Abstract
Continual and rapid mutation of seasonal influenza viruses by antigenic drift necessitates the almost annual reformulation of flu vaccines, which may offer little protection if the match to the dominant circulating strain is poor. S139/1 is a cross-reactive antibody that neutralizes multiple HA strains and subtypes, including those from H1N1 and H3N2 viruses that currently infect humans. The crystal structure of the S139/1 Fab in complex with the HA from the A/Victoria/3/1975 (H3N2) virus reveals that the antibody targets highly conserved residues in the receptor binding site and contacts antigenic sites A, B, and D. Binding and plaque reduction assays show that the monovalent Fab alone can protect against H3 strains, but the enhanced avidity from binding of bivalent IgG increases the breadth of neutralization to additional strains from the H1, H2, H13, and H16 subtypes. Thus, antibodies making relatively low affinity Fab interactions with the receptor binding site can have significant antiviral activity when enhanced by avidity through bivalent interactions of the IgG, thereby extending the breadth of binding and neutralization to highly divergent influenza virus strains and subtypes.
Influenza virus is the etiologic agent responsible for seasonal flu and sporadic pandemics and remains a significant health and economic burden by infecting millions each year. Hemagglutinin (HA), the major surface glycoprotein on influenza virus, facilitates virus entry and infection of host cells by binding sialic acid receptors on the surface of endothelial cells, thereby promoting virus entry into endosomes (1, 2). HA exists in 17 distinct subtypes (primarily in birds), which can be split into two major groups by phylogeny (3, 4) and are classified (H1–H17) by their uniqueness of reactivity against polyclonal antisera. Group 1 is comprised of subtypes H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, and the recently identified H17 (5), whereas the H3, H4, H7, H10, H14, and H15 subtypes form group 2. Annual vaccines against HA are administered as a countermeasure against influenza and are composed of a mixture of representative H1, H3, and influenza B strains that are selected to match the prevailing or anticipated circulating strains. However, the effectiveness of vaccines heavily relies on the match of the dominant circulating virus to the vaccine strains (6). Additionally, the influenza virus rapidly mutates and can escape the host immune response if sufficient viable HA mutations are incorporated to mask the surface from previously elicited antibodies (7, 8). Thus, a vaccine that provides protection by eliciting an antibody response against multiple HA subtypes may potentially combat a much larger range of strains and subtypes of influenza viruses (9).
The HA protein is trimeric in structure and is composed of three identical copies of a single HA0 polypeptide precursor, which upon proteolytic maturation, is cleaved to produce a pH-dependent, metastable intermediate, comprised of HA1 and HA2 subdomains that serve distinct roles in viral infection (10). The membrane distal “head” is composed entirely of HA1 residues and contains the receptor binding site that is used for recognition of sialic acid receptors on host cells (1, 2). The membrane proximal “stem” is assembled from HA2 and some HA1 residues and contains the fusion machinery that is triggered in the low pH environment of late endosomes (11, 12). To inhibit viral infection, antibodies can impede viral attachment to host cells by sterically blocking either receptor binding (13–16), preventing the low pH-induced conformational change (14, 17–19), or interfering with the maturation of HA0 to HA1 and HA2 (18, 20). The HA stem is highly conserved and antibody recognition against this region has been shown to be extremely broad, with neutralization reported against almost all strains within the subtypes from group 1 (17, 21–24), group 2 (18), or both (19, 20). However, eliciting high levels of these stem-directed antibodies by vaccination remains a challenge, either because of poor immunogenicity, mode of immunization, or more restricted access to the HA stem, but recent studies have suggested that such antibodies are produced in some individuals (25, 26) and can be enhanced by DNA prime-boost methods (27). In contrast, HA1 is usually highly immunogenic for most subtypes except H5 (28), although the breadth of neutralization of head-targeted antibodies has generally been poor because of the hypervariability of the residues that surround the receptor binding site (7, 8).
Despite the overall sequence variability of HA1, the receptor binding site is relatively conserved as it is constrained to preserve its receptor-binding function. Broadly neutralizing antibodies that specifically target the receptor binding site have been rare, perhaps in part because of its relatively small footprint. S139/1 was the first antibody to be described with heterosubtypic reactivity, neutralizing strains from multiple subtypes, such as H1, H2, H3, and H13 (29), that cross the HA group barrier. A few other recent reports have described a number of broadly neutralizing antibodies that map to the apex of HA close to or at the receptor binding site (29–32), such as CH65, an antibody specific to the H1 subtype (16), as well as C05, which has activity against multiple subtypes (33). Here, we report the crystal structure of the S139/1 Fab in complex with A/Victoria/3/1975 (H3N2) (Vic75/H3) HA and show that S139/1 achieves heterosubtypic neutralization by targeting the receptor binding site on HA. Furthermore, we show that, although Fab is sufficient for neutralization of H3 isolates, S139/1 is unusually dependent upon avidity for heterosubtypic neutralization. Bivalent binding of the IgG significantly boosts the affinity compared with the Fab and is correlated closely with the antibody’s neutralization potential. These findings suggest that antibodies against the HA receptor binding site may possess much greater cross-reactivity than was previously appreciated, and the use of avidity to extend neutralization breadth may be a general feature of many antibodies targeting highly variable surface glycoproteins of viruses, such as influenza and HIV.
Results
S139/1 IgG Binding Correlates with Neutralization.
Binding of S139/1 Fab and IgG against a panel of 35 HAs within 13 subtypes was assessed by bio-layer interferometry using an Octet RED instrument (n.b. subtypes H8, 9, 11, and the recently identified H17 were not included in this study) (Table 1 and Table S1). The HAs included in the study were selected to be diverse and encompass human viruses that caused significant disease, such as the 1918 and 2009 H1, 1957 H2, and 1968 H3 pandemic strains. Wherever possible, H1 and H3 strains that have been used in human vaccines were selected to approximate the antigenic breadth that circulated in the human population over the last century. The binding data confirm that S139/1 binds all of the previously reported neutralized strains and also A/turkey/Massachusetts/3740/1965 (H6N2) (Mass65/H6), which is recognized with measurable affinity (250 nM) by the bivalent IgG, but not monovalent Fab. Strikingly, this antibody displays an unusually wide range of binding affinities against neutralized viruses, with Kd values ranging from ∼2 µM to ∼10 nM for binding of monovalent Fab to immobilized HA, whereas most reported neutralizing antibodies against HA (including antibodies against the stem) have Fab Kd values in the low nanomolar range (18, 19). In general, bivalent binding of S139/1 IgG increases the apparent affinity to the HA by roughly three orders of magnitudes compared with the Fab.
Table 1.
Binding and neutralization breadth of influenza HA strains by S139/1 Fab and IgG
| Binding |
Neutralization |
|||
| Isolate | Fab Kd (nM) | IgG Kd (nM) | Fab IC50 (µg/mL) | IgG IC50 (µg/mL) |
| A/Aichi/2/1968 (H3N2) | N.T. | N.T. | 0.11 | 0.0034 |
| A/Hong Kong/1/1968 (H3N2) | 14 | 0.4 | N.T. | N.T. |
| A/Victoria/3/1975 (H3N2) | 20 | 0.011 | N.T. | N.T. |
| A/WSN/1933 (H1N1) | 1,800 | 13 | 2.66 | 0.033 |
| A/Adachi/2/1957 (H2N2) | 1,700 | 6.2 | 33.9 | 0.82 |
| A/gull/Maryland/704/1977 (H13N6) | 1,800 | 9.5 | >200 | 1.33 |
| A/black-headed gull/Sweden/5/1999 (H16N3) | N.T. | N.T. | >200 | 4.29 |
| A/turkey/Massachusetts/3740/1965 (H6N2) | >1,000 | 250 | >200 | >100 |
N.T., not tested.
Because the monovalent S139/1 affinity for HA is so variable among neutralized strains, we postulated that bivalent antibody binding (i.e., simultaneous engagement of both Fab arms of an IgG, likely spanning HA trimers on the virus surface) is essential for efficient neutralization, particularly for lower affinity targets. Plaque reduction assays were performed using both S139/1 Fab and IgG against previously known neutralized strains and subtypes, as well as the Mass65/H6 strain. For all strains tested, neutralization by S139/1 IgG was significantly more potent than by S139/1 Fab (the IC50 for IgG was typically ∼100-fold lower than for Fab). A/Aichi/2/1968 (H3N2) (Aichi68/H3), which has high sequence similarity to the A/Hong Kong/1/1968 (H3N2) (HK68/H3) and Vic75/H3 strains in the binding panel (Fig. S1), was neutralized by S139/1 Fab, but more potently neutralized by the IgG (Table 1). On the other hand, all other viruses tested were much more effectively neutralized by the IgG except for the Mass65/H6 strain, which bound with weaker affinity for both Fab and IgG and was not neutralized, suggesting a threshold for neutralization at a Kd of < ∼250 nM for either Fab or IgG. Additionally, we discovered that S139/1 IgG neutralizes A/black-headed gull/Sweden/5/1999 (H16N3) (Sweden/5/99/H16), extending the cross-reactivity of the antibody to the H16 subtype, although no binding was detected to another H16 strain, A/black-headed gull/Sweden/4/1999 (H16N3) (Sweden/4/99/H16), likely because of sequence variation in and around the binding site (Fig. S2).
Structural Characterization of the S139/1-Vic75/H3 Complex.
To investigate the structural basis of S139/1 recognition, the crystal structure of the S139/1 Fab in complex with Vic75/H3 was determined at 2.95 Å resolution (Table 2). The asymmetric unit contains three HA copies that form a biological trimer with one Fab bound to each HA protomer (Fig. 1A). The Vic75/H3 trimer and the variable heavy and light Ig domains of the complex are well ordered and are structurally similar to other unliganded H3 models in the PDB and to the apo S139/1 Fab model that we also determined to 2.05 Å (Table 2). The combining site interface between the HA and the Fab variable domains is structurally conserved between all protomers in the trimer; however, the constant domain positions vary for each Fab molecule because of the flexibility of the Fab elbow linking the variable and constant domains.
Table 2.
X-ray data collection and refinement statistics
| Data collection and refinement | S139/1-Vic75/H3 complex | S139/1 Fab |
| Data collection | ||
| Beamline | APS 23-ID-D | APS 23-ID-D |
| Wavelength (Å) | 1.0333 | 1.0333 |
| Space group | C2 | P3212 |
| Unit cell parameters | a = 225.5, b = 112.9, c = 197.0 | a = b = 106.8, c = 185.5 |
| (Å, °) | α = γ = 90.0, β = 118.8 | α = β = 90.0, γ = 120.0 |
| Resolution (Å) | 50–2.95 (3.05–2.95)* | 50–2.05 (2.15–2.05)* |
| Observations | 343,980 | 564,829 |
| Unique reflections | 91,317 (8,658)* | 76,062 (10,038)* |
| Redundancy | 3.8 (3.8)* | 7.4 (7.2)* |
| Completeness (%) | 99.8 (99.9)* | 99.8 (99.9)* |
| <I/σI> | 17.1 (2.1)* | 16.0 (2.3)* |
| Rsym† | 0.05 (0.46)*† | 0.08 (0.72)*† |
| Rpim† | 0.03 (0.27)*† | 0.03 (0.27)*† |
| Za‡ | 3 | 2 |
| Refinement statistics | ||
| Resolution (Å) | 48.4–2.95 | 46.3–2.05 |
| Reflections (work) | 91,228 | 75,878 |
| Reflections (test) | 4,571 | 4,006 |
| Rcryst (%)§ | 20.8§ | 15.8§ |
| Rfree (%)¶ | 23.3¶ | 20.9ll |
| Protein atoms | 20,687 | 6,594 |
| Carbohydrate atoms | 627 | 42 |
| Waters | 31 | 44 |
| Other | 115 | 10 |
| Average B-value (Å2) | ||
| Overall | 106.2 | 54.1 |
| HA | 85.1 | – |
| Fab | 128.9 | 53.9 |
| Fab variable domain** | 117.1 | 48.4 |
| Fab constant domain†† | 145.1 | 60.3 |
| Carbohydrate | 151.6 | 103.2 |
| Wilson | 98.4 | 41.1 |
| RMSD from ideal geometry | ||
| Bond length (Å) | 0.009 | 0.008 |
| Bond angles (°) | 1.18 | 1.19 |
| Ramachandran statistics (%)‡‡ | ||
| Favored | 96.5 | 96.9 |
| Outliers | 0 | 0 |
| PDB ID code | 4GMS | 4GMT |
*Numbers in parentheses refer to the highest resolution shell.
†Rsym = Σhkl Σi|Ihkl,i − <Ihkl>|/Σhkl Σi Ihkl,I and Rpim = Σhkl [1/(n − 1)]1/2 Σi|Ihkl,i − <Ihkl>|/Σhkl Σi Ihkl,I, where Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l, <Ihkl> is the average intensity for that reflection, and n is the redundancy. Note that despite the high Rsym in the highest resolution shell, the high redundancy enables the average F’s to be well determined (assuming measurement errors are randomly distributed), as reflected by the redundancy-independent measure of the quality of intensity measurements Rpim.
‡Za is the number of HA monomer-Fab complexes or Fabs per crystallographic asymmetric unit.
§Rcryst = Σhkl|Fo – Fc|/Σhkl|Fo| × 100
¶Rfree was calculated as for Rcryst, but on a test set comprising 5% of the data excluded from refinement.
llRfree flags were reassigned for this twinned dataset and was calculated as for Rcryst, but on a test set comprising 5.3% of the data excluded from refinement.
**Residues 1–109 of the light chain and 1–113 of the heavy chain.
††Residues 110–214 of the light chain and 114–214 of the heavy chain.
‡‡Calculated using Molprobity (42).
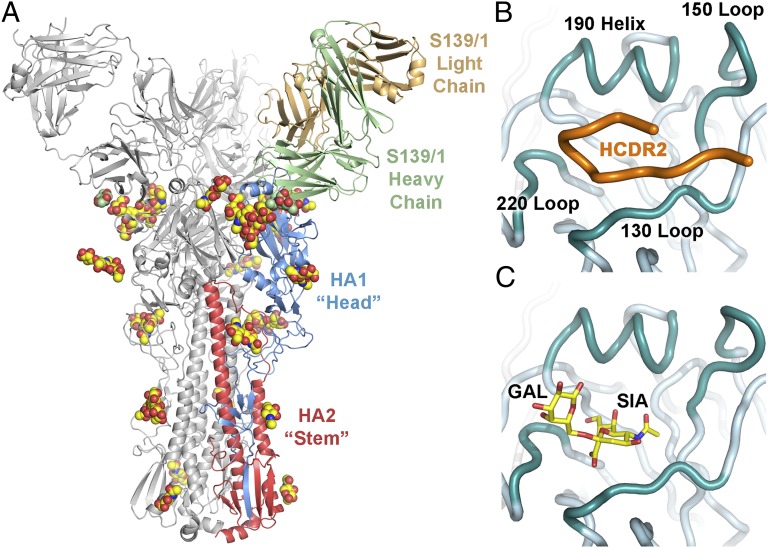
Fig. 1.
S139/1 binds to the receptor binding site of HA. (A) Crystal structure of S139/1 Fab in complex with Vic75/H3 HA. One Fab-HA protomer of the trimeric complex is colored with HA1 in blue, HA2 in red, the Fab heavy chain in green, and the Fab light chain in light orange. The other two Fab-HA protomers are colored gray. The glycans are represented as spheres with carbon in yellow for HA or green for Fab, oxygen in red, and nitrogen in blue. (B) S139/1 inserts its HCDR2 (colored in orange) into the HA receptor binding site and (C) overlaps with the human α2,6 sialoglycan receptor (PDB ID code 1MQN).
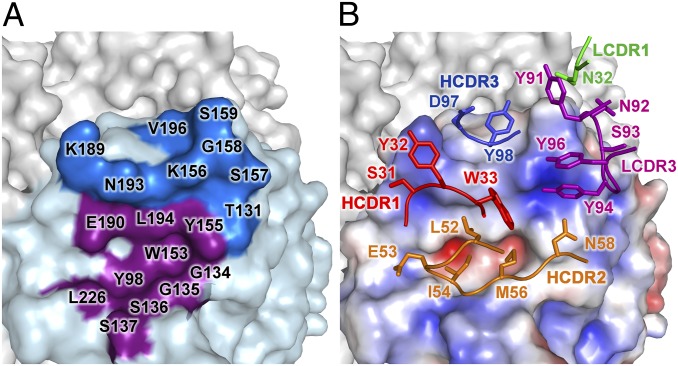
Each S139/1 Fab binds the globular head of a single Vic75/H3 HA protomer at and around the receptor binding site, contacting residues from multiple antigenic sites, A, B and D, as designated for H3 (the equivalent for H1 are Sa and Sb) (7, 8). The receptor binding site is a broad and shallow pocket framed by four loops that form the outer ridges, denoted as the 130 loop, the 150 loop, the 190 helix, and the 220 loop (Fig. 1B). All four of these receptor binding site components are contacted by five of the six complementarity determining regions (CDRs) of S139/1 (Fig. 2). On average, a total of 1,447 Å2 of surface area is buried in the interaction (728 Å2 on HA and 719 Å2 on the Fab), where the heavy and light chains contribute to 78% and 22% to the overall buried surface, respectively. S139/1 has a unique binding mode in that it uses HCDR2 to insert into the receptor binding site to compete with receptor (34) (Fig. 1 B and C) in contrast to C05 (33), CH65 (16), and HC19 (35), which use HCDR3 (Fig. S3). HCDR2 contributes to 43% of the interface, primarily using hydrophobic residues LeuH52, IleH54, and MetH56 to contact HA Trp153 and Leu194 (Fig. 2), which are highly conserved residues involved in receptor binding and are present in 99.9% and 97.5% of HAs from the 17 subtypes [13,627 sequences from the Influenza Virus Resource at the National Center for Biotechnology Information (NCBI) database], respectively. The complex is further stabilized by aromatic residues in LCDR3 and HCDR3, which contact the 150 loop and the 190 helix, respectively. These three CDRs together contribute 80% of the total Fab buried surface area. Correspondingly, the binding site interfaces of HA are dominated by the 150 loop and the 190 helix, which are enveloped by S139/1. Around 75% of buried surface of the epitope on the HA is contributed solely by these two loops, divided evenly, and are therefore the major HA antigenic determinants for S139/1 binding, at least in terms of buried surface.
Fig. 2.
Interaction of S139/1 with Vic75/H3 illustrating the neutralizing epitope. (A) Surface representation of the receptor binding site region with the residues of the HA epitope adjacent to the receptor binding site in dark blue. Residues in the receptor binding site also contacted by S139/1 are colored in purple. (B) The electrostatic potential surface of HA is illustrated (red, negative; blue, positive; white, neutral) in the same orientation as A, with the S139/1 contacting residues in the five CDRs that interact with HA represented as sticks and color-coded by CDR. Interacting residues on the HA and the antibody are labeled by amino-acid letter and residue number.
S139/1 has an N-glycosylation site at Asn72, which is in framework region 3 (FR3) of the heavy chain. In the complex, the S139/1 Fab glycan is positioned away from the combining site, but is proximal to the glycan at HA1 Asn165 on an adjacent HA protomer of the trimer (Fig. S4). However, glycan–glycan interactions do not appear to contribute significantly to binding, because the Kd values measured for glycosylated and unglycosylated S139/1 Fabs are essentially equivalent (Table S2).
Sequence Analysis of the S139/1 Epitope.
To understand the breadth of the cross-reactivity of S139/1, the sequences from the HA panel were analyzed at the antibody contact positions. The major binding determinants arise from both the sequences and conformations of the 130 loop and 150 loop (Table 3 and Table S3). Although the 130 loop does not contribute to a large proportion of the neutralizing interface, its conformation greatly influences binding. A common amino acid insertion at the 133a position (between HA1 residues 133 and 134), which is not present in the Vic75/H3 strain, creates a localized change in the 130 loop conformation around the insertion site, causing it to bulge and clash into S139/1 (Fig. 3A), which is similar in the case of antibodies CH65 and C05 (16, 33). Additionally, the side chain of residue 133a is oriented into the space where the antibody would bind, giving further credence that this insertion would disrupt antibody binding. This insertion exists in ∼75% of H1, ∼94% of H5, and all H6 and H10 strains, and thus enables these strains and subtypes to evade S139/1 recognition.
Table 3.
HA sequence variation at the S139/1 epitope
| Isolate | 133a | 156 | 158 | 150 loop insertion | 193 |
| A/Aichi/2/1968 (H3N2) HA used to raise S139/1 | – | K | G | No | S |
| A/Hong Kong/1/1968 (H3N2) | – | – | – | – | – |
| A/Victoria/3/1975 (H3N2) | – | – | – | – | N |
| A/WSN/1933 (H1N1) | – | – | – | – | – |
| A/Adachi/2/1957 (H2N2) | – | – | – | – | T |
| A/gull/Maryland/704/1977 (H13N6) | – | – | N | – | T |
| A/black-headed gull/Sweden/5/1999 (H16N3) | – | – | K | Yes | N |
| A/turkey/Massachusetts/3740/1965 (H6N2) | S | – | S | Yes | T |
Identical residues in comparison with A/Aichi/2/1968 (H3N2) HA are denoted by a hyphen. Unfavorable residues are shaded in black and tolerable residues are shaded in gray.
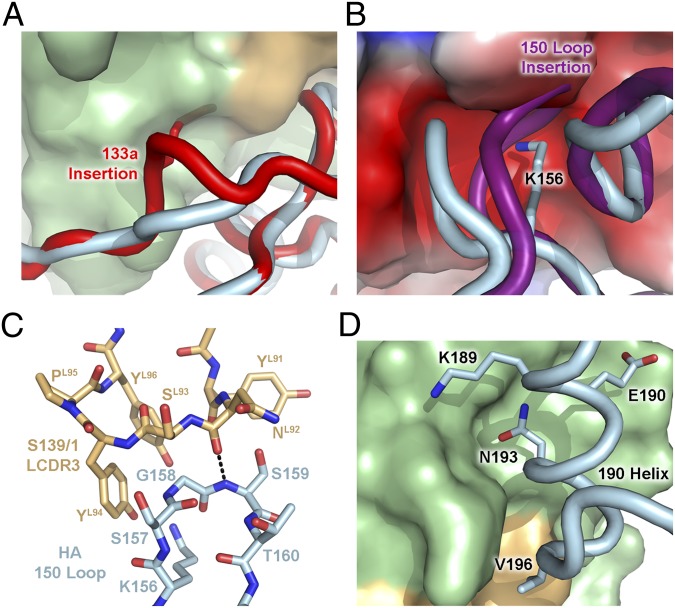
Fig. 3.
Tightly constrained networks of interactions regulate S139/1 binding. (A) Steric effects of the 133a insertion. Receptor binding site residues around position 133 of the A/California/04/2009 (H1N1) HA (PDB ID code 3LZG) are shown as a backbone tube colored in red and aligned with the corresponding residues in Vic75/H3 (light blue). The side chain of the 133a residue is shown as red sticks on the red backbone and would clash with the S139/1 antibody, shown as a surface with the heavy and light chains colored in green and light orange, respectively. (B) Steric effects and electrostatics of the 150 loop. The 150 loop of A/turkey/Italy/2002 (H7N3) HA (PDB ID code 1TI8) is colored in purple and is aligned with Vic75/H3 (light blue), and likely would clash with the S139/1 antibody. The electrostatic surface potential (red, negative; blue, positive; white, neutral) of S139/1 is shown. The Lys156 side chain of Vic75/H3 is depicted as sticks. (C) LCDR3 and the 150 loop are depicted as sticks. The main-chain hydrogen bond between Ser159 and AsnL92 is indicated by dashed lines. (D) The 190 helix packs against the HCDR1-3 loops of S139/1. Interacting residues of Vic75/H3 are shown as sticks.
The sequences and conformation of the 150 loop also determines S139/1 binding. Lys156 of Vic75/H3 forms electrostatic interactions by inserting into an acidic pocket in the Fab formed by GluH35 and GluH50 (Fig. 3B). Evidence for the dependency of this interaction for binding is observed when comparing the A/Japan/305/1957 (H2N2) (Japan57/H2) and A/Adachi/2/1957 (H2N2) (Adachi57/H2) strains, which have identical binding site residues, except for Glu156 or Lys156, respectively. Electrostatic repulsion, as in the case of Japan57/H2, results in no S139/1 binding, whereas Adachi57/H2 is recognized. Another 150-loop binding determinant is Gly158, which is closely stacked on LCDR3 and stabilized by a main-chain hydrogen bond between Ser159 and AsnL92. Any substitution at Gly158 would result in clashes at the antibody interface (Fig. 3C). The most severe example of steric interference with S139/1 binding comes from an N-linked glycan at position 158, which is present in ∼37% of H5 strains (1,021 of 2,738 sequences from the Influenza Virus Resource at the NCBI database). Finally, similar to the conformation changes of the 130 loop caused by 133a, insertions in the 150 loop (around the HA1 158 position) also correlate with poor S139/1 binding. Because the 150 loop is heavily buried by the antibody and the sequences at positions 156 and 158 are critical for antibody recognition, insertions will distort this loop and result in a steric clash with S139/1 (Fig. 3B). The 150-loop insertions are seen in nearly all strains from the H4, H6, H7, H8, H10, H14, and H15 subtypes.
Although the key HA recognition elements can be attributed to the 130 loop and the 150 loop, the 190 helix also plays a major role as it contributes to 37% of the buried surface area. The interacting residues in this helix are quite variable among the HA strains in the panel, but S139/1 overcomes this potential problem by contacting residues with high-sequence conservation and low variability in the 190 helix. For example, residue 190 is a Glu or the isoelectric Asp, which are conserved ∼48% or ∼43% in all HA strains, respectively. Leu194 is highly conserved and contacts hydrophobic residues of HCDR2. S139/1 also evades diversity in the 190 helix by orienting away from variable surface-exposed residue positions, such as Lys189 and Val196 (Fig. 3D). On the other hand, Asn193 is hypervariable and is buried by HCDR1-3 limiting mutational freedom at this position.
Discussion
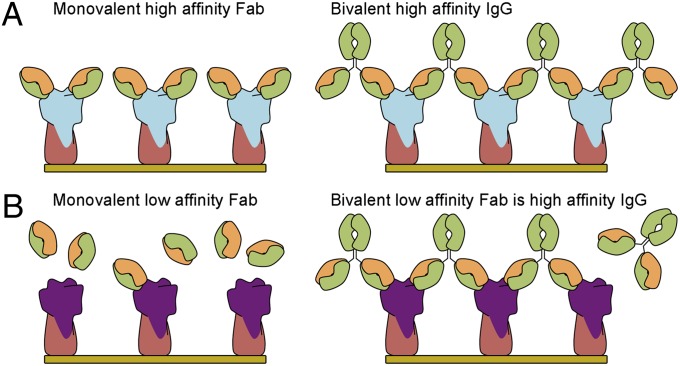
In this study, we show that antibody S139/1 covers the HA receptor binding site and inserts its HCDR2 into the shallow groove where the endogenous sialic acid receptor binds. Avidity appears to be crucial for S139/1 for the recognition and neutralization of influenza virus, in comparison with many other antibodies, not only to HA but also to other antigens, where the monovalent Fab can bind and neutralize almost as effectively as the IgG (17). Increased affinity by multivalency, therefore, plays a very important role for S139/1 (Fig. 4), where the bivalent IgG can enhance binding to HA by roughly three orders of magnitude over the monovalent Fab and directly correlates with the potency of in vitro neutralization. Such a large discrepancy between the binding and neutralizing activities of Fab and IgG has not been observed, for example, with antibodies targeting the stem (17) or the head region away from the receptor binding site (36). Whether or not bivalent recognition can be regarded as a general mechanism for more broadly neutralizing antibodies that target the shallow and relatively small receptor binding site of HA, which is surrounded by hypervariable residues, remains to be seen. The large footprint of an antibody makes it almost inevitable that it will have to bind to some hypervariable residues and, hence, may need to compensate by not acquiring too much specificity and affinity for these positions. Thus, a bivalent interaction can rescue a weaker, but more permissive Fab interaction.
Fig. 4.
Cartoon representation depicting the enhanced avidity by multivalency of receptor binding site targeted IgG over Fab. (A) Monovalent Fab that has high affinity for HA and IgG cross-links between HAs. (B) The Fab now has low affinity for HA, but the bivalent IgG increases the avidity of the low affinity Fab by cross-linking HAs.
The crystal structure of S139/1 in complex with Vic75/H3 reveals that the HCDR2 insertion into the receptor binding site does indeed account for the majority of interactions. These interactions are primarily hydrophobic, where LeuH52, IleH54, and MetH56 contact the highly conserved HA Trp153 and Leu194 residues. The sequence of S139/1 derives from the J558.17 germ-line mouse antibody (IgBlast). Although S139/1 did not originate with hydrophobic residues from the HCDR2 derived from its germ-line sequence (Fig. S5), it acquired hydrophobic residues in HCDR2 through somatic hypermutation, thereby gaining the ability to specifically target the receptor binding site. Hydrophobic HCDR2s are the hallmark of VH1-69 germ-line antibodies, such as the HA stem-targeted CR6261 (17, 22) and F10 (23) antibodies. That a hydrophobic HCDR2 can enter the receptor binding site, as in the case of S139/1, raises the possibility that similar receptor binding site-targeted antibodies from the VH1-69 germ line can be raised in humans (37).
The S139/1 binding mode involves a tightly constrained network of residue contacts and conformations of the outer loops that compose the receptor binding site, including antigenic sites A, B, and D (7). Electrostatics and sterics are important for the overall positioning of the antibody CDRs on the receptor binding site (Fig. 3). In addition, insertions in the 130 loop and the 150 loop in some strains and subtypes alter their respective loop conformations and, thus, abrogate S139/1 binding. Hypervariability around the receptor binding site can lead to HA escape mutants against S139/1, as observed at positions 156, 158, and 193 (29). Although H13 and H16 strains do not fit the same binding profile as H1, H2, and H3 viruses, they can still be bound or neutralized by S139/1 (SI Materials and Methods and Table 3). Furthermore, S139/1 has the potential to be a pan-H2 neutralizing antibody because most of the H2 strains fit the recognition profile. This finding may be of particular interest as immunity in the population diminishes against H2N2 viruses, which were last encountered from 1957 until the subsequent H3N2 pandemic in 1968 (38).
S139/1 was isolated from Aichi68/H3-immunized mice and was selected for cross-reactivity against strains from other HA subtypes (29). Although cross-reactivity of this antibody was achieved by inserting hydrophobic residues into a highly conserved hydrophobic portion of the receptor binding site, HA recognition is also highly dependent on the sequence and conformation of the surrounding loops. Our binding and structural analyses of the S139/1-Vic75/H3 complex raises the possibility that selection of a composite HA as an immunogen that contain residues sampled by a greater representation of different strains and subtypes may lead to a promising vaccine candidate that can offer broader long-lasting protection. Furthermore, the problem of targeting a hypervariable site, in which natural variation or mutations may reduce but not completely abrogate Fab binding, can be overcome by using the antibody bivalency to acquire sufficient binding energy for heterosubtypic neutralization. The importance of bivalent binding may also be particularly pertinent when the target antigen is tightly clustered on the virus surface, as in the case of influenza virus HA (39). In contrast, it may be more difficult for antibodies to bind bivalently to viruses with low glycoprotein density, such as the HIV envelope spike protein (40), where avidity typically plays only a minor role (41).
Materials and Methods
S139/1 Fab was cloned in a pFastBac Dual vector with a C-terminal His6 tag fused to the heavy chain. Fab was produced by baculovirus infection of High Five insect cells and was purified by Ni-NTA, protein G, and MonoS chromatography followed by a final step of gel filtration. HAs were cloned in a pFastBac vector with a C-terminal BirA biotinylation site, thrombin cleavage site, trimerization domain, and a His6 tag. HAs were similarly expressed and purified by Ni-NTA, as previously described (18). For binding studies, HA was biotinylated with BirA and purified by gel filtration. Kd values were measured by bio-layer interferometry using an Octet RED instrument (ForteBio). Neutralization activity of S139/1 Fab and IgG was measured by a plaque reduction assay using Madin-Darby canine kidney cells. Vic75/H3 was treated with trypsin to produce mature HA followed by gel filtration used for crystallization. S139/1 Fab was added to Vic75/H3 HA in a molar ratio of 3.2:1 to achieve three Fabs per HA trimer and the complex was purified from unbound Fab by gel filtration. S139/1-Vic75/H3 crystals were grown using sitting drops and crystals were cryoprotected and flash-cooled in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source (APS) GM/CA-CAT 23-ID-D beamline and the structure was solved by molecular replacement. Further information is described in the SI Materials and Methods and Figs. S6–S9. Sequences of the HAs can be found in Dataset S1.
Supplementary Material
Acknowledgments
We thank Henry Tien of the Robotics Core at the Joint Center for Structural Genomics for automated crystal screening; the staff of the Advanced Photon Source General Medicine/Cancer Institute-Collaborative Access Team (GM/CA CAT) 23-ID-D for beamline support; Robyn Stanfield for assistance with data collection; Xueyong Zhu and Jean-Philippe Julien for assistance with data processing; Marc Elsliger for advice on model refinement; and Megumi Higuchi for cloning the antibody gene. This work was supported in part by Grant AI058113 (to I.A.W.); the Skaggs Institute of Chemical Biology; National Institutes of Health Molecular Evolution Training Program Grant GM080209 (to P.S.L.); the Japan Science and Technology Agency Basic Research Programs; the Japan Initiative for Global Research Network on Infections Diseases; and a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology. The GM/CA CAT has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. This is The Scripps Research Institute manuscript number 21870.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4GMS and 4GMT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212371109/-/DCSupplemental.
References
- 1.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 2.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 3.Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci USA. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobusawa E, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 5.Tong S, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzberg S. The contents of the syringe. Nature. 2008;454:160–161. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 8.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 9.Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. 2012;2:134–141. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 11.Skehel JJ, et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 13.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 14.Barbey-Martin C, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 15.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 21.Kashyap AK, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci USA. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashyap AK, et al. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 2010;6:e1000990. doi: 10.1371/journal.ppat.1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 28.Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med. 2006;354:1343–1351. doi: 10.1056/NEJMoa055778. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida R, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohshima N, et al. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krause JC, et al. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–218. doi: 10.1016/s0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 35.Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol. 1998;5:119–123. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- 36.Fleury D, et al. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat Struct Biol. 1999;6:530–534. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- 37.Krause JC, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. 2012;86:6334–6340. doi: 10.1128/JVI.07158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nabel GJ, Wei CJ, Ledgerwood JE. Vaccinate for the next H2N2 pandemic now. Nature. 2011;471:157–158. doi: 10.1038/471157a. [DOI] [PubMed] [Google Scholar]
- 39.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 41.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.