MicroRNAs play central roles in controlling gene expression in human cells. Sequencing data show that many miRNAs are produced at different levels and as multiple isoforms that can vary in length at their 5′ or 3′ ends, but the biogenesis and functional significance of these RNAs are largely unknown. The authors show here that the human trans-activation response (TAR) RNA binding protein (TRBP), a known molecular partner of the miRNA processing enzyme Dicer, changes the rates of pre-miRNA cleavage in an RNA-structure-specific manner. Furthermore, TRBP can trigger the generation of iso-miRNAs (isomiRs) that are longer than the canonical sequence by one nucleotide. They show that this change in miRNA processing site can alter guide strand selection, resulting in preferential silencing of a different mRNA target. These results implicate TRBP as a key regulator of miRNA processing and targeting in humans.
Keywords: RNAi, Dicer, TRBP, miRNA, post-transcriptional gene silencing
Abstract
MicroRNAs play central roles in controlling gene expression in human cells. Sequencing data show that many miRNAs are produced at different levels and as multiple isoforms that can vary in length at their 5′ or 3′ ends, but the biogenesis and functional significance of these RNAs are largely unknown. We show here that the human trans-activation response (TAR) RNA binding protein (TRBP), a known molecular partner of the miRNA processing enzyme Dicer, changes the rates of pre-miRNA cleavage in an RNA-structure-specific manner. Furthermore, TRBP can trigger the generation of iso-miRNAs (isomiRs) that are longer than the canonical sequence by one nucleotide. We show that this change in miRNA processing site can alter guide strand selection, resulting in preferential silencing of a different mRNA target. These results implicate TRBP as a key regulator of miRNA processing and targeting in humans.
INTRODUCTION
MicroRNAs (miRNAs) are 21- to 24-nt noncoding RNAs that regulate mammalian gene expression post-transcriptionally through sequence complementarity to mRNA targets. Base pairing between a miRNA and a complementary mRNA leads to mRNA degradation (Guo et al. 2010) and/or repression of protein synthesis (Humphreys et al. 2005; Petersen et al. 2006). Beginning in the nucleus, precursor miRNAs are produced by cleavage of primary RNA transcripts and exported in a partially processed pre-miRNA form. Once in the cytoplasm, the endoribonuclease Dicer catalyzes further cleavage of these RNAs to produce the functional guide sequences used to regulate the translation or degradation of specific mRNAs.
Deep sequencing data have revealed that miRNAs are present in different amounts according to tissue type and developmental stage (Morin et al. 2008; Fernandez-Valverde et al. 2010; Lee et al. 2010). Furthermore, many miRNAs are expressed in variant forms called isomiRs that differ by one or more nucleotides at the 5′ or 3′ end of the miRNA sequence (Lu et al. 2005; Sotiropoulou et al. 2009; Fernandez-Valverde et al. 2010; Marti et al. 2010). It is not known how most of these isomiRs are produced or regulated, and how they may contribute to gene regulation.
As the enzyme responsible for processing most miRNAs in the eukaryotic cytoplasm, Dicer must recognize a variety of dsRNA substrates as well as various proteins that are part of the RNA interference (RNAi) machinery. In particular, Dicer binds to the trans-activation response (TAR) RNA binding protein (TRBP) and also interacts with proteins in the Argonaute (Ago) family of endonucleases (Chendrimada et al. 2005; Haase et al. 2005). Dicer processes different kinds of dsRNA substrates at rates that differ by ∼100-fold, and TRBP can enhance the processing rates of these substrates (Chakravarthy et al. 2010). Although it is unknown how miRNA processing kinetics may affect their steady-state concentrations, mutation or truncation of TRBP has been observed in several cancers in which altered miRNA levels are also detected (Melo et al. 2009; Ryan et al. 2010). These data suggest a possible regulatory role of TRBP in miRNA biogenesis.
The large pool of pre-miRNAs predicted to occur in vivo is diverse in both sequence and secondary structure, providing the potential for differential processing by Dicer or Dicer–TRBP complexes. To test this possibility, we measured the kinetics of dsRNA processing using purified Dicer and a Dicer–TRBP complex with a set of natural and systematically designed pre-miRNA substrates. The results of these experiments show that Dicer discriminates among different substrates according to their structure. In addition, TRBP influences substrate recognition and processing by Dicer. We show that for specific substrates, TRBP leads to the production of isomiRs that are 1 nt longer than the miRNAs produced by Dicer alone, which can silence distinct mRNA targets, apparently due to changes in guide-strand selection. These results support a model in which TRBP functions as a key regulator of both dicing kinetics and isomiR generation.
RESULTS
Substrate-specific effects of TRBP on Dicer kinetics
To test the possibility that different pre-miRNAs are processed at distinct rates by Dicer and to determine how these rates are affected by TRBP, we measured in vitro dicing kinetics of several structurally diverse pre-miRNAs including pre-let-7a, pre-miR-21, pre-miR-31, pre-miR-29, pre-miR-200a, pre-miR-34c, pre-miR-16, pre-miR-342, pre-miR-125b-1, and pre-miR-101 (Fig. 1; Supplemental Figs. S1, S2). These pre-miRNAs were selected based on differences in their predicted secondary structures and the functional significance of the mRNAs they are thought to regulate.
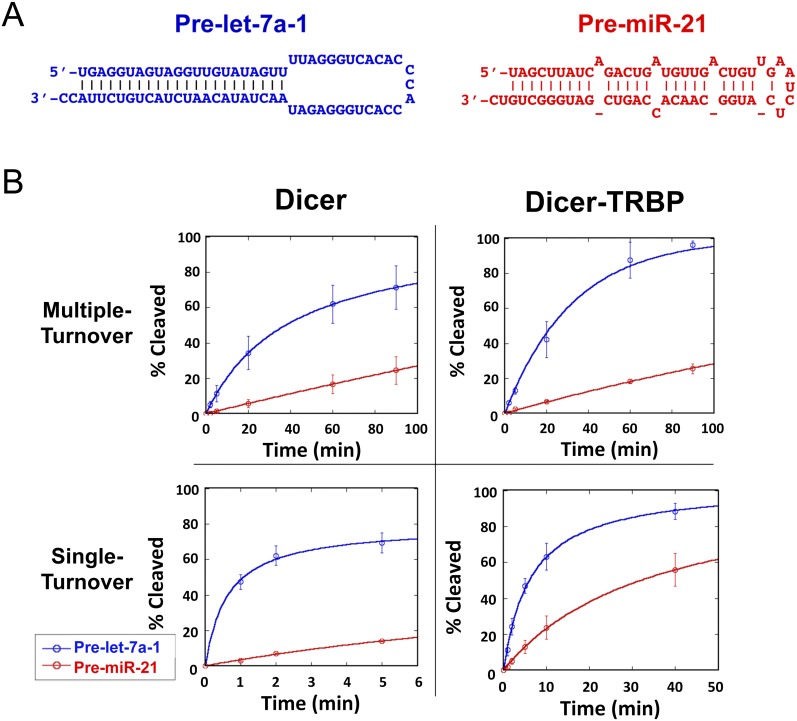
FIGURE 1.
TRBP affects the kinetics of pre-miR processing by Dicer. (A) The predicted secondary structure of pre-let-7a and pre-miR-21. (B) Pre-let-7a and pre-miR-21 processing by Dicer and Dicer–TRBP in single turnover conditions ([Dicer] = 400 nM, [Dicer–TRBP] = 50 nM, [RNA] = 2 nM) and in multiple turnover condition ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM; graphs represent data from three experimental replicates).
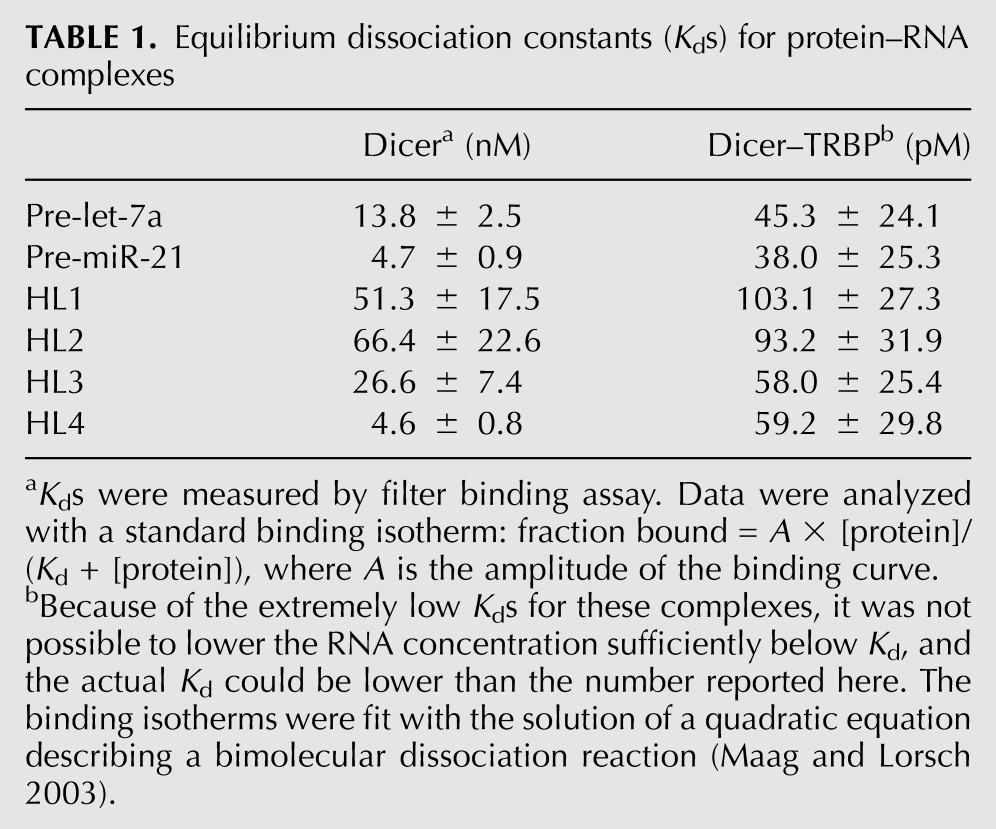
Although all of these substrates yielded the predicted miRNA products when incubated with Dicer, processing rates varied substantially (Supplemental Fig. S1). In particular, pre-let-7a and pre-miR-21 cleavage rates differed by more than fivefold under multiple-turnover conditions and at physiological salt concentrations (Fig. 1A,B). Let-7a is a tumor suppressor miRNA that is abundant in many cells (Long et al. 2009), and miR-21 is a proto-oncogenic miRNA that is commonly overexpressed in cancers (Si et al. 2007). The observed difference in processing kinetics of these two RNAs indicates that Dicer distinguishes between different pre-miRNA substrates, despite similarities in their overall predicted secondary structure. Although pre-miR-21 binds more tightly to Dicer than does pre-let-7a (Table 1), potentially explaining slower turnover on the enzyme, similar differences in processing rates occurred under single-turnover conditions (enzyme in excess over substrate) (Fig. 1B). This implies that substrate discrimination by Dicer occurs downstream from binding. In the presence of TRBP, the difference in processing rates between pre-let-7a and pre-miR-21 is more pronounced, such that pre-miR-21 is processed 11-fold slower than pre-let-7a in multiple turnover dicing assays (Fig. 1B). This finding suggests that the effect of TRBP may vary according to substrate.
TABLE 1.
Equilibrium dissociation constants (Kds) for protein–RNA complexes
Both RNA loop and stem structure affect Dicer–TRBP processing rates
We reasoned that the different dicing kinetics observed for pre-let-7a and pre-miR-21 could result from differences in the secondary structures of these substrates, which vary in primary sequence in both the stem and loop regions. To test this, we designed chimeric substrates in which either the loop or the stem was swapped between the RNAs, or the RNA sequence was changed at or near the Dicer cleavage sites in the stem (Fig. 2A).
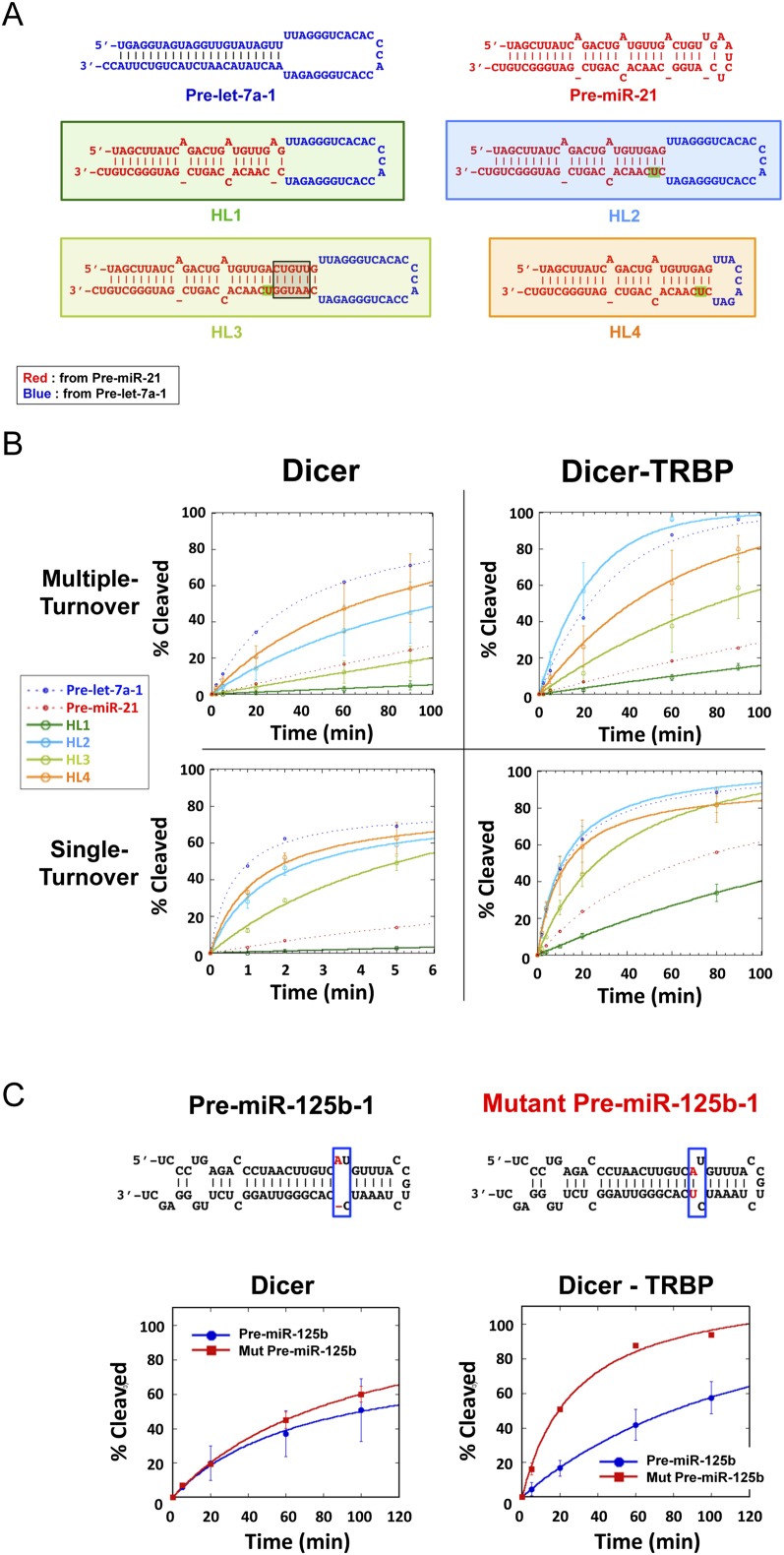
FIGURE 2.
The key structural features of pre-miRs for TRBP-Dicer processing. (A) The predicted secondary structure of chimeric substrates from pre-let-7a, pre-miR-21 representing the effect of cleavage site structure, stem length, and loop size; (red) from pre-miR-21; (blue) from pre-let-7a; (green highlights) indicate where the insertions or mutations were made. HL1 contains the pre-let-7a loop and the pre-miR-21 stem. HL2 has a single nucleotide insertion at the cleavage site of HL1 to create a base pair with the otherwise unpaired nucleotide bulge present in the pre-miR-21 stem. HL3 has five additional base pairs derived from pre-miR-21 inserted onto the end of the HL2 stem. HL4 has a smaller loop than HL2. (B) The effect of stem structure and loop size in Dicer and Dicer–TRBP processing in single turnover conditions ([Dicer] = 400 nM, [Dicer–TRBP] = 50 nM, [RNA] = 2 nM) and in multiple turnover condition ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM; graphs represent data from three experimental replicates). (C) The effect of cleavage site structure for pre-miR-125b processing by Dicer–TRBP complex. Pre-miR-125b and its mutant pre-miR-125b were processed by Dicer and Dicer–TRBP complex ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM; graphs represent data from three experimental replicates).
We first measured equilibrium dissociation constants (Kds) of these substrates for Dicer and a pre-formed Dicer–TRBP complex using a nitrocellulose filter-binding assay (Table 1). As observed previously (Ma et al. 2008; Chakravarthy et al. 2010), the Dicer–TRBP complex binds 102–103 times more tightly to these substrates than is observed for Dicer alone.
We next measured pre-miRNA processing rates of chimeric substrates under both single- and multiple-turnover conditions (Fig. 2B). Under the multiple turnover condition tested (50 nM substrate, 5 nM enzyme), substrate concentration is saturating for Dicer–TRBP, implying that kinetic differences arise from steps that occur downstream from binding (i.e., conformational change or kcat). The results of these experiments revealed structural elements that influence Dicer–TRBP processing rates. Using sets of closely related substrate RNAs, we found that the cleavage site structure is important for pre-miRNA processing by Dicer–TRBP (cf. HL1 and HL2) (Fig. 2B). HL1 contains the pre-let-7a loop and the pre-miR-21 stem and is processed even slower than pre-miR-21 by both Dicer and Dicer–TRBP, suggesting that the stem structure retards processing of pre-miR-21. A single nucleotide insertion at the cleavage site to create a base pair with the otherwise unpaired nucleotide bulge present in the pre-miR-21 stem (HL2) leads to 14-fold and 32-fold increases in processing rates by Dicer and Dicer–TRBP, respectively (Fig. 2B). TRBP-dependent sensitivity to a cleavage site bulge was also observed for processing of pre-miR-125b and its paired cleavage-site mutant, with a threefold rate increase observed for the mutant relative to the native RNA (Fig. 2C).
To test the effect of helical stem length on pre-miRNA processing rates catalyzed by Dicer or Dicer–TRBP, five additional base pairs derived from pre-miR-21 were inserted onto the end of the HL2 stem to form HL3 (Fig. 2A,B). HL3 was cleaved more slowly than HL2 by both Dicer–TRBP and Dicer. Enzyme turnover was hampered only for Dicer, and not for Dicer–TRBP in multiple turnover conditions (Fig. 2B). This suggests that TRBP may facilitate not only substrate binding but also product release after Dicer cleavage in the case of HL3.
To examine the effect of RNA loop size on pre-miRNA processing rates, we prepared a Dicer substrate with a smaller loop than HL2 (HL4) (Fig. 2A). For the Dicer–TRBP complex, the small loop size decreased the processing rate, whereas for Dicer alone the processing rate increased in both single turnover and multiple turnover dicing assays (Fig. 2B). Binding affinities were also affected: For Dicer, the substrate with a smaller loop (HL4) shows 14-fold tighter binding than the substrate with a larger loop (HL2) (Table 1). For Dicer–TRBP, the differences in Kds between substrates are less significant (less than twofold) (Table 1). Because HL2 is processed faster than HL4 by the Dicer–TRBP complex under both single and multiple turnover conditions, it is possible that the large loop is helpful in localizing the substrate in the right position for catalysis and also in releasing products.
Inhibitory effect of TRBP in pre-miR processing
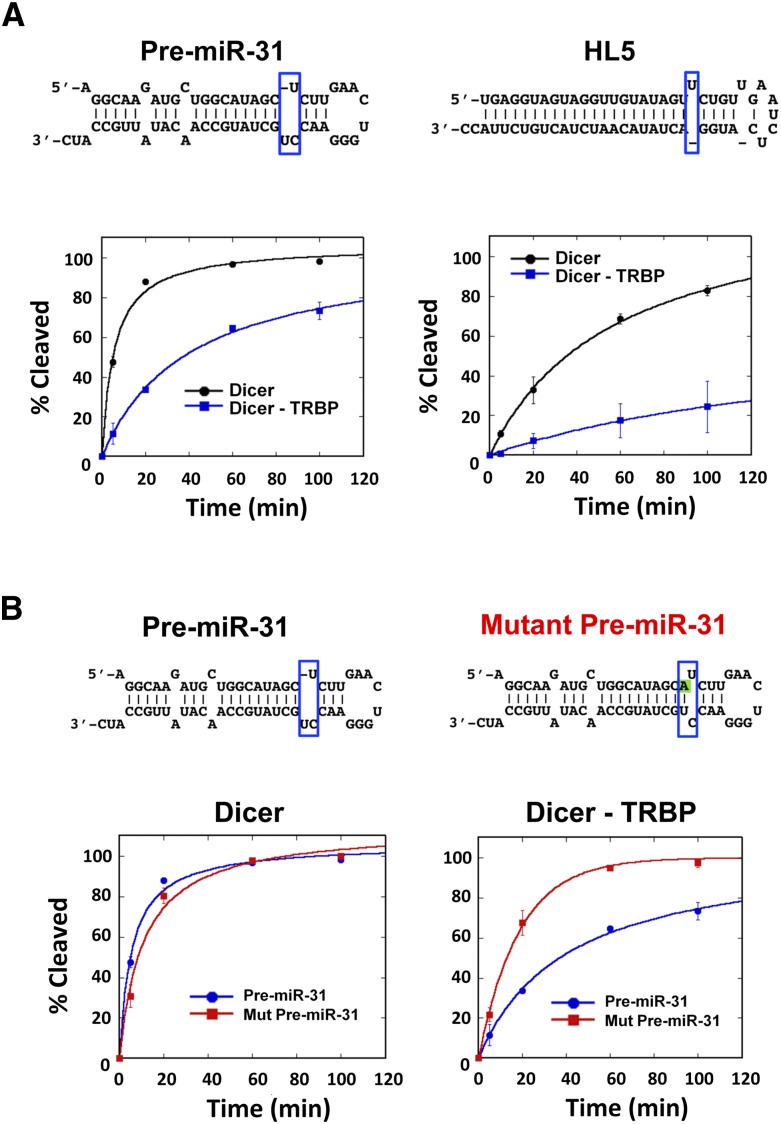
In previous experiments (Chakravarthy et al. 2010) as well as in the experiments described above, TRBP stimulated the rate of pre-miRNA processing to different extents depending on the substrate. However, this effect was not universal. Among the natural pre-miRNAs tested using in vitro dicing assays, pre-miR-31 differed notably from the others in that its processing by Dicer was inhibited fivefold in the presence of TRBP (Fig. 3A). The chimeric substrate HL5 was also cleaved threefold slower by Dicer in the presence of TRBP. Both pre-miR-31 and HL5 have an unpaired nucleotide bulge at the cleavage site in the stem, suggesting a possible role of RNA stem structure in the observed inhibitory effect of TRBP. Consistent with this hypothesis, mutation of pre-miR-31 to create canonical base pairs at the Dicer cleavage site produced an RNA substrate that was processed three times faster than wild-type pre-miR-31 by the Dicer–TRBP complex (Fig. 3B). These results suggest that TRBP is not a general enhancer of Dicer activity, but instead has opposing functions depending on the structure of the dsRNA substrate.
FIGURE 3.
TRBP inhibits processing of some pre-miRs in a structure-dependent way. (A) TRBP inhibits pre-miR-31 and HL5 processing by Dicer ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM). (B) The effect of cleavage site bulge in pre-miR-31 in Dicer–TRBP processing ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM; graphs represent data from three experimental replicates).
TRBP stimulates 5′- and 3′-isomiR production in a substrate-specific manner
Deep sequencing data show that some miRNAs occur in cells as multiple isoforms whose lengths and sequences can vary by one or more nucleotides at the 5′ or 3′ ends (Lee et al. 2010; Marti et al. 2010). In Drosophila, these isomiRs fluctuate during the course of development and may be actively regulated (Fernandez-Valverde et al. 2010). The detection of isomiRs in immunoprecipitated samples of human and mouse Argonaute proteins suggests that these RNAs are functional as gene expression regulators (Meister et al. 2004; Chi et al. 2009; Marti et al. 2010). However, it is unclear how isomiRs are produced and regulated and what their functional significance may be. In particular, 5′ isomiRs, which are less abundant than 3′ isomiRs in nature, could be functionally significant because they can change the seed sequence that is the primary determinant of mRNA target recognition, thereby changing potential target transcripts.
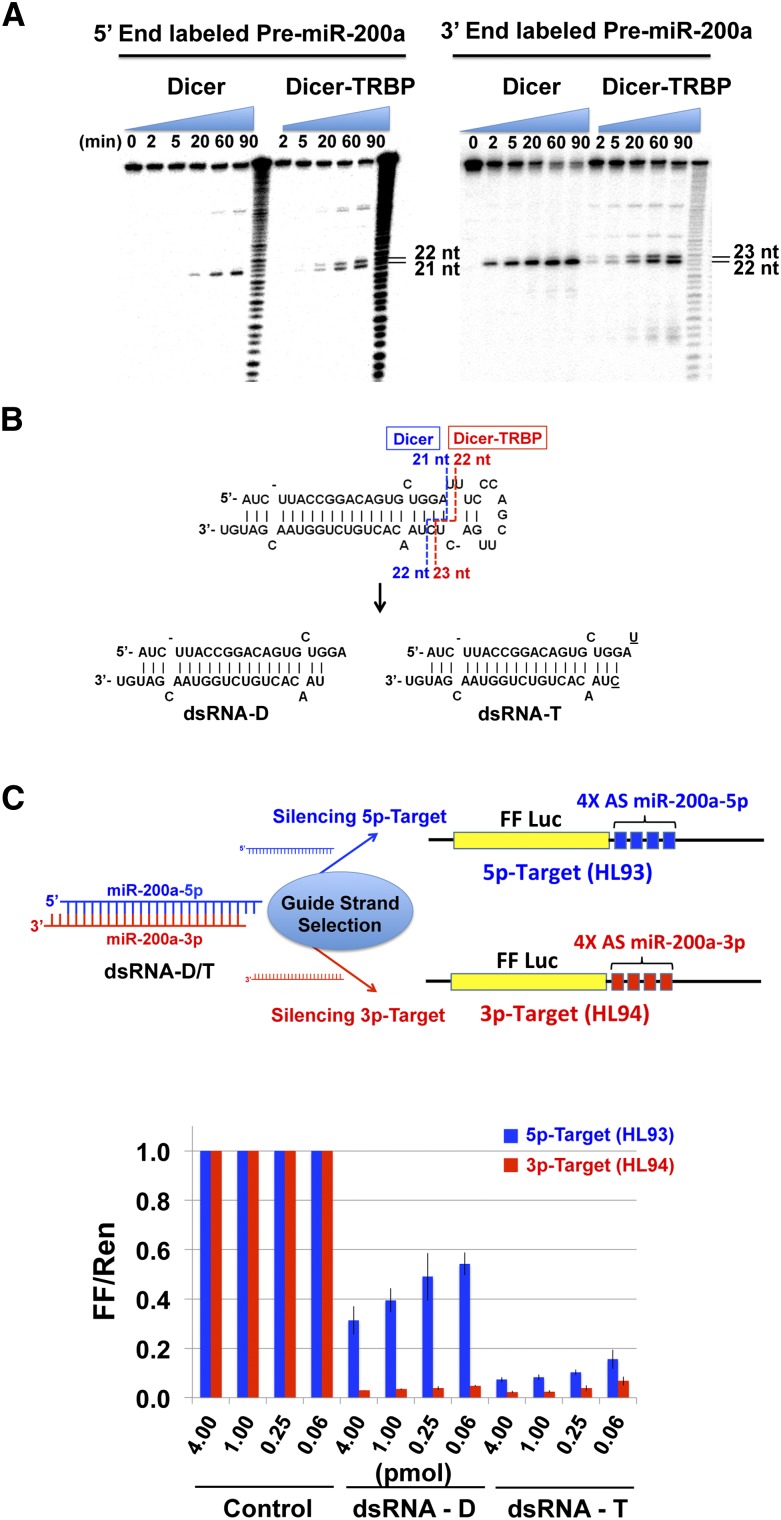
We found that Dicer itself can generate isomiRs in some cases (pre-miR-29 and pre-miR-34c) (Supplemental Fig. S2). Furthermore, we found that TRBP association to Dicer induces or enhances production of isomiRs that are 1 nt longer at the 5′-strand miRNA in the cases of pre-miR-200a, pre-miR-34c, and pre-miR-29 (Fig. 4A; Supplemental Fig. S2). In particular, pre-miR-200a processing by Dicer–TRBP generates 1-nt-longer isomiR-200a (miR-200a-3p) and isomiR-200a* (* indicates a passenger strand, miR-200a-5p) by shifting the cleavage site on the stem 1 nt up for both strands relative to the original cleavage site of Dicer. We validated the cleavage sites of pre-miR-200a by sequencing the products of in vitro pre-miR-200a dicing assays (Fig. 4B). Although the mechanism by which TRBP affects Dicer processing is not understood, it is possible that TRBP alters pre-miR positioning on Dicer by interacting with RNA substrates directly or by changing Dicer conformation.
FIGURE 4.
TRBP contributes to isomiR generation and guide strand selection. (A) Dicer–TRBP generates 1-nt-longer isomiRs for both strands when processing pre-miR-200a ([Dicer], [Dicer–TRBP] = 5 nM, [RNA] = 50 nM). (B) Different dsRNA are produced from Dicer and Dicer–TRBP processing of pre-miR-200a. The exact cleavage sites were identified by sequencing the product small RNA from Dicer and Dicer–TRBP processing reaction. The product dsRNA from Dicer is indicated as dsRNA-D, and the unique product from Dicer–TRBP processing is indicated as dsRNA-T. (C) The effect of differential Dicer cleavage on guide strand selection. dsRNA-D and dsRNA-T show different guide strand selection bias in HEK293 cells. A firefly luciferase construct containing either four tandem 5′-strand miR-200a binding sites in the 3′ UTR (pHL93) or four tandem 3′-strand miR-200a binding sites in the 3′ UTR (HL94) was cotransfected with a control construct that express Renilla luciferase (pRL-TK) (50:1) and with a product dsRNA, either dsRNA-M or dsRNA-I, to HEK293 cells. Dual luciferase assays were performed 36–40 h post-transfection. (Graphs represent data from three experimental replicates.)
Functional effects of differential cleavage upon TRBP association
Following Dicer-catalyzed cleavage, the product dsRNA is expected to be loaded into Argonaute protein complexes such that the guide strand functions in target recognition as part of an active RNA-induced silencing complex (RISC), while the passenger strand is eliminated (Matranga et al. 2005; Rand et al. 2005; Leuschner et al. 2006). Experiments in Drosophila extracts showed that guide strand selection is determined primarily by the thermodynamic stability of the product miRNA duplex ends (Khvorova et al. 2003; Schwarz et al. 2003). For this reason, changes in product dsRNA length can change the relative end stabilities, potentially leading to changes in strand selection during RISC assembly.
To test this possibility in human cells, we used a luciferase reporter assay in HEK293 cells to investigate the silencing activity of RNAs corresponding to the miRNA products produced by cleavage of pre-miR-200a by Dicer or Dicer–TRBP. Two different dsRNAs were used in transfection experiments, one corresponding to the product from Dicer processing (dsRNA-D) and the other, 1 nt longer, corresponding to the unique product from Dicer–TRBP processing (dsRNA-T) (Fig. 4B). Luciferase reporter mRNAs were designed to contain four targeting sites complementary to either the miR-200a-5p (HL93) or miR-200a-3p (HL94) (Fig. 4C). Each luciferase reporter construct was cotransfected together with one of the dsRNAs, dsRNA-D or dsRNA-T, to test the silencing effects of each strand in HEK293 cells (Fig. 4C). Dicer product dsRNA-D shows strong bias for the miR-200a-3p as a guide strand compared with miR-200a-5p: 3p-Target (HL94) containing binding sites for miR-200a-3p was silenced much more efficiently than 5p-Target (HL93) containing miR-200a-5p binding sites (Fig. 4C). This result is explained by the end stability because miR-200a-3p has its 5′ end at the less stable end of the duplex. In the case of Dicer–TRBP product dsRNA-T, both miR-200a-5p and miR-200a-3p are efficient in silencing their respective 5p- and 3p-Targets (HL93 and HL94) (Fig. 4C), suggesting that both strands are working as guide strands to a similar extent. It is possible that the increased thermodynamic stability of the 5′ end of miR-200a-3p enhances selection of miR-200a-5p as a guide strand.
To date, the guide strand selection of miRNAs in vivo has been puzzling because the strand selection varies depending on tissue type (Ro et al. 2007). Our results show that small changes in miRNA sequence have profound effects on miRNA duplex functional asymmetry, altering which strand of a miRNA duplex functions efficiently in mRNA silencing. We find that TRBP-dependent differential cleavage by Dicer could contribute to guide strand selection and target silencing activity of each strand, resulting in differential target recognition.
DISCUSSION
Dicer's RNA-binding partner protein TRBP has been proposed to regulate miRNA processing (Chendrimada et al. 2005; Forstemann et al. 2005; Haase et al. 2005; Paroo et al. 2009), but the molecular mechanisms have been unclear. We find that TRBP, as a molecular binding partner of Dicer, influences pre-miRNA processing by changing both processing kinetics and miRNA product length. These effects may not be unique to the human miRNA processing machinery. Previous findings showed that Loquacious (Loqs), a TRBP homolog in Drosophila, has differential effects on pre-miRNA processing in loqs knockout (KO) flies. The molecular mechanism and the functional significance of the differential effect of Loqs on pre-miR processing have not been determined, but this result suggests that TRBP homologs in other systems may have similar functions during small RNA production (Liu et al. 2007).
Here we show that the RNA hairpin loop and stem structure affect Dicer–TRBP processing with different sensitivity compared with processing by Dicer alone. These differential effects of pre-miR secondary structures on Dicer–TRBP as compared with Dicer alone suggest that TRBP might induce a Dicer conformational change influencing Dicer substrate specificity and kinetics. It is not yet clear how or whether changes in miRNA production rates influence steady-state levels of these RNAs in cells, since other factors could affect miRNA levels in vivo, including transcription rates, miRNA turnover, and the stability of RISC.
Most interestingly, we show here that TRBP affects isomiR production by shifting cleavage sites of Dicer. The effect of TRBP in isomiR formation is substrate-specific and produces unique 5′- and 3′-isomiR populations that are distinct from isomiRs generated by 3′-end modification, including adenylation, uridylation, or truncation of miRNA (Fernandez-Valverde et al. 2010). Generating different pairs of isomiR products changes guide strand selection and target silencing activity in cells and could have similar effects in vivo. In addition, TRBP-induced 3′-strand-isomiR (isomiR-3p) production shifts the seed sequence by one nucleotide, potentially changing the set of target mRNAs that are recognized.
These findings support a direct role for TRBP in regulating miRNA processing kinetics and isoform specificity. It is possible that TRBP affects pre-miRNA processing in vivo in a similar way. However, several regulatory mechanisms of TRBP must be considered, including differential phosphorylation, stabilization by Dicer, and competition with different protein binding partners for access to Dicer, as these will almost certainly influence miRNA biogenesis. For example, four serine phosphorylation sites on TRBP alter processing of a set of pre-miRNAs related to cell growth when fully phosphorylated (Paroo et al. 2009), although the mechanism of this effect is unknown. In addition, proteins other than Dicer, including PKR, PACT, and Merlin, may compete for TRBP binding (Daher et al. 2001; Lee et al. 2004; Laraki et al. 2008). Other pre-miRNA binding proteins such as Lin28, hnRNPA1, and KHSRP (Heo et al. 2008; Trabucchi et al. 2009; Michlewski and Caceres 2010) also influence pre-miR processing in vivo and may change the effects of TRBP. Hence, discovering how TRBP alters pre-miR processing in vivo will be both interesting and challenging to decipher.
In conclusion, our results show the unprecedented role of TRBP in pre-miRNA processing by both affecting kinetics as well as isoform specificity, resulting in altered guide strand selection and target silencing activity. These results highlight the possibility that changes in TRBP levels and post-translational modification could alter dicing efficiency/specificity in vivo and imply that TRBP homologs in other systems may have similar functions during small RNA production.
MATERIALS AND METHODS
Proteins, Dicer–TRBP complex, and RNA substrates preparation
Dicer, TRBP, and Dicer–TRBP complex were prepared as reported before (MacRae et al. 2008). Pre-miRNA substrates were prepared by in vitro transcription using T7 RNA polymerase, and the transcribed RNAs have one ribozyme at each end for homogeneous RNA production. After in vitro transcription, RNAs were gel-purified and end-labeled for the processing assay. For 5′-end labeling, RNA was incubated with T4 polynucleotide kinase (New England Biolabs Inc.) and [γ-32P]ATP for 1 h at 37°C. For 3′-end labeling, RNA was incubated with T4 RNA ligase (New England Biolabs Inc.) and [5′-32P]pCp for overnight at 16°C.
The sequences of RNA substrates used in this study are the following (mutated or inserted nucleotides are underlined in mutant pre-miR sequences):
pre-let-7a, 5′-UGAGGUAGUAGGUUGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUAUACAAUCUACUGUCUUACC-3′;
pre-miR-21, 5′-UAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUC-3′;
HL1, 5′-UAGCUUAUCAGACUGAUGUUGAGUUAGGGUCACACCCACCACUGGGAGAUCCAACACCAGUCGAUGGGCUGUC-3′;
HL2, 5′-UAGCUUAUCAGACUGAUGUUGAGUUAGGGUCACACCCACCACUGGGAGAUCUCAACACCAGUCGAUGGGCUGUC-3′;
HL3, 5′-UAGCUUAUCAGACUGAUGUUGACUGUUGUUAGGGUCACACCCACCACUGGGAGAUCAAUGGUCAACACCAGUCGAUGGGCUGUC-3′;
HL4, 5′-UAGCUUAUCAGACUGAUGUUGAGUUACCAGAUCUCAACACCAGUCGAUGGGCUGUC-3′;
pre-miR-200a, 5′-AUCUUACCGGACAGUGCUGGAUUUCCCAGCUUGACUCUAACACUGUCUGGUAACGAUGU-3′;
pre-miR-200a (3′-1nt) for 3′-end labeling, 5′-AUCUUACCGGACAGUGCUGGAUUUCCCAGCUUGACUCUAACACUGUCUGGUAACGAUG-3′;
pre-miR-29a, 5′-ACUGAUUUCUUUUGGUGUUCAGAGUCAAUAUAAUUUUCUAGCACCAUCUGAAAUCGGUUA-3′;
pre-miR-31, 5′-AGGCAAGAUGCUGGCAUAGCUGUUGAACUGGGAACCUGCUAUGCCAACAUAUUGCCAUC-3′;
mutant pre-miR-31, 5′-AGGCAAGAUGCUGGCAUAGCAUGUUGAACUGGGAACAUGCUAUGCCAACAUAUUGCCAUC-3′;
pre-miR-125b-1, 5′-UCCCUGAGACCCUAACUUGUGAUGUUUACCGUUUAAAUCCACGGGUUAGGCUCUUGGGAGCU-3′;
mutant pre-miR-125b-1, 5′-UCCCUGAGACCCUAACUUGUGAUGUUUACCGUUUAAAUCUCACGGGUUAGGCUCUUGGGAGCU-3′.
Kinetic assays
5′- or 3′-end-labeled RNA was processed with the indicated amount of Dicer or Dicer–TRBP in dicing buffer (20 mM Tris-HCl at pH 6.5, 1.5 mM MgCl2, 25 mM NaCl, 1 mM dithiothreitol, and 1% glycerol). Each time point sample was prepared by being quenched with 1.2 volumes of loading buffer (95% formamide, 18 mM ethylenediaminetetraacetic acid, 0.025% SDS, 0.1% xylene cyanol, and 0.1% bromophenol blue). Samples were heated for 10 min at 70°C before loading to 12%–15% denaturing 7 M urea polyacrylamide gel electrophoresis. The gel was dried, and the amounts of substrates and products were quantified with a PhosphorImager (GE Healthcare).
Luciferase assays
pGL03 control constructs were cut with an XbaI restriction enzyme, and inserts were put into the 3′ UTR to make reporter constructs having miRNA binding sites. pHL93 and pHL94 were prepared by inserting four repeated antisense sequences of miR-200a-5p and miR-200a-3p. The inserted sequences in the pGL03 control vector for pHL93 and pHL94 are the following (XbaI and SpeI restriction sites were used and indicated as italic and underlined):
pHL93: 5′-ACTAGTTCCAGCACTGTCCGGTAAGATGGATCCTCCAGCACTGTCCGGTAAGATTCCAGCACTGTCCGGTAAGATTCCAGCACTGTCCGGTAAGATTCTAGA-3′;
pHL94: 5′-ACTAGTACATCGTTACCAGACAGTGTTAGGATCCACATCGTTACCAGACAGTGTTAACATCGTTACCAGACAGTGTTAACATCGTTACCAGACAGTGTTATCTAGA-3′.
The pGL03 control base construct was cotransfected to HEK293 cells when cells are ∼80%–90% confluent with the pRL-TK construct (with ratio 50:1) and RNA using Lipofectamine 2000. Amounts per one well of a 96-well TC plate were 10 ng of pHL93 or pHL94; 0.2 ng of pRL-TK; 4 pmol (26.7 nM), 1 pmol (6.7 nM), 0.25 pmol (1.7 nM), and 0.06 pmol (0.4 nM) of dsRNA; and 0.25 μL of Lipofectamine 2000 were used. The cells were harvested 36–40 h post-transfection and assayed with the dual luciferase assay (Promega).
Filter binding assay
Dicer or Dicer–TRBP was incubated in buffer (20 mM HEPES (pH 7.5), 25 mM KCl, 5 mM ethylenediaminetetraacetic acid, 1 mM tris(2-carboxyethyl)phosphine, 1% glycerol, 0.01% Igepal 630-CA), with 0.2–20 nM 5′-end 32P-labeled pre-miR substrates for 60 min on ice in 30 μL of total volume.
Samples were applied to a dot-blot apparatus containing two pieces of Whatman filter paper below three membranes: 0.2 μm pore size Tuffryn (Pall Co.), 0.1 μm pore size Protran (Whatman) and Hybond-N (Amersham) using vacuum. After applying the samples, the membranes were washed with 50 μL of binding buffer. The amounts of free RNA (retained on Hybond-N membrane) and protein-bound RNA (retained on Protran membrane) were quantified with a PhosphorImager (GE Healthcare). Kds were determined by fitting the data to binding isotherms with KaleidaGraph (Synergy Software).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
J.A.D. is a Howard Hughes Medical Institute Investigator. This work was funded in part by the National Institutes of Health. We thank all the Doudna laboratory members especially Cameron L. Noland for helpful discussions. We thank O.K. Yoon for help in sequence analysis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035501.112.
REFERENCES
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA 2010. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol 404: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, et al. 2001. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem 276: 33899–33905 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS 2010. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236 doi: 10.1371/journal.pbio.0030236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T 2005. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Laraki G, Clerzius G, Daher A, Melendez-Pena C, Daniels S, Gatignol A 2008. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein–protein interactions. RNA Biol 5: 92–103 [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim H, Ryu CH, Kim JY, Choi BH, Lim Y, Huh PW, Kim YH, Lee KH, Jun TY, et al. 2004. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem 279: 30265–30273 [DOI] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K 2010. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 16: 2170–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q 2007. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 13: 2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long XB, Sun GB, Hu S, Liang GT, Wang N, Zhang XH, Cao PP, Zhen HT, Cui YH, Liu Z 2009. Let-7a microRNA functions as a potential tumor suppressor in human laryngeal cancer. Oncol Rep 22: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA 2008. Autoinhibition of human Dicer by its internal helicase domain. J Mol Biol 380: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Lorsch JR 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol 330: 917–924 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. 2009. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Michlewski G, Caceres JF 2010. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 17: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q 2009. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 139: 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA 2006. Short RNAs repress translation after initiation in mammalian cells. Mol Cell 21: 533–542 [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W 2007. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC 2010. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer 10: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY 2007. miR-21-mediated tumor growth. Oncogene 26: 2799–2803 [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z 2009. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 15: 1443–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]