SUMMARY
Molecular signatures have identified several subsets of Diffuse Large B-Cell Lymphoma (DLBCL) and rational targets within the B-cell receptor (BCR) signaling axis. The OxPhos-DLBCL subset, which harbors the signature of genes involved in mitochondrial metabolism, is insensitive to inhibition of BCR survival signaling, but is functionally undefined. We show that compared with BCR-DLBCLs, OxPhos-DLBCLs display enhanced mitochondrial energy transduction, greater incorporation of nutrient-derived carbons into the TCA cycle and increased glutathione levels. Importantly, perturbation of the fatty acid oxidation program and glutathione synthesis proved selectively toxic to this tumor subset. Our analysis provides evidence for distinct metabolic fingerprints and associated survival mechanisms in DLBCL and may have therapeutic implications.
INTRODUCTION
Tumors often rewire their metabolism to ensure steady supply of intermediary metabolites for synthesis of new biomass, as well as generation of ATP and reducing equivalents (Barger and Plas, 2010; DeBerardinis et al., 2008; Tennant et al., 2010). This metabolic reprogramming is a dynamic process shaped by oncogenes and tumor suppressors (Barger and Plas, 2010; Morrish et al., 2008; Yuneva et al., 2012). One of the first metabolic alterations identified in tumors is elevated glycolysis even in the presence of sufficient oxygen. This program, also known as the Warburg effect or aerobic glycolysis, fulfills important biosynthetic needs (Barger and Plas, 2010; Koppenol et al., 2011; Vander Heiden et al., 2009). The Warburg effect has often been interpreted as an indication of impaired mitochondrial respiration (Koppenol et al., 2011). However, the relevance of mitochondrial respiration in tumors is varied depending on tumor type and evidence for an oxidative class of tumors and tumors with dual capacity for glycolytic and oxidative metabolism exists (Marin-Valencia et al., 2012; Moreno-Sanchez et al., 2009). Moreover, the importance of mitochondria in tumor cell survival and proliferation, including utilization of alternative oxidizable substrates such as glutamine and fatty acids has been increasingly appreciated (Le et al., 2012; Rossignol et al., 2004; Zaugg et al., 2011). The diversity of carbon substrate utilization pathways in tumors is indicative of metabolic heterogeneity that may not only be relevant across different types of cancer but also manifest within a group of tumors that otherwise share a common diagnosis.
Diffuse large B-cell lymphomas (DLBCLs) are a genetically heterogeneous group of tumors and the most common non-Hodgkin lymphomas in adults (Abramson and Shipp, 2005; Lenz and Staudt, 2010). However, the spectrum of fuel utilization pathways and the metabolic fingerprints within DLBCL and other similarly heterogeneous groups of tumors have not been fully elucidated. To date, efforts to capture the molecular heterogeneity of DLBCL have relied on gene expression profiling that has uncovered coordinate signaling and survival paradigms in distinct subsets of DLBCL. In one approach, comparison of the genetic signatures across DLBCLs using genome-wide arrays and multiple clustering algorithms captured tumor-intrinsic distinctions in three separate and reproducible clusters (Monti et al., 2005). Groups of DLBCLs identified by this consensus cluster classification (CCC) scheme are the BCR/proliferation cluster (BCR-DLBCL) displaying up-regulation of genes encoding B-cell receptor (BCR) signaling components, the OxPhos cluster (OxPhos-DLBCL), which is significantly enriched in genes involved in mitochondrial oxidative phosphorylation (OxPhos), and the host response (HR) tumors largely characterized by a brisk host inflammatory infiltrate (Monti et al., 2005). Another classification framework known as cell-of-origin (COO) delineated DLBCL subsets that shared components of their transcriptional profiles with normal B-cell subtypes, including Germinal Center B-cell (GCB)-like and Activated B-cell (ABC)-like (Alizadeh et al., 2000), and a third undefined category, designated “type 3” (Wright et al., 2003). CCC and COO classifications capture largely different molecular aspects of DLBCL (Monti et al., 2005).
Unlike tumors that rely on signaling pathways downstream of the B-cell receptor, OxPhos-DLBCLs do not display active/functional BCR signaling (Chen et al., 2008). However, the nature of survival pathways in this group of tumors is not known and beyond the original CCC assignment, the actual functional attributes of the OxPhos molecular signature have not been fully examined. This signature includes multiple subunits of mitochondrial respiratory chain complexes I (NADH dehydrogenase) and V (mitochondrial ATP synthase) that may suggest alterations in mitochondrial energy transduction. However, given the integrative aspect of cellular metabolism and the requirement of both nuclear and mitochondria-encoded genes for proper functioning of the electron transport machinery, the precise metabolic landscape of this molecular subset could not be predicted.
In the present study, we conducted an integrative analysis to dissect the metabolic fingerprints of DLBCL and to delineate subtype-specific differences that may selectively contribute to growth and survival of DLBCL subsets.
RESULTS
Subtype-Specific Differences in the DLBCL Mitochondrial Proteome
The up-regulation of select genes encoding for subunits of electron transport chain (ETC) complexes in OxPhos-DLBCLs predicts potential differences in mitochondrial oxidative metabolism compared with other DLBCL groups. However, as ETC activity is linked to the supply of carbon substrates and reducing equivalents, the OxPhos signature is likely part of a broader spectrum of changes in mitochondrial nutrient metabolism that may shed light on the actual functional attributes of an “OxPhos” program in this DLBCL subset. To search for additional components of this metabolic program, we initially performed two dimensional differential gel electrophoresis (2D-DIGE) to compare the proteome of mitochondria purified from representative OxPhos- and BCR-DLBCL cell lines Karpas 422 and OCI-Ly1, respectively (Chen et al., 2008). Mitochondrial proteins that were ≥ 2.5 more abundant in the OxPhos cell line were identified by mass spectrometry (Figure S1A). Among 2D-DIGE candidates were subunits of mitochondrial respiratory chain complex I (NADH dehydrogenase), complex II (succinate dehydrogenase, also a TCA cycle enzyme), complex V (ATP synthase), subunits of the pyruvate dehydrogenase (PDH) complex and several other TCA cycle enzymes, mitochondrial ROS detoxification enzyme manganese superoxide dismutase (MnSOD or SOD2), as well as enzymes involved in mitochondrial β-oxidation of fatty acids and metabolism of ketone bodies and amino acids (Figure S1A).
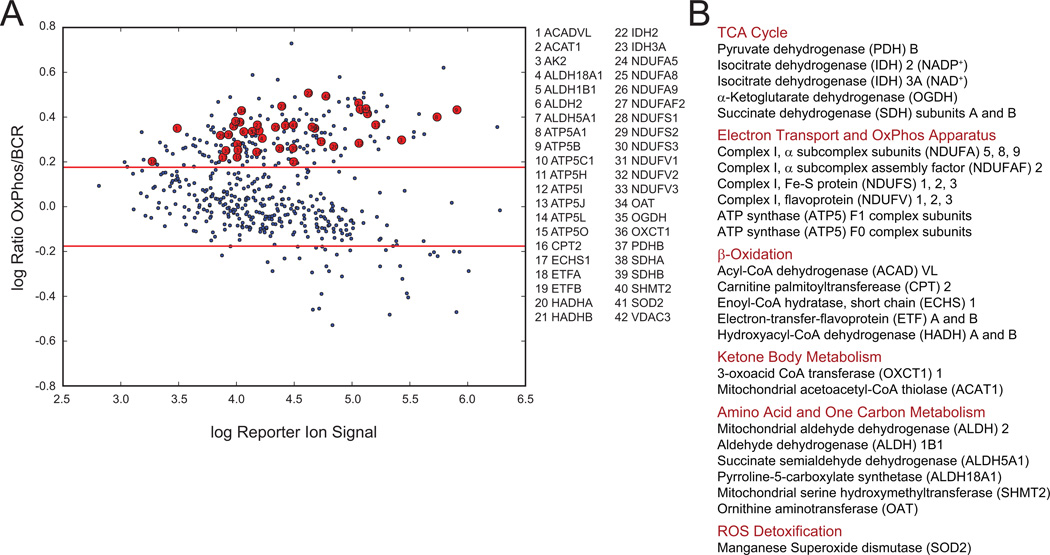
We next wished to interrogate the mitochondrial protein signature defined by 2D-DIGE in a larger panel of OxPhos- and BCR-DLBCL cell lines. This required an independent proteomics approach amenable to multiplexing- isobaric tags for relative and absolute quantification (iTRAQ) (Figure S1B) (Choe et al., 2007; Ross et al., 2004)- for simultaneous quantitative comparison of multiple unique tryptic peptides per candidate mitochondrial protein across three pairs of OxPhos- and BCR-DLBCL cell lines. This analysis confirmed significant quantitative enrichment of mitochondrial β-oxidation enzymes, multiple subunits of respiratory chain complexes, as well as TCA cycle enzymes and MnSOD in OxPhos-DLBCL mitochondria (Figure 1 and Table S1).
Figure 1. Comparison of Mitochondrial Proteome in OxPhos- and BCR-DLBCLs.
(A and B) Multiplex iTRAQ analysis of mitochondria derived from 3 OxPhos- (Karpas 422, Toledo, and Pfeiffer) and 3 BCR- (OCI-Ly1, SU-DHL-4, and SU-DHL-6) DLBCL cell lines. Log-log plots of reporter ions abundance ratios vs. reporter ion intensities for proteins detected in replicate nanoflow LC-MS/MS analyses are shown (A). Proteins within the mitochondrial signature that are enriched in OxPhos-DLBCLs are shown as red circles in (A) and grouped per metabolic pathway in (B). Red lines in A represent global thresholds based on a ±1.5-fold change between OxPhos and BCR subtypes.
See also Figure S1 and Table S1.
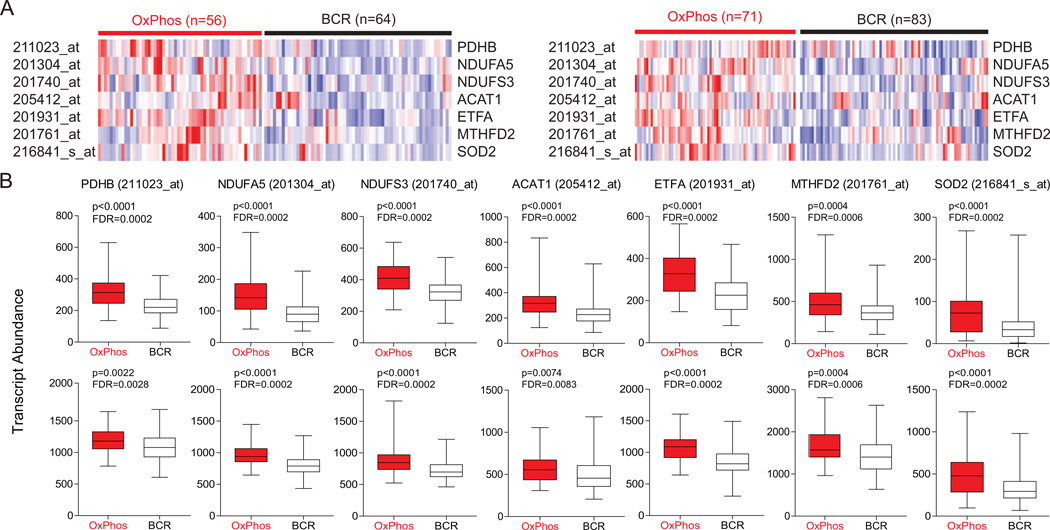
Beyond confirming the differential expression of complex I and V components that were in the original OxPhos-DLBCL cluster signature (Monti et al., 2005), the 2D-DIGE and iTRAQ proteomic analyses identified additional components of the OxPhos-DLBCL mitochondrial signature that were not fully captured by the previous RNA profile analysis of total tumors. To examine whether these observations could also be substantiated in primary OxPhos-DLBCLs, we examined tumor biopsies for differences in the abundance of these newly identified components of the mitochondrial signature at both protein and RNA levels. The small en bloc primary DLBCL biopsies precluded isolation of tumor mitochondria; however, iTRAQ-based quantification in a limited number of primary cases that produced protein samples of suitable purity and grade for LC-MS/MS indicated increased expression of several TCA cycle and mitochondrial β-oxidation enzymes, as well as MnSOD (Table S1). The RNA abundance for candidate components of the mitochondrial proteome signature was also queried in two extensive primary DLBCL expression profile data sets (Lenz et al., 2008; Monti et al., 2005) with CCC designations (Tables S2 and S3) and found to be significantly higher in primary OxPhos- than in BCR-DLBCLs in both data sets (Figure 2). These observations confirm that the differences in mitochondrial signature identified in DLBCL cell lines translate to primary tumor biopsies.
Figure 2. Increased Abundance of Transcripts Encoding Mitochondrial Proteins in Primary DLBCL Tumor Biopsies.
(A) Heat map representation of the relative mRNA levels of genes corresponding to the components of the mitochondrial proteome signature using the Monti et al. (left) and Lenz et al. (right) expression array data sets of primary DLBCL cases with OxPhos and BCR consensus cluster assignments.
(B) Transcript abundance (probe intensity) of the indicated genes in primary OxPhos- and BCR-DLBCLs from the Monti et al. (top) and Lenz et al. (bottom) data sets. Differential expression was determined by a two-sided Mann Whitney test and p values were corrected for multiple hypothesis testing using the false discovery rate (FDR) procedure.
Abbreviations: PDH, pyruvate dehydrogenase; NDUFA5, NADH dehydrogenase (ubiquinone) 1α subcomplex subunit 5; NDUFS3, NADH dehydrogenase (ubiquinone) Fe-S protein 3; ETF, electron-transfer-flavoprotein; ACAT, acetoacetyl-CoA thiolase; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; SOD2, manganese superoxide dismutase.
See also Tables S2 and S3.
The functional categories of proteins in the OxPhos-DLBCL mitochondrial signature predict differences in mitochondrial handling of fatty acids and pyruvate, ETC activity, mitochondrial energy production, and ROS content. We next set out to validate each of these predictions.
Mitochondrial Substrate Oxidation in DLBCL Subsets
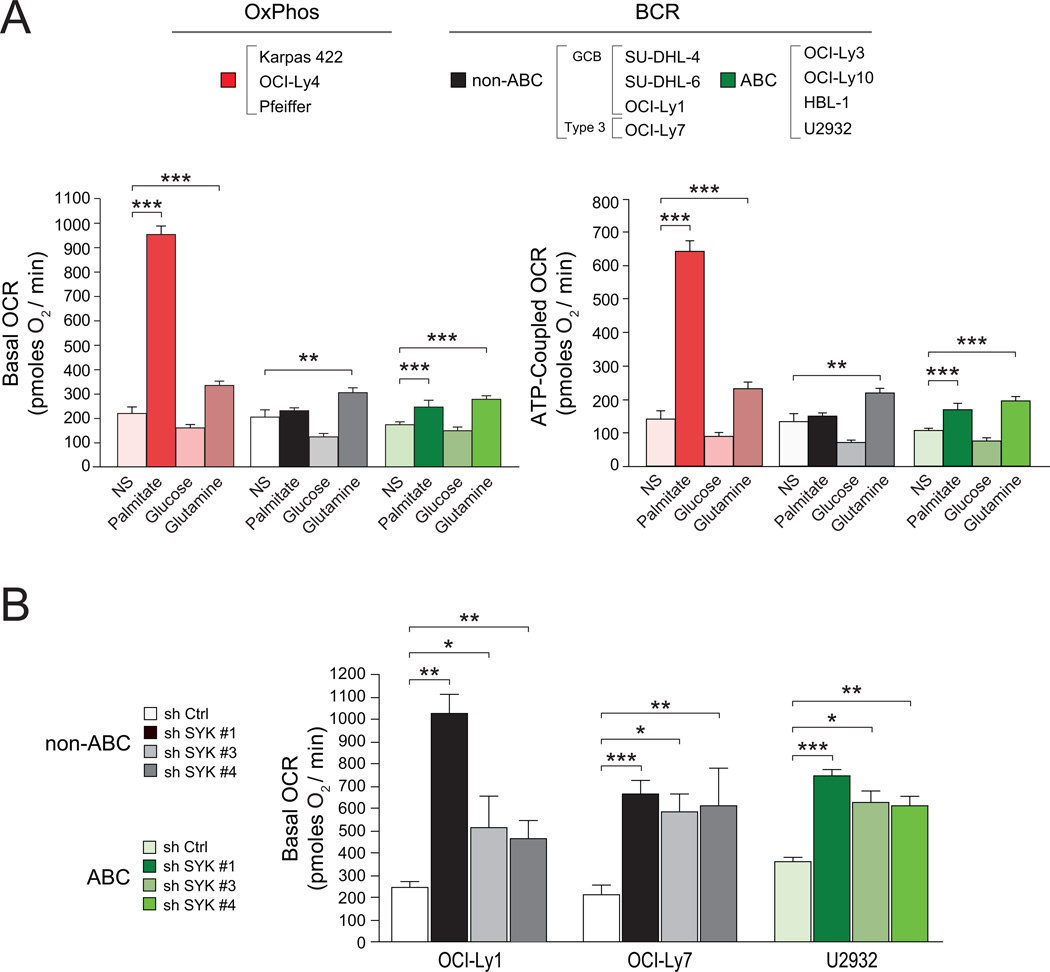
To assess potential differences in mitochondrial carbon substrate utilization, mitochondrial oxygen consumption rate (OCR) was measured in real time in the absence or presence of exogenously added substrates such as glucose, glutamine and palmitate using a panel of 3–4 independent OxPhos and “non-OxPhos” DLBCL cell lines. The “non-OxPhos” panel included cell lines with known CCC assignment as BCR-DLBCLs (Polo et al., 2007) and COO classification as GCB-, ABC-, or Type 3-DLBCLs (Alizadeh et al., 2000; Wright et al., 2003) (see Supplemental Experimental Procedures). Basal OCR in response to exogenously supplied palmitate was significantly higher in OxPhos-DLBCLs relative to “non-OxPhos” DLBCL cell lines [no substrate (NS) vs. palmitate (Figure 3A)]. Basal OCR values per individual DLBCL cell line are shown in Figure S2A. The “non-OxPhos” DLBCLs displayed either no increase in OCR or an increase that was significantly less than that observed in the OxPhos-DLBCLs (Figures 3A and S2A). Importantly, the majority of palmitate-stimulated OCR in OxPhos-DLBCLs was sensitive to the mitochondrial ATP synthase inhibitor oligomycin, indicating mitochondrial fatty acid oxidation (FAO) in this setting is associated with ATP synthesis (Figures 3A and S2A). These observations provide quantitative evidence for increased mitochondrial FAO in OxPhos-DLBCLs. Analysis of other oxidizable substrates showed that none of the DLBCL subsets respire on glucose (NS vs. glucose, Figures 3A and S2A). However, in response to glutamine, all DLBCL subsets exhibited a comparable increase in basal and ATP-associated (oligomycin inhibitable) OCR (NS vs. glutamine, Figures 3A and S2A). Taken together, comparison of the three carbon substrates across and within DLBCL subsets indicates that palmitate is a predominant respiratory fuel in OxPhos-DLBCLs.
Figure 3. Mitochondrial Carbon Substrate Oxidation in DLBCL Subsets and Its Regulation by BCR Signaling.
(A) Basal (left) and ATP-coupled (right) OCR in DLBCL subsets. OCR values shown are average of all cell lines per DLBCL subtype indicated on top. For each cell line, 7–13 independent OCR measurements were taken. NS denotes no substrate added exogenously.
(B) Palmitate-stimulated basal OCR in “non-OxPhos” DLBCL cell lines after acute knockdown of SYK.
Error bars, ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; two-tailed Student’s t-test.
See also Figure S2.
A marked increase in palmitate-induced OCR in OxPhos-DLBCL compared with “non-OxPhos” DLBCL cell lines parallels the absence or presence of functional BCR signaling in these subsets, respectively (Chen et al., 2008). This prompted examination of a potential reciprocal relationship between active BCR signaling and mitochondrial FAO. Such a scenario would predict that inhibition of BCR signaling in non-OxPhos/BCR DLBCLs may manifest in increased mitochondrial FAO. Indeed, acute interference with BCR signaling using multiple independent shRNAs against SYK, an upstream component of this signaling axis, was associated with significant elevation of basal palmitate-induced OCR (Figure 3B). Notably, OCR measurements were taken after 24 hr treatment with shRNAs, which enabled sufficient depletion of SYK (Figure S2B) without affecting cell viability (data not shown). These observations are suggestive of an underlying metabolic plasticity that governs the pattern of fuel oxidation in DLBCL subsets.
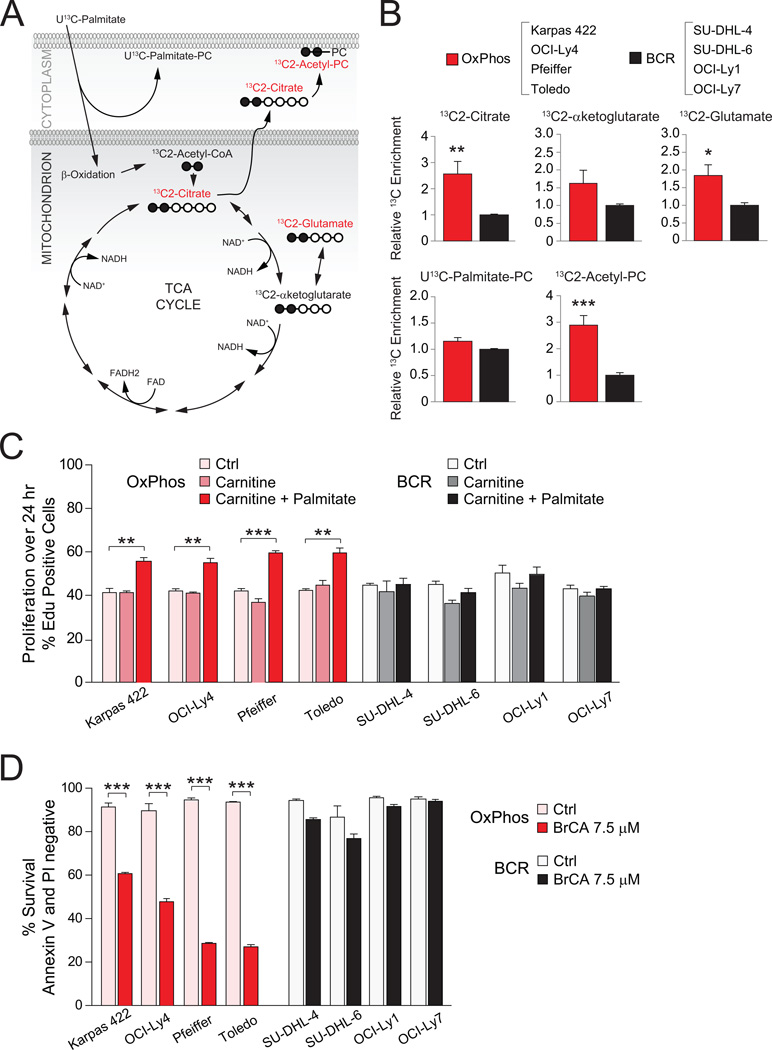
To provide independent and parallel evidence for differential utilization of fatty acids in DLBCL subsets, a targeted 13C isotopomer approach was undertaken. A total of eight cell lines with known designation as OxPhos or BCR were cultured in media containing uniformly labeled 13C-palmitate (U13C-palmitate), and 13C enrichment was assessed in a defined set of intermediates derived from fatty acid metabolism. The complete oxidation of 13C-palmitate by mitochondria yields 8 acetyl units that can donate carbons to the TCA cycle in the form of citrate labeled on two carbons (13C2-citrate), which can in turn lead to the formation of 13C2-α-ketoglutarate that is in isotopic equilibrium with 13C2-glutamate (Figure 4A). The relative level of 13C enrichment in both 13C2-citrate and 13C2-glutamate was significantly higher in the OxPhos-DLBCL cell lines (Figure 4B). This is consistent with greater entry of palmitate carbons into the TCA cycle.
Figure 4. Palmitate Metabolism and Its Effect on DLBCL Proliferation and Survival.
(A–B) 13C isotopomer analysis of uniformly labeled palmitate (U13C-Palmitate). (A) Schematics depicting the number of carbons labeled (filled circles) in a defined set of metabolites derived from palmitate. Metabolites marked in red are selectively elevated in OxPhos-DLBCL cell lines (B). (B) 13C enrichment in palmitate-derived metabolites. For each metabolite, cumulative data obtained from all 4 OxPhos-DLBCL cell lines are shown relative to the mean value of that metabolite in all 4 BCR-DLBCL cell lines listed on top.
(C) Effect of palmitate supplementation on the proliferation of DLBCL cell lines. Control denotes serum-free media containing all amino acids except L-glutamine.
(D) Survival of DLBCL subsets cultured in the absence or presence of BrCA for 24 hr.
Error bars, ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; two-tailed Student’s t-test.
See also Figure S3.
In addition to oxidative metabolism for energy production, palmitate may have important biosynthetic roles, including direct incorporation into phosphatidyl choline (PC) to yield U13C-Palmitate-PC (Figure 4A). Alternatively, palmitate-derived citrate can be exported from mitochondria and its carbons subsequently incorporated into the head group of PC (13C2-Acetyl-PC, Figure 4A). In both DLBCL subtypes, direct incorporation of palmitate into PC (U13C-Palmitate-PC) was observed. In addition, a larger enrichment of palmitate carbons in 13C2-Acetyl-PC was detected in OxPhos cell lines (Figure 4B).
A preferential increase in palmitate utilization in OxPhos-DLBCLs prompted examination of its effect on proliferation in these cells. Supplementation of serum-free media containing amino acids with palmitate and carnitine led to a modest but significant and selective increase in the proliferation of OxPhos-DLBCLs (Figure 4C). In the absence of palmitate, carnitine, which is required for mitochondrial import of long-chain fatty acids, did not influence proliferation (Figure 4C).
To probe the pro-survival benefit of mitochondrial FAO in DLBCL subsets, this program was inhibited using 4-bromocrotonic acid (BrCA), which irreversibly inhibits mitochondrial β-oxidation of both long- and short-chain fatty acids (el-Aleem and Schulz, 1987). Acute treatment of DLBCL cell lines (4 hr) with BrCA interfered with palmitate stimulation of basal OCR in OxPhos-DLBCL cell lines (Figure S3), while longer treatment (24 hr) was selectively toxic to this subset compared with BCR-DLBCLs (Figure 4D). These results suggest that the mitochondrial FAO program provides pro-survival benefits to OxPhos-DLBCLs.
Programmatic Regulation of Fatty Acid Oxidation and Its Relevance to OxPhos-DLBCL Survival
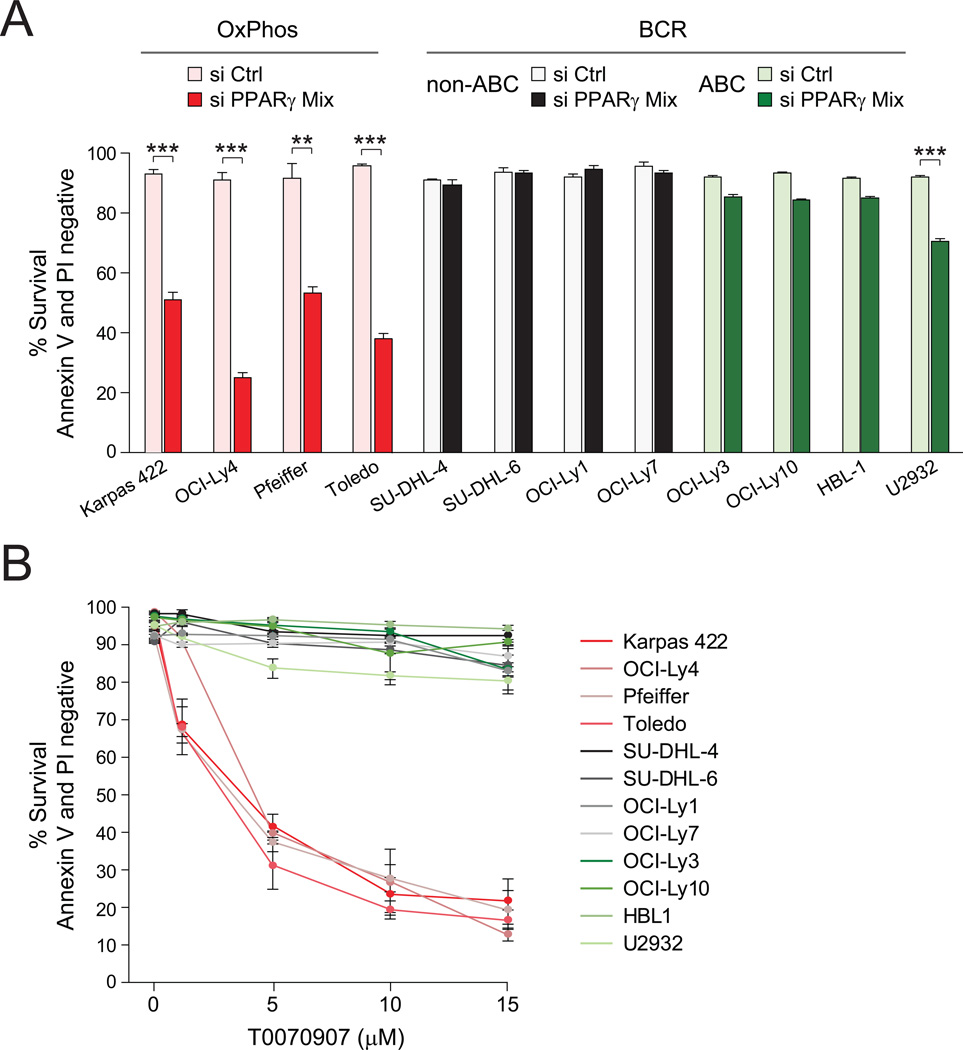
The concomitant increase in the abundance of several mitochondrial FAO enzymes in OxPhos-DLBCLs is consistent, at least in part, with a programmatic increase in the transcriptional regulation of this pathway. These observations warranted systematic interrogation of the primary DLBCL transcript data sets for the prevalence of a defined list of transcriptional regulators of this pathway. Nuclear receptor peroxisome proliferator-activated receptor (PPAR) γ transcripts were found to be significantly more abundant in primary OxPhos-DLBCLs in two independent cohorts of DLBCL tumors (Figure S4A). Importantly, several enzymes identified in the mitochondrial protein signature of OxPhos-DLBCLs such as ETF, ACAD and HADH, as well as multiple subunits of mitochondrial respiratory chain complexes I, II, and ATP synthase are downstream targets of PPARγ (Hsiao et al., 2011). To probe the functional relevance of PPARγ in DLBCL subsets, we tested the effect of its depletion using 3 independent siRNAs and their corresponding mixture (Figures 5A and S4B). The most robust decrease in PPARγ protein levels was achieved using the siRNA mix (Figure S4C) and was accompanied by significant apoptosis in OxPhos-DLBCLs compared with the BCR subset (Figure 5A).
Figure 5. Programmatic Regulation of FAO and Its Relevance to DLBCL Survival.
(A) Effect of siRNA-mediated depletion of PPARγ on the survival of DLBCL subsets.
(B) Survival of the indicated DLBCL cell lines cultured in the absence or presence of increasing concentrations of T0070907 for 96 hr.
Error bars, ± SEM. **p < 0.01; ***p < 0.001; two-tailed Student’s t-test.
See also Figure S4.
To provide a pharmacologic correlate to these findings, we also tested the effect of two selective PPARγ antagonists T0070907 and GW9662 (Lee et al., 2002; Leesnitzer et al., 2002). Short term treatment of DLBCL cell lines with these compounds blocked palmitate stimulation of OCR (data not shown). Upon longer incubation periods (96 hr), these compounds proved selectively toxic to OxPhos-DLBCLs (Figures 5B and S4D). The above genetic and pharmacologic approaches to PPARγ inhibition complement and extend the results shown in Figure 4D that pharmacologic inhibition of mitochondrial β-oxidation program is toxic to OxPhos-DLBCLs, and are collectively congruent with the idea that a sustained mitochondrial FAO program may be relevant for the survival of OxPhos-DLBCLs.
Differential Utilization of Glucose-Derived Carbons in DLBCL Subsets
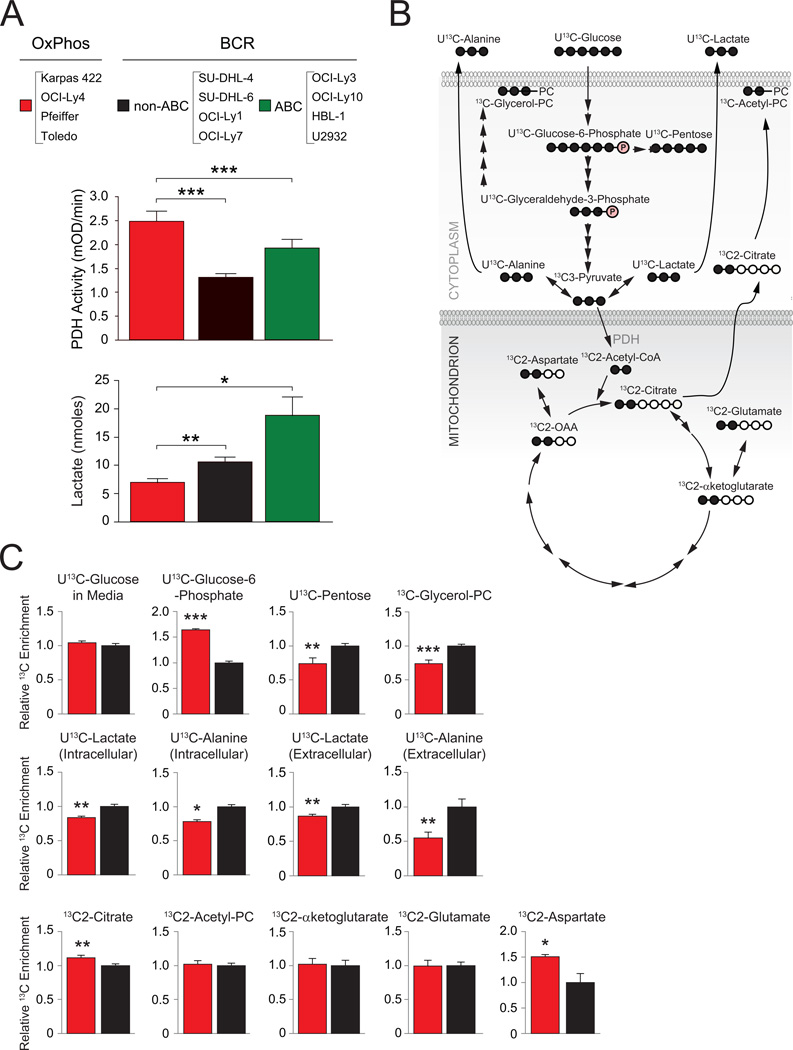
Identification of PDH as a component of the mitochondrial proteome signature in OxPhos-DLBCLs predicts differential mitochondrial handling of pyruvate. Indeed, biochemical analysis of isolated mitochondria derived from a panel of DLBCL cell lines indicated significant increase in PDH enzyme activity in OxPhos-DLBCLs (Figure 6A). Increased PDH activity would further predict diminished availability of glucose-derived pyruvate for lactate synthesis. Biochemical quantification of glucose-derived lactate secreted by OxPhos-DLBCL cell lines compared to “non-OxPhos”/BCR counterparts indicated that this is indeed the case (Figure 6A). These observations prompted a more detailed examination of the fate of glucose carbons in DLBCL subsets.
Figure 6. Utilization of Glucose-Derived Carbons in DLBCL Subsets.
(A) PDH enzyme activity (middle) and lactate production from glucose (bottom) in DLBCL subsets. Data are cumulative from independent DLBCL cell lines listed on top.
(B–C) 13C isotopomer analysis of uniformly labeled glucose (U13C-glucose) (B) Schematics depicting the number of carbons labeled (filled circles) in intermediary metabolites of glucose metabolism. (C) 13C enrichment in glucose-derived metabolites. For each metabolite, cumulative data obtained from all 4 OxPhos-DLBCL cell lines (Karpas 422, OCI-Ly4, Pfeiffer and Toledo; red bars) are shown relative to the mean value of that metabolite in all 4 BCR-DLBCL cell lines (SU-DHL-4, SU-DHL-6, OCI-Ly1 and OCI-Ly7; black bars).
Error bars, ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001; two-tailed Student’s t-test.
Beyond generation of pyruvate, glucose-derived metabolites are central to several biosynthetic pathways (Figure 6B). For example, glucose-6-phosphate can enter the pentose phosphate pathway to yield ribose sugars for nucleic acid synthesis and NADPH generation for lipid synthesis and ROS detoxification (Tennant et al., 2010; Vander Heiden et al., 2009). Dihydroxyacetone phosphate, which is in equilibrium with glyceraldehyde-3-phosphate, is used in glycerol synthesis, providing a necessary backbone for membrane phospholipids. Finally, glucose-derived citrate and aspartate, a surrogate for oxaloacetate (OAA), can be used in lipid and nucleotide synthesis, respectively. To examine differences in the terminal fate of glucose-derived pyruvate and the branch points at which glucose carbons divert to biosynthetic pathways in different DLBCL subsets, we carried out a targeted 13C isotopomer analysis using uniformly labeled 13C-glucose (U13C-glucose).
Glucose uptake was comparable in DLBCL cell lines as evident from the similar levels of remaining U13C-glucose in the media following 8 hr incubation (Figure 6C). The BCR cell lines displayed a larger contribution of glucose-derived carbons to the synthesis of pentose sugars compared with the OxPhos cell lines (U13C-pentose; Figure 6C). This is consistent with reduced levels of U13C-glucose-6-phosphate precursor observed in this subset (Figure 6C) provided that the oxidative branch of the pentose phosphate pathway is utilized to derive pentose sugars. In addition, the contribution of glucose carbons to glycerol incorporation into PC was significantly higher in BCR cell lines (13C-glycerol-PC; Figure 6C).
The overall glycolytic capacity was assessed by comparing both intracellular and extracellular (secreted) U13C-lactate and U13C-alanine (a surrogate for pyruvate). BCR cell lines had significantly higher intracellular and secreted U13C-alanine and U13C-lactate (Figure 6C). These data are consistent with biochemical evidence that glucose-derived lactate is higher in these cells (Figure 6A), suggesting a higher glycolytic flux.
Glucose-derived pyruvate can enter the TCA cycle through a PDH-catalyzed reaction yielding citrate or through a pyruvate carboxylase (PC)-mediated anaplerotic reaction that generates OAA (Fan et al., 2009). The ratio of PDH and PC activities in tumors is variable (DeBerardinis et al., 2007; Fan et al., 2009), suggesting significant diversity and tumor type specificity in the mode of TCA cycle entry of glucose-derived carbons. The PDH reaction can be traced by the pattern of carbon labeling in citrate as 13C2-citrate. The relative 13C enrichment in this metabolite was significantly higher in OxPhos cell lines (Figure 6C), indicating a greater overall diversion of glucose carbons to the TCA cycle. Elevated PDH-catalyzed formation of 13C2-citrate in OxPhos-DLBCLs is consistent with increased levels of PDH and enzyme activity in this subset. Among other TCA cycle intermediates measured in this analysis, 13C enrichment in aspartate (surrogate for OAA) was relatively higher in OxPhos compared with BCR cell lines (13C2-asparate, Figure 6C).
Despite elevated PDH activity in OxPhos-DLBCLs and higher enrichment of glucose carbons in the afore-mentioned TCA cycle intermediates, glucose is not fully oxidized to stimulate mitochondrial OCR in these cells (Figures 3A and S2A). This may be due to greater diversion of glucose-derived TCA intermediates to biosynthetic pathways. Overall, the distinct enrichment patterns of carbons in glucose-derived metabolites indicate relative diminution of lactate production in OxPhos-DLBCL and an attendant increase in the entry of glucose carbons in the TCA cycle via citrate.
Contribution of Mitochondrial Metabolism to Cellular ATP Budget in DLBCL Subsets
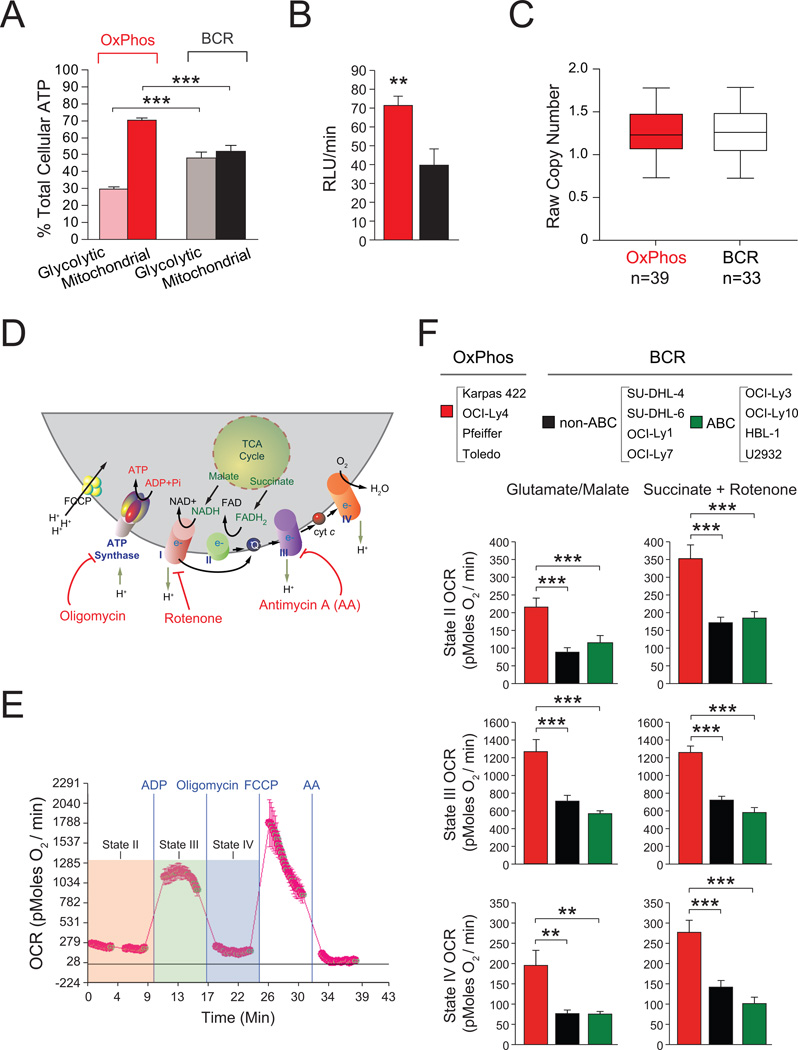
The observed differences in utilization of palmitate- and glucose-derived carbons as well as ATP generation through mitochondrial oxidation of palmitate (ATP-coupled respiration) in DLBCL subsets warranted comparison of mitochondrial and non-mitochondrial contributions to the cellular energy budget. To this end, the portion of total cellular ATP that is sensitive to inhibition of glycolysis versus mitochondrial metabolism was assessed (Guppy et al., 2002). Compared to BCR-DLBCL, the OxPhos subset derives a significantly higher portion of its total energy (~70%) from mitochondrial oxidative metabolism than from glycolysis (Figure 7A). The higher contribution of mitochondria to total cellular ATP in OxPhos-DLBCLs is concordant with increased expression of mitochondrial ATP synthase (complex V) subunits (Figures 1, S1A and Table S1), and an elevated rate of mitochondrial ATP synthesis in OxPhos-DLBCL cell lines (Figure 7B). These observations could be potentially explained by differences in mitochondrial content in DLBCL subtypes. However, assessment of the steady state content of mitochondria in DLBCL cell lines did not reveal any subtype-specific differences (Figure S5A). In addition, mitochondrial SNP copy number analysis in a cohort of primary biopsies with OxPhos or BCR assignments did not reveal any differences (Figure 7C).
Figure 7. Contribution of Mitochondrial Metabolism to Cellular ATP and Energy Transduction in DLBCL Subsets.
(A) Percent contribution of glycolysis and mitochondrial metabolism to total cellular ATP. For each subtype, cumulative data from 4 OxPhos-DLBCL (Karpas 422, OCI-Ly4, Pfeiffer, and Toledo) and 4 BCR-DLBCL (SU-DHL-4, SU-DHL-6, OCI-Ly1, and OCI-Ly7) cell lines are shown.
(B) Mitochondrial ATP synthesis rate. Cumulative data from 4 OxPhos-DLBCL (red bar) and 4 BCR-DLBCL (black bar) cell lines are shown as in (A).
(C) Average copy number of 110 mitochondrial SNP probes for 39 OxPhos-DLBCL and 33 BCR-DLBCL cases. Differences were tested using a Man-Whitney U-test and found to be non-significant.
(D–F) OCR in isolated mitochondria in different respiratory states. (D) Schematics of mitochondrial respiratory complexes and substrates as well as mitochondrial inhibitors used to measure their specific activities. (E) Representative OCR traces in mitochondria isolated from DLBCL cell lines indicating respiratory states examined as described in the Supplemental Experimental Procedures. (F) OCR in isolated mitochondria measured using complex I- or complex II-linked substrates. Data are derived from 4 cell lines per DLBCL subset listed on top.
Error bars in A, B and F, ± SEM. **p < 0.01; ***p < 0.001; two-tailed Student’s t-test.
See also Figure S5.
Increased contribution of mitochondria to total cellular ATP may not only reflect distinct channeling of carbon substrates in mitochondria but also differential activity or efficiency of mitochondrial ETC complexes. Studies in isolated mitochondria derived from DLBCL cell lines enabled direct assessment of mitochondrial respiration at the organelle level independent of the cytosolic processing of carbon substrates.
Mitochondrial respiratory states were assessed using glutamate/malate and succinate as complex I- and II-linked substrates, respectively (Figures 7D and E). When measuring complex II activity, rotenone was included with succinate to inhibit the reverse flow of electrons to complex I. Respiratory rates in the presence of substrate alone (also known as state II respiration) were higher in OxPhos-DLBCL mitochondria (Figure 7F). Addition of ADP, which mimics a state of energy demand driving high rates of respiration (also known as state III respiration) elicited significantly higher OCR in OxPhos-DLBCL mitochondria supplied with either complex I or II substrates (Figure 7F). This increased respiration is used for ATP synthesis and is sensitive to oligomycin. Accordingly, OCR values in the presence of substrate, ADP and oligomycin (state IV respiration) were also significantly higher in OxPhos- compared with BCR-DLBCLs (Figure 7F). These results provide a direct link between the OxPhos signature and an actual quantitative increase in ETC activity in this DLBCL subset. Moreover, independent biochemical measurements of mitochondrial complex I, II and IV in immunocapture assays provided corroborative biochemical evidence for significant elevation of these enzyme activities in OxPhos-DLBCLs (Figure S5B). In aggregate, our observations both at the level of intact cells (Figures 3A, 7A and 7B) and isolated mitochondria (Figures 7F and S5B) demonstrate that the OxPhos signature captures a program of mitochondrial metabolism and energy transduction that is selectively activated in this DLBCL subset.
The Relevance of ROS Content and Glutathione Synthesis in DLBCL Subtypes
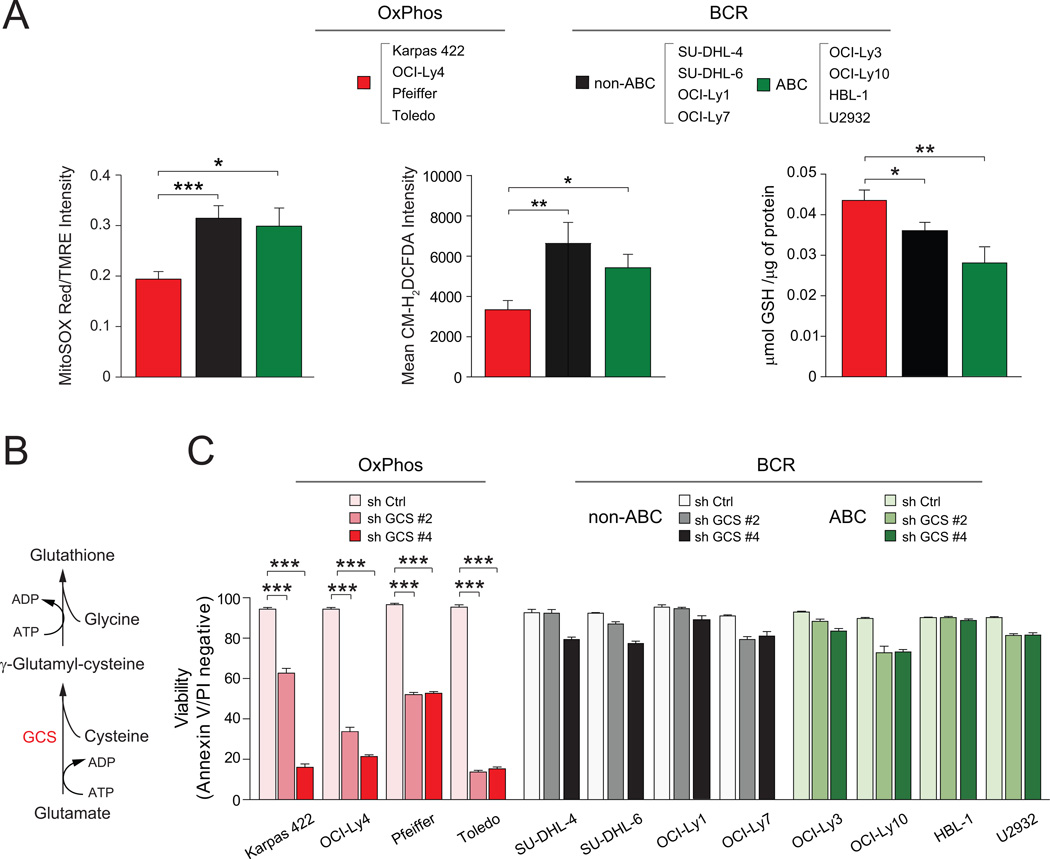
Mitochondria are a predominant source of reactive oxygen species (ROS). While ROS signaling is important for a myriad of cellular functions, excessive mitochondrial superoxide can damage mtDNA, modify proteins and lipids, and inhibit aconitase activity, thus limiting oxidative phosphorylation. Elevated ETC activity in OxPhos-DLBCL, particularly of complex I, predicted increased accumulation of mitochondrial superoxide. However, MnSOD is also concomitantly elevated in this subset (Figures 1, 2, S1A and Table S1), potentially as a mechanism to counteract increased burden of mitochondrial superoxide. Efficient clearance of ROS would also ensure that oxidative phosphorylation could remain elevated in OxPhos-DLBCLs. Assessment of mitochondrial superoxide using MitoSOX Red revealed lower steady state levels in OxPhos-DLBCLs (Figure 8A). These observations were further integrated with the total cellular ROS levels using CM-H2DCFDA, a fluorescent probe that is sensitive to oxidation by peroxyl, alkoxyl, peroxynitrite, NO2˙, CO3˙−, and OH˙ radicals and can thus serve as an indicator of overall “oxidative stress”. The CM-H2DCFDA signal intensity was also significantly lower in OxPhos-DLBCLs (Figure 8A). A potential explanation for these observations is increased diversion of peroxide generated from the SOD-catalyzed reaction into the antioxidant glutathione system. Consistent with this possibility, glutathione (GSH) levels were significantly higher in OxPhos-DLBCLs (Figure 8A), suggesting increased capacity for ROS detoxification.
Figure 8. Differential Contribution of ROS Detoxification to Survival of DLBCL Subsets.
(A) Mitochondrial superoxide (left), total cellular ROS (middle) and GSH (right) levels in DLBCL subtypes. Data are derived from 4 cell lines per DLBCL subset listed on top.
(B–C) De novo GSH synthesis pathway (B) and the effect of GCS depletion on DLBCL survival. Cell viability was assessed 72 hr after knockdown. GCS, γ-glutamyl cysteine synthase.
Error bars, ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001; two-tailed Student’s t-test.
See also Figure S6.
Given the quantitative differences in ROS and GSH content in DLBCL cell lines, we hypothesized that the capacity to maintain a large GSH pool may be required for the survival of OxPhos-DLBCLs. To test this possibility, de novo GSH synthesis was inhibited by shRNA-mediated depletion of γ-glutamyl cysteine synthase (GCS), a rate limiting enzyme that catalyzes the introduction of an amide linkage between the γ-carboxyl group of glutamate and cysteine (Figure 8B). OxPhos-DLBCL cell lines were significantly more sensitive to GCS knockdown compared with the BCR subset (Figures 8C and S6), suggesting that they may be more reliant on GSH for survival.
DISCUSSION
Our integrative analysis using proteomics, mitochondrial respirometry and metabolomics have unraveled metabolic distinctions in DLBCL subsets. We show that compared with “non-OxPhos”/BCR DLBCLs, nutrient and energy metabolism in OxPhos-DLBCL have a significant mitochondrial component, marked by elevated oxidative phosphorylation, increased contribution of mitochondria to total cellular energy budget, greater incorporation of fatty acid- and glucose-derived carbons into the TCA cycle, and increased lipogenesis from these carbon substrates. In comparison, the “non-OxPhos” DLBCLs have greater glycolytic flux. These studies also provide a clear example of heterogeneity in fuel utilization pathways even within the same disease entity.
The differential utilization of glucose- and fatty acid-derived carbons in OxPhos versus “non-OxPhos” DLBCLs appears to parallel the absence or presence of functional BCR signaling, respectively. This is consistent with our observations that acute inhibition of BCR signaling upon SYK depletion is sufficient to enhance palmitate-induced mitochondrial OCR in BCR-DLBCL cell lines. BCR-derived signals are critical for growth and survival of mature B-cells as well as multiple B-cell lymphomas (Kuppers, 2005). These signals also trigger glucose utilization in a PI3K-dependent manner that is marked by an initial increase in lactate production and a subsequent shift to the pentose phosphate pathway during progression to S phase (Doughty et al., 2006). Reduction in glycolytic flux and incorporation of glucose carbons into the pentose pool in OxPhos compared with “non-OxPhos” DLBCLs is consistent with published observations that OxPhos-DLBCLs do not display the full phospho-pattern of signaling intermediates following BCR crosslinking and are insensitive to inhibitors of BCR signaling (Chen et al., 2008).
The common metabolic profile of “non-OxPhos” DLBCLs is striking given that these tumors rely on multiple components of BCR signaling (Chen et al., 2008; Compagno et al., 2009; Davis et al., 2010). This may suggest that proliferation and survival mechanisms in these tumors may converge on glycolysis. The clear distinction between the metabolic profiles of OxPhos- and BCR-DLBCLs suggests that the specific pattern of nutrient metabolism in the former may provide an alternative survival program independent of the BCR network. Moreover, the OxPhos metabolic signature may have broader implications for other tumor types that are independent or have lost dependency on the components of BCR signaling.
Several independent approaches to inhibit the mitochondrial FAO program significantly compromised the survival of OxPhos-DLBCL. Within this context, BrCA-mediated inhibition of mitochondrial FAO and pharmacologic or genetic interference with PPARγ were selectively toxic to OxPhos-DLBCL. The relevance of PPARγ in multiple cancer models and the anti-tumor effects of its inhibition have been reported (Burton et al., 2008). Increased PPARγ activity has been recently implicated in differentiation and stimulation of antibody production in normal human B-lymphocytes (Garcia-Bates et al., 2009). Whether this is accompanied by increased FAO in normal lymphocytes remains to be determined. Nevertheless, it is possible that the metabolic signature in OxPhos-DLBCL represents pathways relevant to B-lymphocytes that are further modified to fulfill nutrient and energy requirements of these tumors.
Findings in other cancer models have suggested that FAO may serve as an alternative survival pathway that is triggered by glucose deprivation or lack of glucose uptake (Barger and Plas, 2010; Schafer et al., 2009). Moreover, a recent report showed increased FAO in solid tumors contributes to rapamycin resistance (Zaugg et al., 2011). The survival-promoting effect of fatty acid metabolism has also been reported in a leukemia model (Samudio et al., 2011). In this model, increased fatty acid metabolism was decoupled from oxidative phosphorylation, which appears to be distinct from enhanced ATP-coupled OCR in response to palmitate in OxPhos-DLBCL.
Concomitant utilization of palmitate-derived acetyl-CoA for ATP production and citrate synthesis in OxPhos-DLBCL suggests FAO and fatty acid synthesis may be concurrent pathways in these cells. This may be surprising in light of the inhibitory effect of citrate-derived malonyl-CoA on mitochondrial transport of long-chain fatty acids through carnitine palmitoyltransferase (CPT)1. However, several possibilities may explain these observations. The potency of inhibition by malonyl-CoA differs between the two CPT1 isoforms and is likely further influenced by their relative abundance (McGarry and Brown, 1997). CPT1A is ~80 fold less sensitive to malonyl-CoA than CPT1B and the ratio of CPT1A to 1B expression is higher in OxPhos-DLBCL. The relative abundance of CPT1A and the possibility that malonyl-CoA may be used rapidly prior to its accumulation may explain how mitochondrial FAO could proceed unhindered in these cells. A scenario for rapid utilization of malonyl-CoA is active fatty acid synthesis as would be expected in a proliferating cell. In addition, fatty acid synthesis may serve as a “sink” for excess acetyl CoA/citrate generated due to enhanced FAO and increased entry of acetyl-CoA into the TCA cycle. Concurrent FAO and fatty acid synthesis in other tumors has been observed (Ookhtens and Baker, 1979; Ookhtens et al., 1984). It is also interesting to note that increased FAO in AML has been linked to a quiescent pool of tumor initiating cells (Samudio et al., 2011). The precise molecular basis for concomitant use of fatty acids for ATP production and citrate synthesis in OxPhos-DLBCL awaits future studies.
Parallel activation of an antioxidant defense mechanism with increased mitochondrial FAO in OxPhos-DLBCL is intriguing. A link between FAO and antioxidant capacity has been previously suggested. For example, mitochondrial FAO may serve as a source of NADPH to regenerate GSH (Pike et al., 2010). On the other hand, a reduced cellular redox state may be important for completion of β-oxidation (Korge and Weiss, 2006; Schafer et al., 2009). It is also possible that elevated GSH in OxPhos-DLBCL is secondary to its increased synthesis from glutamate. This is consistent with higher enrichment of fatty acid-derived glutamate in this subtype. While increased GSH and MnSOD levels are in agreement with lower ROS content, additional mechanisms contributing to ROS handling in OxPhos-DLBCL cannot be excluded.
Our findings provide functional validation of quantifiable metabolic differences associated with transcriptionally-defined subsets of DLBCL. These observations also indicate that the OxPhos molecular signature is a bona fide metabolic program that is selectively activated in these lymphomas, providing distinct growth and survival benefits. Detailed fingerprints of the DLBCL metabolome may uncover important insights into the molecular pathogenesis and underlying heterogeneity of these lymphomas as well additional roadmaps to subtype-specific therapeutic targets.
EXPERIMENTAL PROCEDURES
Primary DLBCL Biopsies
Protein samples were prepared from frozen biopsy specimens of newly diagnosed, previously untreated primary DLBCLs with > 80% tumor involvement according to Institutional Review Board-approved protocols from two institutions (Brigham & Women Hospital and Dana-Farber Cancer Institute). A waiver to obtain informed consent was granted by the IRBs because otherwise discarded tissue was used. The frozen biopsy specimens from which total RNA and high molecular weight DNA were extracted for transcript abundance and SNP analysis have been described in a recent study (Monti et al., 2012).
Sample Preparation and Labeling for iTRAQ Analysis
Mitochondria isolated from DLBCL cell lines or Trizol-purified primary biopsy proteins were solubilized in 7.2 M guanidine hydrochloride, 100 mM ammonium bicarbonate. Protein concentration was determined by the Bradford assay (Bio-Rad, Hercules, CA) and equal amounts of protein were reduced with 10 mM DTT for 30 min at 56°C, and alkylated with 22.5 mM iodoacetamide for 30 min at room temperature in the dark. DTT was added to a final concentration of 20 mM to quench the remaining iodoacetamide. Proteins were digested overnight with trypsin (1:20) at 37°C after addition of 100 mM ammonium bicarbonate solution to dilute the concentration of guanidine HCl to 1 M. Digests were acidified with 10% TFA and desalted by C18. Aliquots of peptides were stored at −80°C.
Mitochondria derived peptides (50 µg) from 3 OxPhos (Karpas 422, Toledo, and Pfeiffer) and 3 BCR (OCI-Ly1, SU-DHL-4, and SU-DHL-6) cell lines were solubilized in 100 µL of 30% 500 mM triethylammonium bicarbonate, pH 8.5/70% ethanol and 1 unit of iTRAQ 8-plex reagent was added to each sample (Karpas 422-113, Toledo-114, Pfeiffer-115, OCI-Ly1-117, SU-DHL-4-118, SU-DHL-6-119; note 116 and 121 reagents were not used). The solution was incubated for 1 hr at room temperature, and the reactions were combined and dried by vacuum centrifugation. Labeled peptides were desalted by C18, dried again by vacuum centrifugation, and stored at −80°C. The same procedure was used to label 100 µg aliquots of primary DLBCL biopsy-derived peptides with iTRAQ 4-plex reagent [biopsy #32 (OxPhos)-114; biopsy #39 (OxPhos)-115; biopsy #42 (BCR)-116)]. iTRAQ-labeled peptide samples were analyzed and data processed as described in supplemental experimental procedures.
Mitochondrial Respirometry
OCR was measured in real time using the XF24 extracellular flux analyzer instrument and the AKOS algorithm v1.5.069 software (Seahorse Bioscience Inc., Chicopee, MA). For whole cell studies, cells were seeded on XF24 V7 plates coated with Cell-Tak (BD Bioscience) at 3×105 cells/well in 600 µl of sodium bicarbonate-free RPMI medium alone or supplemented with 10 mM glucose, 2 mM glutamine or 0.2 mM palmitate. When palmitate was tested, 0.5 mM carnitine was included in the incubation/equilibration medium to ensure import of BSA-conjugated palmitate into mitochondria. The plates were spun at 500 rpm (breaks off) and incubated at 37°C for 10 min to ensure cell attachment. Measurements were taken for a total of 60 min, including a 12 min incubation period prior to starting baseline measurements. Within this assay time, OCR was measured for 3 min periods with 5 intervals between measurements. After baseline measurements, 2.5 µM oligomycin was added in a single 75 µl injection. In all experiments, parallel samples were run in the absence of oligomycin to ensure stable baselines as a quality control parameter for the bioenergetics health of the cells. OCR in isolated mitochondria was determined as detailed in supplemental experimental procedures.
13C Isotopomer-Based Metabolomics Analysis
10×106 cells were incubated for 8 hr at 37°C in glutamine-free un-buffered RPMI medium containing 10 mM malate, 0.5 mM carnitine, 10 mM β-hydroxybutyrate, 10% (v/v) FBS supplemented with 10 mM glucose, 4 mM glutamine, and 0.2 mM BSA-conjugated palmitate. When tracing glucose or palmitate carbons, the media was supplemented with U-13C-glucose or U-13C-palmitate isotopomers (Cambridge Isotope Laboratories) to the final concentrations indicated above. Cell pellets and 1 ml aliquots of media were frozen on dry ice and processed for LC-MS/MS and GC/MS as detailed in supplemental experimental procedures.
Mitochondrial Copy Number Analysis
The mitochondrial copy number was assessed using the mitochondrial SNP probes within the Affymetrix HD-SNP array 6.0 data from an independent series of primary DLBCL samples (Monti et al., 2012, GEO accession number GSE34171). The pre-segmentation data processing was performed according to the SNParray 6.0 analytical pipeline described in (TCGA, 2008). Gene expression profiling of the same samples allowed assignment of consensus cluster subtypes (Monti et al., 2012, GEO accession number GSE34171). Subsequently, the average copy number of 110 mitochondrial SNP probes was computed and visualized using a box plot for the 33 “BCR” and 39 “OxPhos” samples. Differences were tested using a Man-Whitney U-test (Dawson-Saunders and Trapp, 1994).
Statistics
Unless otherwise indicated statistical analysis was performed using two-tailed student’s t-test, assuming unequal variance. Transcript abundance was visualized with box plots (median, line; 25% and 75% quartile, box; whiskers, minimum to maximum).
Supplementary Material
Highlights.
Glucose and fatty acids are differentially utilized in molecular subsets of DLBCL
Mitochondria are predominant in the metabolic signature of BCR-independent DLBCLs
Fatty acid oxidation and glutathione synthesis are relevant to OxPhos-DLBCL survival
Fuel utilization pathways define additional levels of heterogeneity in DLBCL
SIGNIFICANCE.
B-cell receptor (BCR) survival signals are central to pathogenesis of certain DLBCLs. Among transcriptionally-defined groups of DLBCL, the “OxPhos” subset displays increased expression of genes involved in mitochondrial oxidative phosphorylation but lacks an intact BCR signaling network, suggesting dependence on alternative survival mechanisms. However, the functional basis of the OxPhos signature has not been defined. Using an integrative approach, we have carefully dissected the patterns of fuel utilization and energy metabolism in DLBCL with a primary focus on the “OxPhos” subset. We show fatty acid oxidation and glutathione synthesis are distinct metabolic features of OxPhos-DLBCLs selectively required for their survival. Metabolic signatures may provide important insights into the molecular heterogeneity of DLBCL and reveal rational targets in these lymphomas.
ACKNOWLEDGMENTS
We thank Eric Smith for manuscript preparation. We gratefully acknowledge George Rogers, Martin Brand, David Ferrick, Min Wu and Orian Shirihai for advice on respirometry; Wei Jiang and Frank Cook for assistance with LC-MS/MS and GC/MS; and Georg Lenz and Frank Stegmeier for HBL-1 and U2932 DLBCL cell lines. A.U.K was supported by Alexandra Jane Miliotis Fellowship in Pediatric Oncology, the IDEA^2 Program Grant from the Harvard-MIT Division of Health Sciences and Technology and a Medical Student Research Training Fellowship from HHMI. E.N. is supported by a post doctoral fellowship from The Swedish Research Council. This work was supported in part by funding from the Novartis Institutes for Biomedical Research (N.N.D.), NIH PO1 CA092625 (M.A.S) and the German Research Foundation DFG Ch 735/1-1 (B.C.). The authors acknowledge generous support provided through the DFCI strategic research initiative (to J.A.M.). N.N.D. is a recipient of the Burroughs Welcome Fund Career Award in Biomedical Sciences and a consultant for the Novartis Institutes for Biomedical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Experimental Procedures include LC-MS/MS analysis of iTRAQ-labeled samples and data processing, classification of primary tumor biopsies and cell lines based on RNA profiles, mitochondrial isolation and respirometry, metabolomics analysis, knockdown studies, measurements of energy budget, mitochondrial ATP synthesis rate, lactate, PDH activity, ROS and GSH content.
REFERENCES
- Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Barger JF, Plas DR. Balancing biosynthesis and bioenergetics: metabolic programs in oncogenesis. Endocr Relat Cancer. 2010;17:R287–R304. doi: 10.1677/ERC-10-0106. [DOI] [PubMed] [Google Scholar]
- Burton JD, Goldenberg DM, Blumenthal RD. Potential of peroxisome proliferator-activated receptor gamma antagonist compounds as therapeutic agents for a wide range of cancer types. PPAR Res. 2008;2008 doi: 10.1155/2008/494161. 494161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Monti S, Juszczynski P, Daley J, Chen W, Witzig TE, Habermann TM, Kutok JL, Shipp MA. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111:2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson-Saunders B, Trapp R. Basic and Clinical Biostatistics. Norwalk, CT: Appleton & Lange; 1994. Basic and Clinical Biostatistics. 1994. [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, Chiles TC. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood. 2006;107:4458–4465. doi: 10.1182/blood-2005-12-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Aleem SA, Schulz H. Evaluation of inhibitors of fatty acid oxidation in rat myocytes. Biochem Pharmacol. 1987;36:4307–4312. doi: 10.1016/0006-2952(87)90676-9. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bates TM, Baglole CJ, Bernard MP, Murant TI, Simpson-Haidaris PJ, Phipps RP. Peroxisome proliferator-activated receptor gamma ligands enhance human B cell antibody production and differentiation. J Immunol. 2009;183:6903–6912. doi: 10.4049/jimmunol.0900324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. 2002;364:309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao G, Chapman J, Ofrecio JM, Wilkes J, Resnik JL, Thapar D, Subramaniam S, Sears DD. Multi-tissue, selective PPARgamma modulation of insulin sensitivity and metabolic pathways in obese rats. Am J Physiol Endocrinol Metab. 2011;300:E164–E174. doi: 10.1152/ajpendo.00219.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Korge P, Weiss JN. Redox regulation of endogenous substrate oxidation by cardiac mitochondria. Am J Physiol Heart Circ Physiol. 2006;291:H1436–H1445. doi: 10.1152/ajpheart.01292.2005. [DOI] [PubMed] [Google Scholar]
- Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Elwood F, McNally J, Weiszmann J, Lindstrom M, Amaral K, Nakamura M, Miao S, Cao P, Learned RM, et al. T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J Biol Chem. 2002;277:19649–19657. doi: 10.1074/jbc.M200743200. [DOI] [PubMed] [Google Scholar]
- Leesnitzer LM, Parks DJ, Bledsoe RK, Cobb JE, Collins JL, Consler TG, Davis RG, Hull-Ryde EA, Lenhard JM, Patel L, et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- Monti S, Chapuy B, Takeyama K, Rodig S, Hao Y, Yeda K, Inguilizian H, Mermel C, Curie T, Dogan A, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B-cell lymphoma. Cancer Cell. 2012 doi: 10.1016/j.ccr.2012.07.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- Moreno-Sanchez R, Rodriguez-Enriquez S, Saavedra E, Marin-Hernandez A, Gallardo-Perez JC. The bioenergetics of cancer: is glycolysis the main ATP supplier in all tumor cells? Biofactors. 2009;35:209–225. doi: 10.1002/biof.31. [DOI] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookhtens M, Baker N. Fatty acid oxidation to H2O by Ehrlich ascites carcinoma in mice. Cancer Res. 1979;39:973–980. [PubMed] [Google Scholar]
- Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol. 1984;247:R146–R153. doi: 10.1152/ajpregu.1984.247.1.R146. [DOI] [PubMed] [Google Scholar]
- Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2010;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Polo JM, Juszczynski P, Monti S, Cerchietti L, Ye K, Greally JM, Shipp M, Melnick A. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2011;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.