The spliceosomal branch site adenosine of the yeast Saccharomyces cerevisiae, which functions as the nucleophile in the first step of pre-mRNA splicing, adopts an extrahelical conformation in the presence of a phylogenetically conserved pseudouridine (ψ) modification in the U2 snRNA strand. Here, using NMR and fluorescence techniques, the authors show that this extrahelical motif also requires a purineU2 strand-pyrimidineintron strand base pair two residues 5′ to the extrahelical adenosine, with which it forms a base triple between the amino group of the adenosine and the pair's minor groove edge. In addition, they show that the fluorescent adenine analog 2-aminopurine, in which the amino group is in a different location, also adopts an extrahelical conformation, suggesting that the position is not solely dependent upon hydrogen bond formation.
Keywords: base triple, branch site, NMR, pseudouridine, 2-aminopurine fluorescence spectroscopy, RNA
Abstract
The branch site helix from Saccharomyces cerevisiae with pseudouridine (ψ) incorporated in a phylogenetically conserved position of U2 snRNA features an extrahelical branch site adenosine (A) that forms a base triple interaction with the minor groove edge of a widely conserved purineU2 strand-pyrimidineintron strand (RU2-Yintron) base pair two positions upstream. In these studies, NMR spectra of a duplex in which 2-aminopurine (2ap), a fluorescent analog of adenine lacking the proposed hydrogen bond donor, was substituted for the branch site A, indicated that the substitution does not alter the extrahelical position of the branch site residue; thus, it appears that a hydrogen bond between the adenine amino group and the R-Y pair is not obligatory for stabilization of the extrahelical conformation. In contrast, reversal of the orientation of AU2-Uintron to UU2-Aintron resulted in an intrahelical position for the branch site A or 2ap. Fluorescence intensity of 2ap substituted for the branch site A with the original RU2-Yintron orientation (AU or GC) was high, consistent with an extrahelical position, whereas fluorescence in helices with the reversed R-Y orientation, or with a mismatched pair (A-U → G•A or U•C), was markedly quenched, implying that the residue was stacked in the helix. The A 5′ to the branch site residue was not extrahelical in any of the duplexes. These findings suggest that the RU2-Yintron base pair orientation in the ψ-dependent branch site helix plays an important role in positioning the branch site A for recognition and/or function.
INTRODUCTION
As a critical step in the maturation of precursor messenger (pre-m)RNA molecules, intervening sequences, introns, are excised and flanking regions, exons, are ligated to form a contiguous transcript. The spliceosome, which comprises small nuclear (sn)RNAs (U1, U2, U4, U5, U6) and a large number of proteins, is the biomolecular machinery responsible for catalysis of pre-mRNA splicing in eukaryotic nuclei. The first of two transesterification reactions occurs as a result of the nucleophilic attack by the 2′ hydroxyl (2′OH) of a conserved adenosine residue of the intron, called the branch site because of the branched intermediate formed by the intron at the 5′ splice site. The bulged branch site adenosine (A) is positioned by pairing of a consensus region of the intron with a short segment of U2 snRNA.
Because of the crucial role played by RNA components of the branch site region in recognition and catalytic events in splicing, determination of structural features of RNA components of the active site of the spliceosome is important. Solution structures of a short RNA duplex representing the branch site helix from the yeast Saccharomyces cerevisiae were previously solved by NMR in the presence and absence of a post-transcriptional base modification in a phylogenetically conserved location of U2 snRNA (Newby and Greenbaum 2001, 2002a). A duplex that included the conserved pseudouridine (ψ) residue in position 35 of the U2 snRNA strand was characterized by a kinked backbone of the intron strand and an extrahelical conformation of the branch site A, with its 2′OH exposed in the widened major groove. Calculation of electrostatic surface potentials by a nonlinear Poisson-Boltzmann approach identified a region of significant negative potential in the major groove surrounding the 2′OH of the branch site A (Xu et al. 2005) that may contribute to the recognition of the branch site A by other spliceosomal components or its activity in the first step of splicing. In contrast, the branch site A in the unmodified counterpart (U in place of the naturally occurring ψ in the U2 snRNA strand) adopted an intrahelical conformation, and both strands had A-type helical parameters throughout.
Identity of the branch site A is conserved in yeast and mammals (Langford and Gallwitz 1983; Pikielny et al. 1983; Gao et al. 2008). Although the flanking sequence is only loosely conserved in higher eukaryotes (YUNAY, where Y represents a pyrimidine, N is any nucleotide, and the underlined A represents the invariant branch site adenosine) (Gao et al. 2008), the AU2-Uintron base pair associated with the observed base triple is found in ∼75% of the U2-dependent spliceosomes studied to date, whereas a G-C base pair is found in ∼25% of the studied spliceosomes (Lim and Burge 2001; Kol et al. 2005; Corvelo et al. 2010); however, an A-U base pair is found in 100% of the U12-dependent spliceosomes (Padgett and Burge 2003, 2005). Thus, the data suggest that a purineU2-pyrimidineintron (RU2-Yintron) base pair orientation is strictly conserved in yeast, widely conserved in mammals, and likely in all eukaryotes. Splicing assays in which the highly conserved A-U base pair was reversed (i.e., A-U→U-A) resulted in a significant decrease in splicing efficiency (McPheeters and Abelson 1992).
The structure of the branch site duplex suggested formation of a base triple involving the branch site adenosine (A24) (see numbering in Fig. 1) and the A7-U22 base pair (Newby and Greenbaum 2002a). A number of the nine converged structural models imply a hydrogen bond between the exocyclic amino N6H2 of A24 and one or more acceptors in the minor groove of the A7-U22 pair, leading to the conclusion that the extrahelical branch site A may be stabilized by a hydrogen bond. In this study, we examine the need for formation of the postulated hydrogen bond by analysis of duplexes in which 2ap replaces the branch site A, as well as the role of the identity and orientation of the base pair involved in formation of the base triple (Fig. 2). Our results suggest that the exocyclic amino group in the 6 position of adenine may not be essential for stabilization of the extrahelical conformation of the branch site A but that a RU2-Yintron base pair is required. These findings support a model whereby the orientation of the RU2-Yintron base pair in the ψ-dependent branch site maintains a role in positioning and/or stabilizing the branch site nucleophile prior to the first step of splicing.
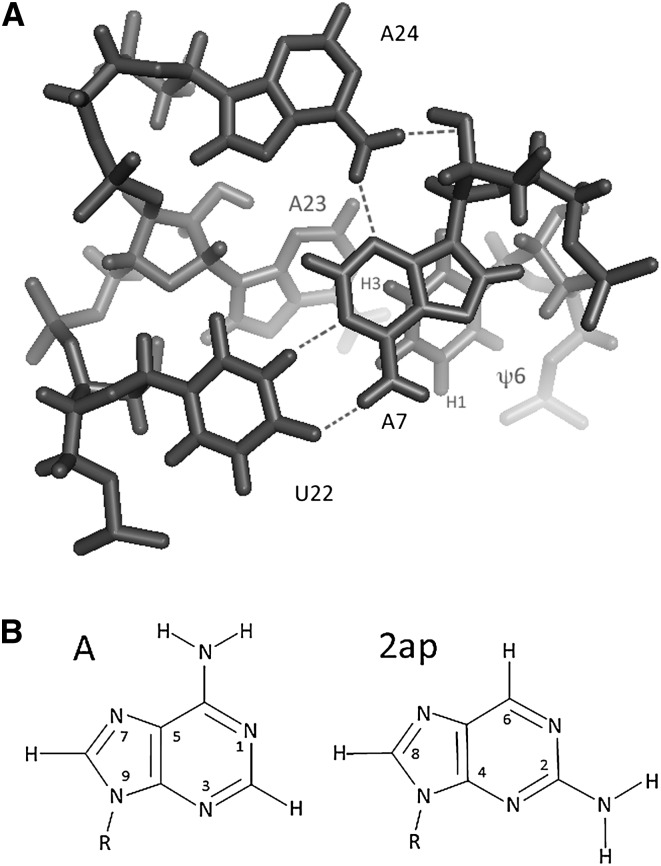
FIGURE 1.
(A) Axial view of the base triple and the adjacent ψ6 and A23 residues in the ψ-modified branch site duplex (ψBP) as seen in the solution structure determined by Newby and Greenbaum (2002a) (1LPW). A7 and U22 participate in a canonical Watson-Crick base pair, while A24, the branch site adenosine residue, adopts an extrahelical conformation and forms a base triple with two hydrogen bonding interactions in the minor group of the A7-U22 pair (with A7 N3 and A7 2′O). (B) Stick structure of adenine and 2-aminopurine (2ap).
FIGURE 2.
RNA duplexes and naming schemes used in these studies. The duplex ψBP, with strands representing the wild-type U2 snRNA (top oligomer) and intron (bottom oligomer) sequences from S. cerevisiae, includes two additional complementary nucleotides added to the 5′ side of the U2 strand and 3′ side of the intron strand to increase thermal stability. Other duplexes include modifications in the base pair A7-U22 that participates in the base triple identified in the solution structure of ψBP; in each case, the mutation from the native yeast sequence is the rectangle. Adenosine residues substituted by the fluorescent analog 2-aminopurine (2ap) are circled. The rationale for testing each of the mutated pairs is explained in the text.
RESULTS
Effect of 2ap substitution on the ψ-dependent branch site duplex
In order to assess the requirement for a hydrogen bond between the amino group of the branch site A and the minor groove edge of the conserved AU2-Uintron pair to stabilize its extrahelical conformation, we examined the conformation of the branch site residue in a duplex in which 2ap replaced the branch site A (ψBP2ap). 2ap, which has an exocylic amino group in position 2 instead of position 6 as found in adenine, forms hydrogen bonds with uracil that are comparable to a typical A-U Watson-Crick base pair but cannot form a hydrogen bond involving the N6H2 exocyclic amino group. We collected one- and two-dimensional NOESY spectra of exchangeable and nonexchangeable protons of ψBP2ap and compared them with those of ψBP, the ψ-containing duplex. Exchangeable protons were assigned from NMR spectra acquired at 7°C in 90% H2O. Assignments of nonexchangeable proton resonances were made from a combination of NOESY and TOCSY experiments acquired in 99.96% D2O at 20°C.
Spectra of ψBP were essentially identical to those acquired previously (Newby and Greenbaum 2001). Proton (1H) resonance chemical shifts and NOEs of ψBP2ap were very similar to corresponding resonances and NOEs in the unsubstituted counterpart (Fig. 3). As was the case for ψBP, no resonance peak was observed for ψ6 H3 resonance under conditions used for these spectra. However, when both temperature and pH were decreased (2°C, pH 5.5, absence of phosphate buffer), we observed a broad peak at ∼10.8 ppm, similar to that observed for ψ H3 of pseudouridine monophosphate or ψ6 H3 of ψBP at −18°C (Schroeder et al. 2005), suggesting that ψ6 H3 in ψBP is not involved in a Watson-Crick base pair. These observations are in contrast to the chemical shift of a Watson-Crick paired ψ6 H3 at ∼13.1 ppm assigned in a fully complementary RNA helix at temperatures up to at least 20°C (Durant and Davis 1999; Newby and Greenbaum 2002b; Schroeder et al. 2005). The absence of a visible peak for ψ6 H3, therefore, led to the conclusion that, as in ψBP, this proton is exchange-broadened beyond detection. Of the exchangeable protons, only the imino 1H of G8 (see Fig. 1 for numbering scheme) displays a slight downfield shift (0.07 ppm) in the spectra of ψBP2ap as compared to the corresponding resonance in ψBP, implying similar base-pairing patterns and helical parameters.
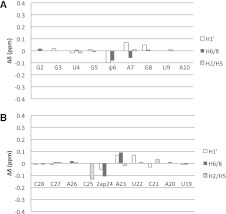
FIGURE 3.
Changes in chemical shifts of nonexchangeable protons of the ψ-dependent branch site duplexes (ψBP and ψBP2ap). Positive values for differences [δ (ψBP2ap) − δ (ψBP)] are associated with downfield shifts and the negative values with upfield shifts. Differences are shown for the anomeric protons H1′, aromatic protons H6/8 (for pyrimidines/purines), and H2/5 (for adenosine/pyrimidines). (A) Nucleotides of the U2 snRNA strand; (B) nucleotides of the intron strand. The slight chemical shift changes in ψBP2ap are consistent with a similar helical structure to the branch site helix ψBP.
Nonexchangeable resonances exhibiting differences in chemical shift >0.10 ppm are 2ap24 H8 and C25 H5, with changes of −0.11 and −0.13 ppm, respectively (where the negative sign represents an upfield shift relative to ψBP). Other resonances with lesser but detectable chemical shift changes (<0.10 ppm) are ψ6 H6 and H1′, A7 H8 and H1′, A23 H8, H1′ and H2, and G8 H1′ and U22 H1′ (Fig. 3). These changes in the chemical shifts were attributed to the altered distribution of electronegative groups on the Watson-Crick face of 2ap24 and, therefore, to differences in shielding effects in the 2ap-substituted duplex ψBP2ap or minor differences in configuration (Figs. 3, 4A). Importantly, the absence of significant chemical shift changes of residues in the immediate vicinity of 2ap24 in ψBP2ap suggests similar chemical environments and is, therefore, consistent with similar conformation of the branch site 2ap to that observed for the branch site A in ψBP.
FIGURE 4.
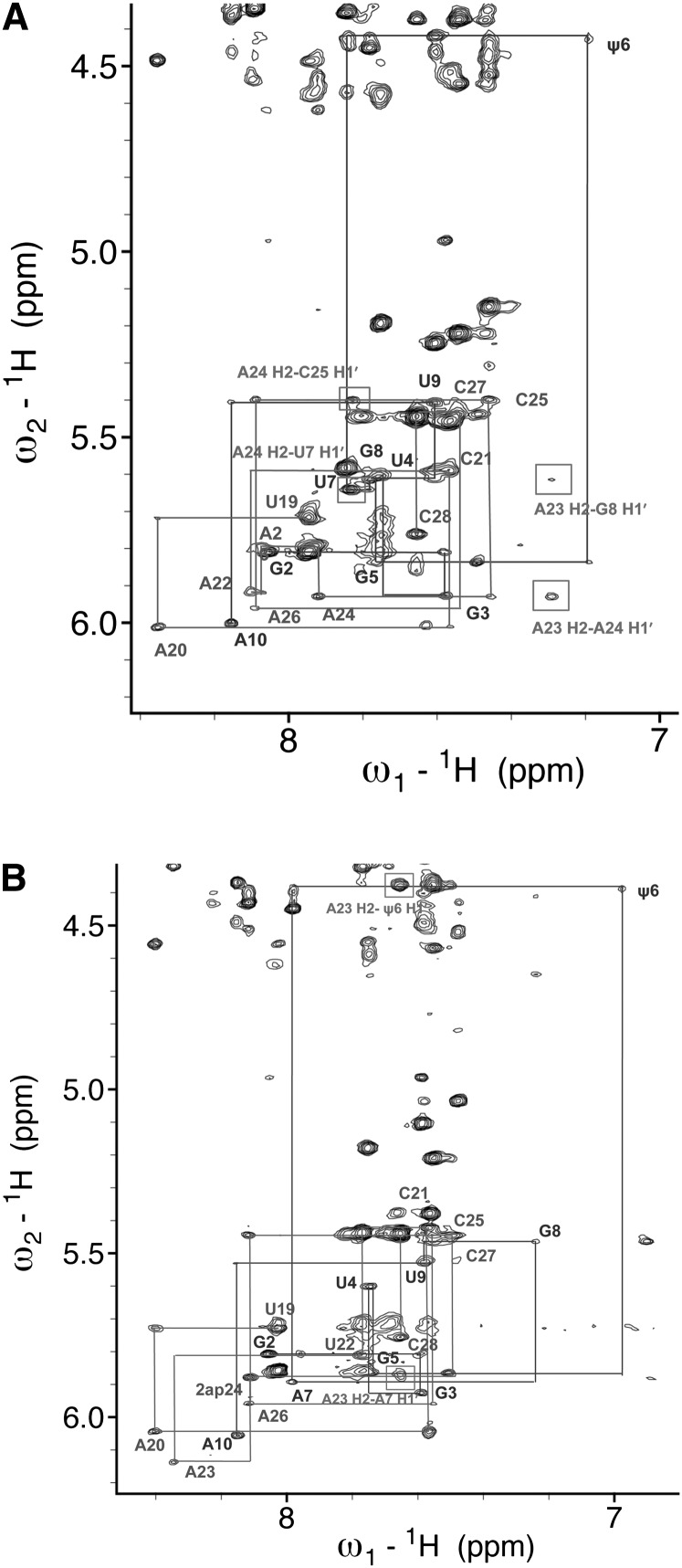
(A) NMR spectrum of the aromatic-anomeric (base-H1′) 1H region of a two-dimensional spectrum of the mutated branch site duplex ψBP2ap. Both spectra shown in A and B were acquired on 0.3 mM RNA, in 99.96% D2O in 50 mM NaCl at 20°C and at 600 MHz with a mixing time of 350 msec. Sequential connectivities between protons H6/H8 and H1′ are marked by lines. Boxed NOEs are those indicating contacts also observed in spectra of ψBP but not in those of uBP (the unmodified duplex in which the branch site A was in an intrahelical conformation) (Newby and Greenbaum 2002a). (B) NMR spectrum of the aromatic-anomeric (base-H1′) 1H region of a two-dimensional spectrum of the mutated branch site duplex (mψBP duplex; 0.3 mM RNA, the same sample conditions as in A). Connectivities between sequential NOEs between protons H6/H8 to H1′ are shown. NOEs involving A24 and A23 H2 protons that confirmed the stacked intrahelical conformation of those residues are marked by rectangles and identified.
We detected no cross-strand NOEs involving A24 H2 in spectra of ψBP that would have been expected if the branch site residue adopted an intrahelical position (e.g., for uBP, the branch site helix with U in place of ψ) (Newby and Greenbaum 2001). We were unable to confirm the A23 H2-C25 H1′ NOE, a marker of the extrahelical A24-conformation, because of chemical shift overlap of C25 H1′ with C28 H5 and A23 H2 with C28 H6. However, in both ψBP and ψBP2ap, A23 H2 exhibited an NOE to A7 H1′, as well as an unusual cross-strand NOE to ψ6 H1′, similarly to the NOEs previously observed in ψBP. These NOEs were not observed in spectra of uBP, in which the branch site A is stacked intrahelically. Also clearly absent were NOEs between A7 H2 and A23 H1′ and between A7 H2 and A24 H2, consistent with extrahelical A in both ψBP and ψBP2ap (Fig. 4A). Moreover, we detected similar patterns of ribose H1′-H2′ correlations in TOCSY spectra of ψBP and ψBP2ap in the current study, as previously observed. Specifically, detection of medium intensity H1′-H2′ TOCSY cross-peaks for riboses of U22, A23, and A24, as well as for terminal residues, implied non-C3′-endo ribose puckers analogous to the pattern observed for ψBP. Chemical shifts of resonances corresponding to these TOCSY cross-peaks were essentially the same for A10, U22, and U19 as noted for ψBP and very similar for A23 and A(2ap)24.
Similarity of chemical shifts, NOE patterns, and ribose H1′-H2′ correlations between ψBPap and ψBP suggest the same extrahelical conformation of the branch site duplex in ψBPap as previously observed for ψBP. Given these findings, we also conclude that the hydrogen bond(s) observed between an amino 1H in position 6 of A in ψBP is not an essential component of stabilization of the extrahelical branch site A.
Conformational changes of the branch site duplex associated with the reversed UU2-Aintron base pair
In order to test the hypothesis that the widely conserved RU2-Yintron orientation is important for stabilization of the extrahelical conformation of the branch site A, we acquired and analyzed NMR spectra of a ψ-modified duplex with reversed orientation of the base pair associated with formation of the observed base triple (mψBP) (shown in Fig. 2). Imino protons were assigned from one- and two-dimensional NMR spectra of mψBP acquired at 7°C in 90% H2O, 10% D2O added for lock, and by similarity to corresponding resonances of ψBP (this report) and uBP (Newby and Greenbaum 2001). The chemical shifts for ψ6 H3 were 12.83 and 13.09 ppm in mψBP and in mψBP2ap (the 2ap-substituted analogous duplex), respectively, similar to the value 13.1 ppm noted for a pseudouridine involved in Watson-Crick pairing (Durant and Davis 1999; Newby and Greenbaum 2002b; Schroeder et al. 2005).
Assignments of nonexchangeable proton resonances were made from a combination of NOESY (Fig. 4B) and TOCSY experiments in 99.96% D2O at 20°C, 350- and 70-msec mixing times, respectively. Chemical shifts for resonances attributed to the helical stem region flanking the branch site region were very similar to those of corresponding protons of uBP and ψBP, although chemical shifts of aromatic/anomeric resonances of U7 and A22 (the pair with reversed orientation) were unique to mψBP (Figs. 4B, 5A). Significantly shifted nonexchangeable proton resonances (|Δδ| > 0.10 ppm) in spectra of mψBP relative to corresponding resonances in the spectrum of ψBP were: ψ6 H6, G8 H8, U9 H1′, C21 H1′, A23 H8, H1′, and H2, as well as A24 H8, with the largest difference in chemical shift observed for G8 H8 (0.55 ppm) (Fig. 5A), suggesting significant structural differences between the two helices.
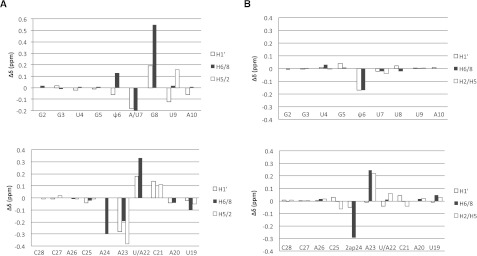
FIGURE 5.
(A) Effects of the reverse UU2-Aintron base pair on the chemical shifts of the modified branch site duplex (mψBP and ψBP). Positive values for differences δ (mψBP) − δ (ψBP) are associated with downfield shifts of resonances in mψBP spectra as compared to those in ψBP spectra, whereas the negative values are associated with upfield shifts of respective resonances (U2 snRNA oligomer) (upper panel) and the intron oligomer (lower panel). Large changes in chemical shifts of mψBP relative to ψBP suggest perturbed helical parameters of the branch site region of mψBP. (B) Changes in chemical shifts of the modified ψ-dependent branch site duplexes as compared to the 2ap-substituted duplex (mψBP and mψBP2ap). Notation and orientation of respective strands are the same as in Figure 3. Changes in chemical shifts of nonexchangeable protons of ψ6, which may form a hydrogen bond with A23 or A24, are consistent with an intrahelical stacked conformation of 2ap24 in mψBP2ap.
NOEs throughout the stem of mψBP were typical of the stacked intrahelical conformation of residues. In particular, NOEs involving the A H2 protons suggested a stacked orientation for A24 and A23, including sequential A H2-H1′i+1 and cross-strand A H2-H1′i−1 NOEs for both A23 and A24. A24 H2 of mψBP displayed NOESY cross-peaks to A23 H2 and C25 H1′, as well as a long-range cross-strand i → i−2 NOE to H1′ of U7, consistent with an intrahelical and stacked conformation of A24. A24 H8-C25 H6 NOE is an additional indicator of the stacked conformation of A24 in mψBP. The NOE between A23 H2 and U7 H1′, as well as long-range G8 H1′ in mψBP are consistent with a stacked intrahelical position of A23 (Fig. 4B).
In contrast to ψBP or ψBP2ap, all riboses of mψBP, with the exception of those of the terminal residues A10 and U19, had very small H1′-H2′ scalar couplings (3JH1′-H2′), as evidenced from the absence of TOCSY cross-peaks, and were thus attributed to a C3′-endo conformation. A phosphorous spectrum of mψBP acquired at 20°C indicated a narrow chemical shift range of −0.11 to −1.04 ppm, typical of A-form helical parameters (data not shown). Thus, all spectral evidence supports the conclusion that the reversal of AU2-Uintron to UU2-Aintron orientation results in a stacked branch site A, a finding that supports the importance of the conserved purineU2-pyrimidineintron orientation in stabilization of the extrahelical branch site conformation.
We also analyzed spectra of an analogous duplex (mψBP2ap), with 2ap replacing the branch site residue. Chemical shifts of mψBP2ap were very similar to those of mψBP (Fig. 5B); the small changes in or near 2ap are attributed to electrostatic differences between 2ap and A. Such changes were seen for ψ6 H1′ and H6, 2ap24 H8, and A23 H5 and H8. The chemical shift changes of ψ6 H1′ and H6, which are likely to pair either with A23 or 2ap24 (such an interaction was observed in the structural model for uBP, the unmodified branch site duplex) (Newby and Greenbaum 2002a), also imply an intrahelical orientation of the 2ap24. Observed NOEs support the conclusion that 2ap of mψBP2ap, like A of mψBP, is in a stacked intrahelical conformation. The observed variations in mψBP2ap are fully consistent with altered electron shielding patterns of protons in the vicinity of the altered groups of 2ap24.
Relative position of adenosines in branch site helices by fluorescence spectroscopy using 2-aminopurine
Based upon similar structural features for duplexes in which the 2ap substituted for the branch site adenine, we exploited the fluorescent properties of 2ap as a probe for conformation of the branch site residue and the 5′ neighboring adenine in a series of fluorescence experiments. 2ap is considered a useful reporter of conformation of an adenine residue because its fluorescence intensity is quenched when 2ap is stacked between neighboring nucleotides but not when involved in a hydrogen bond alone (Rachofsky et al. 2001) and is enhanced considerably when the 2ap fluorophore is exposed to solvent (Millar 1996; Jean and Hall 2001).
As controls, we measured 2ap fluorescence in RNA helices or strands of known, stacked conformation, including the native helix without the ψ modification (uBP2ap), a complementary helix (10 + 102ap), and a single-stranded intron (102ap). Fluorescence signals from the sequences were normalized against that of a duplex containing the conserved ψ modification (ψBP2ap, where 2ap was shown by NMR studies to adopt an extrahelical conformation), which was given an arbitrary value of 1. Sequences are shown in Figure 2.
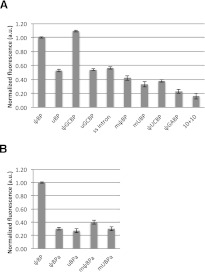
Relative fluorescence intensity for all samples is shown in Figure 6. Consistent with previous results (Newby and Greenbaum 2002a), fluorescence of 2ap substituted for A24 in ψBP2ap is high, whereas 2ap fluorescence in a complementary helix (10 + 102ap) is only ∼15% of the intensity of ψBP2ap. By comparison, fluorescence in the unmodified branch site duplex, uBP2ap, or single-stranded (ss) RNA, 102ap, was ∼53% or ∼57% that of ψBP2ap, respectively, suggesting that A24 was predominantly stacked but (as shown by NMR studies) likely to be conformationally heterogeneous or flexible.
FIGURE 6.
Relative fluorescence intensity profiles of 2ap incorporated in native and mutated branch site constructs. (A) 2ap was incorporated in place of the canonical branch site residue (A24). The native ψ-modified construct (ψBP2ap), single-stranded intron (102ap) and complementary duplex (10 + 102ap) were used as standards; these standards exhibited comparable relative intensities as previously published (Newby and Greenbaum 2002a). Naming conventions are explained in the text (duplex sequences shown in Fig. 2). (B) With the exception of the ψBP2ap control against which fluorescence of all samples was normalized (2ap in the A24 position), 2ap replaced the adenosine residue 5′ to the branch site A (A23) to test for its intra- vs. extrahelical conformation.
In the first set of experiments with altered duplexes, we tested the effects of a base pair switch AU2-Uintron→GU2-Cintron, which maintains the purine-pyrimidine orientation and is present in ∼25% of the studied eukaryotes. Sequences tested in these experiments included a G7-C22 base pair with a ψ-modified and unmodified U2 strand, (ψGCBP2ap and uGCBP2ap), respectively. Fluorescence of ψGCBP2ap was 9% greater than that of ψBP2ap, suggesting that the extrahelical position of A24 was stabilized even more than in the original duplex, whereas fluorescence of uGCBP2ap was ∼53% of the value for ψBP2ap, essentially the same as seen for uBP2ap or the single-stranded intron 102ap, suggesting an intrahelical branch site A.
We then measured fluorescence in a duplex for which we reversed orientation of the native AU2-Uintron base pair to UU2-Aintron (mψBP2ap) (Fig. 2), which is expected to result in a different geometry in the minor groove. We found that 2ap fluorescence of mψBP2ap and of an analogous duplex without the ψ modification (muBP2ap) had emission intensities ∼42% and ∼33%, respectively, that of ψBP2ap, suggesting that the branch site residue was in a predominantly intrahelical conformation in these mutated sequences.
In order to test the structural impact of disrupting the highly conserved Watson-Crick base pair in this position, we replaced the A7-U22 pair with purine-purine (A-U→G•A, ψGABP2ap) and pyrimidine-pyrimidine (A-U→U•C, ψUCBP2ap) pairs, and assessed the fluorescence. We observed that both ψGABP2ap and ψUCBP2ap displayed low fluorescence intensity, comparable to that of the complementary duplex (10 + 102ap; ∼23% of ψBP2ap).
Conformation of adenosine 5′ to the branch site
Biochemical assays have shown that the adenosine adjacent to the branch site (known as 5′ A or, in the numbering scheme used here, A23) may pose as the nucleophile in the first step of splicing in higher organisms like humans, although not in yeast (Query et al. 1994). NMR models of the native yeast sequence indicated that the 5′ A was intrahelical, thus lowering accessibility (Newby and Greenbaum 2002a). In order to determine the relative conformation of the neighboring A for the native and several of the mutated sequences, we detected 2ap fluorescence emission intensities in S. cerevisiae branch site duplexes by site-specific substitution of the neighboring A (2ap23) (Fig. 2) with 2ap in the intron strand. 2ap23 in all duplexes tested, native (AU2-Uintron) ψ-modified (ψBPa) and unmodified (uBPa), as well as in reversed A-U base pair (UU2-Aintron) ψ-modified mψBPa and unmodified muBPa, exhibited low fluorescence intensities, exhibiting ∼30% and ∼27%, and ∼40% and ∼30% the intensity of ψBPap, respectively, indicating that the branch site neighboring residues, 2ap(A)23 were generally stacked in the helix (Fig. 6). It is, therefore, unlikely that the 5′ A would readily substitute for the branch site A in the first step of splicing catalysis for these sequences.
DISCUSSION
The coplanar base triple observed in the branch site helix of S. cerevisiae involving the extrahelical branch site A and the minor groove edge of an AU2-Uintron base pair two positions upstream suggested formation of one or two hydrogen bonds between the exocyclic amino group and acceptors on the base and ribose of the purine of the base pair (Newby and Greenbaum 2002a). This conformation was dependent upon incorporation of a modification conserved in the U2 snRNA strand. It still is not clear how presence of the ψ modification contributes to the extrahelical conformation. Although ψ substitutions in Watson-Crick helices and in multiple sites in tRNA molecules are associated with enhanced thermal stability (Davis and Poulter 1991; Hall and McLaughlin 1992; Arnez and Steitz 1994; Durant and Davis 1999; Yarian et al. 1999) and reduced flexibility in ribosomal RNA (Sakakibara and Chow 2012), inclusion of ψ in certain RNA loops has been shown to be destabilizing (Meroueh et al. 2000). Presence of two conserved pseudouridines at the terminus of a hairpin loop of telomerase RNA provide a stabilizing effect relative to the unmodified molecule but induce markedly different structures in the two hairpins (Kim et al. 2010), similar to the observations by Newby and Greenbaum (2001, 2002a). Formation of a water-mediated hydrogen bond between ψ6 H1 and a phosphate oxygen atom has been proposed to be an important component of pseudouridine-mediated structure stabilization (Davis and Poulter 1991; Hall and McLaughlin 1992); demonstration of hydrogen bond formation was shown in both ψ-substituted complementary helices and the branch site helix (Newby and Greenbaum 2002b). X-ray crystallographic structures of several mammalian branch site duplexes have revealed interactions with tightly bound water molecules involving the ψ and branch site A that support the role of ψ in stabilizing the extrahelical conformation (Lin and Kielkopf 2008).
Based on geometry implying a hydrogen bond between the amino group of the branch site A (A24) and the N3 of the adenine of the base pair in most of the converged structures of ψBP (A7 in our numbering scheme), we hypothesized that hydrogen bond formation was important for stabilization of extrahelical conformation of the branch site A. We tested this premise by NMR-based structural studies of a duplex in which 2ap, an adenine analog with the exocyclic amino group attached to the carbon atom in position 2 instead of position 6. All spectral features supported the conclusion that moving the amino group (the hydrogen bond donor) to a position in which it is not likely to participate in a hydrogen bond did not alter the extrahelical position of the branch site residue, suggesting that, although hydrogen bond formation appears to occur in solution, it is not necessary for stabilization of the extrahelical conformation. Thus, although we cannot rule out the possibility that N2H2 of 2ap could substitute as a direct or water-mediated hydrogen bond donor with acceptors in the minor groove of the RU2-Yintron base pair, it appears that the conformation is at least partially stabilized by other factors, such as energetically preferred stacking interactions.
We next probed the importance of the identity and orientation of the AU2-Uintron base pair with which the branch site residue interacts. Mapping of U2 snRNA and intron sequences in eukaryotes has indicated that a number of complementary base pairs flank the branch site A (Breathnach et al. 1978; Breathnach and Chambon 1981; Dodgson and Engel 1983; Keller and Noon 1984; Padgett et al. 1986). However, the paired nucleotide two positions upstream of the branch site A appears always to be a pyrimidine: U in ∼75% of eukaryotic sequences studied thus far (always opposing an A in the consensus sequence of U2 snRNA) and C in the remaining 25% of the branch site sequences (paired with G) (Lim and Burge 2001; Kol et al. 2005; Gao et al. 2008; Corvelo et al. 2010). Thus, a RU2-Yintron complementary base pair (A7-U22 in the NMR structure and this study) is maintained at this position in all eukaryotes studied to date.
Based on this observation, we used NMR studies to measure the impact of altered minor groove constituents and geometry on the branch site A position. Our measurements indicated unambiguously that reversing the orientation of the AU2-Uintron pair resulted in the branch site A (and also branch site 2ap in analogous experiments) adopting a stacked intrahelical position.
The suitability of 2ap as a structural probe of adenosine conformation has been demonstrated by a number of groups (Sowers et al. 1986; Fagan et al. 1996; Kulinski et al. 1996; Zagorowska and Adamiak 1996; Newby and Greenbaum 2002a). Since our NMR data indicated that 2ap did not perturb the structural features of the branch site in the duplexes examined, we exploited the fluorescent properties of 2ap as a probe of branch site conformation for a number of other duplex sequences. Our earlier data had shown that fluorescence of 2ap substituting for the branch site A in ψBP2ap was high, indicative of extrusion of the residue from the helix (Newby and Greenbaum 2002a). This finding was corroborated by the NMR results reported here.
Based upon similar geometry in the minor groove, we would expect that a GU2-Cintron base pair, which occurs in a minority of eukaryotic sequences, would support similar interactions as seen with the AU2-Uintron base pair. As predicted, replacement of the A-U base pair with a G-C base pair displayed high 2ap fluorescence, consistent with extrahelical position of the branch site A. Interestingly, fluorescence of 2ap in a duplex in which a G-C pair replaced A-U was even greater than for the A-U pair in that position, consistent with even greater exposure to solution. This effect could be the result of greater thermal stability of a G-C pair than for an A-U pair in this position as a result of enhanced hydrogen bonding and stacking interactions (Freier et al. 1981), as well as of slight alterations in helical stacking parameters, such as helical twist and differences in base-base orientation between A-U and G-C base pairs, which may affect the exposure of the fluorophore and, therefore, the intensity of the fluorescent signal.
In contrast, reversal of the positions of the RU2-Yintron pair in the S. cerevisiae sequences (with or without the ψ) resulted in low 2ap fluorescence, implying that a greater proportion of the ensemble reverted to an intrahelical position of the branch point 2ap in the mψBP2ap and muBP2ap duplexes, as was shown for mψBP and mψBP2ap by the NMR studies. Similarly, substitution of RU2•Rintron (G•A) or YU2•Yintron (U•C) pairs for the native RU2-Yintron pair also resulted in relatively low 2ap fluorescence, indicative of a predominantly stacked conformation for the branch site A. Thus, minor groove geometry, and not only presence of a purine in the U2 snRNA strand, appears to be important in stabilization of the extrahelical branch site residue.
We also investigated the impact of base pair identity on the intra- or extrahelical position of the 5′ neighboring A for several of the duplexes. Crystallographic studies of an RNA duplex representing the human U2 snRNA-intron pairing in the absence of the conserved ψ modification indicated that the branch site adenosine was stacked into the helix but that its 5′ neighbor, also an adenosine, was extruded from the helix and was involved in interhelical stacking (Berglund et al. 2001) or, in some cases, surrounded by solvent (Lin and Kielkopf 2008). In some species, although not in yeast (the sequence examined in the study of Berglund et al. [2001]), the neighboring residue can act as the nucleophile in the first step of splicing. For this reason, we also measured fluorescence of helices in which 2ap was substituted for the 5′ neighboring adenosine (2ap23, in these studies). In full agreement with our NMR studies of the ψ-modified helix presented here, as well as our previous studies, our 2ap fluorescence data indicate that the adenosine residue adjacent to the branch site in these constructs is not exposed to solvent. Similar results in each of the mutant sequences implied that, regardless of the intra- or extrahelical conformation of the branch site adenosine, its 5′ neighbor appeared to adopt an intrahelical conformation in solution.
Taken together, our data indicate that, even if hydrogen bond formation is not obligatory, the specific geometry associated with the presence of a RU2-Yintron pair in the conserved position, along with the ψ modification in its phylogenetically conserved location on the U2 snRNA strand across from the branch site, is necessary for the extrahelical position of the branch site residue. Interestingly, in addition to a general conservation of the RU2-Yintron pair interacting with the branch site A, the adjacent pair (three positions 5′ of the branch site) is also very predominantly a RU2-Yintron pair (Lim and Burge 2001). Details of stacking interactions and/or the specific placement of water molecules noted by Lin and Kielkopf (2008) associated with this sequence may contribute to the favorability of the branch site motif.
Mutations associated with components of the branch site motif result in decreased splicing efficiency. Mutations of the branch site adenine suppress splicing, with an A→C mutation causing the least deleterious effect (Query et al. 1996). It is possible that substitution of C may be tolerated (Hornig et al. 1986) because it has an amino group on the Watson-Crick face of the base and may, therefore, be able to form similar hydrogen bonding or other interactions as those formed by the analogous face of A, although with slightly different geometry.
Wide conservation of the AU2-Uintron base orientation suggests that the orientation is critical to the branch site motif formation and function. Splicing assays performed in yeast to effect a reversal of the AU2-Uintron pair to UU2-Aintron (mψBP duplex) show <10% splicing efficiency compared to the wild type (McPheeters and Abelson 1992), a finding that we propose is at least partly the result of the intrahelical position of the branch site A in the presence of the reversed UU2-Aintron base pair. In contrast, however, inhibition of splicing as a result of mutation in the three intron nucleotides 5′ of the branch site A in a SV40/human system were rescued by compensatory mutations in U2 snRNA that maintained complementary pairing, without an apparent effect of orientation of the Rintron-YU2 base pair (Wu and Manley 1989), which may reflect subtle mechanistic differences between the yeast and mammalian spliceosomes. We emphasize, however, that only limited correlations can be made when extrapolating conclusions from a cellular to an in vitro system due to the complexity and dynamic nature of the spliceosomal assembly.
A number of factors may play a role in recognition of the branch site and activity. Among them, electrostatic features of the ψ-modified branch site duplex (Xu et al. 2005) may impact on recognition and function of the branch site. Smith et al. (2009) have recently shown that the branching activity in yeast spliceosomes is also affected by the distance of the branch site A from the U2-U6 helix Ia, suggesting that long range interactions with remote sections of the U2-U6 snRNA complex may play a role in recognition and/or positioning of the branch site adenosine. Specific protein interactions within the branch site region are essential for recruitment of the U2 snRNP and splicing activity (Berglund et al. 1997; Liu et al. 2001). Perturbation of binding between U2 snRNP protein SF3b 155 and sequences flanking the pre-mRNA branch site by a small molecule inhibitor, spliceostatin A, disrupts alternative splicing patterns, perhaps by promoting nonproductive RNA pairing, thus underscoring the role of branch site duplex formation in assembly of the U2 snRNP-branch site complex (Corrionero et al. 2011) that may explain its anti-cancer activity (Kaida et al. 2007; Kotake et al. 2007). Thus, an intricate balance involving multiple recognition motifs and interactions, including conserved base modifications and local RNA interactions as well as protein and long-range RNA interactions, appears to facilitate splicing activity.
MATERIALS AND METHODS
Design and synthesis of RNA helices
A series of RNA duplexes was designed to test the role of the RU2-Yintron base pair and the phylogenetically conserved ψ residue (ψ35 in the S. cerevisiae U2 snRNA sequence) on the conformation of the branch site adenosine or its 5′ neighbor in solution. RNA oligomers were designed to represent fragments of the native U2 snRNA and intron sequences from S. cerevisiae that pair to form the branch site helix; mutant sequences were designed with specific mutations at positions involved in formation of the base triple. 2ap-substituted sequences were designed to test the importance of the hydrogen bond donor in position 6 of the adenine ring on stabilizing the extrahelical branch site A conformation. U2 snRNA strands and intron strands comprised 9 and 10 nt, respectively, including 7 bp from the native duplex plus two additional Watson-Crick pairs added to increase thermal stability. It was shown previously that addition of these base pairs did not alter duplex conformation (Newby and Greenbaum 2001). Duplexes were formed by pairing equimolar amounts of U2 snRNA and intron strands.
We also performed fluorescence studies on duplexes in which the branch site adenosine (A24 in these studies) was replaced with 2ap in the intron and also substituted the adenosine 5′ to the branch site A (A23) with 2ap to test the effect of mutations on the conformation of A23. A complementary duplex of 10 bp (U added to the U2 snRNA strand opposing the branch site A) was used as a control, as was a single-stranded intron sequence. All RNA oligomers were purchased from Dharmacon and were deprotected according to company protocols.
NMR spectroscopy
Samples of the ψBP and 2ap-substituted ψBP2ap, as well as of ψ-modified mutated duplex (mψBP) and the 2ap-substitued version mψBP2ap, were purchased from Dharmacon, lyophilized after deprotection, and resuspended in 50 mM NaCl pH 6.8–7.2 and 0.1 mM EDTA, in 90% H2O/10% 2H2O (D2O). For observation of nonexchangeable protons, samples were lyophilized three times in 99.96% D2O and resuspended in 99.96% D2O, 0.01 mg/mL DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), the latter used as an internal referencing standard (Cambridge Isotope Laboratories, Inc.). NMR data were collected in microvolume NMR tubes (Shigemi, Inc.).
NMR data were acquired on a 600 MHz Bruker Avance spectrometer (Dept. of Chemistry and Biochemistry, Hunter College of CUNY), equipped with the three-channel TXI-cryoprobe. Quadrature detection was achieved by using the States-TPPI method (Marion et al. 1989). Bruker Topspin 2.1 software was used to process two-dimensional experiments. SPARKY was used to visualize spectra and for chemical shift assignments. Spectra were apodized using sine squared function and zero-filled in both the direct and indirect dimensions.
Chemical shift assignments of exchangeable and nonexchangeable protons of the ψBP, ψBP2ap, mψBP, and mψBP2ap duplexes were made from NOESY spectra acquired at mixing times of 50–950 msec. For nonexchangeable protons, phase-sensitive NOESY spectra using a long and weak presaturation pulse for water suppression (NOESYphpr) were acquired at 17°C and 20°C and compared with those of ψBP (Newby and Greenbaum 2001). C2′-endo (or other non-C3′-endo) sugar conformations were identified from the appearance of medium-to-strong H1′-H2′ cross-peaks in TOCSY spectra collected at 70-msec mixing time. A one-dimensional proton-decoupled 31P spectrum of mψBP, also at 20°C, was collected in a Varian Inova 500 MHz spectrometer equipped with an indirect detection 5-mm HCX probe and referenced to 85% phosphoric acid.
Fluorescence studies
Wild-type and variant sequences of U2 snRNA and intron strands were combined in an aqueous buffer of 10 mM NaPi, pH 6.4, 0.1 mM EDTA, and 1 M NaCl. The concentration of each strand was ∼4.5 μM, with a slight (<5%) excess of the non-2ap strand to minimize presence of free 102ap strand in solution. Melting transitions were measured for duplexes containing 2ap and A to confirm that the substitution did not perturb duplex stability. Samples (250 μL) were suspended in 2 mm × 10 mm quartz fluorescence cells (Starna Cells, Inc.). Fluorescence excitation and emission profiles were scanned on a Cary Eclipse fluorescence spectrometer over a range of 200–850 nm. Emission scans were obtained at least five times for each sample using the observed maximum excitation wavelength 308 nm; maximal fluorescence emission was at ∼370 nm. Experiments were repeated on three to eight samples of each duplex. All fluorescence data were processed in Microsoft Excel (Microsoft, Inc.) and Sigma Plot 8.0 for Windows (Microsoft, Inc.).
ACKNOWLEDGMENTS
We thank Dr. Meredith Newby for sharing data; Dr. Faqing Yuan for assistance with Figure 2; the Biochemical Analysis Sequencing & Synthesis Lab and NMR Facility at Florida State University, NMR Facility at Hunter College of CUNY, and the National High Magnetic Field Laboratory (Tallahassee, FL) for access to spectroscopic instrumentation. This research was supported by NIH grant RO1 GM054008 and NSF grant MCB 0929394 to N.L.G. The project described was supported by Grant Number RR003037 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035782.112.
REFERENCES
- Arnez JG, Steitz TA 1994. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567 [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787 [DOI] [PubMed] [Google Scholar]
- Berglund JA, Rosbash M, Schultz SC 2001. Crystal structure of a model branchpoint–U2 snRNA duplex containing bulged adenosines. RNA 7: 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R, Chambon P 1981. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50: 349–383 [DOI] [PubMed] [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P 1978. Ovalbumin gene: Evidence for a leader sequence in mRNA and DNA sequences at the exon–intron boundaries. Proc Natl Acad Sci 75: 4853–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev 25: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvelo A, Hallegger M, Smith CW, Eyras E 2010. Genome-wide association between branch point properties and alternative splicing. PLoS Comput Biol 6: e1001016 doi: 10.1371/journal.pcbi.1001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR, Poulter CD 1991. 1H–15N NMR studies of Escherichia coli tRNAPhe from hisT mutants: A structural role for pseudouridine. Biochemistry 30: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Dodgson JB, Engel JD 1983. The nucleotide sequence of the adult chicken α-globin genes. J Biol Chem 258: 4623–4629 [PubMed] [Google Scholar]
- Durant PC, Davis DR 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 285: 115–131 [DOI] [PubMed] [Google Scholar]
- Fagan PA, Fabrega C, Eritja R, Goodman MF, Wemmer DE 1996. NMR study of the conformation of the 2-aminopurine:cytosine mismatch in DNA. Biochemistry 35: 4026–4033 [DOI] [PubMed] [Google Scholar]
- Freier SM, Hill KO, Dewey TG, Marky LA, Breslauer KJ, Turner DH 1981. Solvent effects on the kinetics and thermodynamics of stacking in poly(cytidylic acid). Biochemistry 20: 1419–1426 [DOI] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36: 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB, McLaughlin LW 1992. Properties of pseudouridine N1 imino protons located in the major groove of an A-form RNA duplex. Nucleic Acids Res 20: 1883–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H, Aebi M, Weissmann C 1986. Effect of mutations at the lariat branch acceptor site on β-globin pre-mRNA splicing in vitro. Nature 324: 589–591 [DOI] [PubMed] [Google Scholar]
- Jean JM, Hall KB 2001. 2-Aminopurine fluorescence quenching and lifetimes: Role of base stacking. Proc Natl Acad Sci 98: 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Keller EB, Noon WA 1984. Intron splicing: A conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci 81: 7417–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NK, Theimer CA, Mitchell JR, Collins K, Feigon J 2010. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res 38: 6746–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol G, Lev-Maor G, Ast G 2005. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet 14: 1559–1568 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3: 570–575 [DOI] [PubMed] [Google Scholar]
- Kulinski T, Bielecki L, Zagorowska I, Adamiak RW 1996. Introductory data on dynamics of RNA bulge duplexes. 2-aminopurine labelled adenosine loops. Collect Czech Chem Commun 61: S265–S267 [Google Scholar]
- Langford CJ, Gallwitz D 1983. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell 33: 519–527 [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB 2001. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci 98: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kielkopf CL 2008. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry 47: 5503–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Marion D, Ikura M, Tschudin R, Bax A 1989. Rapid recording of 2D NMR-spectra without phase cycling—application to the study of hydrogen-exchange in proteins. J Magn Reson 85: 393–399 [Google Scholar]
- McPheeters DS, Abelson J 1992. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71: 819–831 [DOI] [PubMed] [Google Scholar]
- Meroueh M, Grohar PJ, Qiu J, SantaLucia J Jr, Scaringe SA, Chow CS 2000. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res 28: 2075–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar DP 1996. Fluorescence studies of DNA and RNA structure and dynamics. Curr Opin Struct Biol 6: 322–326 [DOI] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2002a. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965 [DOI] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2002b. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci 99: 12697–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA, Burge CB 2003. Splice sites. In Encyclopedia of the human genome (ed. D Cooper), pp. 1134–1145. Wiley, New York [Google Scholar]
- Padgett RA, Burge CB 2005. Splice sites. In Encyclopedia of life sciences. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA 1986. Splicing of messenger RNA precursors. Annu Rev Biochem 55: 1119–1150 [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Teem JL, Rosbash M 1983. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: Implications for U1 RNA and metazoan mRNA splicing. Cell 34: 395–403 [DOI] [PubMed] [Google Scholar]
- Query CC, Moore MJ, Sharp PA 1994. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes Dev 8: 587–597 [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA 1996. Three recognition events at the branch-site adenine. EMBO J 15: 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- Rachofsky EL, Osman R, Ross JBA 2001. Probing structure and dynamics of DNA with 2-aminopurine: Effects of local environment on fluorescence. Biochemistry 40: 946–956 [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Chow CS 2012. Role of pseudouridine in structural rearrangements of helix 69 during bacterial ribosome assembly. ACS Chem Biol 7: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Skalicky JJ, Greenbaum NL 2005. NMR spectroscopy of RNA duplexes containing pseudouridine in supercooled water. RNA 11: 1012–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Konarska MM, Query CC 2009. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell 34: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers LC, Fazakerley GV, Eritja R, Kaplan BE, Goodman MF 1986. Base pairing and mutagenesis: Observation of a protonated base pair between 2-aminopurine and cytosine in an oligonucleotide by proton NMR. Proc Natl Acad Sci 83: 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Manley JL 1989. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev 3: 1553–1561 [DOI] [PubMed] [Google Scholar]
- Xu D, Greenbaum NL, Fenley MO 2005. Recognition of the spliceosomal branch site RNA helix on the basis of surface and electrostatic features. Nucleic Acids Res 33: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF 1999. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA's position 39. Nucleic Acids Res 27: 3543–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorowska I, Adamiak RW 1996. 2-Aminopurine labelled RNA bulge loops. Synthesis and thermodynamics. Biochimie 78: 123–130 [DOI] [PubMed] [Google Scholar]