Abstract
Problem
Decidual macrophages (dMϕ) of the mother and placental macrophages (Hofbauer cells, HC) of the fetus are deployed at a critical location: the feto-maternal interface. This study was conducted to compare DNA methylome of maternal and fetal monocytes, dMϕ, and HC, and thereby to determine the immunobiological importance of DNA methylation in pregnancy.
Methods of Study
Paired samples were obtained from normal pregnant women at term not in labor and their own neonates. Maternal monocytes (MM) and fetal monocytes (FM) were isolated from peripheral blood of mothers and from fetal cord blood, respectively. dMϕ and HC were obtained from the decidua of fetal membranes and placenta, respectively. DNA methylation profiling was done using the Illumina Infinium Human Methylation27 BeadChip. Quantitative real-time PCR and western blot were performed for validation experiments.
Results
1) Significant differences in DNA methylation were found in each comparison (MM vs. FM, 65 loci; dMϕ vs. HC, 266 loci; MM vs. dMϕ, 199 loci; FM vs. HC, 1,030 loci). 2) Many of the immune response-related genes were hypermethylated in fetal cells (FM and HC) compared to maternal cells (MM and dMϕ). 3) Genes encoding markers of classical macrophage activation were hypermethylated and genes encoding alternative macrophage activation were hypomethylated in dMϕ and HC compared to MM and FM, respectively. 4) mRNA expressions of DNMT1, DNMT3A, and DNMT3B were significantly lower in dMϕ than in HC. 5) 5-azacytidine treatment increased expression of INCA1 in dMϕ.
Conclusions
The findings herein indicate that DNA methylation patterns change during monocyte-macrophage differentiation at the feto-maternal interface. It is also suggested that DNA methylation is an important component of biological machinery conferring an anti-inflammatory phenotype to macrophages at the feto-maternal interface.
Keywords: Decidua, DNA methylation, DNA methyltransferase, Epigenetics, Epigenome, Hofbauer cell, Placenta, Pregnancy
Introduction
Macrophages play key roles in immune responses against a wide variety of insults such as infection1–3 and allograft rejection.4, 5 Human pregnancy is considered an immunologic enigma since the semi-allogeneic fetal graft is accepted and tolerated during the course of pregnancy.6–8 The mechanisms proposed for immunological tolerance to the fetus include the expression of non-classical HLA molecules on trophoblasts,9, 10 changes in tryptophan metabolism by indoleamine 2,3-dioxygenase,11, 12 and regulatory T cells.13–21 Placental development, ranging from trophoblast invasion to deportation of trophoblast debris into intervillous maternal circulation, is associated with the changes in the frames of feto-maternal immune interaction.22–27 Therefore, the human placenta is at the epicenter of the feto-maternal interface, and maternal cells of the decidua interact direct with fetal cells, typically trophoblasts, beginning at the time of implantation and continuing throughout pregnancy.15, 28–31 Maternal decidual macrophages (dMϕ) and fetal placental macrophages (Hofbauer cells, HC), are important cell populations at the feto-maternal interface.32–39 They interact continuously with potential immunological signals (microbial or danger signals)40, 41 and cells from the fetus and the mother.42–49 These two types of macrophages (dMϕ and HC) are known to originate from maternal and fetal blood monocytes although chorionic mesenchymal cells have been proposed as a major source of HC before the formation of villous stromal capillaries.50, 51 Changes in the immunophenotype, metabolic characteristics, and distribution of peripheral monocytes and dMϕ have been implicated in the pathogenesis of pregnancy disorders such as preeclampsia,52–56 preterm labor with intact membranes,57 preterm premature rupture of membranes,58 fetal systemic inflammation,2, 59–61 and pyelonephritis.62 HC have been implicated in the pathogenesis of pregnancy complications63 and they are activated in the presence of placental lesions consistent with maternal anti-fetal cellular rejection.64, 65
DNA methylation is a prototypic example of epigenetic regulation of gene expressions along with small non-coding RNA and histone tail modifications.66–69 DNA methylation largely involves the cytosine residues of CpG dinucleotides in mammalian cells, and hypermethylation leads to the decreased expression of the genes as seen in several tumor suppressor genes during carcinogenesis.70, 71 DNA methylation is mediated by a family of DNA methyltransferases (DNMT1, DNMT3A, DNMT3B),72, 73 and is important in various cellular processes such as differentiation,74 transcription of genes,75 and genomic imprinting.76 Recent studies have demonstrated associations between placental DNA methylation patterns and pregnancy disorders. An analysis of 1,505 CpG sites revealed hypomethylation of 34 loci including the promoter region of TIMP3 in early-onset preeclampsia cases compared to control subjects.77 It was also shown that hypomethylation of the H19 promoter region is associated with increased H19 expression in the placentas of fetal growth restriction cases.78 However, considering the heterogeneity of placental cell types and cell type specificity of DNA methylation patterns,79 the procurement of specific cell populations would be important in the comparative analysis of DNA methylation patterns in the placenta.
DNA methylation has been shown to be important in the differentiation of hematopoietic progenitor cells, and a subset of genes involved in differentiation is hypomethylated in monocytes and granulocytes during hematopoiesis.80 Ji et al also have described drastic alterations of DNA methylation with lymphoid and myeloid restriction from their progenitor cells during hematopoiesis.81 These findings indicate that the changes in CpG methylation are very important in lineage-specific hematopoietic cell differentiation. Tissue macrophages are mainly derived from peripheral blood monocytes to meet the local needs.82 We hypothesized that distinct changes in DNA methylation occur during differentiation of maternal monocytes (MMo) into dMϕ and during differentiation of fetal monocytes (FMo) into HC, and differential DNA methylation patterns will give clues for further understanding of the immunological characteristics of these cells during human pregnancy.
In the present study, we have compared the DNA methylome of MMo, FMo, dMϕ, and HC using paired sets of samples and thereby to determine the immunobiological importance of DNA methylation in normal pregnancy.
Materials and Methods
Study Design
The study was designed to compare the DNA methylome of MMo, FMo, dMϕ, and HC. The paired samples were obtained from normal pregnant women at term not in labor and their own babies (n=26) enrolled to the bank of biological materials of the Perinatology Research Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Health, USA. Peripheral blood of mothers was collected by venipuncture when admitted for cesarean section, and fetal cord blood was collected from the umbilical vein during cesarean section. The placentas were used to obtain decidual and villous macrophages. The numbers of paired samples used in the different experiments are as follows: Infinium methylation assay (n=6), pyrosequencing (n=6), quantitative RT-PCR (n=5), immunoblotting (n=4), dMϕ culture with 5-azacytidine treatment (n=5), and BrdU assay (n=4). Samples isolated by flow sorting were used for Infinium methylation assay and pyrosequencing, which require more pure cell populations. Faster cell column isolation was used for quantitative RT-PCR, immunoblotting, and cell culture in order to obtain more fresh samples in spite of relatively lower purity. Participating women provided written informed consent, and the collection and use of the samples and clinical data for research purposes were approved by the Institutional Review Boards of Wayne State University and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services.
Isolation of blood monocytes by flow cytometric cell sorting
Peripheral blood mononuclear cells (PBMCs) were isolated from maternal and fetal cord blood in EDTA by discontinuous density gradient centrifugation using Histopaque 1077 (Sigma-Aldrich, St. Louis, MO, USA). PBMCs were stained with phycoerythrin (PE)-Cy7-conjugated anti-CD14, and allophycocyanin (APC)-conjugated anti-CD15 (BD Biosciences, San Jose, CA, USA; for each). CD14+CD15- monocytes were sorted using a FACS Aria™ Cell Sorter (BD Biosciences), while CD14dim+CD15+ neutrophils and the other unstained cells were negatively selected.
Isolation of tissue macrophages by flow cytometric sorting
For isolation of dMϕ, decidual tissue (7 gm), procured by scraping the decidual side of the chorion and followed by mincing with a razor blade, was incubated in 50 ml of RPMI 1640 (Invitrogen, Carlsbad, CA, USA) containing 10% FBS and Dispase (2 mg/ml; Invitrogen) at 37 °C for 30 min with shaking (175 rpm). Then the samples were centrifuged at 600 × g for 5 min, and the pellets were resuspended and incubated in 50 ml of RPMI 1640 containing type IV collagenase (2 mg/ml; Worthington Biochemical Corporation, Lakewood, NJ, USA), hyaluronidase (2 mg/ml; Sigma-Aldrich), and DNase I (50 μg/ml; Roche, Mannheim, Germany) for 30 min at 37°C with shaking. The digest was homogenized using a syringe with a 20-gauge needle, and incubated for an additional 30 min at 37°C with shaking (175 rpm). After filtering through the gauze and 100-micron nylon mesh, the digests were washed 2 times with DPBS and the cell numbers were counted. Antibody labeling and cell sorting were performed as described above for blood monocytes.
For isolation of HC, the basal plate, the chorionic plate, and grossly visible blood vessels were removed from the placental tissue. After 3 washes with DPBS, 7 gm of villous tissues were used for a four-stage digestion protocol to isolate single cells from villous tissue.83 First, 50 ml of collagenase type 1A (0.624 mg/ml; Sigma-Aldrich) solution containing 0.684 mg/ml of hyaluronidase (Sigma-Aldrich), 0.12 mg/ml of DNase I (Roche), and 1 mg/ml of BSA (Sigma-Aldrich) in DPBS was added to the tissue. Following incubation for 6 min at 37°C with shaking (175 rpm), the tubes were kept on ice for 5 min and supernatant was discarded. For the second stage, 50 ml of trypsin solution (0.25 %, Invitrogen) containing 0.12 mg/ml of DNase I (Roche) and 0.5 mM of EDTA (pH 8.0, Invitrogen) were added and incubated at 37°C for 30 min with shaking (175 rpm). The tissue digest was centrifuged at 600×g for 5 min, and the pellet was then incubated with collagenase type IV (2 mg/ml; Worthington) in 50 ml of RPMI 1640 (10% FBS without antibiotics) containing 2 mg/ml of hyaluronidase (Sigma-Aldrich), and 50 μg/ml of DNase I (Roche) at 37°C for 30 min with shaking (175 rpm). The tissue digest was centrifuged at 600×g for 5 min and the supernatant was removed. Finally, collagenase type IV treatment was repeated. Following the digestion process, tissue digest was homogenized using a syringe with a 20-gauge needle, and incubated for an additional 30 min at 37°C with shaking. This digest was filtered and washed according to the same procedure used for decidual cell isolation as described above. Antibody labeling and cell sorting were performed as described above for blood monocytes.
Monocyte and macrophage isolation using a separation column
Monocytes and macrophages were isolated using cell separation columns. PBMCs of maternal blood and fetal cord blood were isolated by discontinuous density gradient centrifugation using Histopaque 1077 (Sigma-Aldrich). After lysing the red blood cells with sterile 0.2 % NaCl (pH 6.75) for 45 s, the cells were restored with sterile 1.6 % NaCl (pH 6.75) and centrifuged at 300 ×g for 5 min. Monocytes were isolated using Monocyte Isolation Kit II and MS columns (Miltenyi Biotech Inc., Auburn, CA, USA) according to the manufacturer’s instructions. A small fraction of isolated monocytes were stained with APC-conjugated CD14 for the flow cytometric assessment of purity.
For isolation of dMϕ and HC, isolated single cells obtained by the protocols described above were incubated with sterile 0.2 % NaCl (pH 6.75) for 45 s and restored with sterile 1.6 % of NaCl (pH 6.75). Following removal of dead cells using a Dead Cell Removal Kit (Miltenyi Biotech), macrophages were isolated by CD14 microbeads and LS columns (Miltenyi Biotech). A small fraction of isolated macrophages were stained with APC-conjugated CD14 for flow cytometry.
Confirmation of isolated monocytes and macrophages
Each population of positively sorted cells was reanalyzed by flow cytometry to evaluate the purities of monocytes and macrophages. For visualization of the sorted CD14+ monocytes and macrophages in a few representative cases, positively selected PE-Cy7-conjugated anti-CD14+ cells were smeared on silanized slides, and mounted with Prolong Gold Antifade Reagent with DAPI (Invitrogen). The cells were examined using a Leica TCS SP5 spectral confocal system (Leica Microsystems, Wetzler, Germany).
Infinium methylation assay
Genomic DNA of sorted MMo, FMo, dMϕ, and HC from women at term not in labor (n=6) was isolated using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). DNA (500 ng) samples were bisulfite modified using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA), and were hybridized with the Illumina Human Methylation27 BeadChip (Illumina, Inc., San Diego, CA, USA), according to the manufacturer’s instructions. After hybridization, allele-specific single-base extension and x-staining were performed on the BeadChip, and fluorescent signals were acquired with the Bead Array Reader (Illumina). The data from these images were analyzed using BeadStudio Methylation Module (Illumina), and the relative level of methylation (β) was calculated as the ratio of methylated-locus signal to total locus signal intensity.
Bioinformatics analysis
The relative level of methylation (β value) ranges between 0 and 1 with values close to 0 indicating no methylation and values close to 1 indicating presence of methylation. The raw data was quantile normalized84 among the 24 samples and differential methylation was tested using a paired moderated t-test.85 A given locus was deemed significant if its False Discovery Rate86 adjusted p-value (called q-value) was less than 0.05 and the difference in average β values between the two groups was 0.2 β units or more. Differentially methylated gene listswere interpreted using Gene Ontology (GO) terms explaining that they enrich using an over-representation analysis87 implemented in the Bioconductor (www.bioconductor.org) package GOStats.88 All analyses were performed using the R statistical language (www.r-project.org).
Bisulfite DNA pyrosequencing
Based on the results obtained from the Infinium Methylation Assay, the methylation status of LAG3, INCA1, and IL1B promoter regions was validated by bisulfite pyrosequencing. Genomic DNA (500 ng) samples of MMo, FMo, dMϕ, and HC from an independent group of pregnant women at term not in labor (n=6) were treated with bisulfite, and the samples were purified using Zymogen DNA columns (Zymo Research). Bisulfite-treated DNA was eluted in 20 μl of TE buffer (pH 8.0), and 1 μl was used for each PCR. PCR was performed with 0.2 μM of forward and reverse primers, and the PCR product was immobilized to Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden). The Sepharose beads containing the immobilized PCR product were purified, washed and denatured using a 0.2 M NaOH solution and rewashed using the Pyrosequencing Vacuum Prep Tool (Qiagen), according to the manufacturer’s instructions. For pyrosequencing, 0.2 μM of sequencing primers was annealed to the purified single-stranded DNA, and 10 μl of the PCR products were sequenced by Pyrosequencing PSQ96 HS System (Biotage AB, Uppsala, Sweden), according to the manufacturer’s instructions. The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Qiagen).
Real-time quantitative RT-PCR
The total RNA of monocytes and macrophages (n=5) was isolated using the RNeasy Mini Kit (Qiagen). Reverse transcription was done using the ImpromII Reverse Transcription System (Promega, Madison, WI, USA). All PCR analyses were carried out using TaqMan assays (Applied Biosystems, Foster City, CA, USA) for DNMT1 (Hs00154749_m1), DNMT3A (Hs01027166_m1), DNMT3B (Hs00171876_m1), INCA1 (Hs01652223_m1), and RPLPO (4326314E). PCR reactions were done using the 7500 Fast Real-Time PCR System (Applied Biosystems).
Immunoblotting
Total proteins from monocytes and macrophages (n=4) were isolated using RIPA buffer (Sigma-Aldrich) containing a protease inhibitor cocktail (Roche). Twenty μg of proteins were subjected to 10 % SDS-polyacrylamide gel electrophoresis, and electro-transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20, and probed overnight at 4°C with a rabbit polyclonal anti-INCA1 (1:500 dilution; ProSci Inc, Poway, CA, USA) or a mouse monoclonal anti-β-actin (1:5,000 dilution; Sigma-Aldrich). Horseradish peroxidase-conjugated anti-rabbit or mouse IgG (Vector Laboratories, Burlingame, CA, USA) was used as a secondary antibody, and signals were detected by chemiluminescence using ChemiGlow West Reagents (Alpha Innotech Corporation, San Leandro, CA, USA).
Decidual macrophage culture with 5-Azacytidine treatment and BrdU incorporation assay
The dMϕ (n=5) were plated in 6-well plates at the density of 5 × 105 cells per well in RPMI1640 (Invitrogen) media supplemented with 10 % FBS and antibiotics. The cells were treated with 5-azacytidine (Sigma-Aldrich) at the concentration of 5, 10, or 50 μM for 3 days. The cells were then pulsed with BrdU (10 μM) for 1 h, and stained using a BrdU Flow Kit (BD Bioscience), according to the manufacturer’s instructions. Briefly, the cells were fixed and permeabilized with BD Cytofix/Cytoperm buffer, and incubated with BD Cytoperm Plus buffer. The cells were then fixed again with BD Cytofix/Cytoperm Buffer, followed by DNase (0.3 mg/ml) treatment for 1 h at 37°C. After staining with FITC-conjugated anti-BrdU antibody, the cells were analyzed for BrdU uptake using BD LSR II flow cytometry (BD Biosciences) equipped with BD FACSDiva software version 6.0. Total events numbered 3,000.
Statistical analysis
Wilcoxon signed rank tests for related variables and the Mann-Whitney U test for independent variables were performed using the SPSS version 15.0 (SPSS Inc, Chicago, IL, USA). All p-values were two-sided, and a value of p<0.05 was considered to be statistically significant.
Results
Purity of isolated monocytes and macrophages
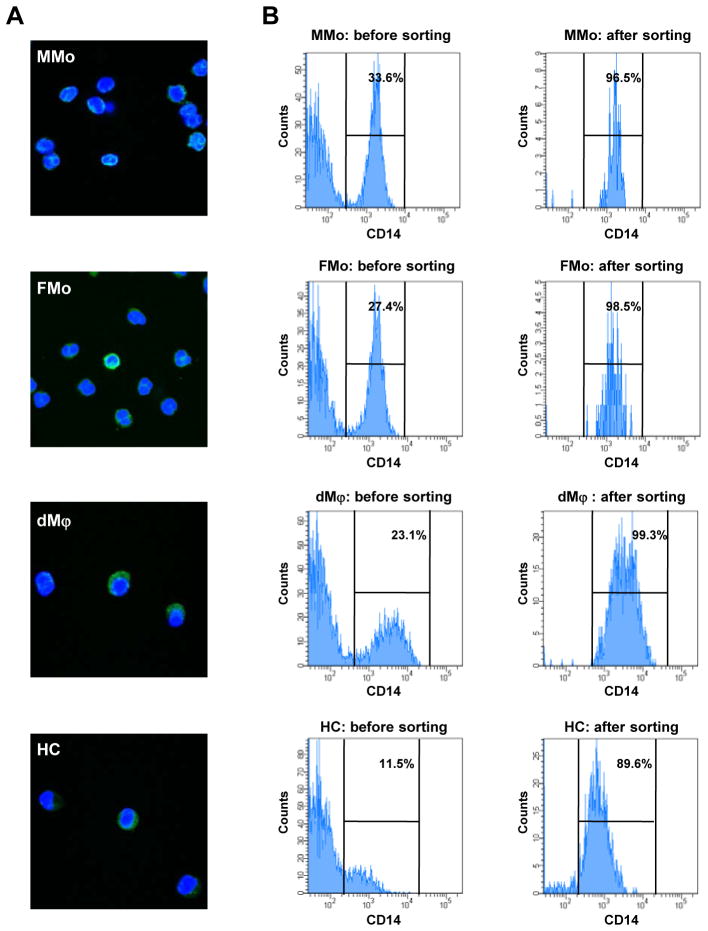
The purity of monocytes and macrophages isolated by cell sorter for microarray and pyrosequencing was higher than 60.0% in all samples analyzed (Figure 1). The median of purity was 99.4% (96.5–100%) for MMo, 98.8% (98.4–99.3%) for FMo, 93.7% (60.0–99.3%) for dMϕ, and 87.5% (69.4–92.3%) for HC, respectively. The median of purity for monocytes and macrophages isolated by MACS column for qPCR, immunoblot, and decidual cell culture was 90.6% (76.7–94.5%) for MMo, 79.1% (46.9–93.3%) for FMo, 70.8% (52.5–85.7%) for dMϕ, and 73.9% (51.1–93.9%) for HC, respectively.
Figure 1.
Identification of isolated monocytes and macrophages. (A) Immunoflourescent staining of CD14 (green) positive sorted maternal monocytes (MMo), Fetal monocytes (FMo), decidual macrophages (dMϕ) and Hofbauer cells (HC). The nuclei are stained with DAPI (blue). Sorted cells show mononuclear morphology. (B) The purity of isolated cells for methylation microarray by flow sorting was more than 80 % in each of MMo, FMo, dMϕ, and HC.
DNA methylation patterns: maternal vs. fetal monocytes and decidual macrophages vs. Hofbauer cells
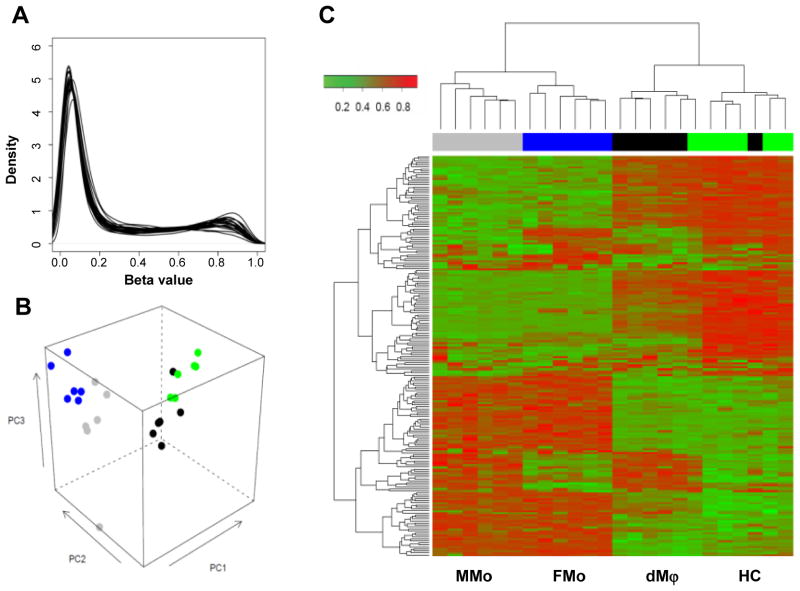
The majority of 27,551 annotated loci on the Illumina Infinium Methylation bead-array showed an absence of methylation (Figure 2A: the left peak near 0.0), with about 9% of all loci showing a methylation level β above 0.8 in each sample (Figure 2A: the right peak). An unsupervised analysis of methylation levels showed clustering of the samples according to the cell populations studied: MMo, FMo, dMϕ, and HC (Figure 2). The basic comparisons of the array data were done according to the host type (Maternal vs. Fetal) and the cell type (Monocytes vs. Macrophages) (Figure 3). The data set is available in www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=tlyrpmsaqwkqwpe&acc=GSE31680.
Figure 2.
Methylation microarray analysis. (A) The distribution of loci methylation β values is shown for each of the 24 samples. The peaks near β=0 illustrates that most of loci are un-methylated in each sample, while the peaks near β=1 show that there is a sizable portion of loci with high methylation β values in each sample. A principal component analysis (PCA) plot (B) and a heat map (C) generated with data from 6 pairs of maternal monocytes (MMo, gray), fetal monocytes (FMo, blue), decidual macrophages (dMϕ, black) and Hofbauer cells (HC, green) show a clear segregation among the cell groups. The PCA plot (B) uses the data from all loci, while the loci chosen for the heat map (C) are the top 200 ones varying the most across all 24 samples (unbiased filtering) as described elsewhere.126
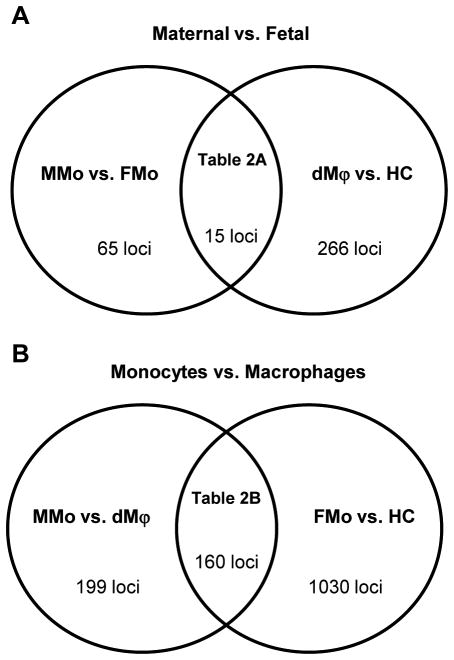
Figure 3.
Venn diagram summary of differentially methylated loci between the groups. Differentially methylated loci were using a FDR cut-off of 5% and average DNA methylation difference (Δβ)>0.2. (A) Division of maternal vs. fetal groups. 65 loci were differentially methylated between maternal monocytes (MMo) and fetal monocytes (FMo), and 266 loci were differentially methylated between decidual macrophages (dMϕ) and Hofbauer cells (HC). Fifteen loci were shared between each comparison. (B) Division of blood monocytes vs. tissue macrophages. One hundred ninety-nine loci were differentially methylated between MMo and dMϕ, and 1,030 loci were listed for differentially methylated genes between FMo and HC. One hundred sixty loci were overlapped between each comparison.
In the comparisons between the mother and the fetus, 65 loci were differentially methylated between MMo and FMo, whereas 266 loci were differentially methylated between dMϕ and HC. Among the 65 differentially methylated loci, 25 loci and 40 loci were hyper- and hypo-methylated in MMo compared to FMo. Interestingly, most of the immune response-related genes such as SP2, LAG3, PGLYRP2, GBP1, and CX3CL1 were hypermethylated in FMo. When the methylation patterns of dMϕ and HC were compared, 145 loci were hypermethylated in dMϕ and 121 loci were hypermethylated in HC. Many of the immune response-related genes such as EDG6, ADA, PGLYRP1, CST7, TRAF1, IL1B, PTGDR, LAG3, and CD79A were found to be hypermethylated in HC. Other differentially methylated loci included the genes related to cell cycle and apoptosis such as INCA1, CASP8, and AIM2 (Table I). Of note, INCA1 and LAG3 were also among those 15 loci whose differential methylation was found both in the comparisons between MMo and FMo and between dMϕ and HC (Table II).
Table I.
Top 30 differentially methylated loci in each comparison
| MMo vs. FMo | dMϕ vs. HC | ||||||
|---|---|---|---|---|---|---|---|
| Gene | q-value | Difference | Direction | Gene | q-value | Difference | Direction |
| CD59 | 2.24E-04 | 0.49 | FMo | LACTB | 4.98E-03 | 0.24 | dMϕ |
| FLJ10945 | 2.24E-04 | 0.29 | FMo | S100A2 | 4.98E-03 | 0.32 | HC |
| LTBR | 3.49E-04 | 0.29 | FMo | PSTPIP1 | 5.42E-03 | 0.25 | HC |
| PRKG2 | 6.95E-04 | 0.57 | FMo | OXCT2 | 5.42E-03 | 0.31 | HC |
| CETP | 6.95E-04 | 0.34 | FMo | CMTM5 | 5.42E-03 | 0.37 | dMϕ |
| ECEL1 | 7.25E-04 | 0.46 | MMo | LTBP3 | 6.66E-03 | 0.30 | HC |
| FLJ10945 | 7.25E-04 | 0.32 | FMo | SLCO4A1 | 6.66E-03 | 0.32 | HC |
| SYDE1 | 1.06E-03 | 0.24 | FMo | HOXB1 | 6.66E-03 | 0.40 | dMϕ |
| OLFML2A | 1.16E-03 | 0.44 | FMo | CDH10 | 6.66E-03 | 0.22 | dMϕ |
| ECEL1 | 1.20E-03 | 0.39 | MMo | CDH9 | 6.66E-03 | 0.21 | dMϕ |
| KLHDC7B | 1.82E-03 | 0.28 | MMo | ADAMTS13 | 6.66E-03 | 0.29 | dMϕ |
| C21orf84 | 1.82E-03 | 0.25 | FMo | KRTHB6 | 7.03E-03 | 0.32 | dMϕ |
| MPI | 3.40E-03 | 0.22 | FMo | IL22RA1 | 7.10E-03 | 0.32 | dMϕ |
| KCNAB3 | 3.40E-03 | 0.74 | MMo | ABCC13 | 7.92E-03 | 0.22 | dMϕ |
| LAG3 | 3.40E-03 | 0.38 | FMo | SMPD3 | 7.92E-03 | 0.22 | dMϕ |
| EDARADD | 5.16E-03 | 0.43 | FMo | TSPYL5 | 7.92E-03 | 0.29 | HC |
| MSRB2 | 5.16E-03 | 0.26 | FMo | TFAP2E | 7.92E-03 | 0.27 | dMϕ |
| POU3F1 | 5.43E-03 | 0.39 | MMo | VDAC1 | 7.92E-03 | 0.22 | dMϕ |
| AKR1C3 | 5.93E-03 | 0.33 | FMo | LRRC4 | 7.92E-03 | 0.27 | dMϕ |
| ZNF710 | 7.20E-03 | 0.29 | MMo | DUOX2 | 7.92E-03 | 0.30 | dMϕ |
| WT1 | 7.20E-03 | 0.27 | MMo | SPARCL1 | 7.92E-03 | 0.20 | dMϕ |
| RUNX2 | 7.54E-03 | 0.29 | FMo | TNFRSF9 | 7.92E-03 | 0.29 | HC |
| NAV1 | 7.54E-03 | 0.42 | MMo | FRMPD2 | 7.92E-03 | 0.36 | dMϕ |
| TFAP2E | 7.64E-03 | 0.22 | MMo | ZNF385 | 8.45E-03 | 0.29 | HC |
| SDS | 7.64E-03 | 0.32 | FMo | SUSD2 | 8.52E-03 | 0.25 | dMϕ |
| RRP22 | 7.78E-03 | 0.25 | MMo | INCA1 | 8.52E-03 | 0.31 | dMϕ |
| SNTB1 | 8.01E-03 | 0.26 | FMo | ACTN3 | 8.84E-03 | 0.30 | dMϕ |
| ACTN3 | 8.01E-03 | 0.30 | MMo | SLC4A3 | 9.55E-03 | 0.27 | dMϕ |
| CHRNE | 9.41E-03 | 0.33 | FMo | CPZ | 9.55E-03 | 0.42 | HC |
| GUCY1B2 | 1.00E-02 | 0.40 | FMo | C15orf2 | 1.03E-02 | 0.24 | dMϕ |
| MMo vs. dMϕ | FMo vs. HC | ||||||
|---|---|---|---|---|---|---|---|
| Gene | q-value | Difference | Direction | Gene | q-value | Difference | Direction |
| LOC283487 | 5.27E-09 | 0.56 | MMo | LOC283487 | 1.39E-09 | 0.61 | FMo |
| CCL14 | 3.96E-08 | 0.45 | MMo | CLDN15 | 4.41E-09 | 0.59 | HC |
| ROBO4 | 9.56E-08 | 0.45 | dMϕ | FLJ38159 | 4.41E-09 | 0.61 | FMo |
| LMO2 | 9.56E-08 | 0.55 | dMϕ | CCL14 | 4.64E-08 | 0.42 | FMo |
| ATF5 | 9.56E-08 | 0.66 | MMo | CDKN2B | 1.11E-07 | 0.68 | HC |
| FLJ38159 | 1.27E-07 | 0.47 | MMo | GIMAP5 | 1.17E-07 | 0.50 | FMo |
| NUMA1 | 4.16E-07 | 0.56 | MMo | BIRC4BP | 1.31E-07 | 0.45 | FMo |
| ICAM3 | 4.16E-07 | 0.48 | dMϕ | ICAM3 | 1.43E-07 | 0.51 | HC |
| CCDC57 | 4.16E-07 | 0.64 | MMo | CDKN2B | 1.43E-07 | 0.63 | HC |
| A2M | 4.16E-07 | 0.34 | MMo | CTNND1 | 1.56E-07 | 0.42 | FMo |
| CLDN15 | 5.33E-07 | 0.40 | dMϕ | ATF5 | 2.50E-07 | 0.59 | FMo |
| KCNE1 | 7.14E-07 | 0.35 | dMϕ | TLR9 | 2.55E-07 | 0.65 | HC |
| GPR21 | 7.14E-07 | 0.51 | dMϕ | HTR2B | 2.70E-07 | 0.38 | FMo |
| ABI3 | 7.30E-07 | 0.56 | MMo | KLHDC7B | 2.87E-07 | 0.50 | HC |
| TLR9 | 7.38E-07 | 0.60 | dMϕ | GPR21 | 2.87E-07 | 0.53 | HC |
| HTR2B | 1.12E-06 | 0.35 | MMo | LTBR | 2.87E-07 | 0.43 | FMo |
| OGFR | 1.12E-06 | 0.40 | dMϕ | IL1B | 2.87E-07 | 0.56 | HC |
| BIRC4BP | 1.25E-06 | 0.36 | MMo | C6orf188 | 3.18E-07 | 0.29 | FMo |
| HYAL2 | 3.09E-06 | 0.27 | MMo | SORBS3 | 3.38E-07 | 0.40 | FMo |
| PTPRCAP | 3.09E-06 | 0.49 | dMϕ | LMO2 | 3.75E-07 | 0.45 | HC |
| NFE2 | 3.09E-06 | 0.43 | dMϕ | SLC39A2 | 3.75E-07 | 0.30 | FMo |
| CDKN2B | 3.45E-06 | 0.49 | dMϕ | ITGB2 | 3.79E-07 | 0.36 | HC |
| ABI3 | 5.81E-06 | 0.52 | MMo | NUMA1 | 4.52E-07 | 0.52 | FMo |
| SORBS3 | 6.84E-06 | 0.32 | MMo | CTSZ | 4.52E-07 | 0.54 | FMo |
| ARL4 | 7.33E-06 | 0.27 | MMo | CD6 | 4.61E-07 | 0.83 | HC |
| XLKD1 | 7.33E-06 | 0.31 | MMo | CSF3R | 5.76E-07 | 0.52 | HC |
| GIMAP5 | 9.26E-06 | 0.34 | MMo | CD6 | 6.49E-07 | 0.74 | HC |
| RNASE1 | 9.38E-06 | 0.51 | MMo | KCNE1 | 8.18E-07 | 0.33 | HC |
| CTNND1 | 1.20E-05 | 0.29 | MMo | CBFA2T3 | 8.25E-07 | 0.55 | HC |
| GPR92 | 1.32E-05 | 0.47 | MMo | RNF36 | 1.29E-06 | 0.34 | FMo |
Difference represents Δβ. q-values represent the False Discovery Rate adjusted p-values
Table II.
Differentially methylated loci between maternal and fetal cells and between monocytes and macrophages
| (A) Maternal vs. Fetal | (B) Monocyte (Mo) vs. Macrophage (Mϕ) | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Gene | Direction | Gene | Direction | Gene | Direction | Gene | Direction |
| ACTN3 | Maternal | A2M | Mo | IL32 | Mo | FAM83F | Mϕ |
| DUOX2 | Maternal | ABI3 | Mo | KRT23 | Mo | FCGR3B | Mϕ |
| INCA1 | Maternal | ABI3 | Mo | LILRB5 | Mo | FGR | Mϕ |
| PLEK2 | Maternal | ANKRD9 | Mo | LOC283487 | Mo | FLJ37396 | Mϕ |
| POU3F1 | Maternal | ARL4 | Mo | LOC92689 | Mo | FXYD1 | Mϕ |
| TFAP2E | Maternal | ATF5 | Mo | MEFV | Mo | GNLY | Mϕ |
| TMEM102 | Maternal | BIRC4BP | Mo | MGC4618 | Mo | GPR21 | Mϕ |
| WT1 | Maternal | BLNK | Mo | NUMA1 | Mo | GPR21 | Mϕ |
| ACOT11 | Fetal | C1QA | Mo | NYX | Mo | GSG1 | Mϕ |
| AKR1C3 | Fetal | C1QA | Mo | PIB5PA | Mo | GUCY1B2 | Mϕ |
| CHRNE | Fetal | C1QC | Mo | PIP | Mo | HP | Mϕ |
| EDARADD | Fetal | C1QC | Mo | PLEKHG5 | Mo | ICAM2 | Mϕ |
| FLJ46365 | Fetal | C1S | Mo | PVALB | Mo | ICAM3 | Mϕ |
| LAG3 | Fetal | C20orf55 | Mo | RNASE1 | Mo | IL12RB2 | Mϕ |
| PRKG2 | Fetal | C3 | Mo | RNASE1 | Mo | IL1B | Mϕ |
| C6orf188 | Mo | RNF36 | Mo | KCNE1 | Mϕ | ||
| CCDC57 | Mo | RUFY3 | Mo | LAIR2 | Mϕ | ||
| CCL13 | Mo | SDC4 | Mo | LCK | Mϕ | ||
| CCL14 | Mo | SLC7A7 | Mo | LMO2 | Mϕ | ||
| CCL5 | Mo | SN | Mo | LMO2 | Mϕ | ||
| CD163 | Mo | SNX8 | Mo | LOC144501 | Mϕ | ||
| CD226 | Mo | SORBS3 | Mo | LOC339789 | Mϕ | ||
| CD300LG | Mo | TADA3L | Mo | LTBP3 | Mϕ | ||
| CD4 | Mo | TBC1D14 | Mo | MFAP4 | Mϕ | ||
| CEBPG | Mo | TIMD4 | Mo | MGC23244 | Mϕ | ||
| CFLAR | Mo | TNFSF12-TNFSF13 | Mo | MKNK2 | Mϕ | ||
| CMKLR1 | Mo | WDR58 | Mo | MPHOSPH9 | Mϕ | ||
| CTNNAL1 | Mo | XLKD1 | Mo | NFE2 | Mϕ | ||
| CTNND1 | Mo | ASGR2 | Mϕ | NKG7 | Mϕ | ||
| CTSK | Mo | AZU1 | Mϕ | NR1I2 | Mϕ | ||
| CTSZ | Mo | BRE | Mϕ | OGFR | Mϕ | ||
| DCHS1 | Mo | C10orf91 | Mϕ | P2RX1 | Mϕ | ||
| DIO1 | Mo | C12orf25 | Mϕ | P2RY2 | Mϕ | ||
| EIF2C4 | Mo | CBFA2T3 | Mϕ | PLAC8 | Mϕ | ||
| ENG | Mo | CCIN | Mϕ | PPP2R4 | Mϕ | ||
| FCGR2A | Mo | CD48 | Mϕ | PRR5 | Mϕ | ||
| FCGR3A | Mo | CD5 | Mϕ | PRTN3 | Mϕ | ||
| FGD4 | Mo | CD6 | Mϕ | PTPN7 | Mϕ | ||
| FLJ20581 | Mo | CD6 | Mϕ | PTPRCAP | Mϕ | ||
| FLJ38159 | Mo | CDKN2B | Mϕ | PTPRCAP | Mϕ | ||
| FOLR2 | Mo | CDKN2B | Mϕ | ROBO4 | Mϕ | ||
| GABARAP | Mo | CDKN2B | Mϕ | RUNX3 | Mϕ | ||
| GAS2L1 | Mo | CDKN2B | Mϕ | S100A5 | Mϕ | ||
| GIMAP5 | Mo | CDKN2B | Mϕ | SCNN1A | Mϕ | ||
| GNA13 | Mo | CDKN2B | Mϕ | SERPINF1 | Mϕ | ||
| GNPDA1 | Mo | CLDN15 | Mϕ | SLC45A1 | Mϕ | ||
| GPNMB | Mo | CLEC3B | Mϕ | SPN | Mϕ | ||
| GPR92 | Mo | CRHBP | Mϕ | TBC1D10C | Mϕ | ||
| GSN | Mo | CRHBP | Mϕ | TBC1D10C | Mϕ | ||
| HAMP | Mo | CSF3R | Mϕ | TLR9 | Mϕ | ||
| HMOX1 | Mo | CST7 | Mϕ | TMC6 | Mϕ | ||
| HNMT | Mo | F12 | Mϕ | TSPAN32 | Mϕ | ||
| HTR2B | Mo | F2RL2 | Mϕ | VAV1 | Mϕ | ||
| HYAL2 | Mo | ||||||
Bold: loci chosen for follow-up study by pyrosequencing.
DNA methylation patterns: maternal monocytes vs. decidual macrophages and fetal monocytes vs. Hofbauer cells
In the comparisons between blood monocytes and tissue macrophages, 199 loci were differentially methylated between MMo and dMϕ, and 1,030 loci were differentially methylated between FMo and HC (Table I). Of note, the majority [160/199 (80%)] of the differentially methylated loci between MMo and dMϕ were also differentially methylated between FMo and HC (Table II).
Gene Ontology analysis of differentially methylated genes revealed enrichment of 215 biological processes for MMo vs. dMϕ comparison and 52 processes for FMo vs. HC (Table III). Most of the biological processes enriched in comparison between FMo vs. HC (51/52) were also enriched in comparison between MMo and dMϕ, and they were related to the variable aspects of immune responses, complement activation, cell adhesion, and angiogenesis. The biological processes enriched in the comparison between MMo and dMϕ included ion homeostasis, coagulation, and aminoglycan metabolic processes.
Table III.
Top 20 biological processes enriched in monocyte vs. macrophage comparison
| Biologic Process | # Enriched Genes/# Genes in GO | q-value | |
|---|---|---|---|
| MMo vs. dMϕ | defense response | 34/477 | 3.80E-14 |
| immune response | 32/522 | 1.46E-11 | |
| immune system process | 35/750 | 1.48E-09 | |
| response to wounding | 25/389 | 2.92E-09 | |
| inflammatory response | 21/268 | 3.25E-09 | |
| acute inflammatory response | 12/65 | 4.05E-09 | |
| response to external stimulus | 29/615 | 5.44E-08 | |
| response to stimulus | 56/2024 | 5.93E-08 | |
| response to stress | 40/1142 | 7.34E-08 | |
| regulation of response to stimulus | 16/215 | 1.16E-06 | |
| positive regulation of immune system process | 12/130 | 7.32E-06 | |
| regulation of immune response | 12/130 | 7.32E-06 | |
| extracellular region | 43/1425 | 1.26E-05 | |
| leukocyte mediated immunity | 10/87 | 1.10E-05 | |
| cell activation | 15/233 | 1.70E-05 | |
| regulation of leukocyte mediated immunity | 7/36 | 2.08E-05 | |
| positive regulation of response to stimulus | 11/121 | 2.32E-05 | |
| lymphocyte mediated immunity | 9/78 | 3.61E-05 | |
| activation of immune response | 8/58 | 3.75E-05 | |
| immune effector process | 11/130 | 4.05E-05 | |
|
| |||
| FMo vs. HC | immune response | 76/522 | 8.10E-09 |
| defense response | 71/477 | 8.10E-09 | |
| immune system process | 94/750 | 4.16E-08 | |
| response to wounding | 58/389 | 3.35E-07 | |
| response to external stimulus | 79/615 | 3.35E-07 | |
| inflammatory response | 45/268 | 5.33E-07 | |
| acute inflammatory response | 18/65 | 2.29E-05 | |
| response to stimulus | 179/2024 | 1.09E-04 | |
| activation of plasma proteins during acute inflammatory response | 12/33 | 1.09E-04 | |
| humoral immune response | 16/61 | 1.95E-04 | |
| complement activation | 11/32 | 5.26E-04 | |
| innate immune response | 22/118 | 8.34E-04 | |
| cell adhesion | 64/572 | 8.36E-04 | |
| biological adhesion | 64/572 | 8.36E-04 | |
| activation of immune response | 14/58 | 1.83E-03 | |
| complement activation, classical pathway | 9/25 | 2.00E-03 | |
| positive regulation of immune response | 17/83 | 2.03E-03 | |
| immune effector process | 22/130 | 2.75E-03 | |
| positive regulation of response to stimulus | 21/121 | 2.75E-03 | |
| leukocyte mediated immunity | 17/87 | 3.14E-03 | |
In the comparison between MMo vs. dMϕ, the genes encoding markers of classical macrophage activation such as TLR9, IL1B, IL12RB2, CD48, and FGR were hypermethylated in dMϕ, while those of alternative macrophage activation markers such as CCL13, CCL14, A2M, HNMT, and IL10 were hypomethylated. The results suggested that the anti-inflammatory property of human dMϕ is partly associated with its DNA methylation pattern. The comparison of FMo vs. HC also demonstrated hypermethylation of the genes encoding markers of classical macrophage activation such as TLR9, IL1B, IL12RB2, CD48, and FGR in HC, whereas the genes encoding alternative macrophage activation markers such as CCL2, CCL13, CCL14, CD209, and A2M were hypomethylated in HC.
Of note, two key enzymes, DNMT1 and DNMT3B, involved in DNA methylation were also among the differentially methylated loci between FMo and HC.
Confirmation of CpG methylation of specific genes: LAG3, INCA1, IL1B
Three genes (LAG3, INCA1, IL1B) were selected for further validation of the methylation status in each type of cells. LAG3 and INCA1 were differentially methylated between maternal and fetal cells (MMo vs. FMo and dMϕ vs HC; Table IIA), and IL1B was found to be differentially methylated between the blood monocyte and the tissue macrophage both in the mother and the fetus (Table IIB). Three CpG sites of each gene for bisulfite pyrosequencing were chosen from the region interrogated by methylation microarray, and the sequencing results of the three genes correlated with the microarray data. As shown in Figure 4A, the methylation percentages of the LAG gene in MMo and dMϕ were lower than those in FMo and HC, respectively (p<0.05 for each). The median of methylation percentages was 59.5% for MMo, 82.8% for FMo, 60.2% for dMϕ, and 82.0% for HC. The degree of methylation across three CpG sites of the INCA1 promoter region was also confirmed to be higher in MMo (61.3%) than in FMo (45.3%), and in dMϕ (35.1%) than in HC (8.7%), respectively (p<0.05 for each, Figure 4B). For IL1B, which was found to be differentially methylated between monocyte and macrophage in the microarray analysis, the median of methylation percentages was 31.1% for MMo, 51.9 % for dMϕ, 30.4% for FMo, and 69.4% for HC, also confirming the microarray data (Figure 4C).
Figure 4.
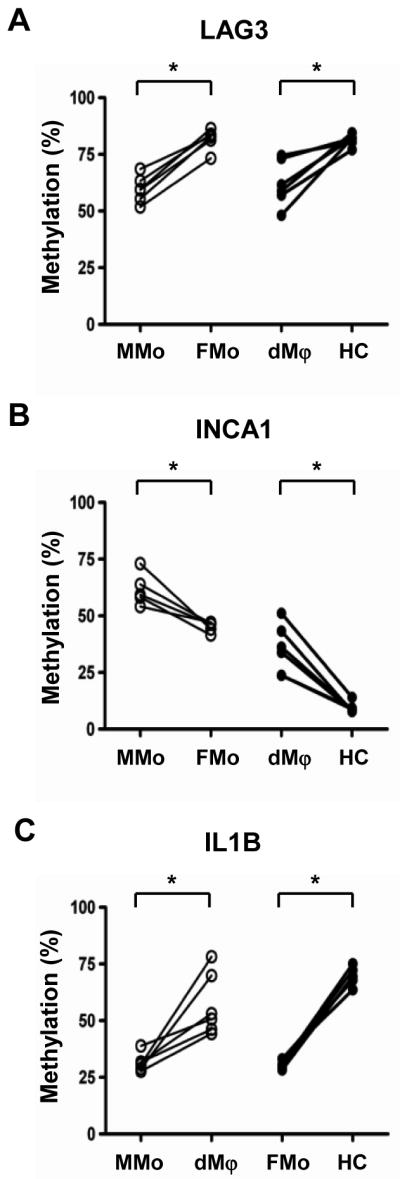
Bisulfite pyrosequencing results of (A) LAG3, (B) INCA1 and (C) IL-1β in 6 samples of genomic DNAs from maternal monocytes (MMo), fetal monocytes (FMo), decidual macrophages (dMϕ) and Hofbauer cells (HC). Bisulfite sequencing was performed across 3 CpG sites of each gene chosen from the region interrogated by methylation microarray, and the average percentages of methylation are well-correlated with microarray data. *p<0.05.
DNMT expression in blood monocytes, decidua, and chorionic villi
As DNMT1 and DNMT3B were among the differentially methylated loci between FMo and HC in the microarray analysis and as they are major methyltransferases, we evaluated mRNA expression levels of DNA methyltransferases (DNMT), DNMT1, DNMT3A, and DNMT3B by quantitative RT-PCR using RNA samples from MMo, FMo, dMϕ, and HC. mRNA expressions of DNMT1, DNMT3A, and DNMT3B were significantly lower in dMϕ when compared to MMo and HC. DNMT1 mRNA expression in MMo was higher than that of FMo (p<0.05 for each, Figure 5). The data suggested that the differences in the methylation patterns of the cells tested are partly associated with the expression of DNMTs.
Figure 5.
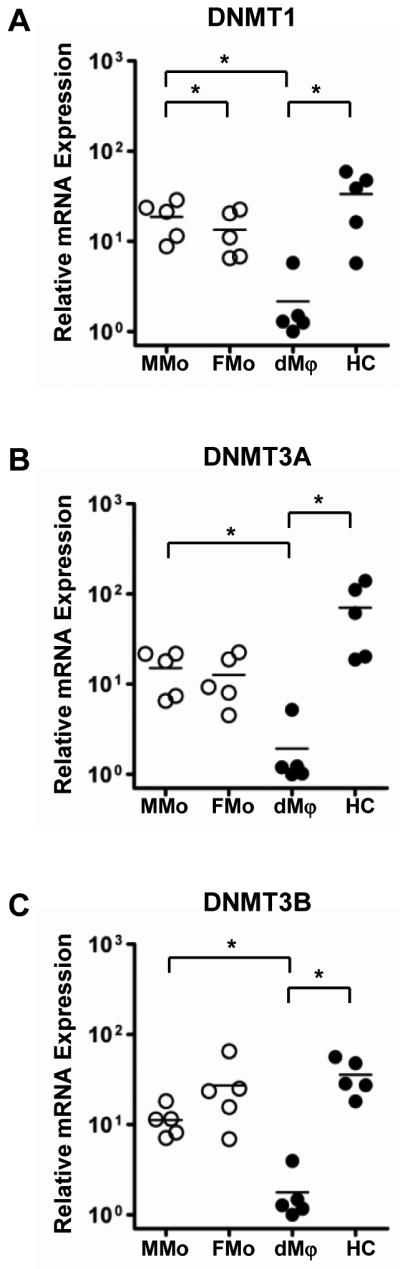
mRNA expressions of DNA methyltransferases by qRT-PCR show distinguished patterns in maternal monocytes (MMo), fetal monocytes (FMo), decidual macrophages (dMϕ), and Hofbauer cells (HC). The DNMTs mRNA expressions were normalized on the content of RPLPO for each. *p<0.05, n=5.
Increased expression of INCA1 mRNAby 5-azacytidine
Based on the microarray and pyrosequencing data, we further studied the effects of 5-azacytidine induced demethylation on the mRNA expression of INCA1 (inhibitor of CDK interacting with cyclin A1). INCA1 was hypermethylated in MMo or dMϕ and hypomethylated in FMo or HC. Quantitative RT-PCR and immunoblotting were used to confirm expression levels of INCA1 in monocytes and macrophages. INCA1 mRNA expression was significantly down-regulated in dMϕ compared to HC (p<0.05); however, there was no significant difference between MMo and FMo (Figure 6A). This seemed to be due to a relatively small difference in the extent of DNA methylation between MMo and FMo. INCA1 protein expression was also significantly lower in dMϕ than in HC as well as in MMo than FMo (Figure 6B).
Figure 6.
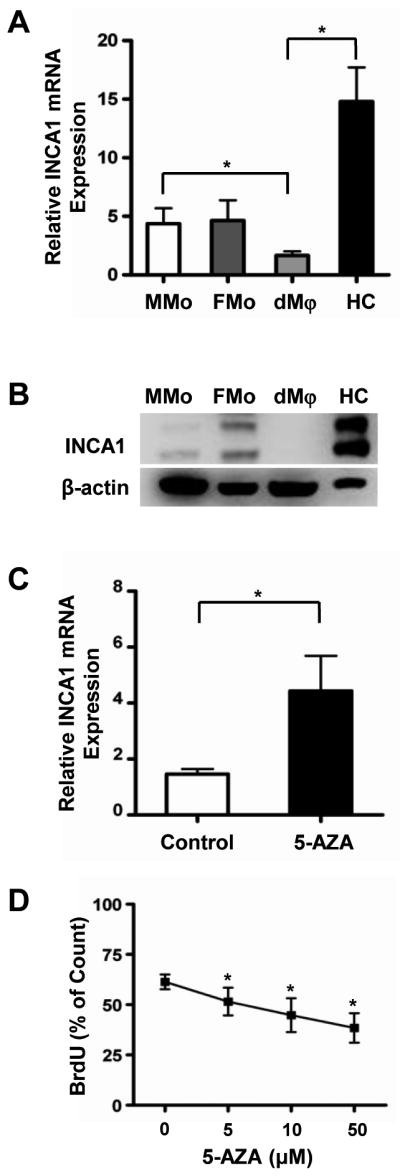
Differential INCA1 expression in monocytes or macrophages by function of DNA methylation. (A) qRT-PCR of INCA1 transcripts of maternal monocytes (MMo), fetal monocytes (FMo), macrophages from decidua (dMϕ), and Hofbauer cells (HC). mRNA expression of INCA1 is significantly down-regulated in dMϕ than in HC. The INCA1 mRNA expressions were normalized on the content of RPLPO. *p<0.05, n=5. (B) Western blot of INCA1 of total proteins from columned MMo, FMo, dMϕ, and HC shows that protein expression of INCA1 is higher in HC than in dMϕ, n=4. (C) qRT-PCR of INCA1 mRNA expression following treatment of 10 μM of 5-Azacytidine for 3 days to dMϕ. mRNA expression of INCA1 was increased in dMϕ exposed to 5-Azacytidine. The INCA1 mRNA expressions were normalized on the content of RPLPO. *p<0.05, n=5. (D) BrdU pulse labeling detection in 5-Azacytidine treated dMϕ. After treatment of 5-Azacytidine (0, 5, 10, or 50 μM), macrophages were pulsed with BrdU (10 μM) for 1 h, and stained with anti BrdU antibody. The percentages of BrdU positive cells were measured by FACS. BrdU labeling decreased with 5-azacytidine treatment in a dose-dependent manner. *p< 0.05 vs. control (0 μM), n=4.
The treatment of isolated dMϕ with 5-azacytidine (10 nM) for 3 days significantly increased INCA1 mRNA expression by 3.1 fold (p<0.05, Figure 6C). Because INCA1 is an inhibitor of cell cycle progression,89, 90 we further studied the effect of the INCA1 methylation on the proliferation of macrophages by BrdU labeling. Following the treatment with 5-azacytidine for 3 days, the dMϕ were pulsed with BrdU. Flow cytometric evaluation demonstrated decreased BrdU labeling in dMϕ with 5-azacytidine treatment in a dose-dependent manner (p<0.05, Figure 6D).
Discussion
Monocytes and macrophages are important in the immunological interaction between the mother and the fetus.91, 92 We investigated the methylome of blood monocytes and tissue macrophages at the feto-maternal interface. The principal findings of this study are: 1) There are distinct differences in the DNA methylation patterns between maternal cells and fetal cells (MMo vs. FMo; dMϕ vs. HC), and the majority of differentially methylated genes related to immune response are hypermethylated in the fetal cells; 2) The methylation patterns of monocytes and macrophages are also quite different (MMo vs. dMϕ; FMo vs. HC), and the differential methylation pattern is more prominent between FMo and HC; 3) Gene Ontology analysis of differentially methylated genes demonstrates significant enrichment of many biological processes associated with immune responses in monocyte vs. macrophage comparisons; 4) Pyrosequencing confirmed microarray results of three genes tested (LAG3, INCA1, IL1B); 5) mRNA expression of DNMT1, DNMT3A, and DNMT3B was significantly lower in dMϕ compared to HC; and 6) Treatment of dMϕ with 5-azacytidine significantly increased mRNA expression of INCA1 with a concomitant decrease in BrdU labeling.
DNA hypermethylation in fetal monocytes relevant to their impaired immune responses
Hypermethylation of the genes related to immune response in FMo compared to MMo indicates that DNA methylation contributes to decreased immune response capacity of FMo compared to MMo. The results are quite consistent with the gene expression data in a recent transcriptome study of adult peripheral monocytes and umbilical cord blood monocytes. Jiang et al have shown differential increases in gene expression between adult monocytes and cord blood monocytes activated by lipopolysaccharides including cytokines, chemokines, transcription factors, signal transduction, apoptotic regulation, and cell structure.93 A previous study on relatively impaired responses of neonatal monocytes and macrophages to multiple Toll-like receptor ligands also supports immature functional capacity of neonatal monocytes compared to that of adult monocytes.94 An additional possible explanation for differential methylation between FMo and MMo may be age-related DNA methylation changes.95 The CD34+ hematopoietic progenitor cells from adult peripheral blood showed differential methylation patterns compared to those from cord blood80 and age-related DNA methylation changes are tissue-specific.95
DNA methylation during monocyte-macrophage differentiation
The present study clearly demonstrates distinct changes of DNA methylation during monocyte-macrophage differentiation. The differences are more profound between FMo and HC. The dMϕ and HC are deployed at the immunologically sensitive location: the feto-maternal interface where an enhanced pro-inflammatory response can be detrimental to the maintenance of pregnancy. Therefore, hypermethylation of certain genes encoding classical activation markers and hypomethylation of a subset of gene-encoding alternative activation markers of macrophages in dMϕ compared to MMo and in HC compared to FMo are biologically quite relevant. DNA methylation seems to contribute to the anti-inflammatory phenotype of dMϕ and HC. Gustafsson et al compared the transcriptome of CD14+ macrophages isolated from first trimester decidual tissue and peripheral blood monocytes. Their study also has shown that the expression of the genes encoding alternative macrophage activation markers such as CCL18 and CD209 is higher in dMϕ. Therefore, dMϕ displays an anti-inflammatory phenotype compared to maternal blood monocytes.96 Of note, their observation of increased expression of alpha-2-macroglobulin (A2M) in dMϕ is consistent with the hypomethylation of this gene in this study, whereas decreased expressions of asialoglycoprotein receptor 2 (ASGR2) and intercellular adhesion molecule 3 (ICAM3) are relevant with the hypermethylation of these genes in dMϕ. In addition to the differential methylation of CpG sites, the M2-macrophage phenotype is epigenetically regulated by chromatin remodeling following reciprocal changes in histone H3 lysine-4 and histone H3 lysine-27 methylation.97 Our data is also consistent with recent observations on the expression of selected molecules associated with macrophage polarization in HC.98, 99 Studies have shown that M2 markers such as DC-SIGN, CD163, and mannose receptor/CD206 are detectable in HC but not in M1 markers (CX3CR1, IL7R, CCR7). Expression of different combinations of M2 markers in dMϕ and HC which do not precisely match the transcriptional profile of in vitro generated M2 macrophages also suggests the potential importance of the tissue microenvironment during monocyte to macrophage differentiation.100, 101
A more profound difference in the DNA methylome between FMo and HC than between MMo and dMϕ has biological implications. The origin of placental HC has been controversial. It has been proposed that they derive from chorionic mesenchymal cells in the villous stroma before the formation of villous capillaries,50, 51 while mobilization of blood monocytes becomes the main route after the development of capillaries102 Therefore, HC might represent a heterogeneous population of cells in the context of histogenesis. Our observation of transdifferentiation of myofibroblasts into macrophages in the chorionic mesoderm of the reflected chorion also supports that HC can differentiate from chorionic mesenchymal cells.103 Furthermore, HC can normally proliferate, or in response to biological stimuli such as inflammation64, 104 and pro-inflammatory cytokines such as IL-6, would confer more chances of DNA methylation by changing gene expression patterns and increasing expression of methyltransferase.105 Hypomethylation of INCA1 in HC compared to dMϕ may also in turn contribute to the difference in the proliferative potential of these fetal and maternal cells.
Differential regulation of DNA methyltransferase expression
This study also reports an interesting feature of the regulation of DNA methyl transferase expression. DNMT1 and DNMT3B were among differentially methylated loci between FMo and HC, and their mRNA expressions in dMϕ were lower than those in MMo and HC, which in turn could modify overall methylation patterns of the cells studied. DNMTs have two types of actions. DNMT1 is responsible for the maintenance of DNA methylation status, while DNMT3A and DNMT3B are important for de novo methylation.72, 73 DNMT1 is expressed ubiquitously whereas DNMT3A and DNMT3B have tissue-specific expression patterns.106 Promoter methylation of DNA methyltransferases themselves may have a broader impact on methylation patterns, and hypomethylation of DNMT3B was demonstrated in glial tumors.107 We also demonstrated that differential DNA methylation patterns have biological significance by showing the effects of INCA1 in dMϕ.
Limitations of this study
There are limitations to this study. First, the samples used for the analysis in the present study were obtained from normal pregnant women at term, and the data related to pathologic pregnancies are not available. Intra-amniotic infection by microbial invasion of the amniotic cavity is a leading cause of preterm birth108–111 and elicits a robust acute inflammatory response of the fetus.2, 60, 61, 112–120 Furthermore, the process is associated with an elevation of the amniotic fluid concentrations of cytokines and chemokines such as macrophage migration inhibitory factor and monocyte chemotactic protein-1.2, 121–123 The fetal chorionic mesenchymal cells also acquire macrophage phenotype in the presence of intra-amniotic infection and chorioamnionitis.103 Therefore, the biological significance of DNA methylation of fetal and maternal monocytes and macrophages during intra-amniotic infection needs to be further studied. The data also might not represent the methylome of dMϕ or HC in the early gestational period. It is well-known that the number of HC decreases with advancing gestation.124 Considering the duration of gestation, it is very likely that methylation patterns of macrophages change across gestation. Houser et al have recently demonstrated that there are two different populations of CD14+ macrophages according to the expression level of CD11c (high vs. low) in the first trimester decidual tissues with different functional implications in lipid metabolism, inflammation, and tissue growth.125 Therefore, it is possible that different subpopulations of macrophages were analyzed together in our analysis.
Conclusions
We report the DNA methylome of maternal and fetal monocytes and macrophages at the feto-maternal interface for the first time. The distinct nature of DNA methylation patterns in each subset of cells strongly suggests that epigenetic regulation of multiple genes is at the core of functional diversification of the critical players in maternal and fetal immune interaction during pregnancy. Particularly, DNA methylation seems to be a part of the machinery conferring an anti-inflammatory phenotype to the macrophages at the feto-maternal interface.
Acknowledgments
This work was supported in part by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest.
References
- 1.Satosar A, Ramirez NC, Bartholomew D, Davis J, Nuovo GJ. Histologic correlates of viral and bacterial infection of the placenta associated with severe morbidity and mortality in the newborn. Hum Pathol. 2004;35:536–545. doi: 10.1016/j.humpath.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179:194–202. doi: 10.1016/s0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 3.Qiu X, Zhu L, Pollard JW. Colony-stimulating factor-1-dependent macrophage functions regulate the maternal decidua immune responses against Listeria monocytogenes infections during early gestation in mice. Infect Immun. 2009;77:85–97. doi: 10.1128/IAI.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langrehr JM, White DA, Hoffman RA, Simmons RL. Macrophages produce nitric oxide at allograft sites. Ann Surg. 1993;218:159–166. doi: 10.1097/00000658-199308000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitchens WH, Chase CM, Uehara S, Cornell LD, Colvin RB, Russell PS, Madsen JC. Macrophage depletion suppresses cardiac allograft vasculopathy in mice. Am J Transplant. 2007;7:2675–2682. doi: 10.1111/j.1600-6143.2007.01997.x. [DOI] [PubMed] [Google Scholar]
- 6.Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates: revention of allogeneic fetal rejection by tryptophan catabolism. Symp Soc Exp Biol. 1953;7:320–338. [Google Scholar]
- 7.Blois SM, Ilarregui JM, Tometten M, Garcia M, Orsal AS, Cordo-Russo R, Toscano MA, Bianco GA, Kobelt P, Handjiski B, Tirado I, Markert UR, Klapp BF, Poirier F, Szekeres-Bartho J, Rabinovich GA, Arck PC. A pivotal role for galectin-1 in fetomaternal tolerance. Nat Med. 2007;13:1450–1457. doi: 10.1038/nm1680. [DOI] [PubMed] [Google Scholar]
- 8.Guleria I, Sayegh MH. Maternal acceptance of the fetus: true human tolerance. J Immunol. 2007;178:3345–3351. doi: 10.4049/jimmunol.178.6.3345. [DOI] [PubMed] [Google Scholar]
- 9.Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, Geraghty DE. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J Immunol. 2003;171:1376–1384. doi: 10.4049/jimmunol.171.3.1376. [DOI] [PubMed] [Google Scholar]
- 10.King A, Burrows TD, Hiby SE, Bowen JM, Joseph S, Verma S, Lim PB, Gardner L, Le Bouteiller P, Ziegler A, Uchanska-Ziegler B, Loke YW. Surface expression of HLA-C antigen by human extravillous trophoblast. Placenta. 2000;21:376–387. doi: 10.1053/plac.1999.0496. [DOI] [PubMed] [Google Scholar]
- 11.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 12.Terness P, Kallikourdis M, Betz AG, Rabinovich GA, Saito S, Clark DA. Tolerance signaling molecules and pregnancy: IDO, galectins, and the renaissance of regulatory T cells. Am J Reprod Immunol. 2007;58:238–254. doi: 10.1111/j.1600-0897.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 13.Mold JE, Michaelsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, Lee TH, Nixon DF, McCune JM. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraccaroli L, Alfieri J, Larocca L, Calafat M, Mor G, Leiros CP, Ramhorst R. A potential tolerogenic immune mechanism in a trophoblast cell line through the activation of chemokine-induced T cell death and regulatory T cell modulation. Hum Reprod. 2009;24:166–175. doi: 10.1093/humrep/den344. [DOI] [PubMed] [Google Scholar]
- 15.Ramhorst R, Fraccaroli L, Aldo P, Alvero AB, Cardenas I, Leiros CP, Mor G. Modulation and Recruitment of Inducible Regulatory T Cells by First Trimester Trophoblast Cells. Am J Reprod Immunol. 2011;67:17–27. doi: 10.1111/j.1600-0897.2011.01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bili H, Fleva A, Pados G, Argyriou T, Tsolakidis D, Pavlitou A, Tarlatzis BC. Regulatory Tau-cell differentiation between maternal and cord blood samples in pregnancies with spontaneous vaginal delivery and with elective cesarian section. Am J Reprod Immunol. 2011;65:173–179. doi: 10.1111/j.1600-0897.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- 17.Ernerudh J, Berg G, Mjosberg J. Regulatory T helper cells in pregnancy and their roles in systemic versus local immune tolerance. Am J Reprod Immunol. 2011;66 (Suppl 1):31–43. doi: 10.1111/j.1600-0897.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- 18.Leber A, Teles A, Zenclussen AC. Regulatory T cells and their role in pregnancy. Am J Reprod Immunol. 2010;63:445–459. doi: 10.1111/j.1600-0897.2010.00821.x. [DOI] [PubMed] [Google Scholar]
- 19.Nevers T, Kalkunte S, Sharma S. Uterine Regulatory T cells, IL-10 and hypertension. Am J Reprod Immunol. 2011;66 (Suppl 1):88–92. doi: 10.1111/j.1600-0897.2011.01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito S, Nakashima A, Shima T, Ito M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am J Reprod Immunol. 2010;63:601–610. doi: 10.1111/j.1600-0897.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 21.Zenclussen ML, Thuere C, Ahmad N, Wafula PO, Fest S, Teles A, Leber A, Casalis PA, Bechmann I, Priller J, Volk HD, Zenclussen AC. The persistence of paternal antigens in the maternal body is involved in regulatory T-cell expansion and fetal-maternal tolerance in murine pregnancy. Am J Reprod Immunol. 2010;63:200–208. doi: 10.1111/j.1600-0897.2009.00793.x. [DOI] [PubMed] [Google Scholar]
- 22.Huppertz B. The feto-maternal interface: setting the stage for potential immune interactions. Semin Immunopathol. 2007;29:83–94. doi: 10.1007/s00281-007-0070-7. [DOI] [PubMed] [Google Scholar]
- 23.Damsky CH, Fitzgerald ML, Fisher SJ. Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J Clin Invest. 1992;89:210–222. doi: 10.1172/JCI115565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas GW, Thomas L, Carr M, Cullen NM, Morris R. Trophoblast in the circulating blood during pregnancy. Am J Obstet Gynecol. 1959;78:960–973. doi: 10.1016/s0002-9378(16)36649-2. [DOI] [PubMed] [Google Scholar]
- 25.Johansen M, Redman CW, Wilkins T, Sargent IL. Trophoblast deportation in human pregnancy--its relevance for pre-eclampsia. Placenta. 1999;20:531–539. doi: 10.1053/plac.1999.0422. [DOI] [PubMed] [Google Scholar]
- 26.Sargent IL, Germain SJ, Sacks GP, Kumar S, Redman CW. Trophoblast deportation and the maternal inflammatory response in pre-eclampsia. J Reprod Immunol. 2003;59:153–160. doi: 10.1016/s0165-0378(03)00044-5. [DOI] [PubMed] [Google Scholar]
- 27.Mor G, Straszewski-Chavez SL, Abrahams VM. Macrophage-trophoblast interactions. Methods Mol Med. 2006;122:149–163. doi: 10.1385/1-59259-989-3:149. [DOI] [PubMed] [Google Scholar]
- 28.Abrahams VM, Kim YM, Straszewski SL, Romero R, Mor G. Macrophages and apoptotic cell clearance during pregnancy. Am J Reprod Immunol. 2004;51:275–282. doi: 10.1111/j.1600-0897.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- 29.Yelavarthi KK, Fishback JL, Hunt JS. Analysis of HLA-G mRNA in human placental and extraplacental membrane cells by in situ hybridization. J Immunol. 1991;146:2847–2854. [PubMed] [Google Scholar]
- 30.Fest S, Aldo PB, Abrahams VM, Visintin I, Alvero A, Chen R, Chavez SL, Romero R, Mor G. Trophoblast-macrophage interactions: a regulatory network for the protection of pregnancy. Am J Reprod Immunol. 2007;57:55–66. doi: 10.1111/j.1600-0897.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 31.Petty HR, Kindzelskii AL, Espinoza J, Romero R. Trophoblast contact deactivates human neutrophils. J Immunol. 2006;176:3205–3214. doi: 10.4049/jimmunol.176.5.3205. [DOI] [PubMed] [Google Scholar]
- 32.Mor G, Koga K. Macrophages and pregnancy. Reprod Sci. 2008;15:435–436. doi: 10.1177/1933719108317253. [DOI] [PubMed] [Google Scholar]
- 33.Nagamatsu T, Schust DJ. The immunomodulatory roles of macrophages at the maternal-fetal interface. Reprod Sci. 2010;17:209–218. doi: 10.1177/1933719109349962. [DOI] [PubMed] [Google Scholar]
- 34.Nagamatsu T, Schust DJ. The contribution of macrophages to normal and pathological pregnancies. Am J Reprod Immunol. 2010;63:460–471. doi: 10.1111/j.1600-0897.2010.00813.x. [DOI] [PubMed] [Google Scholar]
- 35.Svensson J, Jenmalm MC, Matussek A, Geffers R, Berg G, Ernerudh J. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J Immunol. 2011;187:3671–3682. doi: 10.4049/jimmunol.1100130. [DOI] [PubMed] [Google Scholar]
- 36.McIntire RH, Petroff MG, Phillips TA, Hunt JS. In vitro models for studying human uterine and placental macrophages. Methods Mol Med. 2006;122:123–148. doi: 10.1385/1-59259-989-3:123. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton S, Oomomian Y, Stephen G, Shynlova O, Tower CL, Garrod A, Lye SJ, Jones RL. Macrophages Infiltrate the Human and Rat Decidua During Term and Preterm Labor: Evidence That Decidual Inflammation Precedes Labor. Biol Reprod. 2011 doi: 10.1095/biolreprod.111.095505. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Repnik U, Tilburgs T, Roelen DL, van der Mast BJ, Kanhai HH, Scherjon S, Claas FH. Comparison of macrophage phenotype between decidua basalis and decidua parietalis by flow cytometry. Placenta. 2008;29:405–412. doi: 10.1016/j.placenta.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 39.McIntire RH, Hunt JS. Antigen presenting cells and HLA-G--a review. Placenta. 2005;26 (Suppl A):S104–109. doi: 10.1016/j.placenta.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Kim YM, Romero R, Oh SY, Kim CJ, Kilburn BA, Armant DR, Nien JK, Gomez R, Mazor M, Saito S, Abrahams VM, Mor G. Toll-like receptor 4: a potential link between “danger signals,” the innate immune system, and preeclampsia? Am J Obstet Gynecol. 2005;193:921–927. doi: 10.1016/j.ajog.2005.07.076. [DOI] [PubMed] [Google Scholar]
- 41.Romero R, Chaiworapongsa T, Alpay Savasan Z, Xu Y, Hussein Y, Dong Z, Kusanovic JP, Kim CJ, Hassan SS. Damage-associated molecular patterns (DAMPs) in preterm labor with intact membranes and preterm PROM: a study of the alarmin HMGB1. J Matern Fetal Neonatal Med. 2011;24:1444–1455. doi: 10.3109/14767058.2011.591460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIntire RH, Ganacias KG, Hunt JS. Programming of human monocytes by the uteroplacental environment. Reprod Sci. 2008;15:437–447. doi: 10.1177/1933719107314065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elfline M, Clark A, Petty HR, Romero R. Bi-directional calcium signaling between adjacent leukocytes and trophoblast-like cells. Am J Reprod Immunol. 2010;64:339–346. doi: 10.1111/j.1600-0897.2010.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petroff MG, Sedlmayr P, Azzola D, Hunt JS. Decidual macrophages are potentially susceptible to inhibition by class Ia and class Ib HLA molecules. J Reprod Immunol. 2002;56:3–17. doi: 10.1016/s0165-0378(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 45.Toti P, Arcuri F, Tang Z, Schatz F, Zambrano E, Mor G, Niven-Fairchild T, Abrahams VM, Krikun G, Lockwood CJ, Guller S. Focal increases of fetal macrophages in placentas from pregnancies with histological chorioamnionitis: potential role of fibroblast monocyte chemotactic protein-1. Am J Reprod Immunol. 2011;65:470–479. doi: 10.1111/j.1600-0897.2010.00927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulmer JN, Morrison L, Smith JC. Expression of class II MHC gene products by macrophages in human uteroplacental tissue. Immunology. 1988;63:707–714. [PMC free article] [PubMed] [Google Scholar]
- 47.Lidstrom C, Matthiesen L, Berg G, Sharma S, Ernerudh J, Ekerfelt C. Cytokine secretion patterns of NK cells and macrophages in early human pregnancy decidua and blood: implications for suppressor macrophages in decidua. Am J Reprod Immunol. 2003;50:444–452. doi: 10.1046/j.8755-8920.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 48.Rozner AE, Dambaeva SV, Drenzek JG, Durning M, Golos TG. Modulation of cytokine and chemokine secretions in rhesus monkey trophoblast co-culture with decidual but not peripheral blood monocyte-derived macrophages. Am J Reprod Immunol. 2011;66:115–127. doi: 10.1111/j.1600-0897.2010.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atay S, Gercel-Taylor C, Suttles J, Mor G, Taylor DD. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am J Reprod Immunol. 2011;65:65–77. doi: 10.1111/j.1600-0897.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 50.Wynn RM. Derivation and ultrastructure of the so-called Hofbauer cell. Am J Obstet Gynecol. 1967;97:235–248. doi: 10.1016/0002-9378(67)90546-7. [DOI] [PubMed] [Google Scholar]
- 51.Seval Y, Korgun ET, Demir R. Hofbauer cells in early human placenta: possible implications in vasculogenesis and angiogenesis. Placenta. 2007;28:841–845. doi: 10.1016/j.placenta.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Schonkeren D, van der Hoorn ML, Khedoe P, Swings G, van Beelen E, Claas F, van Kooten C, de Heer E, Scherjon S. Differential distribution and phenotype of decidual macrophages in preeclamptic versus control pregnancies. Am J Pathol. 2011;178:709–717. doi: 10.1016/j.ajpath.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renaud SJ, Graham CH. The role of macrophages in utero-placental interactions during normal and pathological pregnancy. Immunol Invest. 2008;37:535–564. doi: 10.1080/08820130802191375. [DOI] [PubMed] [Google Scholar]
- 54.Sacks GP, Studena K, Sargent K, Redman CW. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am J Obstet Gynecol. 1998;179:80–86. doi: 10.1016/s0002-9378(98)70254-6. [DOI] [PubMed] [Google Scholar]
- 55.Kim JS, Romero R, Cushenberry E, Kim YM, Erez O, Nien JK, Yoon BH, Espinoza J, Kim CJ. Distribution of CD14+ and CD68+ macrophages in the placental bed and basal plate of women with preeclampsia and preterm labor. Placenta. 2007;28:571–576. doi: 10.1016/j.placenta.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Gervasi MT, Chaiworapongsa T, Pacora P, Naccasha N, Yoon BH, Maymon E, Romero R. Phenotypic and metabolic characteristics of monocytes and granulocytes in preeclampsia. Am J Obstet Gynecol. 2001;185:792–797. doi: 10.1067/mob.2001.117311. [DOI] [PubMed] [Google Scholar]
- 57.Gervasi MT, Chaiworapongsa T, Naccasha N, Blackwell S, Yoon BH, Maymon E, Romero R. Phenotypic and metabolic characteristics of maternal monocytes and granulocytes in preterm labor with intact membranes. Am J Obstet Gynecol. 2001;185:1124–1129. doi: 10.1067/mob.2001.117681. [DOI] [PubMed] [Google Scholar]
- 58.Gervasi MT, Chaiworapongsa T, Naccasha N, Pacora P, Berman S, Maymon E, Kim JC, Kim YM, Yoshimatsu J, Espinoza J, Romero R. Maternal intravascular inflammation in preterm premature rupture of membranes. J Matern Fetal Neonatal Med. 2002;11:171–175. doi: 10.1080/jmf.11.3.171.175. [DOI] [PubMed] [Google Scholar]
- 59.Kim SK, Romero R, Chaiworapongsa T, Kusanovic JP, Mazaki-Tovi S, Mittal P, Erez O, Vaisbuch E, Gotsch F, Pacora P, Yeo L, Gervasi MT, Lamont RF, Yoon BH, Hassan SS. Evidence of changes in the immunophenotype and metabolic characteristics (intracellular reactive oxygen radicals) of fetal, but not maternal, monocytes and granulocytes in the fetal inflammatory response syndrome. J Perinat Med. 2009;37:543–552. doi: 10.1515/JPM.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gotsch F, Romero R, Kusanovic JP, Mazaki-Tovi S, Pineles BL, Erez O, Espinoza J, Hassan SS. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- 61.Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim JS, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naccasha N, Gervasi MT, Chaiworapongsa T, Berman S, Yoon BH, Maymon E, Romero R. Phenotypic and metabolic characteristics of monocytes and granulocytes in normal pregnancy and maternal infection. Am J Obstet Gynecol. 2001;185:1118–1123. doi: 10.1067/mob.2001.117682. [DOI] [PubMed] [Google Scholar]
- 63.Tang Z, Abrahams VM, Mor G, Guller S. Placental Hofbauer cells and complications of pregnancy. Ann N Y Acad Sci. 2011;1221:103–108. doi: 10.1111/j.1749-6632.2010.05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JS, Romero R, Kim MR, Kim YM, Friel L, Espinoza J, Kim CJ. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. 2008;52:457–464. doi: 10.1111/j.1365-2559.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, Lee DC, Draghici S, Gotsch F, Kusanovic JP, Hassan SS, Kim JS. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. 2009;182:3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Janson PC, Winqvist O. Epigenetics--the key to understand immune responses in health and disease. Am J Reprod Immunol. 2011;66 (Suppl 1):72–74. doi: 10.1111/j.1600-0897.2011.01050.x. [DOI] [PubMed] [Google Scholar]
- 69.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62:78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 72.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 73.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 74.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, Ferreira JC, Pinto D, Scherer SW, Shaffer LG, Coullin P, Caniggia I, Beyene J, Slim R, Bartolomei MS, Weksberg R. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res. 2011;21:465–476. doi: 10.1101/gr.111922.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuen RK, Penaherrera MS, von Dadelszen P, McFadden DE, Robinson WP. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 2010;18:1006–1012. doi: 10.1038/ejhg.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koukoura O, Sifakis S, Zaravinos A, Apostolidou S, Jones A, Hajiioannou J, Widschwendter M, Spandidos DA. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32:51–57. doi: 10.1016/j.placenta.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 79.Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6:368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 81.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wetzka B, Clark DE, Charnock-Jones DS, Zahradnik HP, Smith SK. Isolation of macrophages (Hofbauer cells) from human term placenta and their prostaglandin E2 and thromboxane production. Hum Reprod. 1997;12:847–852. doi: 10.1093/humrep/12.4.847. [DOI] [PubMed] [Google Scholar]
- 84.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 85.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 86.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 87.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 88.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 89.Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 90.Diederichs S, Baumer N, Ji P, Metzelder SK, Idos GE, Cauvet T, Wang W, Moller M, Pierschalski S, Gromoll J, Schrader MG, Koeffler HP, Berdel WE, Serve H, Muller-Tidow C. Identification of interaction partners and substrates of the cyclin A1-CDK2 complex. J Biol Chem. 2004;279:33727–33741. doi: 10.1074/jbc.M401708200. [DOI] [PubMed] [Google Scholar]
- 91.Mor G, Abrahams VM. Potential role of macrophages as immunoregulators of pregnancy. Reprod Biol Endocrinol. 2003;1:119. doi: 10.1186/1477-7827-1-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Southcombe J, Tannetta D, Redman C, Sargent I. The immunomodulatory role of syncytiotrophoblast microvesicles. PLoS One. 2011;6:e20245. doi: 10.1371/journal.pone.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang H, Van De Ven C, Satwani P, Baxi LV, Cairo MS. Differential gene expression patterns by oligonucleotide microarray of basal versus lipopolysaccharide-activated monocytes from cord blood versus adult peripheral blood. J Immunol. 2004;172:5870–5879. doi: 10.4049/jimmunol.172.10.5870. [DOI] [PubMed] [Google Scholar]
- 94.Levy O, Zarember KA, Roy RM, Cywes C, Godowski PJ, Wessels MR. Selective impairment of TLR-mediated innate immunity in human newborns: neonatal blood plasma reduces monocyte TNF-alpha induction by bacterial lipopeptides, lipopolysaccharide, and imiquimod, but preserves the response to R-848. J Immunol. 2004;173:4627–4634. doi: 10.4049/jimmunol.173.7.4627. [DOI] [PubMed] [Google Scholar]
- 95.Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gustafsson C, Mjosberg J, Matussek A, Geffers R, Matthiesen L, Berg G, Sharma S, Buer J, Ernerudh J. Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS One. 2008;3:e2078. doi: 10.1371/journal.pone.0002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Joerink M, Rindsjo E, van Riel B, Alm J, Papadogiannakis N. Placental macrophage (Hofbauer cell) polarization is independent of maternal allergen-sensitization and presence of chorioamnionitis. Placenta. 2011;32:380–385. doi: 10.1016/j.placenta.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 99.Bockle BC, Solder E, Kind S, Romani N, Sepp NT. DC-sign+ CD163+ macrophages expressing hyaluronan receptor LYVE-1 are located within chorion villi of the placenta. Placenta. 2008;29:187–192. doi: 10.1016/j.placenta.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 100.Liu H, Shi B, Huang CC, Eksarko P, Pope RM. Transcriptional diversity during monocyte to macrophage differentiation. Immunol Lett. 2008;117:70–80. doi: 10.1016/j.imlet.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 102.Takahashi K, Naito M, Katabuchi H, Higashi K. Development, differentiation, and maturation of macrophages in the chorionic villi of mouse placenta with special reference to the origin of Hofbauer cells. J Leukoc Biol. 1991;50:57–68. doi: 10.1002/jlb.50.1.57. [DOI] [PubMed] [Google Scholar]
- 103.Kim SS, Romero R, Kim JS, Abbas A, Espinoza J, Kusanovic JP, Hassan S, Yoon BH, Kim CJ. Coexpression of myofibroblast and macrophage markers: novel evidence for an in vivo plasticity of chorioamniotic mesodermal cells of the human placenta. Lab Invest. 2008;88:365–374. doi: 10.1038/labinvest.3700749. [DOI] [PubMed] [Google Scholar]
- 104.Castellucci M, Celona A, Bartels H, Steininger B, Benedetto V, Kaufmann P. Mitosis of the Hofbauer cell: possible implications for a fetal macrophage. Placenta. 1987;8:65–76. doi: 10.1016/0143-4004(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 105.Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001;276:39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 106.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 107.Rajendran G, Shanmuganandam K, Bendre A, Mujumdar D, Goel A, Shiras A. Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol. 2011;104:483–494. doi: 10.1007/s11060-010-0520-2. [DOI] [PubMed] [Google Scholar]
- 108.Romero R, Espinoza J, Chaiworapongsa T, Kalache K. Infection and prematurity and the role of preventive strategies. Semin Neonatol. 2002;7:259–274. doi: 10.1016/s1084-2756(02)90121-1. [DOI] [PubMed] [Google Scholar]
- 109.Romero R, Gotsch F, Pineles B, Kusanovic JP. Inflammation in pregnancy: its roles in reproductive physiology, obstetrical complications, and fetal injury. Nutr Rev. 2007;65:S194–202. doi: 10.1111/j.1753-4887.2007.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 110.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McElrath TF, Hecht JL, Dammann O, Boggess K, Onderdonk A, Markenson G, Harper M, Delpapa E, Allred EN, Leviton A. Pregnancy disorders that lead to delivery before the 28th week of gestation: an epidemiologic approach to classification. Am J Epidemiol. 2008;168:980–989. doi: 10.1093/aje/kwn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Berry SM, Romero R, Gomez R, Puder KS, Ghezzi F, Cotton DB, Bianchi DW. Premature parturition is characterized by in utero activation of the fetal immune system. Am J Obstet Gynecol. 1995;173:1315–1320. doi: 10.1016/0002-9378(95)91378-5. [DOI] [PubMed] [Google Scholar]
- 113.Chaiworapongsa T, Romero R, Berry SM, Hassan SS, Yoon BH, Edwin S, Mazor M. The role of granulocyte colony-stimulating factor in the neutrophilia observed in the fetal inflammatory response syndrome. J Perinat Med. 2011;39:653–666. doi: 10.1515/JPM.2011.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jarvis JN, Deng L, Berry SM, Romero R, Moore H. Fetal cytokine expression in utero detected by reverse transcriptase polymerase chain reaction. Pediatr Res. 1995;37:450–454. doi: 10.1203/00006450-199504000-00011. [DOI] [PubMed] [Google Scholar]
- 115.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, Ghezzi F, Berry SM, Qureshi F, Jacques SM, Kim JC, Kadar N, Romero R. Funisitis and chorionic vasculitis: the histological counterpart of the fetal inflammatory response syndrome. J Matern Fetal Neonatal Med. 2002;11:18–25. doi: 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 116.Romero R, Espinoza J, Goncalves LF, Gomez R, Medina L, Silva M, Chaiworapongsa T, Yoon BH, Ghezzi F, Lee W, Treadwell M, Berry SM, Maymon E, Mazor M, DeVore G. Fetal cardiac dysfunction in preterm premature rupture of membranes. J Matern Fetal Neonatal Med. 2004;16:146–157. doi: 10.1080/14767050400009279. [DOI] [PubMed] [Google Scholar]
- 117.Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179:186–193. doi: 10.1016/s0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 118.Romero R, Maymon E, Pacora P, Gomez R, Mazor M, Yoon BH, Berry SM. Further observations on the fetal inflammatory response syndrome: a potential homeostatic role for the soluble receptors of tumor necrosis factor alpha. Am J Obstet Gynecol. 2000;183:1070–1077. doi: 10.1067/mob.2000.108885. [DOI] [PubMed] [Google Scholar]
- 119.Romero R, Savasan ZA, Chaiworapongsa T, Berry SM, Kusanovic JP, Hassan SS, Yoon BH, Edwin S, Mazor M. Hematologic profile of the fetus with systemic inflammatory response syndrome. J Perinat Med. 2011 doi: 10.1515/JPM.2011.100. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Romero R, Soto E, Berry SM, Hassan SS, Kusanovic JP, Yoon BH, Edwin S, Mazor M, Chaiworapongsa T. Blood pH and gases in fetuses in preterm labor with and without systemic inflammatory response syndrome. J Matern Fetal Neonatal Med. 2011 doi: 10.3109/14767058.2011.629247. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Esplin MS, Romero R, Chaiworapongsa T, Kim YM, Edwin S, Gomez R, Mazor M, Adashi EY. Monocyte chemotactic protein-1 is increased in the amniotic fluid of women who deliver preterm in the presence or absence of intra-amniotic infection. J Matern Fetal Neonatal Med. 2005;17:365–373. doi: 10.1080/14767050500141329. [DOI] [PubMed] [Google Scholar]
- 122.Chaiworapongsa T, Romero R, Espinoza J, Kim YM, Edwin S, Bujold E, Gomez R, Kuivaniemi H. Macrophage migration inhibitory factor in patients with preterm parturition and microbial invasion of the amniotic cavity. J Matern Fetal Neonatal Med. 2005;18:405–416. doi: 10.1080/14767050500361703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Romero R, Chaiworapongsa T, Espinoza J, Gomez R, Yoon BH, Edwin S, Mazor M, Maymon E, Berry S. Fetal plasma MMP-9 concentrations are elevated in preterm premature rupture of the membranes. Am J Obstet Gynecol. 2002;187:1125–1130. doi: 10.1067/mob.2002.127312. [DOI] [PubMed] [Google Scholar]
- 124.Vinnars MT, Rindsjo E, Ghazi S, Sundberg A, Papadogiannakis N. The number of CD68(+) (Hofbauer) cells is decreased in placentas with chorioamnionitis and with advancing gestational age. Pediatr Dev Pathol. 2010;13:300–304. doi: 10.2350/09-03-0632-OA.1. [DOI] [PubMed] [Google Scholar]
- 125.Houser BL, Tilburgs T, Hill J, Nicotra ML, Strominger JL. Two unique human decidual macrophage populations. J Immunol. 2011;186:2633–2642. doi: 10.4049/jimmunol.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]