A detailed functional analysis of the CRISPR-associated Thermus thermophilus Cas5d protein identifies it as a sequence-specific pre-crRNA processing endonuclease. High-resolution X-ray structural studies of a second Cas5d ortholog highlight similarities with previously characterized CRISPR processing enzymes. A combination of structural and functional analyses based on these results allow a modeling of the interaction of Cas5d with its RNA substrate and suggest that it is a member of a larger conserved family of CRISPR RNA endonucleases.
Keywords: CRISPR, RNA endonuclease, RNA processing
Abstract
Small RNAs derived from clustered, regularly interspaced, short palindromic repeat (CRISPR) loci in bacteria and archaea are involved in an adaptable and heritable gene-silencing pathway. Resistance to invasive genetic material is conferred by the incorporation of short DNA sequences derived from this material into the genome as CRISPR spacer elements separated by short repeat sequences. Processing of long primary transcripts (pre-crRNAs) containing these repeats by a CRISPR-associated (Cas) RNA endonuclease generates the mature effector RNAs that target foreign nucleic acid for degradation. Here we describe functional studies of a Cas5d ortholog, and high-resolution structural studies of a second Cas5d family member, demonstrating that Cas5d is a sequence-specific RNA endonuclease that cleaves CRISPR repeats and is thus responsible for processing of pre-crRNA. Analysis of the structural homology of Cas5d with the previously characterized Cse3 protein allows us to model the interaction of Cas5d with its RNA substrate and conclude that it is a member of a larger family of CRISPR RNA endonucleases.
INTRODUCTION
Individual CRISPR loci in bacterial and archaeal genomes feature a cluster of repeats separated by short spacer elements. They also typically include a set of CRISPR-associated (cas) genes that code for proteins, a number of which have been shown to be necessary for CRISPR-based gene interference. Specific subtypes of CRISPR systems are defined by the structure and organization of the repeats, as well as by the particular set of accompanying protein-coding cas genes (for review, see Deveau et al. 2010; Horvath and Barrangou 2010; Marraffini and Sontheimer 2010).
Three major types of CRISPR systems that may be further divided into 10 subtypes have been described (Makarova et al. 2011a). In the type I system of Escherichia coli, the products of the cas1 and cas2 genes are not required to effect CRISPR-based immunity using existing CRISPR spacers, and are therefore believed to function in the acquisition of new repeats (Brouns et al. 2008; Makarova et al. 2011a). The Cas3 protein, endowed with both helicase and HD nuclease domains, has been implicated as the ultimate effector protein in gene silencing, with evidence suggesting that it targets mature crRNAs to invading DNA sequences (Beloglazova et al. 2011; Mulepati and Bailey 2011; Sinkunas et al. 2011).
Analysis of both wild-type and mutant E. coli strains suggests that an initial pre-crRNA is processed to yield mature crRNA by endonucleolytic cleavage at the base of the repeat hairpin RNA structure (Brouns et al. 2008). The enzymatic activity required for this processing has been shown to reside in the Cse3 protein, a component of the multiprotein CRISPR-associated complex for antiviral defense (Cascade) complex containing Cse1–Cse4 and Cas5e (Jore et al. 2011; Wiedenheft et al. 2011).
Type II CRISPR systems, which have been shown to target phage and plasmid DNA, feature a single large protein, Cas9, which is required for both generation of effector RNAs and target cleavage (Deltcheva et al. 2011; Makarova et al. 2011a; Jinek et al. 2012). Maturation of type II pre-crRNA has been shown to involve formation of a duplex between a single-stranded CRISPR repeat and trans-encoded tracrRNA, followed by cleavage of the repeat by RNase III (Deltcheva et al. 2011). Finally, in the type III system, for which there is evidence of the targeting of both DNA and RNA (Marraffini and Sontheimer 2008; Hale et al. 2009; Hatoum-Aslan et al. 2011), the processing of pre-crRNA occurs in two steps, the first of which, endonucleolytic cleavage of a repeat RNA, resembles the type I processing mechanism and is performed by the cas6 protein product (Carte et al. 2008, 2010; Wang et al. 2011).
The type I CRISPR system is the most well-understood from a structural and mechanistic perspective. The structure and activity of the effector Cas3 HD nuclease have recently been reported (Beloglazova et al. 2011; Mulepati and Bailey 2011). Additionally, the high-resolution X-ray structures of the pre-crRNA processing Cse3, alone and complexed to hairpin substrate RNA, have been described (Gesner et al. 2011; Sashital et al. 2011). Recently, cryo-electron microscopy was used to determine the subnanometre resolution structures of the Ecoli Cascade complex (containing the proteins Cse1–4, Cse5e, and crRNA) both before and after target binding (Wiedenheft et al. 2011).
The diversity of CRISPR systems complicates the analysis of individual pathways. For example, CRISPR repeats in the type I Dvulg CRISPR system are predicted to form stable RNA hairpins (Kunin et al. 2007) analogous to that described for the Cse3 substrate RNA, but the identity of the pre-crRNA processing activity has been unclear. Here we report the high-resolution X-ray structure of a hypothetical protein from Mannheimia succiniciproducens that shares strong sequence similarity with the Cas5d family of Dvulg-type Cas proteins. We show that the Cas5d ortholog from Thermus thermophilus is an RNA endonuclease that specifically binds and cleaves pre-crRNA, and establish the mechanism of RNA cleavage. Comparison of Cas5d by structural alignment with Cse3 allows us to model the interaction of Cas5d with its hairpin RNA substrate, and argues for a conservation of RNA recognition between diverse CRISPR RNA processing enzymes.
RESULTS
Crystal structure and homology modeling of Cas5d
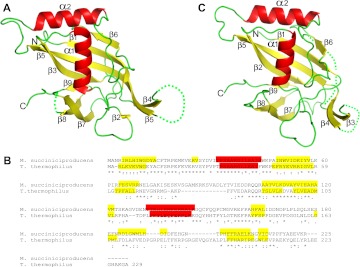
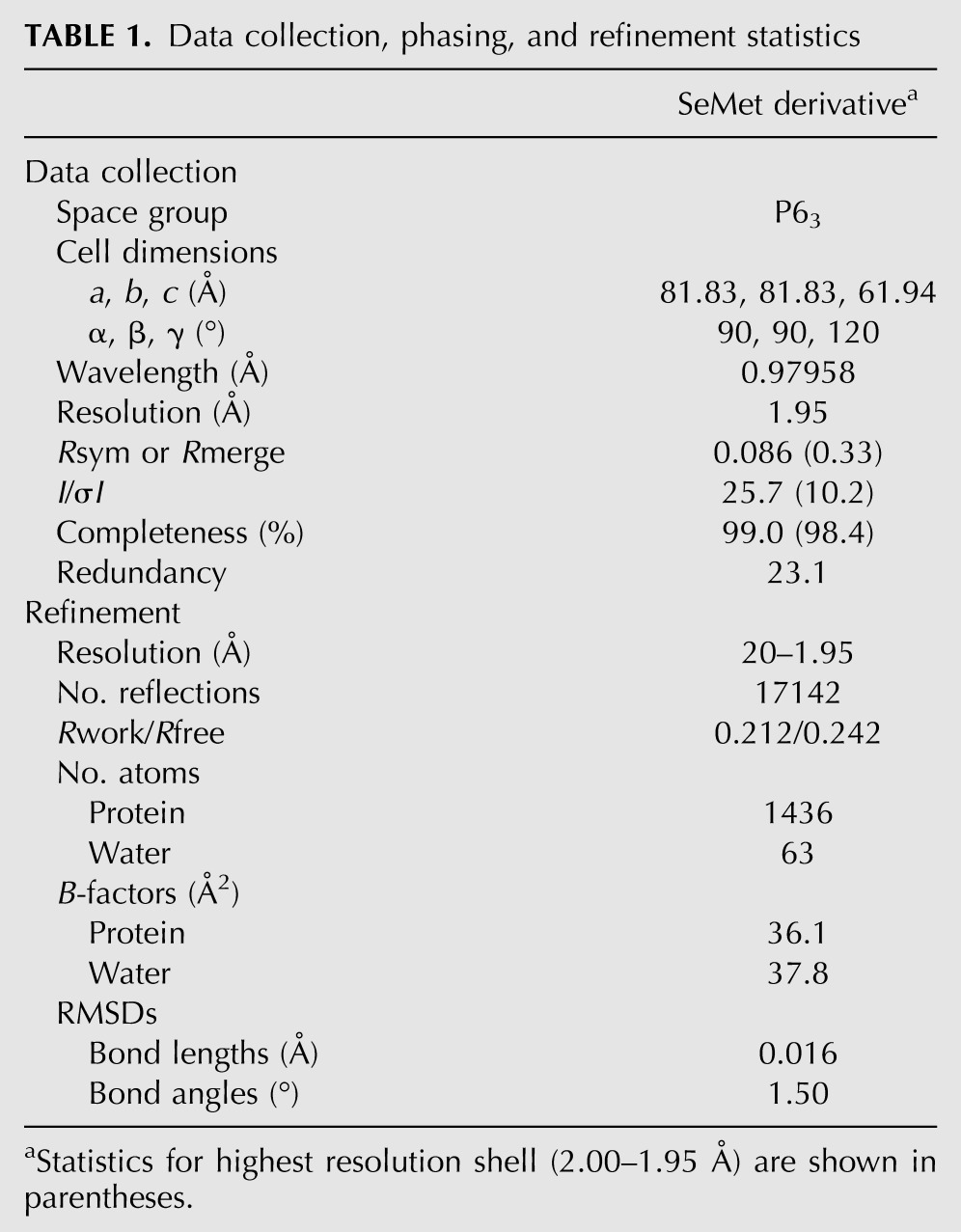
The ms0988 gene codes for a hypothetical protein predicted from the genomic sequence of the capnophilic (favored growth in the presence of CO2) Gram-negative bacterium M. succiniciproducens cultured from bovine rumen (Hong et al. 2004). The X-ray structure of recombinant MS0988 was determined using the single anomalous dispersion (SAD) method and refined to a resolution of 1.95 Å (Fig. 1A; Table 1). Inspection of the structure revealed the presence of an embedded modified ferredoxin-like fold with the canonical β-α-β-β-α-β arrangement of secondary structure elements, a feature common to many Cas proteins (Makarova et al. 2011b). Notable in the structure was an extensive disordered region (amino acids 71–103) representing a loop between β4 and β5.
FIGURE 1.
Structural analysis of the CRISPR RAMP domain family member Cas5d. (A, top) Ribbon diagram derived from the X-ray structure of M. succiniciproducens MS0988. α-helices and β-strands are colored red and yellow, respectively. (B) Alignment of MS0988 and T. thermophilus TTP0133 primary sequence and secondary structure diagrams depicting α-helices (red) and β-strands (yellow) derived from pdb and homology modeling by Protein Homology/analogY Recognition Engine (Phyre; http://www.sbg.bio.ic.ac.uk/phyre2) (Kelley and Sternberg 2009). Amino acids 71–103, 196–199, and 221–235 are disordered in the MS0988 structure. Also indicated are sequence identity (*), conservation (:), and partial conservation (.). (C, top) Structural model of T. thermophilus Cas 5d based on submission of the primary sequence of TTP0133 to Phyre.
TABLE 1.
Data collection, phasing, and refinement statistics
BLAST analysis of the ms0988 sequence revealed a strong homology with the Cas5 family of CRISPR-associated proteins, and specifically with the Cas5d class characteristic of the Dvulg CRISPR subtype (Haft et al. 2005; see Supplemental Material). Cas5 was first described as a family of proteins about 250 amino acids in length with a conserved 43-amino acid N-terminal region (Haft et al. 2005). It was originally identified in five separate CRISPR subtypes, with sequence outside the conserved N-terminal region specific for each subtype. The family includes the hypothetical Cas5d protein coded for by ttp0133 upstream of the CRISPR-5 locus of the T. thermophilus HB27 megaplasmid pTT27 (Henne et al. 2004). The ttp0133 sequence is embedded within an operon that includes putative cas3, cas5d, cas8c, cas7, and cas4 genes corresponding to the Dvulg CRISPR subtype (Haft et al. 2005). These are located ∼100 nucleotides (nt) upstream of seven copies of a CRISPR repeat element (5′-GTTGCACCGGCCCGAAAGGGCCGGTGAGGATTGAAAC-3′) (Grissa et al. 2007a,b) similar to the hairpin repeats associated with the Dvulg CRISPR subtype (Kunin et al. 2007). Alignment of primary sequences revealed that TTP0133 (hereafter Cas5d) is 40% identical and 65% similar to that of MS0988 (Fig. 1B), suggesting that the structure of the latter forms an excellent basis for homology modeling of the structure of the T. thermophilus Cas5d (Fig. 1C).
A distinguishing feature of the Dvulg CRISPR subtype is the lack of a gene coding for an ortholog of Cas6, the pre-crRNA processing RNA endonuclease found in type I and III CRISPR systems. It has been proposed, based on a bioinformatic analysis, that Cas5d or Cas7 might process the pre-crRNA in this system (Makarova et al. 2011a,b). Three pre-crRNA processing endonucleases have been extensively characterized structurally and functionally (Carte et al. 2008, 2010; Haurwitz et al. 2010; Gesner et al. 2011; Sashital et al. 2011; Wang et al. 2011; Sternberg et al. 2012). Both the Pyrococcus furiosus Cas6 and T. thermophilus Cse3 feature tandem ferredoxin-like folds, although their mode of interaction with their RNA substrates is clearly distinct. A recent structural analysis of Cas6 RNA interaction suggests that the 5′ end of a single-stranded repeat is anchored in a groove between the ferredoxin-like folds, and traverses the protein to position the site of cleavage at the active site on the opposing surface of the protein (Wang et al. 2011). In contrast, the interaction of T. thermophilus Cse3 with its hairpin RNA substrate is similar to the RNA-binding mode of Csy4, the Cse3 functional homolog from Pseudomonas aeruginosa. Csy4 features a single N-terminal ferredoxin-like fold and a separate C-terminal domain that includes two α-helices joined to the main body by extended linker sequences (Haurwitz et al. 2010). Both RNA–protein interfaces feature major-groove RNA recognition of an incomplete turn of an A-form helix. In the former case this involves sequence-specific interactions by a short β-hairpin extending from the C-terminal ferredoxin-like fold; in the latter, an α-helix of the C-terminal domain plays a similar role in major groove recognition.
CRISPR repeat binding and cleavage by Cas5d
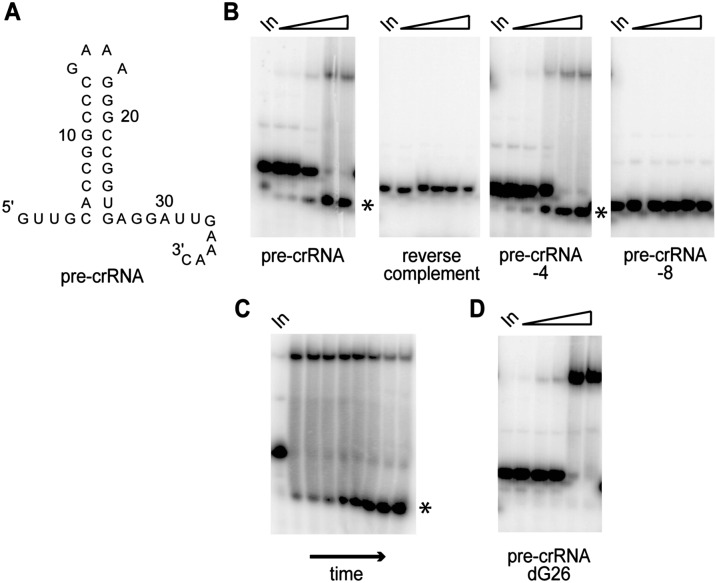
In order to elucidate the function of Cas5d in the CRISPR pathway, we cloned, expressed, and purified T. thermophilus TTP0133. We examined the RNA-binding properties of Cas5d by electrophoretic mobility shift assay (EMSA) using a panel of RNAs based on the T. thermophilus CRISPR repeat (Fig. 2A). Cas5d tightly bound (KD ∼50 nM) the 37-nt RNA representing the downstream repeat element, but even at high concentrations of protein (1 μM) no affinity was observed for the reverse complement of the repeat RNA (Fig. 2B).
FIGURE 2.
Specific pre-crRNA binding and cleavage by Cas5d. (A) Secondary structure of T. thermophilus Dvulg repeat RNA. (B) Gel mobility-shift assays for RNA binding by Cas5d with increasing concentrations of protein (2, 30, 40, 500, 1000 nM). (Left to right) A 37-nucleiotide model of pre-crRNA as shown in A, reverse complement of A RNA, 4- and 8-nt truncations of pre-crRNA. Cleaved RNA is indicated (*). (C) Time course for gel mobility-shift assay of pre-crRNA binding by 100 nM of Cas5d. (Left to right) 1, 2, 5, 10, 15, 30, 45, 60 min. Cleaved RNA is indicated (*). (D) Gel mobility-shift assays for binding of pre-crRNA substituted with dG at position 26 by Cas5d with increasing concentrations of protein (2, 30, 40, 500, 1000 nM).
In the EMSA experiment, we noted the concentration-dependent generation of a specific lower mobility species that we concluded to be a Cas5d cleavage product (Fig. 2B). Analysis of both the upper and lower bands from this experiment by denaturing PAGE revealed that the upper species included both full-length and cleaved RNA, while the lower species corresponded to the discrete cleavage product (data not shown). In a time course of binding, we noted that binding of Cas5d to substrate RNA was rapid, with all of the RNA bound within a minute, and dissociation occurring as a function of RNA cleavage over time (Fig. 2C). These results strongly suggest that Cas5d is a sequence-specific RNA endonuclease responsible for processing pre-crRNAs in the CRISPR pathway.
We prepared modified repeat RNAs truncated by four and eight nucleotides at the 3′ end in order to examine the effect of these modifications on RNA binding and cleavage by Cas5d. In the former case, there was no significant impairment of RNA binding or cleavage, while in the latter, no significant RNA binding or cleavage was observed (Fig. 2B). Thus, a portion of the 3′ tail of the substrate RNA is important for both RNA binding and cleavage; nevertheless, the enzyme showed no affinity for the 3′ cleavage product (data not shown).
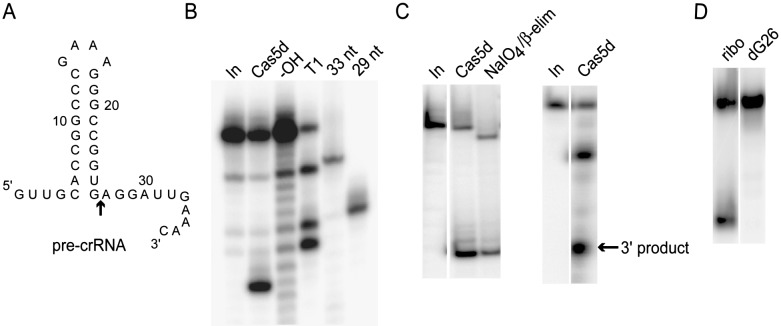
We mapped the pre-crRNA cleavage site by comparison with RNase T1 and base hydrolysis ladders, and determined it be 3′ to G26 at the base of the predicted RNA hairpin (Fig. 3A,B). The 5′ cleavage product was resistant to oxidation with periodate and base-mediated elimination, indicating a 3′ end lacking 2′ and/or 3′ hydroxyls. The 3′ product could be 5′ end-labeled, indicating the presence of a 5′ hydroxyl (Fig. 3C). The generation of functionalized 2′/3′ and free 5′-hydroxyl termini on the longer and shorter products, respectively, is consistent with a cleavage mechanism involving attack of the G26 2′-hydroxyl group on the scissile phosphate, as observed in both protein and RNA-catalyzed RNA cleavage. Also consistent with this mechanism, substitution of a 2′-deoxy residue at the G26 position abolished cleavage in the presence of the enzyme, but the affinity of Cas5d for this RNA was unimpaired (Figs. 2D, 3D). Previously characterized Cse3 CRISPR processing also proceeds via this mechanism (Gesner et al. 2011; Jore et al. 2011). As observed with the type I Cse3 (Gesner et al. 2011; Sashital et al. 2011) and Csy4 (Haurwitz et al. 2010) endonucleases, and the type III Cas6 endonuclease (Carte et al. 2008), pre-crRNA cleavage by Cas5d was found to be metal independent (data not shown).
FIGURE 3.
Characterization of cleavage products and mechanism of RNA cleavage by Cas5d. (A) Secondary structure of T. thermophilus Dvulg repeat RNA. Position of RNA cleavage is indicated by arrow. (B) Mapping of cleavage location by denaturing PAGE showing comparison to RNase T1 and base hydrolysis ladders. Also shown are 29- and 33-nt RNA size markers corresponding to 3′ truncations of the pre-crRNA. (C, left) Periodate/base elimination of pre-crRNA treated with Cas5d affects the mobility of the full-length pre-crRNA, but not that of the 5′ product, indicating that the latter has a modified 2′/3′ terminus. (C, right) The smaller of two RNAs produced upon Cas5d cleavage of pre-crRNA is labeled by 32P upon treatment with T4 polynucleotide kinase, indicating the presence of a free 5′ hydroxyl on the 3′ product. (D) A modified pre-crRNA substrate containing a dG residue at position 26 is not cleaved by Cas5d supporting a model whereby RNA cleavage occurs by intramolecular attack of the G26 2′ hydroxyl on the scissile phosphate.
Following the experiments described above, we tested the activity of recombinant MS0988 protein. Although we could not measure binding by EMSA of MS0988 to a 32-nt RNA derived from the repeat associated with the ms0988 locus (data not shown), we were able to observe MS0988-mediated cleavage of this RNA and mapped the cleavage to the base of a predicted hairpin structure (see Supplemental Material). This cleavage is specific since MS0988 did not cleave the T. thermophilus repeat described above, just as the T. thermophilus Cas5d ortholog was not observed to cleave the MS0988 substrate (data not shown).
Modeling CRISPR RNA recognition by Cas5d
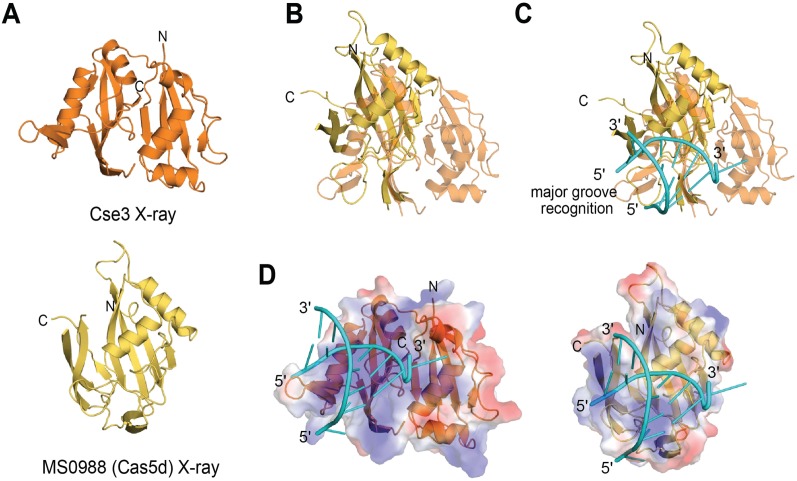
The fact that Cas5d recognizes an RNA substrate structurally similar to that bound by Csy4 and Cse3 suggests that the mode of Cas5d RNA recognition might be similar to that observed with those proteins. As both MS0988 Cas5d and Cse3 feature ferredoxin-like folds (Fig. 4A), we used iSuperpose from the Mobyle suite (Neron et al. 2009) to determine the structural overlap between MS0988 Cas5d and Cse3; the result of this calculation is a superimposition of the Cas5d structure on the C-terminal RNA recognition domain of Cse3 (Fig. 4B) (PDB 3QRQ). We then examined this superposition within the context of the Cse-3•RNA X-ray structure. This model suggests that as with Cse3 and Csy4, the face of Cas5d obverse to the β-sheet of the ferredoxin-like fold is most proximal to the bound RNA (Fig. 4C; Haurwitz et al. 2010; Gesner et al. 2011; Sashital et al. 2011). This is consistent in both cases with the overall surface electrostatics of the Cse3 and Cas5d proteins, in which a significant basic patch corresponds to the actual and modeled RNA-binding surfaces (Fig. 4D). Strikingly, in the model, the loop connecting β2 and α1 of Cas5d is predicted to function similarly to the major groove recognition elements of Cse3 and Csy4. It occupies a space analogous to the major groove-recognition hairpin of Cse3, but does not overlay with the major groove-recognition helix of Csy4.
FIGURE 4.
Homology modeling of Cas5d as a member of the larger pre-crRNA RNA endonuclease family. (A) X-ray structures of (top) Cse3 (PDB 3QRQ) and (bottom) MS0988 (PDB 3KG4). (B) Overlay of the X-ray structures of MS0988 (yellow) and Cse3 (orange) as determined by iSuperpose (Neron et al. 2009). (C) Overlay of the X-ray structure of MS0988 (yellow) with that of a Cse3•RNA complex (PDB 3QRQ). Cse3 and RNA are depicted in yellow and cyan, respectively. (D) Surface electrostatics for Cse3 and MS0988 within the actual and modeled protein RNA complexes. (Blue) Basic; (red) acidic; (white) neutral.
We performed site-directed mutagenesis to examine the functional importance of several conserved amino acid residues within the loop connecting β2 and α1 that is predicted in the model to be involved in RNA recognition by Cas5d. Tyr27 is a conserved residue found at the end of this loop; Glu23 is also conserved, but predicted to be a surface-exposed residue within the loop (see Supplemental Material). We examined cleavage of the fully ribo repeat as well as binding to the deoxyG-substituted repeat RNA. While the conservative Y27F mutation had no affect on either RNA binding or cleavage by Cas5d, mutation of Glu23 to Gln abolished RNA cleavage, likely due to the fact that this mutation also abolished binding of Cas5d to the RNA (Fig. 5; Supplemental Material).
FIGURE 5.
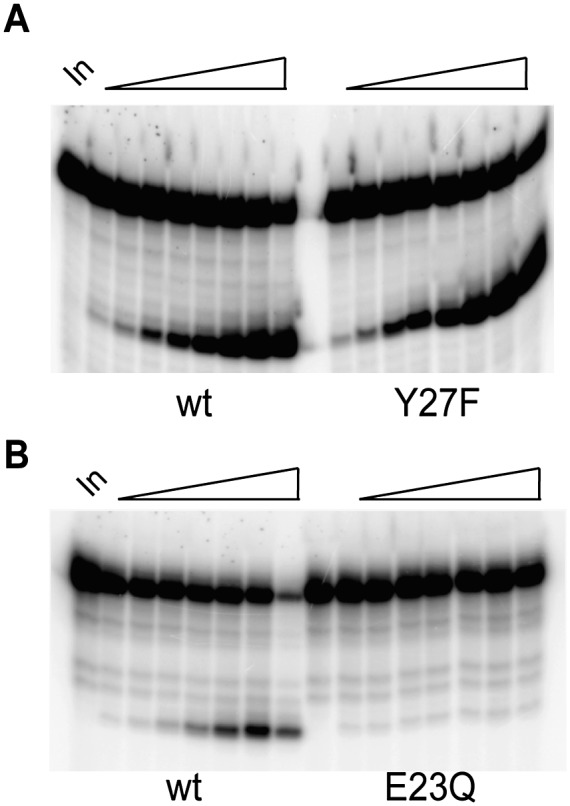
Functional analysis of Cas5d mutants. (A) Comparison of RNA endonucleolytic activity mediated by wild-type and Y27F Cas5d. (B) Comparison of RNA endonucleolytic activity mediated by wild-type and E23Q Cas5d. Denaturing PAGE analysis of 32P end-labeled repeat RNA incubated with 100 nM wild-type, Y27F, and E23Q Cas5d over a (left to right) 1-, 2-, 5-, 10-, 15-, 30-, 45-, 60-min time course.
DISCUSSION
In diverse CRISPR systems, the processing of pre-crRNAs to generate mature effector RNAs involves the activity of an incompletely defined set of endoribonucleases that recognize and cleave distinct substrates. Those characterized to date include Cas6 that interacts with and cleaves single-stranded RNA (Carte et al. 2008, 2010), as well as Cse3 and Csy4 that bind and cleave hairpin RNA repeats (Haurwitz et al. 2010; Gesner et al. 2011; Sashital et al. 2011). Here we have reported the structure of a Cas5d ortholog (Figs. 1A, 4A) that consists of a single ferredoxin-like fold and also demonstrated that T. thermophilus Cas5d specifically binds and cleaves the hairpin RNA of a Dvulg CRISPR repeat. The finding that the M. succiniciproducens ortholog cleaves its cognate RNA provides further support for the identification of Cas5d as the processing endonuclease in the Dvulg CRISPR subtype.
Analysis of the MS0988 Cas5d structure (Fig. 1; Supplemental Material) reveals that the N-terminal homology sequence that defines that the Cas5 family does not consist of a defined domain including, as it does, portions of β-1 as well as all of α-1 and β-3. The elements of this sequence certainly contribute structurally to the overall ferredoxin-like fold; the predicted RNA-recognition loop between β2 and α1 is also contained within this sequence.
RNA binding and cleavage properties of Cas5d
The results reported here are consistent with Cas5d promoting RNA cleavage by catalysis of the intramolecular attack of a 2′-hydroxyl on the scissile phosphodiester to generate a 2′/3′ cyclic phosphodiester and free 5′ hydroxyl in the 5′ and 3′ products, respectively. In this respect, the enzymatic mechanism of pre-crRNA cleavage resembles that involving Cse3 (Gesner et al. 2011; Jore et al. 2011; Sashital et al. 2011) and is distinct from the hydrolytic cleavage of type II CRISPR repeats by RNase III that generate products with a free 3′ hydroxyl and 5′ phosphate (Deltcheva et al. 2011).
The weak affinity between Cas5d and the cleaved RNA substrate contrasts with the examples of Cse3 and Csy4 in which the cleavage product remains tightly bound to the enzyme (Haurwitz et al. 2010; Gesner et al. 2011; Sternberg et al. 2012). The former observation is consistent with the cryo-em structure in which Cse3 is an integral component of the targeting Cascade complex (Jore et al. 2011; Wiedenheft et al. 2011). It will be of interest to determine whether Cas5d is organized in a complex similar to Cascade, and whether product release following cleavage informs the targeting mechanism in this case.
The Cas5d substrate RNA is predicted to form a double-stranded 9-bp hairpin structure capped by a GNRA tetraloop. The high G-C content and presence of the tetraloop suggest that the hairpin structure is very stable. Both Csy4 and Cse3 recognize the major groove of their RNA substrates, suggesting that a similar mechanism of RNA recognition might be the basis for Cas5d substrate interaction; at least 6 bp of RNA could be recognized in this manner, based on analogy with the Cse3-RNA and Csy4-RNA complexes (Haurwitz et al. 2010; Gesner et al. 2011; Sashital et al. 2011).
Overall features of RNA recognition and catalysis
The results of site-directed mutagenesis support the model suggesting that the loop between β2 and α1 is involved in RNA recognition. Although Tyr27 is an absolutely conserved residue, the side-chain does not appear to be involved in either binding or catalysis, consistent with its predicted location in the structure; the conservation of this residue must be a consequence of its contribution to the stability of the overall Cas5d fold. In contrast, the effects on RNA-binding upon mutation of Glu23 argues for the importance of this residue in mediating Cas5d–RNA interaction; although not common, a variety of modes of interaction of Glu with nucleotide bases in protein–nucleic acid complexes have been documented (Kondo and Westhof 2011).
The proposed RNA recognition loop is found within the region of conserved amino acids that originally defined the Cas5 family (Haft et al. 2005) and our model suggests that the active site may lie outside of this region. Identification of the Cas5d active site will require further functional and structural analysis including high-resolution structural analysis of a Cas5d–RNA complex. Given the lack of conserved, potentially catalytic residues among Cas5d homologs and considering the model proposed here, Cas5d may promote RNA cleavage by appropriate orientation of the pre-crRNA substrate.
Together, the structural and functional analyses presented here suggest that Cas5d is a member of a larger family of CRISPR RNA endonucleases that recognize structurally similar RNA substrates through related mechanisms. Cas5d resembles both Cse3 and Csy4 in recognition and cleavage of hairpin RNA CRISPR repeats (Haurwitz et al. 2010; Gesner et al. 2011; Sashital et al. 2011). The strong sequence specificity of binding suggests that Cas5d may recognize the major groove of an A-form RNA helix in a manner similar to both Cse3 and Csy4. We previously discussed the modular nature of these proteins in terms of separate RNA-binding and catalytic functions (Gesner et al. 2011). While the structural models of Cas5d and the Cas5d–RNA complex suggest similarities to both of these systems, the Cas5d endonuclease appears to represent a minimalist solution to both of these functions embodied in a single domain.
MATERIALS AND METHODS
Cloning, expression, and purification of M. succiniciproducens MS0988
The target gene for NYSGXRC-10400b (M. succiniciproducens strain MBEL55E, gene ms0988, amino acids 2–225) was codon optimized and synthesized (Codon Devices), amplified via PCR, and inserted into pET26b vector (modified for TOPO directed cloning), which drives the expression of protein with a noncleavable C-terminal hexa-histadine tag (Invitrogen). BL21(DE3)-Codon+RIL cells (Stratagene) were transformed with this vector and grown overnight in 1 L of HY medium (Medicilon, Inc.) at 37°C until OD600 reached ∼1. The temperature was reduced to 22°C for 20 min and SeMet buffer (Medicilon, Inc.) was added. Growth was continued for 20 min and expression was induced by addition of 1 mM IPTG. After an additional 21 h of growth, cells were harvested and frozen at –80°C.
The cells were resuspended and lysed by sonication; the lysate was clarified by centrifugation at 38,900g for 30 min. The protein solution was applied to a Ni-NTA column (Qiagen), washed with Buffer A (50 mM Tris-HCl at pH 7.8, 500 mM NaCl, 10 mM imidazole, 10 mM methionine, 10% glycerol), and eluted with buffer A containing 500 mM imidazole. The solution was concentrated by Amicon ultrafiltration (Millipore), and run on an S75 gel-filtration column (buffer 10 mM Hepes at pH 7.5, 150 mM NaCl, 10 mM methionine, 10% glycerol, 5 mM DTT). A yield of 40.7 mg of protein was obtained and analyzed by SDS-PAGE; mass spectrometric analysis revealed intact, fully labeled SeMet protein of interest. The final sample was concentrated to 10 mg/mL.
Crystallization and structure determination
Single crystals were obtained by mixing 1 μL of the protein at 10 mg/mL with 1 μL of precipitant (100 mM Bis-Tris at pH 5.5, 28% PEG 3350, 200 mM ammonium sulfate), followed by vapor diffusion equilibration against 100 μL of the same precipitant at room temperature. Following cryoprotection with 20% DMSO, the crystals were immersed in liquid nitrogen. Single wavelength anomalous diffraction data extending to 1.95 Å resolution were collected at the selenium peak wavelength at the Argonne National Laboratory Advanced Photon Source Sector 31-ID. Diffraction from these crystals was consistent with space group P63 (a = b = 81.83, c = 61.94), with one molecule per asymmetric unit.
Five selenium sites were located using SHELXD, and density-modified phases were calculated with SHELXE (Pape and Schneider 2004). Following rounds of automated and manual model building, with ARP/wARP (Perrakis et al. 1999) and Coot (Emsley and Cowtan 2004), refinement with Refmac (Murshudov et al. 1997) converged at Rwork = 21.1% and Rfree = 24.2%. The final model consists of N-terminal cloning artifacts SerLeu, amino acids Ala2–Ala220 (disordered regions correspond to 71–103, 196–199, 221–225, and C-terminal cloning artifact EGHHHHHH) and 64 waters.
Cloning, expression, and purification of Thermus thermophilus Cas5d
The T. thermophilus Cas5d gene ttp0133 (NC_005838.1) was codon-optimized for expression in E. coli and synthesized by Bio Basics and cloned into the pACYC-duet vector (Novagen) using EcoRI and SalI sites. Site-directed mutagenesis was performed by PCR using a mutagenic primer and the cloned Cas5d as a template. E. coli BL-21 Gold cells were transformed with the appropriate plasmid, grown to an OD600 of ∼0.8 and induced with 1 mM IPTG for 12 h at 22°C. Cells were lysed at 4°C for 60 min (100 mM NaCl, 20 mM Tris-HCL at pH 8.0, 1 mM 2-mercaptoethanol, 20 mM imidazole, 1 mM PMSF) and then sonicated. Lysate was cleared by centrifugation at 40,000g for 20 min, heated to 55°C for 30 min, and centrifuged again at 40,000g for 20 min. The supernatant was bound to Ni Sepharose 6 Fast Flow resin (GE Healthcare) and eluted with lysis buffer containing 200 mM imidazole. The resultant His6-tagged Cas5d fusion proteins were purified by Superdex 75- and cation exchange chromatography. Protein was dialyzed into and stored at −20°C in buffer containing 100 mM NaCl, 10 mM Tris-HCL (pH 8.0), 0.5 mM 2-mercaptoethanol, 0.5 mM EDTA, and 15% glycerol.
RNA preparation
RNAs purchased from IDT were designed to model a full or partial CRISPR repeat:
5′-GUUGCACCGGCCCGAAAGGGCCGGUGAGGAUUGAAAC-3′ (modeling full repeat),
5′-GUUGCACCGGCCCGAAAGGGCCGGUdGAGGAUUGAAAC-3′ (noncleavable repeat),
5′-GUUUCAAUCCUCACCGGCCCUUUCGGGCCGGUGCAAC-3′ (reverse complement of the repeat),
5′-GUUGCACCGGCCCGAAAGGGCCGGUGAGGAUUG-3′ (4-nt truncation), and
5′-GUUGCACCGGCCCGAAAGGGCCGGUGAGG-3′ (8-nt truncation).
Gel mobility shift and cleavage assays
5′-32P-radiolabeled RNA substrate was incubated in 10-μL reaction solutions with 0–10 μM Cas5d protein (100 mM NaCl, and 10 mM Tris-HCL at pH 8.0) for 30 min at 55°C and immediately loaded onto a 6% tris-glycine (w/v) polyacrylamide gel. For cleavage assays, reactions were quenched at various time points in an equal volume of loading dye containing 2% SDS and 7 M urea before resolution on a 20% 29:1 (w/v) 8 M urea-sequencing PAGE. Gels were exposed to a phosphor screen (Molecular Dynamics), scanned with a Typhoon (GE Healthcare) PhosphorImager, and analyzed using ImageQuant software (Molecular Dynamics).
Identification of cleavage site
For RNaseT1 digestion, 5′-32P-radiolabeled RNA substrate was incubated in a buffer containing 20 mM sodium citrate, 7 M urea, 1 mM EDTA, 80 mM HCl, and 0.2 μg/μL yeast tRNA successively for 2 min at 95°C, 2 min at 4°C, and 2 min at 55°C. A total of 30 units of RNase T1 was added to reactions and incubated at room temperature for 1.5 min, then quenched with 400 μL of 300 mM sodium acetate and 100 μL of phenol. Following phenol-chloroform extraction, reactions were precipitated at −20°C in 70% ethanol for 20 min and pelleted by centrifugation at 15,000 rpm at 4°C for 20 min. Base hydrolysis of 5′-32P-radiolabeled RNA was carried out in 50 mM sodium carbonate (pH 9), 1 mM EDTA, and 2 mg/mL yeast tRNA at 55°C for 15 min. Reactions were quenched by addition of 400 μL of 300 mM sodium acetate and 100 μL of phenol. Reactions were phenol/chloroform extracted and ethanol precipitated as described above and resolved on a 20% 8 M urea 29:1 polyacrylamide sequencing gel.
Characterization of Cas5d-dependant 3′ RNA cleavage product
The 37-nt RNA was incubated with 30 ng of Cas5d as described above for 30 min at 55°C, followed by phenol/chloroform extraction and ethanol precipitation. RNA was radiolabeled using T4 kinase (Invitrogen) and [γ-32P]ATP (PerkinElmer) according to the manufacturer's instructions and analyzed as described above.
Characterization of Cas5d-dependent 5′ RNA cleavage product
RNA was digested by Cas5d as described above, phenol/chloroform extracted and ethanol precipitated, and resuspended in 50 μL of water. Oxidation-elimination reaction experiments were conducted essentially as described (Igloi and Kossel 1985). Resuspended reactions were incubated in 60 mM borate-boric acid buffer (pH 6.8) and 25 mM sodium periodate at 0°C for 60 min. Reactions were quenched with 10 μL of glycerol before phenol/chloroform extraction and ethanol precipitation. Resuspended reactions were incubated in 1 M lysine-HCL (pH 9.3) for 90 min at 45°C and then subjected to phenol/chloroform extraction, ethanol precipitation, and analysis on a 20% 29:1 (w/v), 8 M urea sequencing gel.
DATA DEPOSITION
Protein Data Bank: Coordinates for the M. succiniciproducens Cas5d have been deposited under accession code 3KG4.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Mark Glover and Steven Chaulk for helpful advice. This work was supported by a Discovery Grant to A.M.M. from the Natural Sciences and Engineering Research Council of Canada (NSERC). The NYSGXRC was supported by NIH Grant U54 GM074945. We gratefully acknowledge the efforts of all members of the NYSGXRC, past and present. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the beam line facilities at Sector 31 of the Advanced Photon Source was provided by SGX Pharmaceuticals, Inc. (now Eli Lilly) who constructed and operate the facility.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033100.112.
REFERENCES
- Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF 2011. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J 30: 4616–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16: 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64: 475–493 [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, MacMillan AM 2011. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 18: 688–692 [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007a. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35: W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007b. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BCM Bioinformatics 8: 172 doi: 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60 doi: 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA 2011. Mature clustered regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci 108: 21218–21222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329: 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, et al. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22: 547–553 [DOI] [PubMed] [Google Scholar]
- Hong SH, Kim JS, Lee SY, In YH, Choi SS, Rih JK, Kim CH, Jeong H, Hur CG, Kim JJ 2004. The genomic sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol 22: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Igloi GL, Kossel H 1985. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Applications of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res 13: 6881–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Dounda JA, Charpentier E 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE 2009. Protein structure prediction on the web: A case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kondo J, Westhof E 2011. Classification of pseudo pairs between nucleotide bases and amino acids by analysis of nucleotide–protein complexes. Nucleic Acids Res 39: 8628–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61 doi: 10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin A, et al. 2011a. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV 2011b. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 6: 38 doi: 10.1186/1745-6150-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S 2011. Structural and biochemical analysis of the nuclease domain of the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J Biol Chem 286: 31896–31903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neron B, Menager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C 2009. Mobyle: A new full web bioinformatics framework. Bioinformatics 25: 3005–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Schneider TR 2004. HKL2MAP: A graphical user interface for phasing with SHELX programs. J Appl Crystallogr 37: 843–844 [Google Scholar]
- Perrakis A, Morris R, Lamzin VS 1999. Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 18: 680–687 [DOI] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Haurwitz RE, Doudna JA 2012. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: Recognition and cleavage. Structure 19: 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]