes.
It is interesting that RBP42 appears to be similar to the mammalian protein G3BP (Fig. 1). Both RBP42 and G3BP are cytoplasmic, associate with polysomes (Angenstein et al. 2002; Atlas et al. 2007), and bind mRNAs (Atlas et al. 2004; Taniuchi et al. 2011). Whereas G3BP–mRNA interactions have been limited to studies of c-Myc, Tau, and b-F1-ATPase mRNAs, here we identified RBP42–mRNAs targets using an unbiased, global approach. Our findings that RBP42 binds F1-ATPase mRNAs and many more mRNAs involved in energy metabolism in trypanosomes may help facilitate the discovery of additional G3BP targets in metazoans. Indeed, G3BP is altered in several cancer cells, and its binding to F1-ATPase and c-Myc mRNAs has been speculated to play a role in the metabolic changes that occur during altered cell growth (Guitard et al. 2001; Tourriere et al. 2001; Ortega et al. 2010; Shim et al. 2010; Cairns et al. 2011). It will be interesting to see whether RBP42 also plays a role in adjusting the bioenergetics in trypanosomes, possibly in response to the constantly changing nutritional environment during the parasite life cycle.
How does RBP42 binding to its target mRNAs contribute to metabolic adaptation in trypanosomes? Our findings that translational rates are decreased when cells are depleted of RBP42 are suggestive of RBP42 functioning as a translational regulator. It is possible that RBP42 promotes the translation of target mRNAs by helping to recruit initiation or elongation factors to polysomes to facilitate protein production. Alternatively, RBP42 may stall ribosomes, slowing down or abrogating the translation of enzymes utilized in energy metabolism in the cell. Expression of amyloid precursor protein (APP) in human neuronal cells relies on a combination of both positive and negative translational regulation (Lee et al. 2010). The hnRNP C protein binds to the coding region of APP and promotes translation, whereas FMRP, the fragile X-mental retardation protein, binds to the same region and stalls ribosome movement (Brown et al. 2001; Lee et al. 2010; Darnell et al. 2011). In trypanosomes, RBP42 targeting of mRNAs may have either a stimulatory or inhibitory effect as dictated by the environment within different transcripts.
As RBP42 was originally identified by its interaction with the RPB4 subunit of RNA polymerase II, its cytoplasmic localization was surprising. Although RBP42 may function in the nucleus as well as the cytoplasm, our immunofluorescence microscopy data and cell fractionation analysis show that the bulk of RBP42 functions in the cytoplasm in association with polysomes. Accordingly, we speculate that the copurification of RBP42 with RPB4 indicates that trypanosomal RPB4 functions in the cytoplasm. In yeast, Rpb4p functions in the cytoplasm, participating in post-transcriptional reactions as an Rbp4/Rbp7 dimer, which is independent of the core RNA polymerase II complex (Selitrennik et al. 2006; Harel-Sharvit et al. 2010). The Rbp4/Rpb7 complex regulates the decay of mRNAs whose products are involved in protein synthesis, including ribosomal proteins such as RPL25 and translation factors such as TIF1 (Pillai et al. 2003; Lotan et al. 2005). Regulation of this specific group of mRNAs requires Rbp4/Rbp7 to interact directly with the mRNA decay machinery. In addition, Rpb4/Rpb7 stimulates translation initiation of mRNAs, probably through a direct interaction with the translation initiation factor eIF3 (Harel-Sharvit et al. 2010). The dual roles of RBP4 in mRNA decay and translation suggest that it possesses a dynamic set of protein partners. We speculate that a similar scenario occurs in trypanosomes. Specifically, trypanosomal RBP4 may participate in post-transcriptional gene control, partnering with RBP42 to stimulate translation, modulate mRNA decay, or possibly pause translation under certain environmental conditions.
In summary, RBP42 is a new member of the growing family of RNA-binding proteins that target the coding region of mRNAs. By taking a panoramic approach to RNA target discovery, we can begin to define the networks in which RBP42 participates. The studies described herein show that RBP42 is integrated into the vitally important energy metabolic network in the procyclic life cycle stage of T. brucei. How RBP42 and other RNA-binding proteins coordinate cellular processes in an organism heavily reliant on post-transcriptional control remains to be discovered.
MATERIALS AND METHODS
Trypanosome strains and transformations
Wild-type T. brucei Lister 427 procyclic (tsetse midgut form) parasites were grown in SDM-79 medium, supplemented with 10% fetal bovine serum (FBS) at 27°C in a humidified chamber containing 5% CO2. TbRPB4-tagged cells (described by Das et al. 2006) contain a NEO marker (neomycin resistance gene) in place of one RPB4 allele and a BLE marker (phleomycin resistance gene) associated with an in-frame C-terminal tandem affinity protein (TAP) tag within the second RPB4 allele. The transgenes are maintained in TbRPB4-tagged cells with 15 μg/mL G418 and 2.5 μg/mL phleomycin. (See Table 1 for strains.)
TABLE 1.
Strains
T. brucei Lister 427 cell line 29-13 (Wirtz et al. 1999) constitutively expressing T7 RNA polymerase and tetracycline repressor was transfected with an RBP42-RNAi construct, in which a pZJM (BLE) derivative contained a region of the RBP42 ORF. The clonal cell lines produced are named RBP42-clones 1–4 and grew in the presence of 2.5 μg/mL phleomycin. RBP42-clone 3 was used to create the AM1-5 cell lines in which a PUR cassette replaced one of the RBP42 alleles under non RNAi-induced conditions. Double-stranded RNAi was induced by adding tetracycline (2 μg/mL).
Plasmid constructions
T. brucei RBP42 (Gene ID Tb927.6.4440) was previously designated as A7 in Table 2 of Das et al. (2006). The RBP42-RNAi construct was completed in pZJM by inserting a 486 bp DNA sequence containing the 55- to 540-bp region of the RBP42 ORF. Primers AD277 (5′-ACAAAGCTTTTGCTACGGCTCCTGAAAATC-3′) and AD278 (5′-ACACTCGAGCAGGTTTCTTATTTGGGGCAC-3′) contain HindIII and XhoI recognition sequences to facilitate insertion into pZJM and were used to PCR-amplify the insert from T. brucei genomic DNA. The DNA sequences of the recombinant plasmids were verified.
Antibodies
Anti-RBP42 antibodies were produced after injecting rabbits with recombinant RBP42 protein made in Escherichia coli as a GST-fusion protein. GST-tagged RBP42 was purified using glutathione-Sepharose beads, according to the manufacturer's protocol (GE Healthcare), and soluble RBP42, separated from its GST domain by Precision protease, was used to raise polyclonal antibodies by the Lampire Corporation. Anti-DRBD3 antibodies were raised in rabbits against an N-terminal 83-amino-acid polypeptide from DRBD3 (Estevez 2008). Anti-GRASP antibodies were a gift from Graham Warren; anti-BiP antibodies were a gift from James Bangs; and anti-ribosomal protein L5 and anti-PABP1 antibodies were gifts from Noreen Williams. Anti-α tubulin antibody was from Sigma Corporation.
Antibody specificities were assayed by incubating parasite extracts, made in RIPA buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% Na-deoxycholate, 2.5 mM Na-pyrophosphate, 1 mM Β-glycerophosphate, 1 mM Na3VO4, 1 μg/mL leupeptin), with anti-RBP42 or anti-DRBD3 antibodies bound to Dynabeads protein G (Invitrogen). Immunodepleted samples were analyzed by immunoblotting using ECL reagents from GE Healthcare.
Cellular fractionation and polysome purification
The cellular fractionation protocol was adapted from the method described by Michaeli et al. (1993). Briefly, 109 procyclic T. brucei cells were disrupted in 5 mL hypotonic buffer (10 mM Tris-HCl at pH 7.5, 10 mM KCl, 2 mM Mg2Cl, 1 mM DTT, complete protease inhibitor; Roche) containing 0.2% NP-40 by 15–20 strokes in a Dounce homogenizer. Nuclei were isolated by pelleting through a 3 mL 0.8 M sucrose cushion (8000g, 10 min). The cytoplasmic extracts, collected from the top of the sucrose cushion, were further separated into S100 sup and pellet by centrifugation (100,000g, 1 h) in an SW55 rotor.
For polysome analysis, cellular extracts were made in polysome buffer (20 mM Tris-HCl at pH 7.5, 120 mM KCl, 2 mM MgCl2, 1 mM DTT, complete protease inhibitors) containing 1% NP-40. Total lysate was cleared by centrifugation (10,000g, 10 min), and post-nuclear supernatants were collected. To isolate purified polysomes, 3 mL of cytoplasmic extract was layered over a 7 mL 30% sucrose cushion and centrifuged (33,000 rpm, 2.5 h) in a SW41 rotor. The polysome pellet was resuspended in polysome buffer and stored at −80°C. Approximately 90 OD260 units of polysomes were recovered from 1010 cells. Polysome profiles were generated by fractionating cytoplasmic extracts on 5 mL 15%–50% sucrose gradients (41,000 rpm, 2 h) in a SW55 rotor. Ten 0.5 mL gradient fractions were collected manually from the bottom of the tube by punching a hole, and each fraction was analyzed for RNA and proteins.
In vivo crosslinking and immunocapture of protein–RNA complexes
Wild-type T. brucei procyclic cells were harvested from mid-log-phase cells (4 × 106 to 5 × 106 cells/mL), washed once with cold phosphate buffered saline (PBS) and resuspended in cold PBS to a concentration of 5 × 108/mL. Cell suspensions (10 mL) were transferred to a 100-mm Petri dish, placed on an ice tray, and UV-irradiated twice with 400 mJ/cm2 in a Stratalinker 1800 (Stratagene) at 4–5 cm distant from the UV source (each irradiation step takes 1.5 min). Cells were rapidly pelleted, snap frozen in liquid N2, and stored at −80°C in small aliquots.
Immunocapture of crosslinked protein–RNA complexes was carried out according to the method described by Ule et al. (2003). Cellular extracts were made by concentrating UV-irradiated cells by centrifugation and resuspending them in buffer A (PBS, 0.1% SDS, 0.5% Na-deoxycholate, 0.5% NP-40). Extracts were treated with RQ1 DNase (15 min, 37°C) to reduce viscosity and then incubated with titrated amounts of RNase T1 to fragment the protein-bound RNAs. The protein-bound RNA samples were clarified by centrifugation (25 min, 90K) in a Beckman TLA120.2 rotor, and specific protein–RNA complexes were captured by incubation (2 h, 4°C) with anti-RBP42 or anti-DRBD3 antibodies attached to Dynabeads Protein G beads. Captured protein–RNA complexes were washed extensively, twice with buffer A and twice with buffer B (buffer A supplemented with 5× PBS), and analyzed by immunoblot or processed for CLIP.
Purification of poly(A)+ RNA–protein complexes
Cytoplasmic extracts, collected as described above, were adjusted to 0.5% SDS and 1% 2-mercaptoethanol and incubated for 5 min at 90°C. The heated extracts were quickly chilled on ice by incubating for 5 min, and then LiCl was added to a final concentration of 0.5 M. Subsequently, poly(A)+ RNA–protein complexes were isolated by two subsequent rounds of selection over oligo-d(T) cellulose (type 7, GE Healthcare) and collected in elution buffer containing 10 mM Tris-HCl (pH 7.4), 0.05% SDS. The protein components of the isolated poly(A)+ RNA–protein complexes were analyzed by immunoblotting with specific antibodies.
RBP42 and DRBD3 HITS-CLIP
The cDNA libraries from the protein-bound RNAs were made according to the method described by Licatalosi et al. (2008). After capturing the protein–RNA complexes with anti-RBP42 and anti-DRBD3 antibodies, an RNA linker (RL3; 5′P-GUGUCAGUCACUUCCAGCGG-3′-puromycin) was ligated to the 3′ end of the protein-bound-RNAs using T4 RNA ligase, and the 5′ ends were phosphorylated with T4 polynucleotide kinase while suspended on beads. To purify the protein–RNA complexes, a small amount of 32P-γ-ATP was used as a tracer in the T4 PNK reaction. Protein–RNA complexes were resolved on PAGE-SDS, transferred to nitrocellulose membrane, and excised after detection on X-ray film. RNAs were extracted by treating the excised nitrocellulose filters with proteinase K and purified by ethanol precipitation. A 5′ RNA linker (RL5D; 5′-AGGGAGGACGAUGCGGNNNNG-3′) was ligated to the 5′ end of the purified RNA using T4 RNA ligase. The random assortment of 4 nucleotides (nt) at all four positions at the 3′ end of the 5′ linker resulted in a set of 256 unique 5′ linkers. During a subsequent analysis, these linker sequences serve as indices to distinguish individual RNAs from PCR-generated amplifications. First-strand cDNA was produced from these linker-ligated RNAs using Superscript (Invitrogen) and a primer complementary to the 3′ linker sequence (DP3; 5′-CCGCTGGAAGTGACTGACAC-3′). Libraries of cDNA were generated by PCR amplification of the first-strand cDNA using 5′ primer (DP5; 5′-AGGGAGGACGATGCGG-3′) and DP3. As a quality-control measure, a fraction of these PCR-generated cDNAs were inserted in TOPO plasmids and transformed into bacteria. The presence of inserts was confirmed by restriction analysis of plasmid DNAs isolated from 20 randomly selected bacterial colonies. Capillary sequencing of these 20 clones confirmed a heterogeneous pool of cDNA inserts of the desired 30- to 60-bp length. To prepare the cDNA libraries for sequencing in an Illumina sequencer, the cDNAs from the RBP42 and DRBD3–bound RNAs were reamplified with two primers (DSFP5; 5′-AATGATACGGCGACCACCGACTATGGATACTTAGTCAGGGAGGACGATGCGG-3′ and 5′-CAAGCAGAAGACGGCATACGACCGCTGGAAGTGACTGACAC-3′) for 15 and 25 cycles, respectively.
Mapping short reads to T. brucei genome
High-quality reads (after signal filtering from Illumina) were aligned to the genome of T. brucei strain 927 using CASAVA (version 1.7). The last 10 bp of each read has been masked during the mapping to accommodate those RNA sequences whose fragment length is shorter than read length. In CASAVA1.7, the mapping algorithm (ELAND version 2) allows multi-seed and gapped alignment with two mismatches at each seed. The reads with identical index sequence at their 5′ end were further collapsed in order to minimize PCR-based amplification artifacts and generate the unique reads set for downstream peak analysis. The reads were then annotated and grouped into five categories based on their mapped locations: coding region, intercoding region, abundant RNAs (rRNA, tRNA), anti-sense RNAs, or other (i.e., ncRNA, snRNA). The read distribution among these groups was compared between RBP42-bound and DRBD3-bound RNAs.
Peak profiling and Gene Ontology analysis
F-Seq (version 1.84), an algorithm based on kernel density estimation, was run to identify candidate regions (peaks) with significant enrichment of RBP42 (or DRBD3)-bound RNA sequence tags (Boyle et al. 2008). The fragment size, f, was set to 40. The threshold, s, was 6, instead of 4 (default). The generated list of peaks was then annotated. To select a list of significant genes with high peaks, we plotted the distribution of F-Seq enrichment score (ES) for all peaks and performed extensive visual inspection of top-ranked peaks using the IGV browser (Integrative genomic browser; version 1.5 www.broadinstitute.org/igv). An empirical cutoff (ES ≥ 25) was used to generate a list of significant genes, which were categorized by Gene Ontology (GO) analysis. GO analysis was performed using GOToolBox (Martin et al. 2004) using a nonredundant gene list and a GOSlim set from Alsford et al. (2011). GO associations with a P-value <0.001 were considered overrepresented GO categories.
Fluorescence light microscopy
Cells were fixed in 2% paraformaldehyde for 15 min at room temperature, adhered to slides coated with poly L-lysine, stained with DAPI, and visualized under a 100×/1.30 oil-immersion objective (combined with a 10× ocular for a magnification of 1000×) using a UplanF1 objective. An Olympus BX61 was used in combination with DAPI and FITC-sensitive filters. Vectashield (Vector Laboratories) was used as anti-fade. Intact cells were detected using bright field optics. Images were captured using a Hamamatsu ORCA-ER camera and acquired at 0.065-μM × 0.065-μM pixel resolution.
RBP42 immunolocalization was performed using anti-RBP42 antibodies. After fixing and adhering to poly-L-lysine coated slides, cells were permeabilized using 0.2% NP40 for 5 min at room temperature. Z-stacks were obtained with 0.2-μM spacing between each stack. A total of 15 stacks were generated per field.
Protein alignments
Multiple alignments of RBP42 proteins from T. brucei, Trypanosoma cruzi, Leishmania major, and Leptomonas seymouri were performed using CLUSTALW. Alignments were refined manually and visualized using GeneDoc (http://www.psc.edu/biomed/genedoc).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank James Bangs, Graham Warren, and Noreen Williams for antibodies. We thank Rose Garcia for administrative assistance and Han Wu and Edgar Garcia for assistance with graphics. We thank Michael Mathews, Andrew Parrott, and members of the R. Darnell laboratory for extremely helpful discussions. This work was supported in part by National Institutes of Health grants RO1 29478 and RO1 53835 and a Foundation of UMDNJ award to V.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033829.112.
REFERENCES
- Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, Roditi I, Englund PT 2001. The surface coat of procyclic Trypanosoma brucei: Programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci 98: 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21: 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenstein F, Evans AM, Settlage RE, Moran ST, Ling SC, Klintsova AY, Shabanowitz J, Hunt DF, Greenough WT 2002. A receptor for activated C kinase is part of messenger ribonucleoprotein complexes associated with polyA-mRNAs in neurons. J Neurosci 22: 8827–8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas R, Behar L, Elliott E, Ginzburg I 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem 89: 613–626 [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Sapoznik S, Ginzburg I 2007. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J Neurosci Res 85: 173–183 [DOI] [PubMed] [Google Scholar]
- Bandziulis RJ, Swanson MS, Dreyfuss G 1989. RNA-binding proteins as developmental regulators. Genes Dev 3: 431–437 [DOI] [PubMed] [Google Scholar]
- Barnes CJ, Li F, Mandal M, Yang Z, Sahin AA, Kumar R 2002. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res 62: 1251–1255 [PubMed] [Google Scholar]
- Barquilla A, Crespo JL, Navarro M 2008. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci 105: 14579–14584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besteiro S, Barrett MP, Riviere L, Bringaud F 2005. Energy generation in insect stages of Trypanosoma brucei: Metabolism in flux. Trends Parasitol 21: 185–191 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS 2008. F-Seq: A feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Riviere L, Coustou V 2006. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol Biochem Parasitol 149: 1–9 [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487 [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95 [DOI] [PubMed] [Google Scholar]
- Coustou V, Biran M, Breton M, Guegan F, Riviere L, Plazolles N, Nolan D, Barrett MP, Franconi JM, Bringaud F 2008. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J Biol Chem 283: 16342–16354 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Li H, Liu T, Bellofatto V 2006. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol Biochem Parasitol 150: 201–210 [DOI] [PubMed] [Google Scholar]
- De Gaudenzi J, Frasch AC, Clayton C 2005. RNA-binding domain proteins in kinetoplastids: A comparative analysis. Eukaryot Cell 4: 2106–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez AM 2008. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res 36: 4573–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moya SM, Estevez AM 2010. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA 1: 34–46 [DOI] [PubMed] [Google Scholar]
- Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, et al. 2011. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol 176: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitard E, Parker F, Millon R, Abecassis J, Tocque B 2001. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett 162: 213–221 [DOI] [PubMed] [Google Scholar]
- Hammarton TC 2007. Cell cycle regulation in Trypanosoma brucei. Mol Biochem Parasitol 153: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M 2010. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143: 552–563 [DOI] [PubMed] [Google Scholar]
- Ho HH, He CY, de Graffenried CL, Murrells LJ, Warren G 2006. Ordered assembly of the duplicating Golgi in Trypanosoma brucei. Proc Natl Acad Sci 103: 7676–7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K, Stirling R, Hume D, Kennedy D 2004. Rasputin, more promiscuous than ever: A review of G3BP. Int J Dev Biol 48: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM 2007. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP). Oncogene 26: 4209–4215 [DOI] [PubMed] [Google Scholar]
- Kramer S, Carrington M 2011. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol 27: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M 2010. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci 123: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, et al. 2010. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol 17: 732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Spath G, Roditi I 2011. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS ONE 6: e22463 doi: 10.1371/journal.pone.0022463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B 2004. GOToolBox: Functional analysis of gene datasets based on Gene Ontology. Genome Biol 5: R101 doi: 10.1186/gb-2004-5-12-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Goldring A, Agabian N 1993. Identification of mRNA-binding proteins of the lower trypanosomatid Leptomonas collosoma. Exp Parasitol 76: 308–313 [DOI] [PubMed] [Google Scholar]
- Ortega AD, Willers IM, Sala S, Cuezva JM 2010. Human G3BP1 interacts with β-F1-ATPase mRNA and inhibits its translation. J Cell Sci 123: 2685–2696 [DOI] [PubMed] [Google Scholar]
- Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B 1996. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol 16: 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M 2004. Glycosomes: Parasites and the divergence of peroxisomal purpose. Mol Microbiol 53: 717–724 [DOI] [PubMed] [Google Scholar]
- Pillai B, Verma J, Abraham A, Francis P, Kumar Y, Tatu U, Brahmachari SK, Sadhale PP 2003. Whole genome expression profiles of yeast RNA polymerase II core subunit, Rpb4, in stress and nonstress conditions. J Biol Chem 278: 3339–3346 [DOI] [PubMed] [Google Scholar]
- Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K 1999. Evidence for novel cell cycle checkpoints in trypanosomes: Kinetoplast segregation and cytokinesis in the absence of mitosis. J Cell Sci 112: 4641–4650 [DOI] [PubMed] [Google Scholar]
- Riviere L, van Weelden SW, Glass P, Vegh P, Coustou V, Biran M, van Hellemond JJ, Bringaud F, Tielens AG, Boshart M 2004. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei: Gene identification and role in carbohydrate metabolism. J Biol Chem 279: 45337–45346 [DOI] [PubMed] [Google Scholar]
- Selitrennik M, Duek L, Lotan R, Choder M 2006. Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot Cell 5: 2092–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Su ZY, Chae JI, Kim DJ, Zhu F, Ma WY, Bode AM, Yang CS, Dong Z 2010. Epigallocatechin gallate suppresses lung cancer cell growth through Ras-GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev Res 3: 670–679 [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38: 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Gunasekera K, Cross GA, Ochsenreiter T 2011. Gene expression in Trypanosoma brucei: Lessons from high-throughput RNA sequencing. Trends Parasitol 27: 434–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 27: 2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S 2009. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA 15: 648–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K, Nishimori I, Hollingsworth MA 2011. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol Cancer Res 9: 856–866 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J 2001. RasGAP-associated endoribonuclease G3Bp: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21: 7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Walrad P, Paterou A, Acosta-Serrano A, Matthews KR 2009. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog 5: e1000317 doi: 10.1371/journal.ppat.1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
- Zikova A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD 2009. The F0F1-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog 5: e1000436 doi: 10.1371/journal.ppat.1000436 [DOI] [PMC free article] [PubMed] [Google Scholar]