Throughout the world’s oceans, the sediments that cover the seafloor, and the cracks and fissures of the basaltic ocean crust, are permeated by microbial life (1, 2). As these bacteria and archaea are mostly not available in laboratory culture and thus elude direct physiological characterization, Jørgensen et al. correlate geochemical regimes in subseafloor sediments with in situ abundance of specific groups of uncultured bacteria and archaea to infer their habitat preferences, to develop specific hypotheses about their metabolism, and to design promising cultivation strategies (3).
The abundance of microbial cells in subseafloor sediments correlates with sedimentation rate and distance from land; cell densities in seafloor sediment generally increase toward continental margins and shelves, and decline toward the open ocean (1). This pattern is consistent with the observation that bacteria and archaea in the subsurface assimilate buried organic carbon as heterotrophic anaerobes (4). In principle, more specific information about carbon and energy sources of the subsurface biosphere could be obtained by culturing representative species and genera of subsurface bacteria and archaea, followed by study of defined enrichments and pure cultures in the laboratory. However, selectively enriching and isolating subsurface microorganisms in the absence of specific hypotheses that could guide such an effort remains an ongoing challenge. Although some progress has been made, the major evolutionary lineages of subsurface bacteria and archaea have so far remained uncultured or contain very few cultured representatives; therefore, the specific metabolic activities and biogeochemical roles of most subsurface microorganisms are obscure (5, 6). Given these limitations, a systematic study of the in situ geochemical conditions of a microbial subsurface environment should reveal much about the physiological preferences and capabilities of its inhabitants, and offer clues about cultivation conditions and strategies. As specific groups of bacteria or archaea use only specific electron donors and acceptors, they should occur predominantly in sediment horizons that match these physiological preferences (Fig. 1).
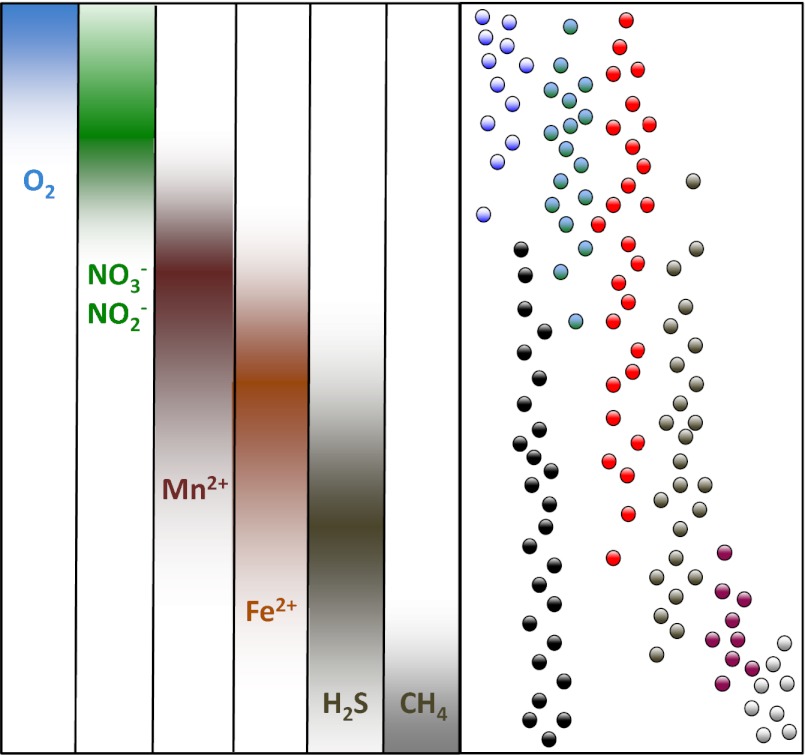
Fig. 1.
Simplified scheme of stratified microbial populations correlated to chemical zonation as result of microbial respiration (blue, aerobic respiration; green, nitrate reduction; maroon, manganese reduction; rust color, iron reduction; black-gray, sulfate reduction; light gray, methanogenesis). The dots on the right show typical downcore distributions of different microorganisms. The oxygen- and nitrate-respiring physiologies of the blue and green microbes are apparent. However, the red microbe appears to be a generalist that thrives throughout the oxic to the iron-reducing zone, whereas the sulfate reducer (in dark gray) also thrives in the metal-reducing zone. The uncultured microorganism in purple appears to be a sulfate-dependent anaerobic methane oxidizer, sandwiched between the sulfate reducers and the methanogens (light gray). Finally, the uncultured subsurface archaeon in black avoids oxygen, but is otherwise ubiquitous. These examples have actual precedents and illustrate the challenges of assigning metabolism based on geochemical stratification.
In their study of Arctic sediment microbiota, Jørgensen et al. (3) apply this approach in unprecedented thoroughness: pore water concentration gradients of major microbial electron acceptors that are essential for microbial respiration (nitrate, oxidized metals, sulfate), and electron donors that function as microbial energy sources (ammonia, sulfide, organic C), were systematically examined for correlations with the abundance of major subsurface bacterial and archaeal phylum-level groups. As phylogenetically specific counts of bacterial and archaeal cells in sediments run into methodological limitations, a molecular proxy was used: the relative abundance of bacterial and archaeal DNA fragments in high-throughput sequencing analyses. Among the microbial groups detected in this way, many have resisted cultivation in their entirety, such as the bacterial Japan Sea group I (7) and the Deep Sea Archaeal Group (8). Their names do not reflect any physiological characteristics, but instead refer to their predominant habitat or the location of their discovery. Both are assumed to be heterotrophic anaerobes; they require sedimentary buried organic matter as a carbon source, and avoid oxygen and do not require it for respiration. Other study subjects fall into what could be called the just barely cultivated category. The Marine Group I archaea constitute the predominant archaea in the marine water column; their cultured representatives gain energy by oxidizing ammonia to nitrite with oxygen as the physiological electron acceptor, and build up cell biomass by autotrophically assimilating inorganic dissolved carbon (9, 10).
The relative abundance profiles of different microbial groups in the sediment column should reflect the chemical microenvironments of subsurface sediments, as they change in a generally predictable downward sequence: oxygen, the most desirable electron acceptor with a high respiratory energy yield, is depleted first, followed by nitrate, and then by a procession of less familiar, less energy-rich but still microbially acceptable electron acceptors, most importantly oxidized metals, sulfate, and finally CO2 (Fig. 1). How are the observed depth profiles of subsurface bacteria and archaea correlated to the availability and utilization of specific electron acceptors?
Predictions about habitat range and physiology were flamboyantly confounded by the Marine Group I archaea. Although the few cultured representatives of this group are aerobes, Marine Group I archaea extend downward into the nitrate-reducing and metal-reducing zones of the sediment column. Their abundance in the pyrosequencing surveys correlates with the quantity of nitrate that persists absorbed to mineral particles, even after pore water nitrate is depleted. Thus, geochemical logic dictates that the Marine Group I archaea in the sediment column should respire anaerobically with nitrate. Nitrate reduction has been suggested previously, based on clone library detection of Marine Group I archaea in anoxic, nitrate-containing sediments (11), but Jørgensen et al. (3) go further and establish a quantitative link between nitrate availability and Marine Group I abundance. Interestingly, anaerobic, nitrate-respiring Marine Group I archaea are not only physiologically but also phylogenetically distinct from their aerobic counterparts in the water column, and form their own branches within the Marine Group I lineage—a beautiful demonstration that critical biogeographic patterns are often only accessible at higher resolution, and will go unnoticed by phylum-level molecular surveys.
Another interesting correlation was observed for the Deep Sea Archaeal Group, also known by its synonym Marine Benthic Group B (12). This widespread, apparently heterotrophic, archaeal group occurs predominantly in organic-rich marine sediments and has a checkered history of theories what it is supposed to be: Links to sulfate-dependent anaerobic methane oxidation have been proposed but remain unverified. Here, relative deep-sea archaeal group abundance in the pyrosequencing survey mirrors the relative contributions of total organic carbon to all sedimentary carbon, and of ferric iron oxide (i.e., Fe2O3) to total sediment weight, suggesting iron reducing degradation of organic matter as a likely metabolism—an eminently testable hypothesis.
Within the bacterial domain, this study substantiates several phylum-level trends: downcore, the relative abundance of the Chloroflexi and Planctomycetes increases from the oxic surface sediment toward anoxic sediment horizons with nitrate and
This study provides geochemically informed, carefully calibrated inferences on microbial metabolism for specific subsurface lineages.
with reduced iron and manganese. Refining the observed correlations with higher phylogenetic resolution, especiallywithin the highly diversified Chloroflexi and Planctomycetes (13), should provideadditional clues about the manner in which environmental controls and preferred geochemical niches partition specific subgroups within these bacterial phyla. In deeper sediment layers that contain sulfide, either as pore water sulfide or in solid phase as pyrite, the Japan Sea I group appears, and shows distribution patterns that appear linked to specific subgroups of the sulfate reducers within the Deltaproteobacteria, and the sulfur-oxidizing Epsilonproteobacteria. Thus, involvement of the Japan Sea I group in the sulfur cycle appears likely, and is consistent with the widespread occurrence of this group in reduced marine sediments (7).
This study (3) provides geochemically informed, carefully calibrated inferences on microbial metabolism for specific subsurface lineages, and, by the same token, generates testable hypotheses about cultivation strategies. Equipped with defensible working hypotheses, microbiologists are now in a better position to choose rational enrichment and isolation strategies for specific groups of subsurface bacteria and archaea. Those who have tried their hands at targeted enrichments and isolations appreciate that this means essentially placing bets on their favorite metabolic theory regarding uncultured or poorly cultured phyla. However, the odds improve with a better basis of well informed observations. The authors of this study have to be thanked for energizing the game.
Footnotes
References
- 1.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109(40):16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev. 2011;75(2):361–422. doi: 10.1128/MMBR.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jørgensen SL, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci USA. 2012;109:E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103(10):3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol. 2008;66(2):181–196. doi: 10.1111/j.1574-6941.2008.00566.x. [DOI] [PubMed] [Google Scholar]
- 6.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2008;2(1):3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 7.Webster G, Parkes RJ, Fry JC, Weightman AJ. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol. 2004;70(9):5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152(4):1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 10.Hallam SJ, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4(4):e95. doi: 10.1371/journal.pbio.0040095. and erratum (2006) 4(12):e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin AM, Teske A. Sediment-associated microdiversity within the marine group I Crenarchaeota. Environ Microb Rep. 2010;2:693–703. doi: 10.1111/j.1758-2229.2010.00163.x. [DOI] [PubMed] [Google Scholar]
- 12.Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65(10):4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durbin AM, Teske A. Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ Microbiol. 2011;13(12):3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x. [DOI] [PubMed] [Google Scholar]