Abstract
Although the protooncogene c-Jun plays a critical role in cell proliferation, cell death, and malignant transformation, DNA microarray screens have identified only a few human cancer types with aberrant expression of c-Jun. Here, we show that c-Jun accumulation is robustly elevated in human glioblastoma and that this increase contributes to the malignant properties of the cells. Most importantly, the increase in c-Jun protein accumulation occurs with no corresponding increase in c-Jun mRNA or the half-life of the c-Jun protein but, rather, in the translatability of the transcript. The c-Jun 5′UTR harbors a potent internal ribosomal entry site (IRES) with a virus-like IRES domain that directs cap-independent translation in glioblastoma cells. Accumulation of c-Jun is not dependent on MAPK activity but can be stimulated by a cytoskeleton-dependent pathway. Our findings provide evidence that human c-Jun is an IRES-containing cellular transcript that contributes to cancer development through translational activation. This previously undescribed mechanism of c-Jun regulation might also be relevant to other types of human cancer and offers unique potential targets for therapy.
The c-Jun protein is a transcription factor that forms a variety of dimeric complexes, collectively termed activator protein-1 (AP-1), and positively regulates cell proliferation and tumor progression. The c-Jun protein stimulates cell cycle progression through two main mechanisms: (i) induction of genes coding for components of the cell cycle machinery, such as cyclin D1, and (ii) repression of tumor suppressor genes, such as p53 (1). In addition, the c-Jun protein activates several events required for tumor progression, including the expression of matrix metalloproteinases (MMPs), proteolytic enzymes that facilitate growth, invasion, and metastasis of cancer cells (2). Reduction of c-Jun/AP-1 activity using dominant-negative c-Jun (TAM67) or conditional inactivation of the c-Jun gene causes cell arrest (3), interferes with tumor development (4, 5), suppresses the invasive ability in keratinocytes (6) and fibroblasts (7), and blocks papilloma-to-carcinoma conversion (8). Although the oncogenic activity of c-Jun has been convincingly demonstrated by multiple lines of evidence in human cell lines and mouse models, there are only few examples of elevated c-Jun expression in human cancer (9–11).
Expression of c-Jun is markedly elevated on exposure of cells to various extracellular stimuli, including growth factors, cytokines, cellular stress, and UV irradiation (12). These external stimuli cause a rapid and dramatic increase in c-Jun gene transcription, mainly through activation of the MAPK family of serine/threonine kinases, particularly by JNKs and p38, which phosphorylate the transcription factors c-Jun, ATF2, and MEF2C, and thereby activate transcription of the c-Jun gene (13–15). The MAPK pathway can also contribute to the stability of the c-Jun protein. Phosphorylation by JNK protects c-Jun from ubiquitination and subsequent degradation (16), whereas ERK increases c-Jun stability via inactivation of glycogen synthase kinase 3 (9).
In addition to external stimuli, expression of c-Jun can be regulated by cell-cell contacts (17–20). Recent studies have shown that loss of cell-cell contacts, by means of cell separation or functional inhibition of the adhesion molecule E-cadherin, causes a marked and sustained increase in c-Jun protein accumulation and that this increase is not transcriptionally but rather translationally controlled (18). Cell contact control of c-Jun translation appears to be mediated by the cytoskeletal network: Depolymerization of the cytoskeleton by overexpression of cofilin 1 (21) or addition of cytoskeleton disrupting agents (18, 22, 23) mimics the effect of cell separation and causes a dramatic increase in c-Jun accumulation, whereas Taxol inhibits the cell contact-dependent increase (18). As in the case of cell contacts, the cytoskeletal-dependent increase is not accompanied by an increase in c-Jun mRNA or in the half-life of the c-Jun protein. The increase in c-Jun accumulation is translationally regulated and is mediated by the UTRs of the c-Jun transcript, particularly by the 5′UTR (22).
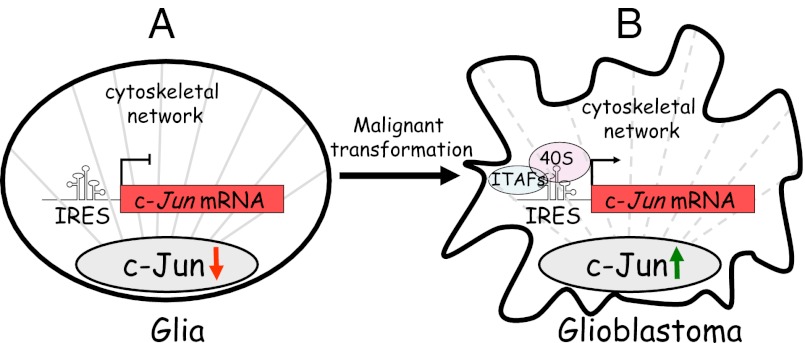
Considering that tumor promotion and progression are often accompanied by loss of adhesion molecules and/or restructuring of the cytoskeleton, we decided to examine whether expression of c-Jun in tumor cells is translationally activated. Here, we examined the expression of c-Jun in astrocytomas, tumors of glial origin that arise in the brain. The most malignant form of these tumors, glioblastoma multiforme (grade IV), is one of the most aggressive human cancers, with a median survival of less than 1 y (24). We show that accumulation of c-Jun in these tumors increases with the grade of malignancy and that this increase contributes to the malignant properties of the cells. Most importantly, we demonstrate that accumulation of c-Jun is translationally regulated by a cap-independent mechanism. Translation of c-Jun is mediated by the internal ribosomal entry site (IRES), is not dependent on MAPK activity, and can be activated by a cytoskeleton-dependent pathway. These findings provide evidence for translational activation of c-Jun in cancer cells and for the presence of an IRES element in human c-Jun mRNA.
Results
High Accumulation of c-Jun in Human Glial Tumors by Posttranscriptional Activation.
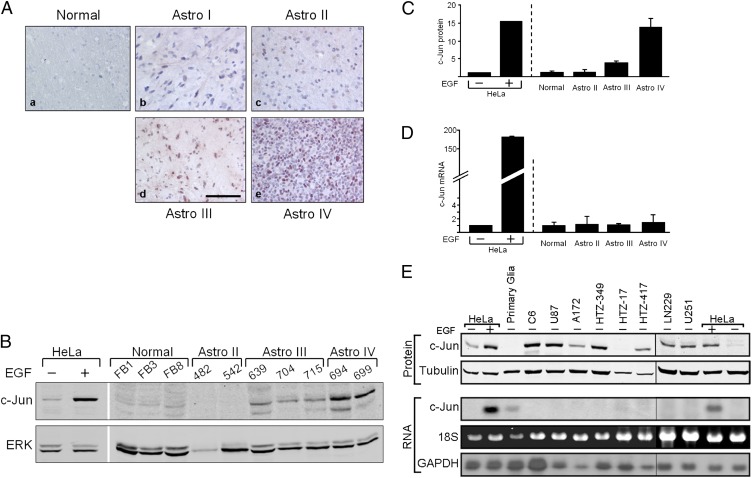
To examine whether c-Jun expression in human glial tumors is elevated, we immunostained brain sections from normal and tumor tissues that were defined by histological criteria as astrocytoma grade I (pilocytic astrocytoma), II (diffuse astrocytoma), III (anaplastic astrocytoma), or IV (glioblastoma). The results clearly showed that accumulation of c-Jun in normal tissue was low but increased with the grade of malignancy (Fig. 1A). In tumors of patients with glioblastoma, high and nuclear-specific expression of c-Jun was observed in almost all tumor cells. Expression of c-Jun was also examined by Western blot and quantitative RT-PCR. HeLa cells, untreated or treated with epidermal growth factor (EGF), a known stimulator of c-Jun transcription, were used as a control. Similar to the immunostaining results, Western blot analysis showed that c-Jun protein accumulation increased with the grade of malignancy, demonstrating a low level in normal tissue and in astrocytoma of grade II, and a threefold or 13-fold increase in astrocytoma of grade III or IV, respectively (Fig. 1 B and C). However, RNA analysis revealed that unlike the concomitant increase in c-Jun protein and mRNA in EGF-treated HeLa cells, the increase in c-Jun protein accumulation in astrocytoma was not accompanied by a corresponding increase in c-Jun mRNA (Fig. 1D). This latter finding is consistent with previous DNA microarray data, which failed to identify a significant increase in c-Jun mRNA associated with astrocytoma grade, progression, or patient survival (25–27).
Fig. 1.
Up-regulation of c-Jun in human glioblastoma is posttranscriptionally controlled. (A) Immunohistochemical staining of c-Jun in tissue sections of human normal brain and grade I to IV astrocytomas (Astro) shows strong and nuclear specific staining of c-Jun in almost all cells of astrocytoma grade IV. (Scale bar: 40 μm.) Tissue samples of normal brain and grade II to IV astrocytomas were assayed for both c-Jun protein and mRNA expression. HeLa cells treated with EGF or untreated were used as controls. (B) Protein expression was assayed by Western blotting using anti–c-Jun and anti-ERK antibodies. (C) Blot was scanned, and the intensity of the c-Jun band was calculated relative to ERK. (D) RNA expression was assayed by quantitative RT-PCR. Data are the mean ± SD. (E) Western blot (Upper) and Northern blot (Lower) analyses show c-Jun expression in rat primary glia, in rat (C6) and human (U87, A172, HTZ349, HTZ17, HTZ417, LN229, and U251) glioblastoma cell lines, and in HeLa cells treated with EGF or untreated. Tubulin was used as a loading control in Western blots, and 18S rRNA and GAPDH were used as loading controls in Northern blots.
Similar results were also obtained when rat (C6) and human (U87, A172, HTZ349, HTZ17, HTZ417, LN229, and U251) glioblastoma cell lines were assayed. Western blot analysis showed that rat primary glia accumulated a low, hardly detectable level of the c-Jun protein, whereas in a glioblastoma cell line of rat and most cell lines of humans, the level of c-Jun was high, similar to that in EGF-treated HeLa cells (Fig. 1E, Upper). Northern blot analysis showed that the increase in c-Jun protein in glioblastoma was not accompanied by an increase in c-Jun mRNA (Fig. 1E, Lower). These findings strongly suggest that up-regulation of c-Jun in glioblastoma is posttranscriptionally controlled.
Accumulation of c-Jun Contributes to the Malignant Properties of Glioblastoma Cells.
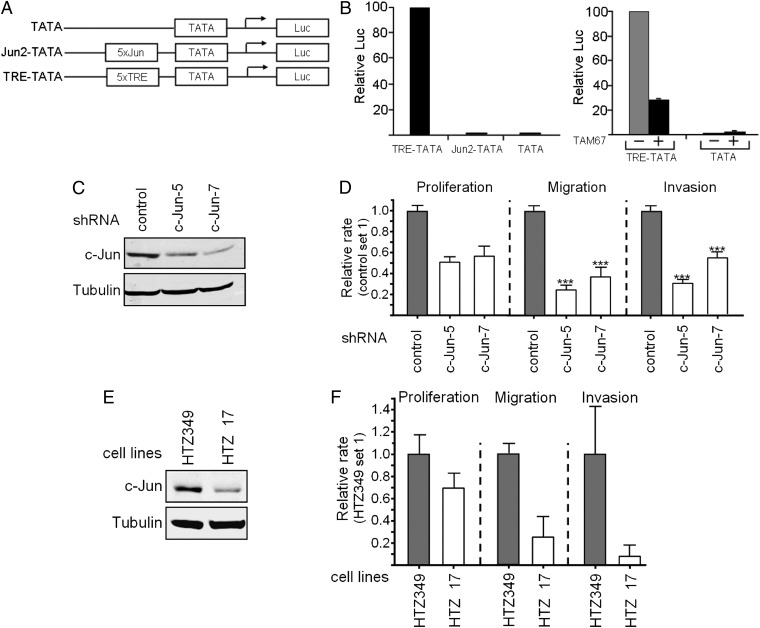
The c-Jun protein is known to autoregulate its own transcription via an AP-1 site in the regulatory region of the gene (1). The finding that the increase in c-Jun protein is not accompanied by an increase in c-Jun mRNA raised the possibility that in glioblastoma cells, the c-Jun protein is transcriptionally inactive. To examine the transcription activity of c-Jun, we transfected U87 cells with reporter constructs that contain a minimal TATA box attached to five copies of the AP-1 sequence from the c-Jun (Jun2-TATA) or MMP1 (TRE-TATA) promoter. A reporter construct that lacks the AP-1 sequence was used as a control (Fig. 2A). Analysis of reporter gene expression revealed a marked difference between the expression levels of the two AP-1–containing constructs. Whereas expression of the Jun2-TATA construct was low, similar to that of the control construct, expression of the TRE-TATA construct was 100-fold higher (Fig. 2B). This high level declined considerably on cotransfection of TAM67, a dominant-negative form of the c-Jun protein. This finding suggests that the accumulated c-Jun protein is functional but incapable of activating its own promoter.
Fig. 2.
c-Jun protein is transcriptionally active and contributes to the malignant properties of glioblastoma cells. (A) Schematic representation of luciferase reporter constructs used in this study. (B) Promoter activity was assayed using U87 cells transfected with indicated reporter constructs with or without the dominant-negative c-Jun construct TAM67 (+) or empty pCDNA3 vector (−). Transfection efficiency was controlled by cotransfection of CMV-Rnl. Luciferase activity obtained with TRE-TATA was given the arbitrary value of 100 and used to normalize all other results. Values are the mean ± SD of three separate experiments. (C) Cellular level of c-Jun in U87 cells stably transfected with c-Jun–directed shRNA (c-Jun-5 or c-Jun-7) or with control shRNA was assayed by Western blotting. Tubulin was used as a loading control. (D) Proliferation, migration, and invasion of the stably transfected c-Jun-5 and c-Jun-7 cells vs. control cells. Migration and invasion assays were performed in a Boyden chamber. Experiments were repeated at least three times. Data are given as the mean ± SEM. ***P < 0.05. (E) Comparison between two human glioblastoma cell lines that express a high (HTZ349) or low (HTZ17) level of c-Jun by Western blotting, using tubulin as a loading control. (F) Proliferation, migration, and invasion of HTZ17 cells vs. HTZ349 cells. Experiments were repeated at least three times. Data are given as the mean ± SEM.
To evaluate the functional activity of c-Jun in glioblastoma further, we assayed whether down-regulation of c-Jun affects the malignant properties of the cells. Stable transfection of U87 cells with c-Jun shRNA, c-Jun-5 or c-Jun-7, reduced the expression of c-Jun by about 50% or 70%, respectively (Fig. 2C). Analysis of cell proliferation of the stable transfectants revealed a decreased proliferation rate after knockdown of c-Jun in comparison to control cells (Fig. 2D). Furthermore, the migration and invasion capacity was significantly reduced after silencing of c-Jun (Fig. 2D). Similar results were obtained when glioblastoma cell lines with high (HTZ349) and low (HTZ17) levels of endogenous c-Jun were assayed (Fig. 2E). HTZ349 cells proliferated at a higher rate and exhibited an increased capacity of migration and invasion compared with HTZ17 cells (Fig. 2F). Taken together, our results suggested that the accumulated c-Jun protein is transcriptionally active and contributes to the malignant properties of glioblastoma cells.
Constitutive c-Jun Accumulation Is Not Due to an Increase in Protein Stability and Is Independent of MAPK Activity.
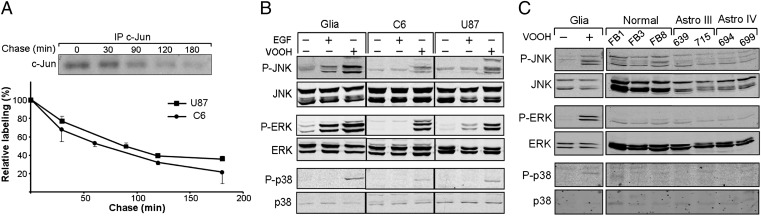
Given that accumulation of cellular proteins may reflect an increase in protein stability, we measured the half-life of c-Jun in glioblastoma cells. Pulse–chase analysis revealed that the half-life of c-Jun in U87 and C6 cells was about 90 min and 70 min, respectively (Fig. 3A), somewhat shorter than the reported half-life of c-Jun in EGF-treated cells (130 min) and similar to that in untreated cells (90 min) (16, 28, 29). Thus, accumulation of c-Jun in glioblastoma cells is not due to an increase in stability of the c-Jun protein.
Fig. 3.
Accumulation of the c-Jun protein is not due to an increase in c-Jun stability and is not dependent on MAPK activity. (A) To estimate the half-life of c-Jun, U87 and C6 cells were metabolically pulse-labeled with 35[S]methionine/35[S]cysteine and chased for the indicated periods of time. The intensities of the immunoprecipitated (IP) c-Jun bands were determined by scanning and calculated using EZQuant-Gel software. The c-Jun signal intensities are expressed as a percentage of that present at the end of the labeling pulse. The data shown are of two independent experiments. (B) Protein samples from primary glia and C6 and U87 cells untreated (−) or treated (+) with EGF or VOOH were analyzed by Western blotting using the indicted antibodies. (C) Protein samples from primary glia, untreated (−) or treated (+) with VOOH, or from tissue samples of normal brain and grade III and IV astrocytomas were analyzed by Western blotting using the indicated antibodies.
Stability of the c-Jun protein and transcription of the c-Jun gene are known to be mainly controlled by the MAPKs, particularly JNK, ERK, and p38. We examined the activation state of the MAPK pathway in glioblastoma cells using specific antibodies that recognize phosphorylated (and therefore activated) JNK, ERK, and p38. EGF or peroxovanadate (VOOH), a general phosphatase inhibitor, was used as a control. The results clearly showed that the MAPK pathway is not activated in glioblastoma cells: The level of phospho-JNK (P-JNK), phospho-ERK (P-ERK), or phospho-p38 (P-p38) in U87 and C6 cells was low, similar to that in primary glia (Fig. 3B). Similar results were obtained when normal and tumor brain samples were assayed. Here too, the level of P-JNK, P-ERK, or P-p38 in astrocytoma of grade III or IV was similar to that in normal tissue (Fig. 3C). Thus, consistent with the above results, aberrant accumulation of c-Jun in glioblastoma is independent of MAPK activity.
High Accumulation of c-Jun in Primary Glia by Cytoskeleton-Dependent Pathway.
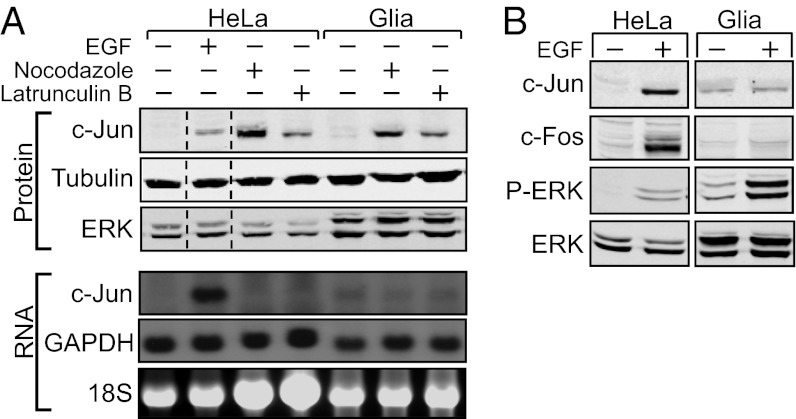
Although expression of c-Jun is known to be mainly activated by the MAPK pathway, recent studies have shown that c-Jun expression can also be activated by a cytoskeleton-dependent pathway (18, 22, 23). This latter pathway does not affect the transcription of the c-Jun gene or the stability of the c-Jun protein but, rather, the translatability of its transcript (22). We examined whether both pathways can stimulate the expression of c-Jun in primary glia. Indeed, addition of latrunculin B, which depolymerizes the actin network, or nocodazole, which depolymerizes the microtubules, caused a marked increase in c-Jun protein accumulation in primary glia (Fig. 4A, Upper). Northern blot analysis revealed that the increase in c-Jun protein accumulation was not accompanied by a corresponding increase in c-Jun mRNA (Fig. 4A, Lower), indicating that cytoskeletal control of c-Jun expression is posttranscriptional. Unexpectedly, analysis of c-Jun expression in EGF-treated cells revealed that activation of the MAPK pathway induced the expression of c-Jun and c-Fos (another target gene of the MAPK pathway) in HeLa cells but not in primary glia (Fig. 4B). These findings suggest that the cellular context of glial cells facilitates the cytoskeleton-dependent increase in c-Jun expression but lacks components essential for MAPK-mediated induction.
Fig. 4.
Cytoskeletal-dependent pathway activates the expression of c-Jun in primary glia. (A) Western blot (Upper) and Northern blot (Lower) analyses show c-Jun expression in HeLa cells and rat primary glia untreated (−) or treated (+) with EGF, nocodazole, or latrunculin B. Tubulin and ERK were used as loading controls in Western blots, and 18S rRNA and GAPDH were used as loading controls in Northern blots. (B) Western blot analysis shows the expression of c-Jun and c-Fos and the phosphorylation of ERK in HeLa cells and rat primary glia untreated or treated with EGF.
Expression of c-Jun Is Translationally Activated.
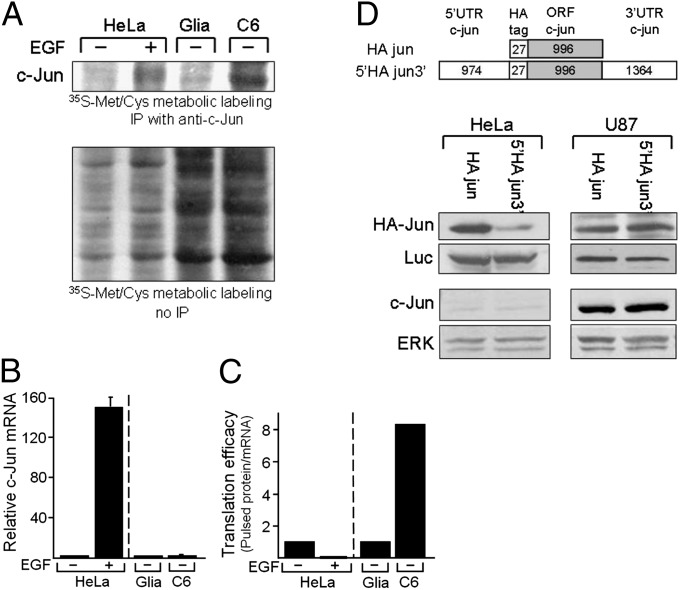
The possibility that accumulation of c-Jun in glioblastoma is translationally controlled was assayed by measuring the rate of de novo c-Jun synthesis in primary glia and glioblastoma cells (C6) or in HeLa cells untreated or treated with EGF. Protein extracts from cells labeled metabolically with 35S-Met/Cys were immunoprecipitated with anti–c-Jun antibodies (Fig. 5A). In parallel, the cellular amount of c-Jun mRNA was quantitatively determined by RT-PCR (Fig. 5B). Translation efficiency was calculated as the ratio between the rate of de novo synthesis of the c-Jun protein and the cellular amount of the c-Jun mRNA. The results clearly demonstrated that translation efficiency of c-Jun was elevated by eightfold in glioblastoma cells but not in EGF-treated HeLa cells (Fig. 5C).
Fig. 5.
Accumulation of c-Jun in glioblastoma cells is translationally controlled. (A) Protein extracts of EGF-treated (+) or untreated (−) HeLa cells, primary glia, or C6 cells, metabolically labeled with 35[S]methionine/35[S]cysteine, were fractionated by electrophoresis, before or after immunoprecipitation (IP) with anti–c-Jun antibodies, and visualized by autoradiography. De novo synthesis was calculated by scanning the intensity of the c-Jun band. (B) Parallel cultures were used to measure the c-Jun mRNA by quantitative RT-PCR. Experiments were repeated three times. Data are the mean ± SD. (C) Translation efficiency was calculated as the ratio between the rate of de novo synthesis of the c-Jun protein and the cellular amount of the c-Jun mRNA. The calculated ratio in untreated HeLa cells or in primary glia was assigned the arbitrary value of 1 and was used to normalize the rate in EGF-treated HeLa or C6 cells, respectively. (D, Upper) Schematic representation of the HA-Jun reporter construct used in this study. (D, Lower) HeLa and U87 cells were transfected with the indicated HA-Jun constructs together with the luciferase construct, pjLuc, to control for transfection efficiency. Expression of the transfected HA-Jun and luciferase constructs and the endogenous c-Jun and ERK was assayed by Western blotting.
Translational control is often mediated by the UTRs of the transcript. In the case of c-Jun, these regions are exceptionally long (the 5′UTR and 3′UTR have 974 and 1,364 bases, respectively) and GC-rich, and they have the potential of forming stable secondary structures. Previous studies have shown that these regions can confer repression of protein translation in HeLa cells, cells that accumulate a low basal level of the c-Jun protein (22). Given the observed increase in translation efficiency in glioblastoma cells, we decided to examine whether the c-Jun UTRs affect translation in these cells too. We used an HA-tagged c-Jun construct that contains (5′HAjun3′) or does not contain (HAjun) the c-Jun 5′UTR and 3′UTR. The constructs were transfected into HeLa and U87 cells, together with a firefly luciferase (FL) construct as a control. The levels of the endogenous c-Jun and exogenous HA-Jun were assayed. As expected, expression of the 5′HAjun3′ construct in HeLa cells was considerably lower than that of the HAjun. By contrast, in U87 cells, the two constructs were expressed at a similar high level (Fig. 5D). This finding indicates that the c-Jun UTRs do not repress translation in glioblastoma cells.
Translation of c-Jun in Glioblastoma Is Cap-Independent and IRES-Mediated.
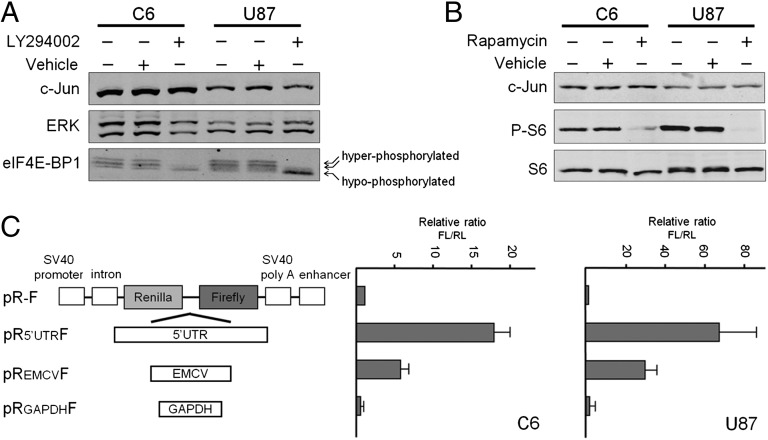
Initiation of translation in eukaryotic cells can occur by means of at least two distinct mechanisms: cap-dependent scanning and internal ribosome entry (30). To examine which mechanism underlies the increase in c-Jun translation, we treated glioblastoma cells with LY294002, a potent PI3K inhibitor that affects eIF4E-BP1 (4E-BP1) phosphorylation and prevents cap-dependent translation (31). In the absence of LY294002, 4E-BP1 was highly phosphorylated in both C6 and U87 cells, as evidenced by the prevalence of the slow-migrating hyperphosphorylated forms of the protein (Fig. 6A). As expected, 4E-BP1 phosphorylation was strongly inhibited by LY294002, as the hypophosphorylated form became more prominent. Strikingly, despite hypophosphorylation of 4E-BP1, enhancement of c-Jun translation was maintained (Fig. 6A). Similar results were obtained with rapamycin, another inhibitor of cap-dependent translation (31). Exposure of C6 or U87 cells to rapamycin abolished the phosphorylation of ribosomal S6, a downstream effector of rapamycin, but did not affect the accumulation of c-Jun (Fig. 6B). These findings indicate that c-Jun can be expressed under conditions in which cap-dependent translation is impeded. To examine whether translational activation occurs via an IRES-mediated mechanism, we inserted the c-Jun 5′UTR into a bicistronic vector (pR-F), which contains Renilla luciferase (RL) and FL, in the first and second cistrons, respectively. Two negative controls were used: an empty vector that contains the multiple cloning site in the intercistronic region (pR-F) and a vector containing a segment from the coding region of human GAPDH (pRGAPDHF). A plasmid containing the encephalomyocarditis virus IRES (pREMCVF) was used as a positive control (32). The constructs were transfected into C6 and U87 cells, and luciferase activity was assayed. The ratio of FL/RL activity was calculated and normalized to the value of the control vector pR-F, arbitrarily set at 1. The results, presented in Fig. 6C, suggested that the c-Jun 5′UTR contains a potent IRES that can direct a marked increase in expression of the downstream cistron.
Fig. 6.
Translation of c-Jun is cap-independent and IRES-mediated. (A) Pattern of c-Jun expression and 4E-BP1 phosphorylation in C6 or U87 cells untreated (−) or treated (+) with LY294002. (B) Pattern of c-Jun expression and S6 phosphorylation in C6 and U87 cells untreated or treated with rapamycin. (C) Bicistronic reporter plasmids, schematically represented (Left), were transfected into C6 or U87 cells. EMCV, encephalomyocarditis virus. RL and FL activities were determined. The ratio of FL/RL in the empty pR-F plasmid was assigned the arbitrary value of 1 and used to normalize all other results. The data shown are the mean ± SEM of at least three separate experiments.
Translational Activation Is Driven by a Virus-Like IRES Domain.
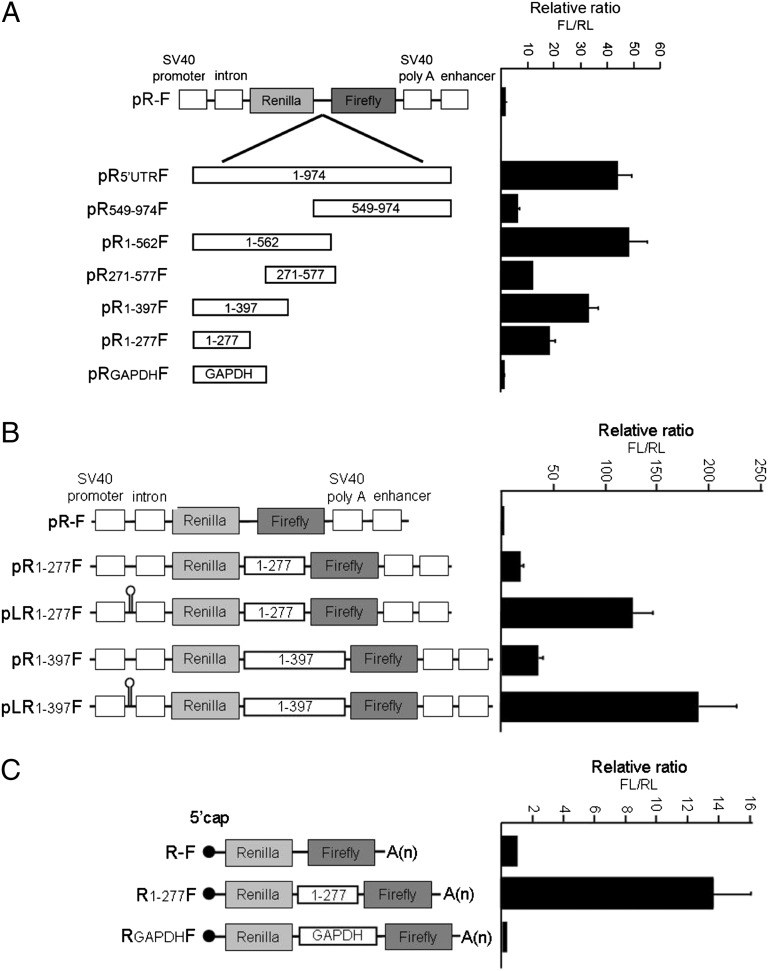
Deletion analysis revealed that IRES activity is mainly located within the first 562 nt of the 5′UTR (Fig. 7A). Subsequent truncations of this region to generate constructs that contain the first 397 nt (pR1-397F) or 277 nt (pR1-277F) of the 5′UTR resulted in an almost stepwise decrease in the FL/RL ratio, suggesting that the deleted sequences contribute, structurally or functionally, to IRES activity. To rule out the possibility that translation of the downstream cistron was a result of ribosome reinitiation, we inserted at the transcription start site of the pR1-277F and pR1-397F constructs a stable hairpin structure that inhibits cap-dependent translation (33). The hairpin strongly inhibited RL expression but not the expression of FL (Fig. S1). This resulted in an increase in the FL/RL ratio, indicating that the two cistrons were independently translated (Fig. 7B). In addition to reinitiation of ribosomes, increased activity of FL in a bicistronic construct can be generated through cryptic promoter activity or cryptic splicing. Because Northern blot analysis (Fig. S2) could not eliminate the possible contribution of cryptic promoter or splicing activity, we transfected cells with mRNA instead of plasmid DNA. The bicistronic constructs were subjected to in vitro transcription followed by capping and poly (A) tailing reactions (Fig. S3 A and B). The mRNA was transfected into U87 cells, and the resulting RL and FL activities were measured. A small but measurable amount of FL activity was produced from the R-F or the RGAPDHF-negative control mRNAs. Insertion of the first 277 bases of the c-Jun 5′UTR resulted in a 14-fold increase in FL/FR ratio over the negative controls (Fig. 7C). These findings clearly indicate that the c-Jun 5′UTR harbors IRES activity.
Fig. 7.
Mapping of IRES activity in the c-Jun 5′UTR. U87 cells were transfected with bicistronic reporter plasmids that contain the entire c-Jun 5′UTR or fragments thereof (A), with or without a hairpin structure at the transcription start site (B) or with capped and poly(A)-tailed bicistronic RNA, obtained by in vitro transcription (C). Luciferase activities were determined, and the ratio of FL/RL activity in the empty pR-F construct was assigned the arbitrary value of 1 and used to normalize all other results. The data shown are the mean ± SEM of at least three separate experiments.
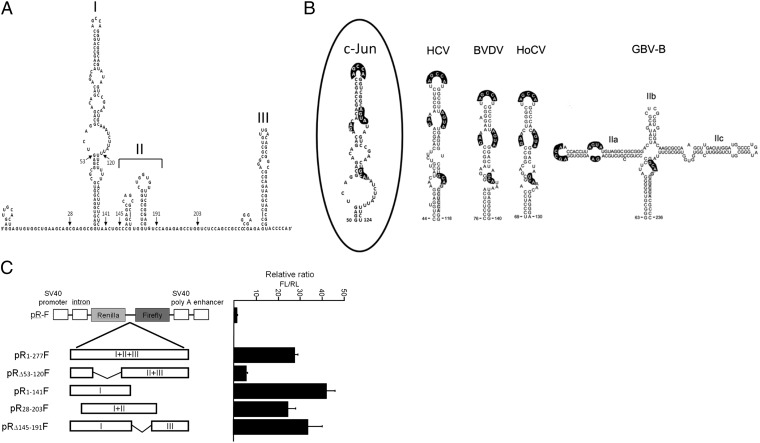
Analysis by the MFold prediction algorithm (34) revealed that the first 277 bases of the c-Jun 5′UTR might form a stable secondary structure (ΔG = −99.3 kcal/mol) that contains several stem-loop domains, designated as domains I to III (Fig. 8A). Sequence inspection revealed that domain I has striking homology to a conserved and functionally essential stem-loop structure that has been identified in IRES elements of four genetically diverse flaviviruses: hepatitis C virus (HCV), bovine viral diarrhea virus (BVDV), hog cholera virus (HoCV), and hepatitis GB virus B (GBV-B) (35). The viral stem-loop structure has conserved and functionally required primary nucleotide sequences within the terminal loop and internal bulge loops (35), and these sequences are also present in domain I of the c-Jun 5′UTR (highlighted in Fig. 8B). To assess the contribution of the different c-Jun domains to IRES activity, we generated bicistronic constructs that contain the following combination of domains in the intercistronic region: I and II (pR28-203F), I and III (pRΔ145-191F), II and III (pRΔ53-120F), or only I (pR1-141F). The constructs were transfected into U87 cells, and luciferase activity was assayed. The results showed that deletion of domain II, domain III, or both domains II and III had almost no effect on IRES activity. By contrast, deletion of domain I reduced the activity strongly, indicating that IRES activity is mainly directed by domain I (Fig. 8C). Taken together, our results show that IRES-mediated translation of c-Jun is driven by a virus-like IRES domain and constitutes a primary mechanism for up-regulation of c-Jun in glioblastoma cells.
Fig. 8.
Virus-like IRES domain drives translational activation. (A) Predicted secondary structure of the first 277 bases of the c-Jun 5′UTR. Structural domains are labeled I to III. (B) Domain I of c-Jun IRES and a conserved IRES domain identified in four genetically diverse flaviviruses: heptatis C virus (HCV), bovine viral diarrhea virus (BVDV), hog cholera virus (HoCV), and hepatitis GB virus B (GBV-B). Bases shown with heavy highlighting are conserved among the IRES domains. (Adapted with permission from ref. 35.) (C) U87 cells were transfected with bicistronic reporter plasmids that contain a combination of the IRES domains, as indicated. Luciferase activities were determined, and the ratio of FL/RL activity in the empty pR-F construct was assigned the arbitrary value of 1 and used to normalize all other results. The data shown are the mean ± SEM of at least three separate experiments.
Discussion
Recent studies using comparative genomic and proteomic profiling of cells have suggested that translational control is more important in the regulation of gene expression than often assumed and that this mechanism might play a major role in tumor progression. In line with this notion, this study shows that expression of the protooncogene c-Jun in tumor cells is not transcriptionally but, rather, translationally controlled. Analysis of human glial tumors revealed that accumulation of the c-Jun protein increases with the grade of malignancy and that this increase is not accompanied by a corresponding increase in c-Jun mRNA. Robust accumulation of c-Jun is also observed in rat and human glioblastoma cell lines, and there too, expression of c-Jun is posttranscriptionally controlled. This finding is supported by transfection experiments that assayed the expression of reporter constructs that contain the AP-1 sequence from the c-Jun or the MMP1 promoter. The results showed that the c-Jun protein is transcriptionally active in glioblastoma cells but incapable of activating its own promoter. The transcription activity of c-Jun is executed by forming AP-1 complexes that consist of homo- or heterodimers with members of the Jun, Fos, and ATF protein subfamilies. These c-Jun/AP-1 complexes display subtle but important variations in DNA binding specificity, and their formation depends on the relative abundance of each of the Jun, Fos, and ATF proteins in the cell (36–38). Thus, the observed differences in transactivation of reporters that contain the AP-1 sequence from the c-Jun or MMP1 promoter suggest that in glioblastoma cells, the cellular context facilitates the formation of c-Jun/AP-1 complexes that can interact with the AP-1 sequence of the MMP1 promoter (and thereby activate the expression of proteolytic enzymes that contribute to the invasive capability of cancer cells) but not with that of c-Jun.
The MAPK pathway constitutes a major signaling cascade that controls the transcription of the c-Jun gene and the stability of the c-Jun protein. Pulse–chase analysis showed that accumulation of c-Jun is not due to an increase in c-Jun protein stability, and, consistently, no significant increase in phosphorylation, and therefore activation of the MAPK family members, JNK, ERK, or p38, was observed in glioblastoma cell lines and tumor brain samples. The finding that accumulation of c-Jun in glioblastoma is independent of MAPK activity was corroborated by the unexpected results, which showed that activation of the MAPK pathway by EGF fails to induce the expression of c-Jun in primary glia. Glial cells appear to lack downstream components essential for MAPK-mediated transcriptional activation of the c-Jun gene. This finding is of particular interest because the EGF receptor (EGFR) is reportedly a primary contributor to glioblastoma initiation and progression (39). Amplification of the EGFR is one of the highly specific genetic events associated with glioblastoma and is often accompanied by genetic alterations that result in a constitutively active receptor protein. The oncogenic role of EGFR has been functionally validated in cell culture and animal models, but the mechanistic basis of its function is still unclear. Our results suggest that in glioblastoma, the tumorigenic activity of EGFR is not mediated by the c-Jun pathway. However, the c-Jun protein is an important driver of glial malignancy. Consistent with its role in cell proliferation, apoptosis, and tumor promotion, knockdown of c-Jun by stable transfection of shRNA decreased proliferation, migration, and invasion capacity of glioblastoma cells. The increase in c-Jun accumulation might be triggered by a signaling pathway that is activated at early stages of malignancy. Nevertheless, accumulation of c-Jun in human brain tumors may serve as a hallmark of tumor progression and provide a molecular target for tumor prevention.
We measured the rate of de novo c-Jun synthesis in primary glia and glioblastoma cells and in HeLa cells untreated or treated with EGF. Our results clearly showed that unlike EGF, which causes an increase in c-Jun protein accumulation by stimulating the transcription of the c-Jun gene, the increase in c-Jun accumulation in glioblastoma cells is translationally controlled. However, treatment with rapamycin or LY294002 revealed that accumulation of c-Jun can also occur under conditions in which cap-dependent translation is impeded. In line with the structural properties of the human c-Jun 5′UTR, which is exceptionally long (974 bases) and GC-rich and has the potential of forming stable secondary structures, transfection of bicistronic DNA and RNA constructs revealed the presence of a potent IRES in the c-Jun 5′UTR. The 5′UTR of avian c-Jun, which is considerably shorter (301 bases), also contains IRES activity (40). Translation of the other human Jun family members, Jun D and Jun B, is apparently cap-dependent (41, 42). Deletion analysis showed that in the human c-Jun transcript, the first 562 bases, but not the last 425 bases, harbor IRES activity and that the first 277 bases are sufficient to direct IRES-mediated translation. This region is predicted to form a stable secondary structure with several stem-loop domains. Subsequent deletion analysis revealed that domain I, which is located at the 5′ border of the c-Jun transcript, highly contributes to translational activation. This domain has striking homology to a phylogenetically conserved sequence and secondary structure in flavivirus IRESs (35). In viral IRESs, mutation analysis showed that the conserved primary nucleotide sequences within the terminal loop and internal bulge loops are functionally essential. Considering that the flaviviruses infect very different host species, it has been suggested that the conserved loop sequences interact with elements of the host translational machinery that are broadly conserved among different mammalian species. Indeed, cryoelectron microscopy studies have shown that the apical half of this conserved IRES domain makes direct contact with the 40S ribosomal subunit (43). This contact may be an important determinant not only of viral IRES function but in the translation activation of c-Jun in glioblastoma cells.
IRES elements are found in the 5′UTR of several oncogenes, growth factors, and proteins involved in cancer (44). Cellular IRESs are active under physiologically relevant conditions that are important in cancer, and when cap-dependent translation is compromised (e.g., during mitosis and tumor stress responses, such as hypoxia and nutrient deprivation). The precise molecular mechanism of cellular IRES-directed translation is not completely understood. In several cases, IRES-directed translation has been attributed to the activity of auxiliary proteins, known as IRES trans-acting factors, although the requirement for these proteins is not absolute and seems to be IRES-specific. Although the mechanism that underlies the translational activation of c-Jun in glioblastoma cells has yet to be unraveled, our results suggest that cytoskeleton dynamics might constitute an important component in this process. We showed that although activation of the MAPK pathway failed to induce the expression of c-Jun, depolymerization of the actin or microtubule network elevated the expression of c-Jun markedly. Restructuring of the cytoskeleton is imperative in the process of cell proliferation and in migration and invasion of cancer cells. In glioblastoma, control of cytoskeletal dynamics has been attributed to the orchestrated activity of several signaling pathways, including Rac1, RhoA, and RhoC (45–47). Interestingly, the two latter pathways have also been implicated in c-Jun regulation (48, 49). Recent evidence points to a functional interaction between the cytoskeleton and the translation machinery (50). The microtubule and actin networks are associated with polysomes and translation effectors, such as aminoacyl-tRNA synthetases and translational initiation and elongation factors, are involved in the targeting and transport of mRNA molecules and can actively regulate global and local translation. It is possible that cytoskeleton dynamics trigger a signaling pathway that functionally contributes to IRES-mediated translation of c-Jun. Our findings provide evidence that human c-Jun is an IRES-containing cellular transcript and that, similar to some previously identified IRES-containing transcripts, it contributes to cancer development through cap-independent translation. This previously undescribed mechanism of c-Jun regulation offers unique potential targets for therapy.
Materials and Methods
Reagents and Plasmids.
Nocodazole, VOOH, and rapamycin were purchased from Sigma. Latrunculin B was a gift from Y. Kashman (Tel Aviv University, Tel Aviv, Israel). EGF was purchased from R&D Systems. LY294002 was purchased from A.G. Scientific. The reporter constructs 5XcollTRE-TATA-Luc (TRE-TATA), 5Xjun2TRE-TATA-Luc (Jun2-TATA), and TATA-Luc (TATA) (37) were gifts from P. Angel (German Cancer Research Center, Heidelberg, Germany). The expression vector for dominant-negative c-Jun, pEGP-TAM67 (3), was kindly provided by R. F. Hennigan (University of Cincinnati, Cincinnati, OH). The pR-F, pLRp27F, and pRGAPDHF bicistronic reporter constructs (33) were gifts from L. Hengst (Innsbruck Medical University, Innsbruck, Austria), and pREMCVF (32) was a gift from G. J. Goodall (Center for Cancer Biology, Adelaide, Australia). The HA-Jun expression vectors (pHAjun and p5′HAjun3′) and the luciferase reporter, pjLuc, have been described previously (22). Generation of bicistronic pR-F plasmids containing the c-Jun 5′UTR (pR5′UTRF) or fragments thereof (pR549-974F, pR1-562F, pR271-577F, pR1-397F, pR1-277F, pRΔ53-120F, pR1-141F, pR28-203F, and pRΔ145-191F) and plasmids with a hairpin structure at the transcription start site (pLR1-277F and pLR1-397F) is described in SI Materials and Methods and primers used for cloning are shown in Table S1. A panel of shRNA constructs for c-Jun and a control vector encoding a noneffective 29-mer cassette were purchased from OriGene Technologies. shRNA constructs with the strongest effect on c-Jun (c-Jun 5 and c-Jun 7) were used for further experiments. CMV-Rnl (Promega) and pCDNA3 (Clontech) are both commercial vectors.
Tissue Samples and Immunohistochemical Analysis.
All tissue samples were obtained in accordance with the ethical guidelines of the University of Regensburg Medical Center and approved by the ethical committee of the University of Regensburg (application number 09/101). For protein and RNA analysis, the samples were collected from surgical specimens, quick-frozen immediately in precooled isopentane, and stored at −80 °C until further analysis. Histological diagnosis of the tumor samples was performed by an independent pathologist. Each tissue sample was divided in two and processed for RNA or protein preparation. For immunohistochemistry, paraffin-embedded sections were deparaffinized, rehydrated, and subsequently incubated with primary rabbit anti–c-Jun antibody (Santa Cruz Biotechnology) overnight at 4 °C. The secondary biotin-labeled anti-rabbit antibody (DAKO) was incubated for 30 min at room temperature, followed by incubation with streptavidin-POD (DAKO) for 30 min. Antibody binding was visualized using AEC-solution (DAKO). Finally, the sections were counterstained with hemalum solution (DAKO). The evaluation of the staining was performed semiquantitatively by light microscopy.
Cell Culture.
Rat primary glia cultures were prepared from cerebral cortices of 1- to 2-d-old Sprague–Dawley rat pups, as previously described (51). The experiments were conducted in accordance with regulations and guidelines of the animal care and use committee of Tel-Aviv University. The detailed protocol is included in SI Materials and Methods. Glioblastoma human (U87, A172, HTZ349, HTZ17, HTZ417, LN229, and U251) or rat (C6) cell lines and HeLa cells were grown at 37 °C in DMEM supplemented with 10% (vol/vol) FBS, in a humidified atmosphere containing 5% CO2. Cells were treated with drugs at the following end concentrations and periods of time: Noc (30 μg/mL) and latrunculin B (4 μg/mL) for 18 h, EGF (100 ng/mL) for 30 min, VOOH (0.1 mM) for 15 min, and rapamycin (100 nM) and LY294002 (20 μM) for 24 h. Cell proliferation was assessed using the Cell Proliferation Kit II (XTT; Roche Applied Science) according to the protocol supplied.
Migration and Invasion Assay.
Migration and invasion assays were performed as previously described (52, 53). Briefly, migration was assessed in Boyden chambers containing polycarbonate filters with a pore size of 8 μm (Costar) coated with gelatin. The lower compartment was filled with fibroblast-conditioned medium used as a chemoattractant, and the filter was placed above. Cells were harvested by trypsinization and resuspended in DMEM without FCS. Cell suspensions (800 μL) at a density of 3 × 104 cells/mL were placed in the upper compartment of the chambers. After incubation at 37 °C for 4 h, filters were removed and cells adhering to the lower surface were fixed, stained, and counted. For invasion assays, 2.5 × 105 cells/mL were used and filters were coated with a commercially available reconstituted basement membrane (Matrigel, diluted 1:3 in H2O; Becton Dickinson). Each condition was assayed in triplicate, and assays were repeated at least twice.
Protein Preparation and Western Blot Analysis.
Cellular protein extracts were prepared by sonication of the cells in passive lysis buffer (Promega) containing a mixture of protease inhibitors (Roche Diagnostics). For analysis of LY294002, rapamycin, and MAPK activity, a mixture of phosphatase inhibitors (Roche Diagnostics) was added. The lysate was centrifuged at 20,000 × g for 15 min at 4 °C. Equal protein samples (20–40 μg) were separated on 10% (wt/vol) or 15% (wt/vol) for analysis of 4E-BP1) SDS-polyacrylamide gels and analyzed by Western blotting using Odyssey Blocking Buffer (LI-COR Biosciences) and the following antibodies: anti–c-Jun (Transduction Laboratories); anti–HA-tag (Covance); anti-FL (Chemicon International); antitubulin, anti-phospho ERK, and anti-ERK (Sigma); anti-JNK, anti-p38, anti–phospho-c-Jun, and anti–c-Fos (Santa Cruz Biotechnology); anti–phospho-p38, anti–phospho-JNK, anti-S6, and anti–phospho-S6 (Cell Signaling Technology); and anti–4E-BP1 (Abcam). Anti-mouse or anti-rabbit IgG coupled to IRDye 800CW (LI-COR Biosciences) was used as a secondary antibody, and protein bands were visualized by the Odyssey infrared imaging system (LI-COR Biosciences). Bend intensity was determined using Odyssey software (LI-COR Biosciences).
Isolation and Quantification of RNA.
Total RNA was isolated from tissue samples using the RNAeasy Mini Kit (Qiagen) and from cell cultures using the EZ-RNA reagent (Biological Industries) according to the manufacturers’ instructions. RNA was analyzed by Northern blot and quantitative RT-PCR as previously described (18). The detailed protocol is included in SI Materials and Methods.
In Vitro Transcription.
The bicistronic plasmids pR-F, pR1-277F, and pRGAPDHF (containing T7 promoter upstream to the Renilla cistron) were linearized using BamHI. Capped and polyadenylated transcripts were synthesized using the T7 mScript mRNA Production System (Epicentere) according to the protocol supplied. RNAs were purified by LiCl precipitation. An aliquot of each RNA was run on an agarose gel to verify RNA quality.
DNA and RNA Transfection and Luciferase Assay.
For DNA or RNA transfection, cells (7.5 × 105 per well) were seeded into six-well plates 24 h before transfection. DNA (3 μg) was transfected to U87 cells using the ProFection mammalian transfection system (Promega) and to C6 or HeLa cells using jetPEI (Polyplus transfection) according to the protocols supplied. Protein extracts for immunoblotting were prepared 48 h after transfection. Clones of U87 cells, stably transfected with c-Jun or control shRNA, were selected in the presence of puromycin (0.65 μg/mL; Sigma). RNA (4 μg) was transfected to U87 cells using the TransIT-mRNA transfection kit (Mirus) according to the manufacturer’s recommendations. FL and RL activities were assayed 48 h after DNA transfection and 10 h after RNA transfection, using the Dual-Luciferase Reporter Assay Systems (Promega) according to the manufacturer’s instructions and were recorded by a luminometer (LKB).
Pulse–Chase Analysis.
Pulse–chase analysis was performed as described before (22). Briefly, C6 and U87 cells were pulse-labeled with 200 μCi/mL [35S]methionine and [35S]cysteine (PerkinElmer Life and Analytical Sciences) in methionine-free medium for 60 min and chased in medium containing 2 mM unlabeled methionine for the indicated periods. Total cell extracts were immunoprecipitated with protein A/G-Sepharose (Santa Cruz Biotechnology) bound to anti c-Jun antibodies (Transduction Laboratories). Bound proteins were separated by SDS/PAGE, transferred to nitrocellulose, and analyzed by autoradiography. Bend intensities were determined using EZQuant-Gel software (EZQuant, Israel). The identity of the c-Jun protein was verified by immunoblotting.
Measurement of Translation Efficiency.
To measure the rate of de novo c-Jun synthesis, primary glia and C6 cells or HeLa cells, untreated or treated with EGF, were pulse-labeled with 200 μCi/mL [35S]methionine and [35S]cysteine in methionine-free medium for 30 min. Total cell extracts were immediately prepared, and the c-Jun protein was immunoprecipitated and analyzed as described above. In parallel, total RNA was prepared from duplicated cell cultures, and the cellular amount of c-Jun mRNA was determined by real-time RT-PCR as described above. Translation efficiency was calculated as the ratio between the rate of de novo synthesis of the c-Jun protein and the cellular amount of the c-Jun mRNA.
Supplementary Material
Acknowledgments
We thank Drs. P. Angel, R. F. Hennigan, L. Hengst, and G. Goodall for providing plasmids and Dr. Y. Kashman for the donation of latrunculin B. This research was supported by the Israel Cancer Association, the Israel Ministry of Health, and Israel Science Foundation Grant 425/08.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 16770 (volume 109, number 42).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203659109/-/DCSupplemental.
References
- 1.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: An overview. Mol Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 3.Hennigan RF, Stambrook PJ. Dominant negative c-jun inhibits activation of the cyclin D1 and cyclin E kinase complexes. Mol Biol Cell. 2001;12:2352–2363. doi: 10.1091/mbc.12.8.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young MR, et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eferl R, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 6.Dong Z, et al. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Lamb RF, et al. AP-1-mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol Cell Biol. 1997;17:963–976. doi: 10.1128/mcb.17.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper SJ, et al. Expression of dominant negative c-jun inhibits ultraviolet B-induced squamous cell carcinoma number and size in an SKH-1 hairless mouse model. Mol Cancer Res. 2003;1:848–854. [PubMed] [Google Scholar]
- 9.Lopez-Bergami P, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spangler B, Vardimon L, Bosserhoff AK, Kuphal S. Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res. 2011;24:148–164. doi: 10.1111/j.1755-148X.2010.00787.x. [DOI] [PubMed] [Google Scholar]
- 11.Mathas S, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002;21:4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 13.van Dam H, et al. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 16.Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 17.Lallemand D, et al. Stress-activated protein kinases are negatively regulated by cell density. EMBO J. 1998;17:5615–5626. doi: 10.1093/emboj/17.19.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knirsh R, et al. Loss of E-cadherin-mediated cell-cell contacts activates a novel mechanism for up-regulation of the proto-oncogene c-Jun. Mol Biol Cell. 2009;20:2121–2129. doi: 10.1091/mbc.E08-12-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reisfeld S, Vardimon L. Cell to cell contacts control the transcription activity of the glucocorticoid receptor. Mol Endocrinol. 1994;8:1224–1233. doi: 10.1210/mend.8.9.7838155. [DOI] [PubMed] [Google Scholar]
- 20.Vardimon L, Ben-Dror I, Oren A, Polak P. Cytoskeletal and cell contact control of the glucocorticoid pathway. Mol Cell Endocrinol. 2006;252:142–147. doi: 10.1016/j.mce.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Rüegg J, Holsboer F, Turck C, Rein T. Cofilin 1 is revealed as an inhibitor of glucocorticoid receptor by analysis of hormone-resistant cells. Mol Cell Biol. 2004;24:9371–9382. doi: 10.1128/MCB.24.21.9371-9382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polak P, et al. The cytoskeletal network controls c-Jun translation in a UTR-dependent manner. Oncogene. 2006;25:665–676. doi: 10.1038/sj.onc.1209114. [DOI] [PubMed] [Google Scholar]
- 23.Oren A, et al. The cytoskeletal network controls c-Jun expression and glucocorticoid receptor transcriptional activity in an antagonistic and cell-type-specific manner. Mol Cell Biol. 1999;19:1742–1750. doi: 10.1128/mcb.19.3.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furnari FB, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 25.Rickman DS, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 26.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 27.van den Boom J, et al. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163:1033–1043. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 29.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 30.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 32.Bert AG, Grépin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1alpha and other cellular 5′ UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 37.van Dam H, et al. Autocrine growth and anchorage independence: Two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 1998;12:1227–1239. doi: 10.1101/gad.12.8.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dam H, Castellazzi M. Distinct roles of Jun : Fos and Jun : ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 39.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal A, Briggs J, Rinehart-Kim J, Basso J, Bos TJ. The chicken c-Jun 5′ untranslated region directs translation by internal initiation. Oncogene. 2000;19:2836–2845. doi: 10.1038/sj.onc.1203601. [DOI] [PubMed] [Google Scholar]
- 41.Staber PB, et al. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- 42.Short JD, Pfarr CM. Translational regulation of the JunD messenger RNA. J Biol Chem. 2002;277:32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 44.Stoneley M, Willis AE. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 45.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 46.Yan B, Chour HH, Peh BK, Lim C, Salto-Tellez M. RhoA protein expression correlates positively with degree of malignancy in astrocytomas. Neurosci Lett. 2006;407:124–126. doi: 10.1016/j.neulet.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 47.Panopoulos A, Howell M, Fotedar R, Margolis RL. Glioblastoma motility occurs in the absence of actin polymer. Mol Biol Cell. 2011;22:2212–2220. doi: 10.1091/mbc.E10-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spangler B, et al. ETS-1/RhoC signaling regulates the transcription factor c-Jun in melanoma. Int J Cancer. 2012;130:2801–2811. doi: 10.1002/ijc.26277. [DOI] [PubMed] [Google Scholar]
- 49.Marinissen MJ, et al. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol Cell. 2004;14:29–41. doi: 10.1016/s1097-2765(04)00153-4. [DOI] [PubMed] [Google Scholar]
- 50.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 51.Mor E, et al. Species-specific microRNA roles elucidated following astrocyte activation. Nucleic Acids Res. 2011;39:3710–3723. doi: 10.1093/nar/gkq1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothhammer T, et al. The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell Mol Life Sci. 2004;61:118–128. doi: 10.1007/s00018-003-3337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albini A, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]