Abstract
Problem
CD300a is an immunomodulatory molecule of the immunoglobulin receptor superfamily expressed in the leukocytes of myeloid and lymphoid lineages. However, its biological function on CD8+ T lymphocytes remains largely unknown. This study was conducted to assess the biological significance of CD300a expression in T lymphocytes and to determine whether its expression in peripheral T lymphocytes changes in pregnant women presenting with anti-fetal rejection.
Methods of Study
Microarray analysis was performed using total RNA isolated from peripheral CD300a+ and CD300a− T lymphocytes. Flow cytometric analysis of the peripheral blood samples of pregnant women and pathologic examination of the placentas were conducted.
Results
A large number of genes (N = 1,245) were differentially expressed between CD300a− and CD300a+ subsets of CD8+ T lymphocytes, which included CCR7, CD244, CX3CR1, GLNY, GZMB, GZMK, IL15, ITGB1, KLRG1, PRF1, and SLAMF7. Gene Ontology analysis of differentially expressed genes demonstrated enrichment of biological processes such as immune response, cell death, and signal transduction. CD300a expression in CD8+ T lymphocytes was coupled to a more cytotoxic molecular signature. Of note, the proportion of CD300a+CD8+ T lymphocytes increased in pregnant women with chronic chorioamnionitis (anti-fetal rejection of the chorioamniotic membranes; P < 0.05).
Conclusion
The findings of this study strongly suggest an increase of systemic T lymphocyte-mediated cytotoxicity in pregnant women with chronic chorioamnionitis as a manifestation of maternal anti-fetal rejection.
Keywords: placenta, maternal anti-fetal rejection, prematurity, preterm birth, transcriptome
Introduction
Tolerance of the fetus by the mother during pregnancy is essential for viviparity.1, 2 Biological characteristics such as separation of maternal circulation from fetal circulation3, 4 and expression of non-polymorphic HLA-G on trophoblasts5 are well-established mechanisms protective of the fetus. In addition, a large body of evidence indicates that the maternal immune system also undergoes alterations to avoid rejection of the semi-allogeneic fetus, and a role for regulatory T lymphocytes has been emphasized recently.6–8 As T lymphocytes play a key role in immune response, many investigators have studied changes of peripheral T lymphocyte subsets in the context of maternal tolerance of the fetus.9–14 Several studies have noted that the proportion of peripheral CD8+ T lymphocytes either increases or does not change during pregnancy,9–11, 13 while the absolute number of CD3+, CD4+, and CD8+ T lymphocytes decreases during pregnancy.12–14 There are also other reports showing that the expression of activation markers such as HLA-DR and CD25 in T lymphocytes changes during pregnancy.9, 12 These observations overall strongly suggest that pregnancy is associated with changes involving peripheral T lymphocyte numbers and activation status.
CD300a is a leukocyte surface molecule of the immunoglobulin receptor (IgR) superfamily. It has extracellular V-type Ig-like domains and a cytoplasmic domain with immunoreceptor tyrosine-based inhibitory motifs (ITIM). CD300a can be detected on the surface of various leukocyte populations such as monocytes, macrophages, granulocytes, dendritic cells, NK cells, and T and B lymphocytes.15, 16 Among CD4+ T lymphocytes, CD300a expression is mainly restricted to Th1 memory and effector cells.17 IFNγ-producing CD4+ T lymphocytes are enriched in CD300a+ expressing cells, and stimulated CD4+ T lymphocytes producing TNFαα and IL-2 are also largely positive for CD300a. Marked up-regulation of T-box transcription factor Eomesodermin (Eomes) is another characteristic of stimulated CD300a+CD4+ T lymphocytes.18 However, the biological significance of CD300a expression on CD8+ T lymphocytes remains largely unknown.
Villitis of unknown etiology is a destructive inflammation of the placenta and a consequence of maternal anti-fetal rejection.19 Infiltration of maternal CD8+ T lymphocytes into the fetal chorionic villi is its characteristic feature, and we have found increased expression of CD300a in villitis of unknown etiology. Increased expression of CD300a in villitis of unknown etiology led us to think that CD300a-expressing CD8+ cells represent a functionally distinct subset of T lymphocytes involved in maternal immune response against the fetus during pregnancy.
This study was performed to assess the biological meaning of CD300a expression in CD8+ T lymphocytes using gene expression profiling. A further flow cytometric analysis of peripheral blood samples from pregnant women was conducted to determine whether the expression of CD300a in T lymphocytes changes in women presenting with anti-fetal rejection.
Materials and methods
Patient Samples
Whole peripheral blood was collected by venipuncture from healthy non-pregnant women (N = 11) and normal pregnant women (N = 40). All normal pregnant women eventually delivered babies at term (37+0 ~ 41+6 weeks) without any maternal and neonatal complications such as intra-amniotic infection, hypertensive disorders, diabetes mellitus, and small-for-gestational-age neonates. Cord blood (N = 17) was collected from the umbilical vein during delivery; both maternal and cord blood samples were obtained in two cases. All patients provided written informed consent, and the collection and use of the samples were approved by the Institutional Review Boards of the participating institutions.
Cell Isolation/Sorting
Peripheral blood mononuclear cells (PBMCs) were isolated from samples of the peripheral blood of healthy non-pregnant women by discontinuous density gradient centrifugation using Histopaque 1077 and Histopaque 1119 (Sigma-Aldrich, St. Louis, MO, USA), and labeled with CD300a-PE (Beckman Coulter, Miami, FL, USA), CD3-FITC (BD Biosciences, San Jose, CA, USA), CD4-APC (BD Biosciences), and CD8-APC-CY7 (BD Biosciences). CD4+CD300a-, CD4+CD300a+, CD8+CD300a-, and CD8+CD300a+ T lymphocyte subsets were sorted using a FACSAria III cell sorter (BD Biosciences). The purity of sorted cells ranged from 80.3 to 98.4%.
Microarray Analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. DNase-treated total RNA (500 ng) was amplified and biotin-labeled with the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA). Labeled cRNAs were hybridized to Illumina’s HumanHT-12 Expression BeadChip (Illumina, San Diego, CA, USA). BeadChips were imaged using a BeadArray Reader, and raw data were obtained with BeadStudio Software V.3.4.0 (Illumina). Gene expression data was quantile normalized,20 probes non-detected in at least three samples were removed, and differential expression was tested using a paired moderated t-test.21 A threshold of 5% on the False Discovery Rate22 adjusted P values (called q-values) together with a minimum of 1.5-fold change was used to determine significance. Gene ontology analysis was performed using enrichment methods previously described23, 24 while pathway analysis was conducted with the Signaling Pathway Impact Analysis (SPIA) method.25, 26 The complete data set is available in a MIAME (Minimal Information about a Microarray Experiment) compliant format in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) database (entry ID: E-TABM-902).
Real-time Quantitative RT-PCR (qRT-PCR)
Reverse transcription of total RNA was done using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo(dT) primers. qRT-PCR analyses were performed with TaqMan gene expression assays (CCL5, Hs00174575_m1; CCR7, Hs99999080_m1; CD244, Hs00175569_m1; CD300a, Hs00381974_m1; CX3CR1, Hs00365842_m1; GNLY, Hs00246266_m1; GZMB, Hs00188051_m1; GZMK, Hs00157878_m1; INFG, Hs99999041_m1; IL15, Hs99999039_m1; ITGB1, Hs01127543_m1; KLRG1, 00929964_m1; PRF1, Hs00169473_m1; SLAMF7, Hs00221793_m1; Applied Biosystems, Austin, TX, USA) using a Biomark System (Fluidigm, South San Francisco, CA, USA). The human ribosomal protein, large, P0 (RPLP0; Applied Biosystems) was used for normalization.
Flow Cytometry
Peripheral blood in EDTA (100 µl) was incubated with a combination of CD300a-PE, CD3-FITC, CD45RA-FITC (BD Biosciences), CCR7-PE-CY7 (BD Biosciences), CD4-APC, and CD8-APC-CY7. Analyses were performed with the BD FACS LSR II flow cytometer and BD FACSDiva software (BD Biosciences). For intracellular staining of granzyme A, granzyme B, and perforin, 100 µl of whole blood were first labeled with antibodies against surface antigens [CX3CR1-FITC (Biolegend, San Diego, CA, USA), CD300a-PE, CD3-PE-CY7 (BD Biosciences), and CD4-APC-CY7 (Biolegend) or CD8-APC-CY7]. After lysis with 1X FACS Lysing Solution (BD Biosciences) and fixation/permeabilization with Fixation/Permeablization solution (BD Biosciences), the cells were stained with Granzyme A-Alexa Fluor 700 (Biolegend), Granzyme B-Alexa Fluor 647 (Biolegend), and Perforin-PerCp-Cy5.5 (Biolegend).
Placental Pathology
Four hematoxylin-and-eosin stained sections (two full-thickness sections of the placental disc, one umbilical cord section, and one roll of chorioamniotic membranes) were reviewed for the presence of placental pathologic findings according to the criteria proposed by the Perinatal Section of the Society for Pediatric Pathology. Placental histological changes were categorized as those associated with amniotic fluid infection (acute chorioamnionitis), maternal vascular underperfusion, and fetal vascular obstruction.27–30 Villitis of unknown etiology, chronic deciduitis, and chronic chorioamnionitis were diagnosed according to previously described criteria.19, 31, 32
Statistical Analysis
For continuous variables, distributions were examined for normality using Kolmogorov-Smirnov tests. When data were far from normality, the Mann-Whitney U test was performed for independent variables, and the Wilcoxon signed rank test was used for related variables. Statistical analyses were performed using SPSS Version 19.0 (SPSS, Inc., Chicago, IL, USA). All P values were two-sided, and a value of P < 0.05 was considered to be statistically significant.
Results
Transcriptome of CD300a+ and CD300a− T Lymphocytes
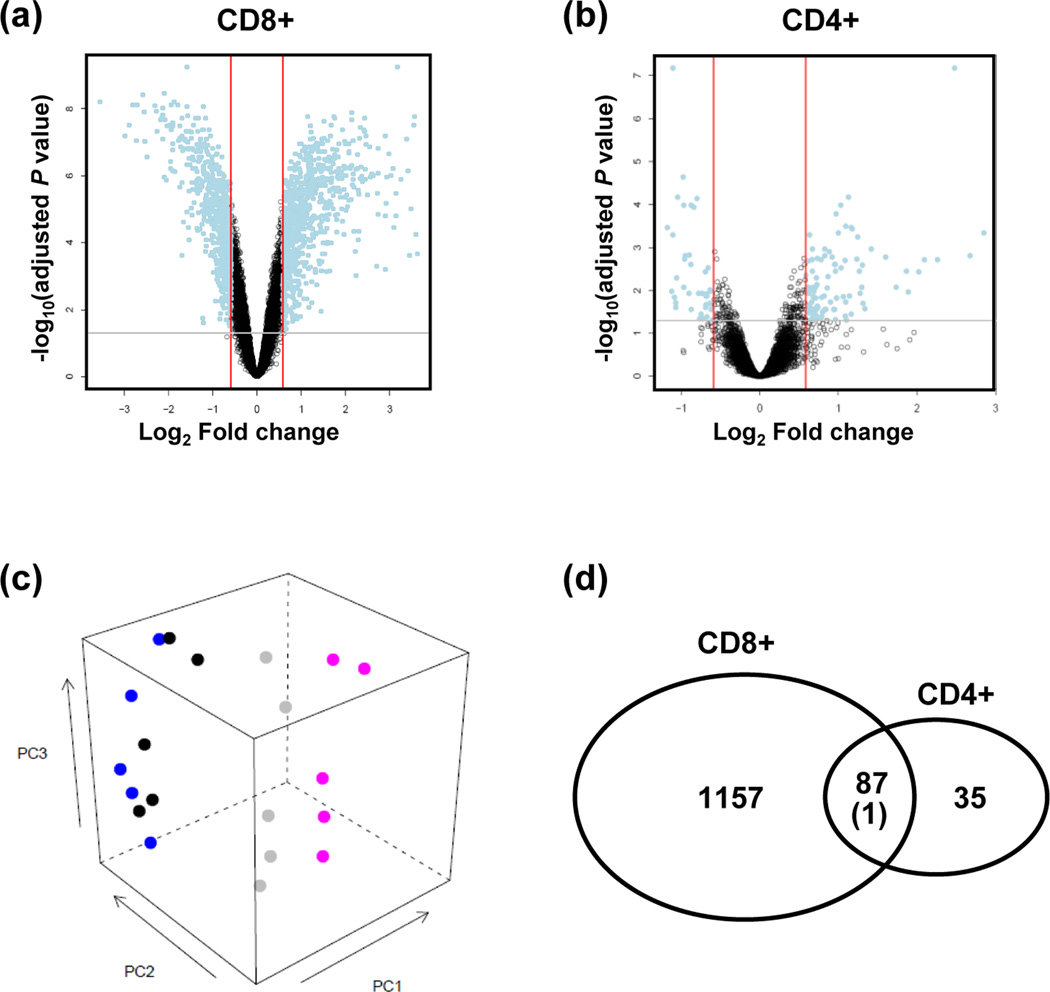
Microarray analyses of CD300a+CD8+ T lymphocytes and CD300a−CD8+ T lymphocytes from peripheral blood of five healthy adult women revealed differential regulation of 1,245 genes out of more than 25,000 annotated genes on the array (adjusted P value < 0.05, fold change > 1.5) (Figure 1a, 1c). Gene Ontology (GO) analysis showed enrichment of 141 biological processes, largely related to immune process, cell death, and signal transduction (Table I). One hundred thirty-six genes were differentially expressed among 613 genes associated with the immune system process (Table II). The impact evidence captured by SPIA from signaling perturbations and enrichment revealed eight pathways such as cytotoxicity and cytokine-cytokine receptor interaction (Table I).
Figure 1.
The transcriptome of CD300a+ and CD300a− T lymphocytes. (a, b) Volcano plots showing the relationship between the FDR-corrected P values of the genes (−log 10 of) and their fold changes (log2 of). Blue dots outside the two red lines and above the gray line have fold changes >1.5 and FDR-corrected P values < 0.05. Positive values on the x-axis indicate genes whose expression increased in CD300+ T lymphocytes, while negative values indicate genes down-regulated in CD300a+ T lymphocytes. (a: CD8+ T lymphocytes; b: CD4+ T lymphocytes) (c) A principal component analysis plot generated with data from five pairs of samples (CD300+CD8+ (pink), CD300a−CD8+ (gray), CD300a+CD4+ (black), CD300a−CD4+ (blue) T lymphocytes), showing a more clear segregation between CD300a+CD8+ and CD300a−CD8+ than between CD300a+CD4+ and CD300a−CD4+ T lymphocytes. (d) Among the differentially expressed genes between CD300a+CD8+ and CD300a−CD8+ T lymphocytes, 87 genes were regulated in the same direction as those expressed differentially between CD300a+CD4+ and CD300a−CD4+ T lymphocytes. Far more significantly enriched genes (N = 1,157) were identified in the CD8+ T lymphocytes with CD300a expression, while only 35 more genes were differentially regulated in the CD4+ T lymphocytes with CD300a expression.
Table I.
Top 10 biological processes and 8 pathways enriched in CD300a+CD8+ T lymphocytes compared to CD300a-CD8+ T lymphocytes
| No. Enriched Genes / No. Total Genes | Adjusted P value | ||
|---|---|---|---|
| Biologic process | Immune system process | 136/613 | 6.43E-16 |
| Immune response | 103/410 | 1.31E-15 | |
| Defense response | 83/353 | 1.52E-10 | |
| Death | 123/690 | 1.71E-07 | |
| Cell death | 122/687 | 2.10E-07 | |
| Response to stimulus | 244/1678 | 2.83E-07 | |
| Programmed cell death | 114/650 | 1.31E-06 | |
| Apoptosis | 113/646 | 1.59E-06 | |
| Signal transduction | 293/2154 | 3.00E-06 | |
| Cell communication | 312/2340 | 6.13E-06 | |
| Pathway | Natural killer cell mediated cytotoxicity | 35/104 | 7.19E-10 |
| Graft-versus-host disease | 21/37 | 8.47E-10 | |
| Cytokine-cytokine receptor interaction | 40/146 | 1.51E-09 | |
| Antigen processing and presentation | 24/68 | 3.54E-06 | |
| Allograft rejection | 14/30 | 3.62E-05 | |
| Asthma | 10/20 | 0.00070 | |
| Type I diabetes mellitus | 13/33 | 0.00172 | |
| Autoimmune thyroid disease | 12/35 | 0.00604 | |
Table II.
Top 20 differentially expressed genes related to immune system process in CD300a+CD8+ lymphocytes
| Gene Symbol | Gene Name | Fold Change | Adjusted P value |
|---|---|---|---|
| CD300A | CD300a molecule | 9.00 | 5.86E-10 |
| CD79A* | CD79a molecule, immunoglobulin-associated alpha | 2.98 | 5.86E-10 |
| KIR2DL3 | killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 3 | 11.82 | 1.71E-08 |
| PLCG2 | phospholipase C, gamma 2 (phosphatidylinositol-specific) | 4.40 | 2.16E-08 |
| HLA-DOA* | major histocompatibility complex, class II, DO alpha | 3.82 | 4.26E-08 |
| CCL3 | chemokine (C-C motif) ligand 3 | 3.81 | 4.26E-08 |
| TNFSF14 | tumor necrosis factor (ligand) superfamily, member 14 | 3.17 | 6.53E-08 |
| PRKCA* | protein kinase C, alpha | 3.35 | 7.78E-08 |
| CCL3L3 | chemokine (C-C motif) ligand 3-like 3 | 3.95 | 9.28E-08 |
| SLAMF7 | SLAM family member 7 | 1.99 | 1.10E-07 |
| IL15 | interleukin 15 | 2.53 | 1.16E-07 |
| FYB* | FYN binding protein (FYB-120/130) | 1.84 | 1.37E-07 |
| CD55* | CD55 molecule, decay accelerating factor for complement (Cromer blood group) | 2.99 | 1.53E-07 |
| VIPR1* | vasoactive intestinal peptide receptor 1 | 3.20 | 1.84E-07 |
| TBK1 | TANK-binding kinase 1 | 2.60 | 1.92E-07 |
| APOBEC3F | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F | 2.88 | 1.94E-07 |
| CLCF1 | cardiotrophin-like cytokine factor 1 | 2.90 | 3.25E-07 |
| DLL1* | delta-like 1 (Drosophila) | 2.70 | 3.46E-07 |
| TGFBR2* | transforming growth factor, beta receptor II (70/80kDa) | 2.02 | 3.79E-07 |
| MAPK1 | mitogen-activated protein kinase 1 | 2.26 | 4.09E-07 |
down-regulated
On the contrary, a relatively smaller number of genes (123 genes) was found to be differentially expressed between CD300a+ and CD300a−CD4+ T lymphocytes than in CD8+ T lymphocytes (Figure 1b, 1c). On GO analysis, 63 biological processes were enriched including signal transduction, immune response, and cell communication, which were similar to those for CD300a+CD8+ T lymphocytes (Table III). Among the genes related to the immune process, 16 genes were differentially expressed in CD300a+CD4+ T lymphocytes (Table IV). Cytokine-cytokine receptor interaction was found to be enriched by the SPIA pathway analysis.
Table III.
Top 10 biological processes and the pathway enriched in CD300a+CD4+ T lymphocytes compared to CD300a−CD4+ T lymphocytes
| No. Enriched Genes / No. Total Genes | Adjusted P value | ||
|---|---|---|---|
| Biologic process | Signal transduction | 41/2154 | 0.00449 |
| Immune response | 15/410 | 0.00449 | |
| Negative regulation of smooth muscle cell proliferation | 3/6 | 0.00449 | |
| Smooth muscle cell proliferation | 4/17 | 0.00449 | |
| Regulation of smooth muscle cell proliferation | 4/17 | 0.00449 | |
| Cell communication | 42/2340 | 0.00455 | |
| Response to external stimulus | 15/438 | 0.00455 | |
| Response to stimulus | 33/1678 | 0.00731 | |
| Response to wounding | 11/267 | 0.00888 | |
| Muscle cell proliferation | 4/24 | 0.00950 | |
| Pathway | Cytokine-cytokine receptor interaction | 9/146 | 4.76E-05 |
Table IV.
Differentially expressed genes related to immune system process in CD300a+CD4+ lymphocytes
| Gene Symbol | Gene Name | Fold Change | Adjusted P value |
|---|---|---|---|
| CD300A | CD300a molecule | 5.57 | 6.53E-08 |
| CTLA4* | cytotoxic T-lymphocyte-associated protein 4 | 1.79 | 1.17E-04 |
| IL15 | interleukin 15 | 1.96 | 1.68E-04 |
| CCL5 | chemokine (C-C motif) ligand 5 | 7.21 | 4.58E-04 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 2.00 | 4.58E-04 |
| TNFRSF4 | tumor necrosis factor receptor superfamily, member 4 | 2.05 | 2.97E-03 |
| OSM | oncostatin M | 1.95 | 3.53E-03 |
| CD226 | CD226 molecule | 1.64 | 8.41E-03 |
| GZMA | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 3.70 | 1.11E-02 |
| GPR44 | G protein-coupled receptor 44 | 1.64 | 1.51E-02 |
| IFNG | interferon, gamma | 2.13 | 1.86E-02 |
| TNFSF14 | tumor necrosis factor (ligand) superfamily, member 14 | 1.55 | 2.17E-02 |
| APOBEC3G | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G | 1.76 | 2.95E-02 |
| TNFSF13B | tumor necrosis factor (ligand) superfamily, member 13b | 1.54 | 3.33E-02 |
| CFH | complement factor H | 1.73 | 4.08E-02 |
| CTSW | cathepsin W | 1.61 | 4.98E-02 |
down-regulated
Among the differentially expressed genes between CD300a+CD4+ and CD300a−CD4+ T lymphocytes, 87 genes were regulated in the same direction as in the comparison between CD300a+CD8+ and CD300a−CD8+ T lymphocytes. The gene FCRL3, down-regulated in CD300a+CD4+ T lymphocytes compared to CD300a−CD4+ T lymphocytes, was found to be up-regulated in CD300a+CD8+ compared to CD300a-CD8+ T lymphocytes (Figure 1d).
Differential mRNA Expression of Immune-related Genes between CD300a+ and CD300a− CD8+ T Lymphocytes
Chemokines/Cytokines
Among chemokine genes, CCL3, CCL5, and CCL20 showed an increased mRNA expression in CD300a+CD8+ T lymphocytes, whereas mRNA expression of CXCL16 was decreased in CD300a+CD8+ T lymphocytes compared to CD300a-CD8+ T lymphocytes. mRNA expression of some cytokines of the hematopoietin family, including interleukins, were up-regulated in CD300a+CD8+ T lymphocytes. These included IL15 sharing certain biological activities with IL-2 such as T and NK cell activation and proliferation; IL29, a member of the IL-10 family; and CLCF1, a member of the IL-6 family. mRNA expression of several members of the TNF family (TNF, TNFSF9, TNFSF10, TNFSF12, TNFSF14, FASLG, and CD40LG) was up-regulated, while mRNA expression of another member of the TNF family, LTB, was down-regulated in CD300a+CD8+ T lymphocytes compared to CD300a−CD8+ T lymphocytes. Among the members of the interferon family, mRNA expression of interferon-gamma (IFNG), interferon gamma-inducible protein 16 (IFI16), and interferon-induced protein with tetratricopeptide repeat 2 (IFIT2), was up-regulated in CD300a+CD8+ T lymphocytes, while mRNA expression of interferon-induced transmembrane protein 3 (IFITM3) was down-regulated.
Chemokine/Cytokine Receptors
Among chemokine receptor genes, mRNA expression of CXCR3 (a receptor for CXCL9, −10, and −11), CXCR6 (a receptor for CXCL16), CX3CR1 (a receptor for CX3CL1), and CCR6 (a receptor for CCL20) was up-regulated in CD300a+CD8+ T lymphocytes compared to CD300a− cells, while mRNA expression of CCR7 (a receptor for CCL19 and -21) was down-regulated. Regarding interleukin receptor genes, mRNA expression of IL10RA, IL12RB1, IL18R1, IL18RAP, and IL28RA was up-regulated in CD300a+CD8+ T lymphocytes, while mRNA expression of IL6R and IL11RA were down-regulated. Among TNF receptor genes, mRNA up-regulation of TNFRSF1B, TNFRSF4, FAS, and TNFRSF9 was observed in CD300+CD8+ T lymphocytes, whereas mRNA expression of CD27 (TNFRSF7), TNFRSF25, and ectodysplasin A receptor (EDAR) was down-regulated. mRNA expression of interferon gamma receptor 2 (IFNGR2) and TGFBR2 was down-regulated in the CD300+ subset.
Effector Molecules for Cytotoxicity
One of the major effector functions of cytotoxic T lymphocytes is secretion of cytotoxic granules to induce apoptosis of target cells. mRNA expression of the major components of the granzyme genes (GZMA, GZMB, and GZMK) was up-regulated in CD300+CD8+ T lymphocytes. mRNAs of granulysin (GNLY) and perforin (PRF1) were also highly expressed in CD300+CD8+ T lymphocytes.
Killer Cell Receptors
CD300+CD8+ T lymphocytes showed the enriched mRNA expression of killer cell receptors. A subset of killer cell immunoglobulin-like receptors (KIR2DL1, −2, −3, −4, −5A, KIR2DS5, KIR3DL1, −2, and −3) and killer cell lectin-like receptors (KLRA1, −B1, −C1, −C3, −D1, −F1, and −G1) showed increased mRNA expression in CD300a+CD8+ T lymphocytes. Among inhibitory killer cell receptors, mRNA expression of KIR3DL1, KIR3DL2, KIR2DL, and KLRC1/KLRD1 (CD94) were up-regulated. Among activating killer cell receptors, mRNA expression of KIR2DS and KLRC3/KLRD1 (CD94) were up-regulated. mRNA expression of NCR3 (NKp30) and CD244, among other membrane receptors, was also upregulated.
Differential mRNA Expression of Immune-related Genes between CD300a+CD4+ and CD300a−CD4+ T Lymphocytes
Among genes related to the immune process, 16 genes were differentially expressed in CD300a+CD4+ T lymphocytes (Table IV). Among the differentially expressed immune-related genes between CD300a+CD4+ and CD300a−CD4+ T lymphocytes, 12 genes were regulated in the same direction as those expressed differentially between CD300a+CD8+ and CD300a−CD8+ T lymphocytes: CD300a, IL15, CCL5, TIMP1, TNFRSF4, CD226, GZMA, IFNG, TNFSF14, APOBEC3G, CFH, and CTSW. Four genes differentially expressed only in CD300a+CD4+ T lymphocytes, but not in CD300a+CD8+ T lymphocytes, were CTLA4, OSM, GPR44, and TNFSF13B. CTLA4 was down-regulated but the others were up-regulated in CD300a+CD4+ T lymphocytes compared to CD300a−CD4+ T lymphocytes.
Validation of Differential Expression of Immune-related Genes in CD300a+ T Lymphocytes
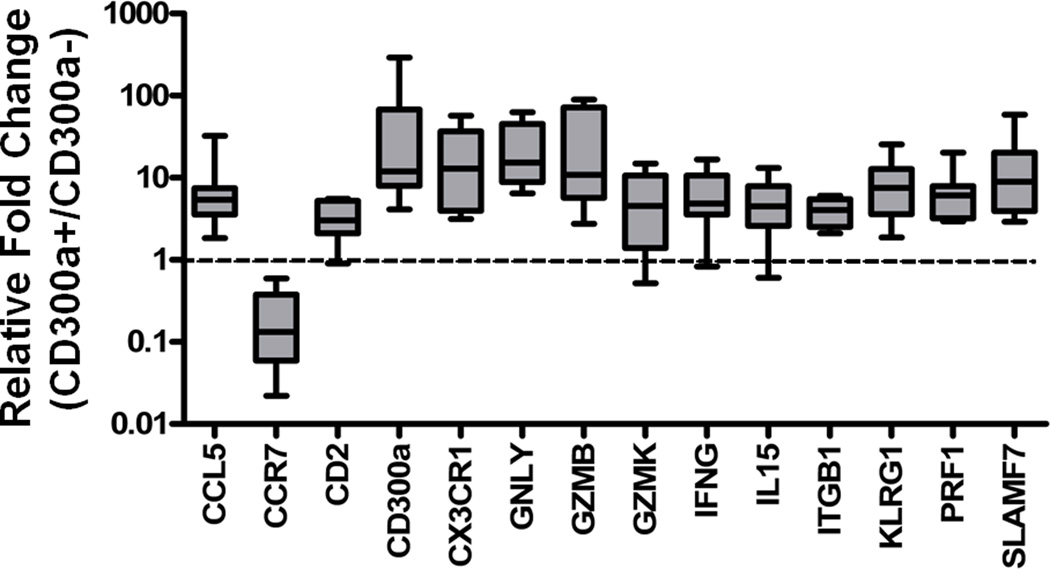
For validation of the microarray results, the differential mRNA expressions of the selected genes (CCL5, CCR7, CD244, CD300a, CX3CR1, GLNY, GZMB, GZMK, IFNG, IL15, ITGB, KLRG1, PRF1, and SLAMF7), related to cytotoxicity and cytokine-cytokine receptor interaction, between peripheral CD300a+ and CD300a−CD8+ T lymphocytes (N = 9), were analyzed by qRT-PCR. All genes other than CCR7 were expressed significantly higher in CD300a+CD8+ T lymphocytes than in CD300a−CD8+ T lymphocytes, while the expression of CCR7 was lower in CD300a+CD8+T lymphocytes than in CD300a−CD8+ T lymphocytes, which confirmed the microarray analysis (Figure 2).
Figure 2.
Validation of microarray analysis to compare gene expression between CD300a+CD8+ and CD300a−CD8+ T lymphocytes. Differential mRNA expression of the genes related to cytotoxicity and cytokines-cytokine receptor interaction between CD300a+ and CD300a−CD8+ T lymphocytes was confirmed by quantitative real-time-RT-PCR. CCR7 mRNA expression was lower, whereas the others were higher in CD300a+ than in CD300a− CD8+ T lymphocytes.
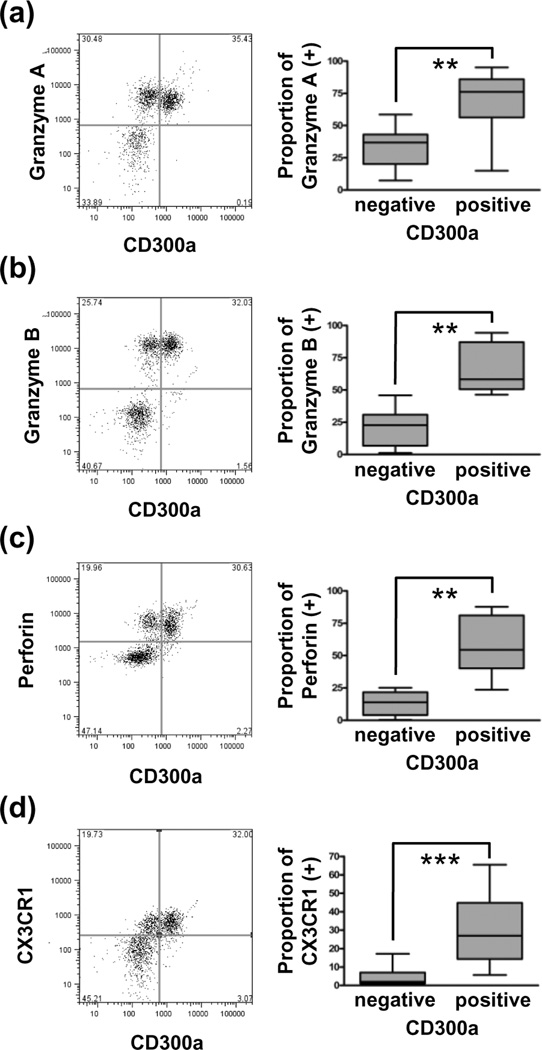
Granzyme A, granzyme B, perforin, and CX3CR1 protein expressions in peripheral CD300a+ and CD300a−CD8+ T lymphocytes (N = 30) were also assessed by flow cytometry, all of which were expressed higher in the CD300a+ sub-population than in the CD300a- sub-population (Figure 3).
Figure 3.
Expression of cytotoxicity effector proteins in CD300a+CD8+ peripheral T lymphocytes. Granzyme A (a), granzyme B (b), perforin (c), and CX3CR1 (d) were expressed higher in CD300a+ than CD300a− CD8+ T lymphocytes. **, P < 0.01; ***, P < 0.001.
CD300a+ T Lymphocytes in Peripheral Blood of Pregnant Women
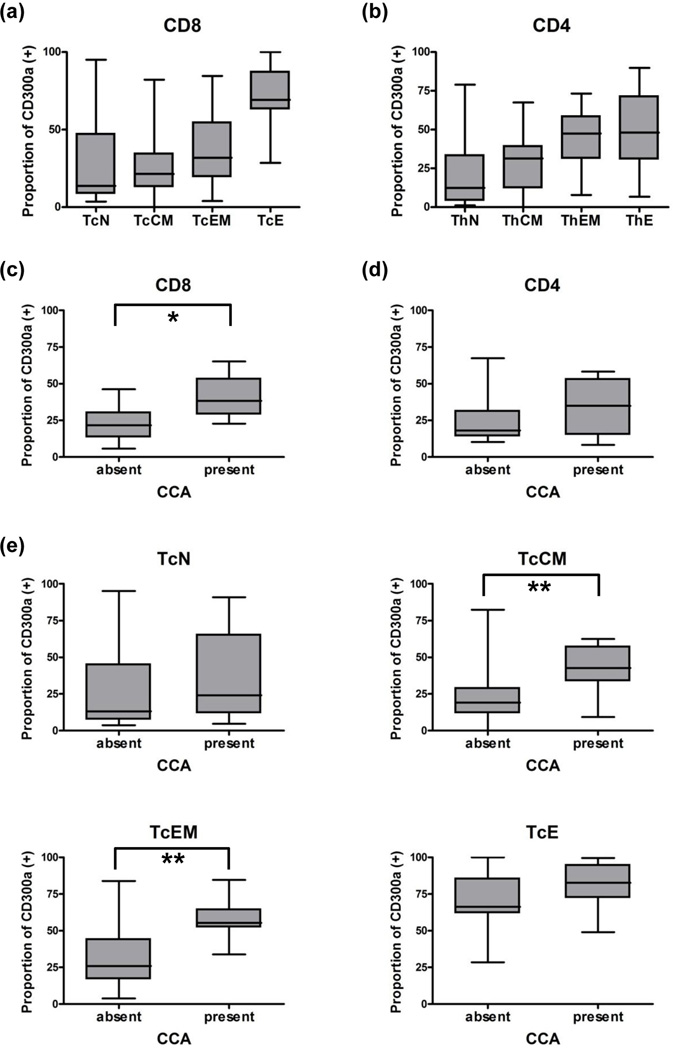
The demographic characteristics of the study population are shown in Table V. On flow cytometric analysis of peripheral blood from pregnant women (N = 19), CD300a was detected in 26.6% (5.9–65.2%) of CD8+ lymphocytes and in 18.3% (8.4–67.3%) of CD4+ T lymphocytes. When evaluated in subsets of Tc and Th lymphocytes by co-labeling for CD45RA and CCR7,33 the proportion of CD300a+ cells was greater in the order of effector, effector memory, central memory, and naïve T lymphocytes (N = 40, Figure 4a, 4b). The proportion of CD300a+ T lymphocytes was not different between preterm and term pregnant women [preterm (N = 7) vs. term (N = 12): CD8+, 30.4% (11.7–41.6%) vs. 19.7% (5.9–65.2%); and CD4+, 21.9% (14.0–67.3%) vs. 18.3% (8.4–48.1%)]. The proportions of CD300a+ T lymphocytes in cord blood of neonates (N = 17) were quite small [CD8+, 7.3% (1.4–17.9%); CD4+, 6.1% (1.3–18.1%)], compared to those in adults (P < 0.001).
Table V.
Demographics and clinical characteristics of pregnant women
| Maternal age (years) (median, range) | 25 (15–40) |
| Gestational age at sampling (weeks) (median, range) | 39.0 (13.3 – 41.7) |
| Gestational age at delivery (weeks) (median, range) | 39.7 (37.0 – 41.9) |
| Labor at sampling (present/absent/unknown) | 14/25/1 |
| Race (African American /others/unknown) | 35/4/1 |
| Para (primiparity/multiparity) | 13/27 |
| Baby gender (male/female) | 17/23 |
| Smoking (yes/no) | 3/37 |
| Interval between sampling and delivery (days) (median, range) | 1 (0–194) |
Figure 4.
The proportion of CD300a+ T lymphocytes in peripheral blood by flow cytometry. (a,b) The naïve, central memory, effector memory, and effector subsets of CD8+ T lymphocytes (a) and CD4+ T lymphocytes (b) showed the gradual increase of CD300a expression. (c,d) The increase of CD300a expression in pregnant women with chronic chorioamnionitis compared to the expression in those without was detected in CD8+ T lymphocytes. (c: CD8+ T lymphocytes; d: CD4+ T lymphocytes) (e) Central memory and effector memory CD8+ T lymphocytes showed higher expression of CD300a in cases with chronic chorioamnionitis than in those without. *, P < 0.05; **, P < 0.01; TcN, naïve cytotoxic T lymphocytes, TcCM, central memory cytotoxic T lymphocytes; TcEM, effector memory cytotoxic T lymphocytes; TcE, effector cytotoxic T lymphocytes.
We analyzed the proportion of the CD300a+ subset of T lymphocytes in samples of the peripheral blood of pregnant women according to placental pathology findings: intra-amniotic infection, maternal vascular underperfusion, fetal vascular thrombo-occlusive diseases, and chronic placental inflammation (villitis of unknown etiology, chronic chorioamnionitis, and chronic deciduitis). The median sampling to delivery interval was 1 day (range: 0–194 days). The proportion of CD300a+CD8+ T lymphocytes was higher in cases with chronic chorioamnionitis than in those without chorioamnionitis (Figure 4c). When evaluated in subsets of Tc and Th lymphocytes by co-labeling for CD45RA and CCR7,33 the proportion of CD300a+ cells was greater in the effector memory and the central memory subsets of CD8+ T lymphocytes but not in either effector or naïve subsets (Figure 4e). The other placental findings were not associated with changes in CD300a+ peripheral T lymphocytes.
Discussion
This study reports the transcriptome of CD300a+CD8+ and CD300a+CD4+ T lymphocytes for the first time. The principal findings of this study are: 1) CD300a expression defines a distinct sub-population of CD8+ T lymphocytes; 2) the gene expression profile of CD300a+CD8+ T lymphocytes is consistent with a more cytotoxic phenotype than that of CD300a-CD8+ T lymphocytes; and 3) the proportion of CD300a+ peripheral Tc lymphocytes is significantly increased in pregnant women with chronic chorioamnionitis (anti-fetal cellular rejection of the chorioamniotic membranes) than in women without chronic chorioamnionitis.
Significance of CD300a Expression in CD8+T Lymphocytes
CD300a is constitutively expressed in granulocytes, NK cells, and dendritic cells, in which it functions as an inhibitory receptor.34–39 Co-ligation of CD300a exerts an inhibitory effect on CD32a–mediated signaling in neutrophils.34 In mast cells, CD300a inhibits kit-mediated SCF-induced signaling to cause impairment of differentiation, survival, and activation.35 CD300a also suppresses eotaxin-dependent transmigration of eosinophils and the anti-apoptotic effects of IL-5 and GM-CSF.39 CD300a cross-linking inhibits the cytotoxicity of NK cells.36, 38 In dendritic cells, CD300a inhibits TNF-α production from activated plasmacytoid dendritic cells and up-regulates type I IFN production.37 In contrast to non-lymphocytic leukocytes, only a subset of B and T lymphocytes was found to express CD300a.16–18
In this study, the majority of CD300a+CD8+ T lymphocytes were memory and effector cells as was the case with CD4+ T lymphocytes.17 Fetal T lymphocytes, most of which are of a naïve subset, were almost negative for CD300a. Interestingly, far more genes were differentially expressed between CD300a+CD8+ and CD300a−CD8+ T lymphocytes than between CD300a+CD4+ and CD300a−CD4+ T lymphocytes. This suggests that CD300a is biologically more significant in CD8+ T lymphocytes than in CD4+ T lymphocytes. This is in contrast to the comparison of the transcriptional profiles between activated CD4+ T lymphocytes and CD8+ T lymphocytes, each group of which showed similar numbers of differentially expressed genes.40
CD300a+CD8+ T lymphocytes had differential expressions of cytokine/chemokine and related genes. CCL3 and CCL20 were up-regulated upon T lymphocyte activation and responsible for the pro-inflammatory response of T lymphocytes, while CCL5 was highly expressed in resting T lymphocytes, suggesting its importance in the homeostasis of resting T lymphocytes.40 CXCL16 and CXCR6 are examples of co-expression of ligand and its receptor in the same cell, and activation of T lymphocytes is known to lead to marked down-regulation of CXCL16.41 In addition, IFNG, a major effector cytokine of T lymphocytes, is up-regulated in CD300a+CD8+ T lymphocytes. Several key effector molecules for cytotoxicity were also up-regulated in CD300a+CD8+ T lymphocytes. CD300a+CD8+ T lymphocytes expressed GZMA, GZMB, and GZMK. GZMB is specific for effector-type cells, while GZMK is found mainly in a subset of memory-type cells. GZMA is highly expressed on primed CD8+ T lymphocytes regardless of differentiation status. High expression of CX3CR1 is also compatible with the features of peripheral cytotoxic effector CD8+ T lymphocytes with intracellular perforin and granzyme B.42
Differential Expression of Immune-related Genes in CD300a+CD4 T Lymphocytes
A certain number of the genes (N = 87) shared by CD300a+CD4+ and CD300a+CD8+ T lymphocytes reflect common cellular events shared by CD4+ and CD8+ T lymphocytes in the CD300a+ sub-population. Four immune process-related genes differentially expressed only in CD300a+CD4+ T lymphocytes, but not in CD300a+CD8+ T lymphocytes, were CTLA4, OSM, GPR44, and TNFSF13B. CTLA4 is a co-stimulatory molecule expressed by activated T lymphocytes and binds to B7-1 and B7-2 on antigen-presenting cells. CTLA4 transmits an inhibitory signal to T lymphocytes.43 Oncostatin M encoded by OSM is a member of the IL-6 family acting as a growth regulator44 and as an inflammatory mediator.45 Th1 lymphocytes regulate hematopoietic progenitor cell homeostasis by production of oncostatin M.46 GPR44 encoding G-protein-coupled receptor 44 is selectively expressed in Th2 cells.47 TNFSF13B encodes a cytokine of the tumor necrosis factor (TNF) ligand family, which is a ligand for receptors TNFRSF13B/TACI, TNFRSF17/BCMA, and TNFRSF13C/BAFFR.
Changes in T Lymphocyte Immune Responses during Pregnancy
Most studies about T lymphocyte tolerance of the fetal allograft have been done mainly about T-helper cells.48, 49 In earlier studies, maternal tolerance toward the fetal allograft was explained by the predominant Th2 immunity over Th1 immunity.50 The Th1/Th2 theory is now blending into the Th1/Th2/Th17/Treg paradigm, and the role of Treg cells is increasingly emphasized.6, 51, 52
In contrast, studies about the changes of cytotoxic T lymphocytes during normal pregnancy are limited.9–14 Alteration of cytotoxic T lymphocytes in pregnancy-related complications such as implantation failures and preeclampsia supports the significance of cytotoxic T lymphocytes for the successful maintenance of normal pregnancy.53–55 Yang et al. showed increased peripheral CD8+T lymphocyte activation in women with multiple implantation failures and recurrent pregnancy losses. The proportion of activated T lymphocytes was inversely correlated with IFNγ/IL-10 producing CD3+/CD4+ T lymphocyte ratios.54 De Groot et al. reported that women with preeclampsia had a higher partner-specific cytotoxic T-lymphocytic precursor frequency when compared to women with uncomplicated pregnancies, suggesting an increased cytotoxic T-cell response to paternal antigens in women with preeclampsia.53
Chronic chorioamnionitis is a common placental lesion found in cases of late preterm birth, and represents maternal anti-fetal cellular rejection involving the chorioamniotic membranes.32, 56, 57 We demonstrated in this study that CD300a+CD8+ T lymphocytes are mainly cytotoxic effector cells and memory cells, and the proportion of CD300a+CD8+ T lymphocytes in peripheral blood was higher in pregnant women who subsequently delivered placentas showing chronic chorioamnionitis than in those who did not. It suggests that peripheral CD8+ T lymphocytes of pregnant women with chronic chorioamnionitis have increased systemic cytotoxic capacity as a manifestation of maternal anti-fetal rejection. It is intriguing that the pregnant women with villitis of unknown etiology (anti-fetal cellular rejection involving the chorionic villi) did not show a significant increase of CD300a+ T lymphocytes in peripheral blood.19 Further analysis of more cases would be necessary to address this issue.
Conclusion
We report the transcriptome of CD300a+ peripheral T cells for the first time. The findings herein strongly suggest a role for CD300a+CD8+ T cells in the maternal anti-fetal immune response. There is a significant increase in the proportion of CD300a+CD8+ T cells in women presenting with anti-fetal cellular rejection in the chorioamniotic membranes.
Acknowledgements
This work was supported in part by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services.
Footnotes
Disclosure of Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Koch CA, Platt JL. T cell recognition and immunity in the fetus and mother. Cell Immunol. 2007;248:12–17. doi: 10.1016/j.cellimm.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fest S, Aldo PB, Abrahams VM, Visintin I, Alvero A, Chen R, Chavez SL, Romero R, Mor G. Trophoblast-macrophage interactions: a regulatory network for the protection of pregnancy. Am J Reprod Immunol. 2007;57:55–66. doi: 10.1111/j.1600-0897.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 3.Leiser R, Kaufmann P. Placental structure: in a comparative aspect. Exp Clin Endocrinol. 1994;102:122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- 4.Redline RW. The structural basis of maternal-fetal immune interactions in the human placenta. Curr Top Microbiol Immunol. 1997;222:25–44. doi: 10.1007/978-3-642-60614-4_2. [DOI] [PubMed] [Google Scholar]
- 5.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 6.Saito S, Nakashima A, Shima T, Ito M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am J Reprod Immunol. 2010;63:601–610. doi: 10.1111/j.1600-0897.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 7.Fraccaroli L, Alfieri J, Larocca L, Calafat M, Mor G, Leiros CP, Ramhorst R. A potential tolerogenic immune mechanism in a trophoblast cell line through the activation of chemokine-induced T cell death and regulatory T cell modulation. Hum Reprod. 2009;24:166–175. doi: 10.1093/humrep/den344. [DOI] [PubMed] [Google Scholar]
- 8.Ramhorst R, Fraccaroli L, Aldo P, Alvero AB, Cardenas I, Leiros CP, Mor G. Modulation and Recruitment of Inducible Regulatory T Cells by First Trimester Trophoblast Cells. Am J Reprod Immunol. 2011 doi: 10.1111/j.1600-0897.2011.01056.x. 10.1111/j.1600-0897.2011.01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhnert M, Strohmeier R, Stegmuller M, Halberstadt E. Changes in lymphocyte subsets during normal pregnancy. Eur J Obstet Gynecol Reprod Biol. 1998;76:147–151. doi: 10.1016/s0301-2115(97)00180-2. [DOI] [PubMed] [Google Scholar]
- 10.Kumru S, Boztosun A, Godekmerdan A. Pregnancy-associated changes in peripheral blood lymphocyte subpopulations and serum cytokine concentrations in healthy women. J Reprod Med. 2005;50:246–250. [PubMed] [Google Scholar]
- 11.Luppi P, Haluszczak C, Trucco M, Deloia JA. Normal pregnancy is associated with peripheral leukocyte activation. Am J Reprod Immunol. 2002;47:72–81. doi: 10.1034/j.1600-0897.2002.1o041.x. [DOI] [PubMed] [Google Scholar]
- 12.Mahmoud F, Abul H, Omu A, Al-Rayes S, Haines D, Whaley K. Pregnancy-associated changes in peripheral blood lymphocyte subpopulations in normal Kuwaiti women. Gynecol Obstet Invest. 2001;52:232–236. doi: 10.1159/000052981. [DOI] [PubMed] [Google Scholar]
- 13.Tallon DF, Corcoran DJ, O'Dwyer EM, Greally JF. Circulating lymphocyte subpopulations in pregnancy: a longitudinal study. J Immunol. 1984;132:1784–1787. [PubMed] [Google Scholar]
- 14.Watanabe M, Iwatani Y, Kaneda T, Hidaka Y, Mitsuda N, Morimoto Y, Amino N. Changes in T, B, and NK lymphocyte subsets during and after normal pregnancy. Am J Reprod Immunol. 1997;37:368–377. doi: 10.1111/j.1600-0897.1997.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 15.Daish A, Starling GC, McKenzie JL, Nimmo JC, Jackson DG, Hart DN. Expression of the CMRF-35 antigen, a new member of the immunoglobulin gene superfamily, is differentially regulated on leucocytes. Immunology. 1993;79:55–63. [PMC free article] [PubMed] [Google Scholar]
- 16.Silva R, Moir S, Kardava L, Debell K, Simhadri VR, Ferrando-Martinez S, Leal M, Pena J, Coligan JE, Borrego F. CD300a is expressed on human B cells, modulates BCR-mediated signaling, and its expression is down-regulated in HIV infection. Blood. 2011;117:5870–5880. doi: 10.1182/blood-2010-09-310318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark GJ, Rao M, Ju X, Hart DN. Novel human CD4+ T lymphocyte subpopulations defined by CD300a/c molecule expression. J Leukoc Biol. 2007;82:1126–1135. doi: 10.1189/jlb.0107035. [DOI] [PubMed] [Google Scholar]
- 18.Narayanan S, Silva R, Peruzzi G, Alvarez Y, Simhadri VR, Debell K, Coligan JE, Borrego F. Human Th1 cells that express CD300a are polyfunctional and after stimulation up-regulate the T-box transcription factor eomesodermin. PLoS One. 2010;5:e10636. doi: 10.1371/journal.pone.0010636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, Lee DC, Draghici S, Gotsch F, Kusanovic JP, Hassan SS, Kim JS. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. 2009;182:3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 21.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 23.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 24.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 25.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarca AL, Romero R, Draghici S. Analysis of microarray experiments of gene expression profiling. Am J Obstet Gynecol. 2006;195:373–388. doi: 10.1016/j.ajog.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redline RW, Ariel I, Baergen RN, Desa DJ, Kraus FT, Roberts DJ, Sander CM. Fetal vascular obstructive lesions: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2004;7:443–452. doi: 10.1007/s10024-004-2020-x. [DOI] [PubMed] [Google Scholar]
- 28.Redline RW, Boyd T, Campbell V, Hyde S, Kaplan C, Khong TY, Prashner HR, Waters BL. Maternal vascular underperfusion: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2004;7:237–249. doi: 10.1007/s10024-003-8083-2. [DOI] [PubMed] [Google Scholar]
- 29.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, Vogler C. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2003;6:435–448. doi: 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 30.Redline RW, Heller D, Keating S, Kingdom J. Placental diagnostic criteria and clinical correlation--a workshop report. Placenta. 2005;26(Suppl A):S114–S117. doi: 10.1016/j.placenta.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Khong TY, Bendon RW, Qureshi F, Redline RW, Gould S, Stallmach T, Lipsett J, Staples A. Chronic deciduitis in the placental basal plate: definition and interobserver reliability. Hum Pathol. 2000;31:292–295. doi: 10.1016/s0046-8177(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 32.Kim CJ, Romero R, Kusanovic JP, Yoo W, Dong Z, Topping V, Gotsch F, Yoon BH, Chi JG, Kim JS. The frequency, clinical significance, and pathological features of chronic chorioamnionitis: a lesion associated with spontaneous preterm birth. Mod Pathol. 2010;23:1000–1011. doi: 10.1038/modpathol.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez Y, Tang X, Coligan JE, Borrego F. The CD300a (IRp60) inhibitory receptor is rapidly up-regulated on human neutrophils in response to inflammatory stimuli and modulates CD32a (FcgammaRIIa) mediated signaling. Mol Immunol. 2008;45:253–258. doi: 10.1016/j.molimm.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachelet I, Munitz A, Berent-Maoz B, Mankuta D, Levi-Schaffer F. Suppression of normal and malignant kit signaling by a bispecific antibody linking kit with CD300a. J Immunol. 2008;180:6064–6069. doi: 10.4049/jimmunol.180.9.6064. [DOI] [PubMed] [Google Scholar]
- 36.Cantoni C, Bottino C, Augugliaro R, Morelli L, Marcenaro E, Castriconi R, Vitale M, Pende D, Sivori S, Millo R, Biassoni R, Moretta L, Moretta A. Molecular and functional characterization of IRp60, a member of the immunoglobulin superfamily that functions as an inhibitory receptor in human NK cells. Eur J Immunol. 1999;29:3148–3159. doi: 10.1002/(SICI)1521-4141(199910)29:10<3148::AID-IMMU3148>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 37.Ju X, Zenke M, Hart DN, Clark GJ. CD300a/c regulate type I interferon and TNF-alpha secretion by human plasmacytoid dendritic cells stimulated with TLR7 and TLR9 ligands. Blood. 2008;112:1184–1194. doi: 10.1182/blood-2007-12-127951. [DOI] [PubMed] [Google Scholar]
- 38.Lankry D, Simic H, Klieger Y, Levi-Schaffer F, Jonjic S, Mandelboim O. Expression and function of CD300 in NK cells. J Immunol. 2010;185:2877–2886. doi: 10.4049/jimmunol.0903347. [DOI] [PubMed] [Google Scholar]
- 39.Munitz A, Bachelet I, Eliashar R, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL-5, GM-CSF, and eotaxin on human peripheral blood eosinophils. Blood. 2006;107:1996–2003. doi: 10.1182/blood-2005-07-2926. [DOI] [PubMed] [Google Scholar]
- 40.Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics. 2008;9:225. doi: 10.1186/1471-2164-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shashkin P, Simpson D, Mishin V, Chesnutt B, Ley K. Expression of CXCL16 in human T cells. Arterioscler Thromb Vasc Biol. 2003;23:148–149. doi: 10.1161/01.atv.0000043906.61088.4b. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura M, Umehara H, Nakayama T, Yoneda O, Hieshima K, Kakizaki M, Dohmae N, Yoshie O, Imai T. Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J Immunol. 2002;168:6173–6180. doi: 10.4049/jimmunol.168.12.6173. [DOI] [PubMed] [Google Scholar]
- 43.Magistrelli G, Jeannin P, Herbault N, Benoit De Coignac A, Gauchat JF, Bonnefoy JY, Delneste Y. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur J Immunol. 1999;29:3596–3602. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Rose TM, Bruce AG. Oncostatin M is a member of a cytokine family that includes leukemia-inhibitory factor, granulocyte colony-stimulating factor, and interleukin 6. Proc Natl Acad Sci U S A. 1991;88:8641–8645. doi: 10.1073/pnas.88.19.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modur V, Feldhaus MJ, Weyrich AS, Jicha DL, Prescott SM, Zimmerman GA, McIntyre TM. Oncostatin M is a proinflammatory mediator. In vivo effects correlate with endothelial cell expression of inflammatory cytokines and adhesion molecules. J Clin Invest. 1997;100:158–168. doi: 10.1172/JCI119508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broxmeyer HE, Bruns HA, Zhang S, Cooper S, Hangoc G, McKenzie AN, Dent AL, Schindler U, Naeger LK, Hoey T, Kaplan MH. Th1 cells regulate hematopoietic progenitor cell homeostasis by production of oncostatin M. Immunity. 2002;16:815–825. doi: 10.1016/s1074-7613(02)00319-9. [DOI] [PubMed] [Google Scholar]
- 47.Nagata K, Tanaka K, Ogawa K, Kemmotsu K, Imai T, Yoshie O, Abe H, Tada K, Nakamura M, Sugamura K, Takano S. Selective expression of a novel surface molecule by human Th2 cells in vivo. J Immunol. 1999;162:1278–1286. [PubMed] [Google Scholar]
- 48.Jin LP, Fan DX, Zhang T, Guo PF, Li DJ. The Costimulatory Signal Upregulation is Associated with Th1 Bias at the Maternal-Fetal Interface in Human Miscarriage. Am J Reprod Immunol. 2011 doi: 10.1111/j.1600-0897.2011.00997.x. 10.1111/j.1600-0897.2011.00997.x. [DOI] [PubMed] [Google Scholar]
- 49.Kheshtchin N, Gharagozloo M, Andalib A, Ghahiri A, Maracy MR, Rezaei A. The expression of Th1- and Th2-related chemokine receptors in women with recurrent miscarriage: the impact of lymphocyte immunotherapy. Am J Reprod Immunol. 2010;64:104–112. doi: 10.1111/j.1600-0897.2010.00829.x. [DOI] [PubMed] [Google Scholar]
- 50.Wegmann TG, Lin H, Guilbert L, Mosmann TR. Bidirectional cytokine interactions in the maternal-fetal relationship: is successful pregnancy a TH2 phenomenon? Immunol Today. 1993;14:353–356. doi: 10.1016/0167-5699(93)90235-D. [DOI] [PubMed] [Google Scholar]
- 51.Toldi G, Rigo J, Jr, Stenczer B, Vasarhelyi B, Molvarec A. Increased Prevalence of IL-17-Producing Peripheral Blood Lymphocytes in Pre-eclampsia. Am J Reprod Immunol. 2011;66:223–229. doi: 10.1111/j.1600-0897.2011.00987.x. [DOI] [PubMed] [Google Scholar]
- 52.Liu YS, Wu L, Tong XH, Wu LM, He GP, Zhou GX, Luo LH, Luan HB. Study on the relationship between Th17 cells and unexplained recurrent spontaneous abortion. Am J Reprod Immunol. 2011;65:503–511. doi: 10.1111/j.1600-0897.2010.00921.x. [DOI] [PubMed] [Google Scholar]
- 53.de Groot CJ, van der Mast BJ, Visser W, De Kuiper P, Weimar W, Van Besouw NM. Preeclampsia is associated with increased cytotoxic T-cell capacity to paternal antigens. Am J Obstet Gynecol. 2010;203:496, e1–e6. doi: 10.1016/j.ajog.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 54.Yang KM, Ntrivalas E, Cho HJ, Kim NY, Beaman K, Gilman-Sachs A, Kwak-Kim J. Women with multiple implantation failures and recurrent pregnancy losses have increased peripheral blood T cell activation. Am J Reprod Immunol. 2010;63:370–378. doi: 10.1111/j.1600-0897.2010.00811.x. [DOI] [PubMed] [Google Scholar]
- 55.Darmochwal-Kolarz D, Saito S, Rolinski J, Tabarkiewicz J, Kolarz B, Leszczynska-Gorzelak B, Oleszczuk J. Activated T lymphocytes in pre-eclampsia. Am J Reprod Immunol. 2007;58:39–45. doi: 10.1111/j.1600-0897.2007.00489.x. [DOI] [PubMed] [Google Scholar]
- 56.Gersell DJ, Phillips NJ, Beckerman K. Chronic chorioamnionitis: a clinicopathologic study of 17 cases. Int J Gynecol Pathol. 1991;10:217–229. [PubMed] [Google Scholar]
- 57.Jacques SM, Qureshi F. Chronic chorioamnionitis: a clinicopathologic and immunohistochemical study. Hum Pathol. 1998;29:1457–1461. doi: 10.1016/s0046-8177(98)90016-8. [DOI] [PubMed] [Google Scholar]