Abstract
The parameters in a complex synthetic gene network must be extensively tuned before the network functions as designed. Here, we introduce a simple and general approach to rapidly tune gene networks in Escherichia coli using hypermutable simple sequence repeats embedded in the spacer region of the ribosome binding site. By varying repeat length, we generated expression libraries that incrementally and predictably sample gene expression levels over a 1,000-fold range. We demonstrate the utility of the approach by creating a bistable switch library that programmatically samples the expression space to balance the two states of the switch, and we illustrate the need for tuning by showing that the switch’s behavior is sensitive to host context. Further, we show that mutation rates of the repeats are controllable in vivo for stability or for targeted mutagenesis—suggesting a new approach to optimizing gene networks via directed evolution. This tuning methodology should accelerate the process of engineering functionally complex gene networks.
Keywords: gene network optimization, evolvability, synthetic biology
Engineering reliable and predictable synthetic gene networks presents unique challenges because genetic parts such as promoters, ribosome binding sites, and protein coding regions often behave unexpectedly when used in novel designs. Noise (1, 2), metabolic load (3), poorly characterized interactions with the host (4), and general uncertainty about the detailed functionality of parts conspire to limit the complexity of synthetic gene networks to a small number of interacting genes (5, 6). As a result, a complex gene network that has been predicted analytically to perform well may in fact perform poorly, if it works at all, when ultimately implemented in cells. Furthermore, even if a gene network can be made to perform well in a particular strain and environment, there is no guarantee that the same network will perform well if ported to a new strain or environment (7). More generally, synthetic networks often operate in substantially different parameter regimes than expected during the design process and must, therefore, be tuned (2, 8–11) before they function properly, and even retuned when used in new environments or with different hosts.
One way to improve the performance of a poorly tuned gene network is to introduce focused variability into the design, generating a library of circuits (12–15) with the same genetic components and connectivity, but with each member of the library operating in a different parameter regime. In bacteria, for example, promoters (16, 17), ribosome binding sites (RBS) (18, 19), RNA stability (20, 21), protein stability (22), and other biochemical details such as transcription factor regulation (23) or enzyme catalysis (24) can be varied to sample different regions of parameter space. Furthermore, sensitivity analysis (25) can guide the designer to parameters that, when tuned, are most likely to result in improved performance. Subject to screening or selection, the effectiveness of a tuning library is proportional to the range of parameters it samples and is inversely proportional to its size. More specifically, a useful tuning approach explores the parameter space over a large range with high resolution; results in a predictable relationship between the genetic sequence and the values of the corresponding parameters; and is scalable to complex networks. Ideally, a good tuning method should also be evolvable, forcing the host organism to focus mutations on highly tunable elements in the network, such that it complements directed evolution techniques (26, 27) by more frequently sampling mutations that enhance functionality.
Here, we introduce a tuning mechanism for gene networks in Escherichia coli that couples the straightforward tunability of translation initiation rates via the RBS spacer region (18, 28, 29) with the high mutation rate and strong bias for insertion/deletion mutations inherent to simple sequence repeats (SSR) (30). We implement this mechanism by embedding mono- or di-nucleotide SSRs between the Shine–Dalgarno sequence and the start codon of target genes. We call this sequence motif the rbSSR.
We describe multiple methods to generate libraries of rbSSR sequences that vary in repeat number, using them to evaluate this tuning approach against the criteria above. We found that these libraries incrementally and predictably sample gene expression levels over a 1,000-fold range, and that the range of expression can be expanded by coarsely tuning promoter strength. We demonstrate the utility of the approach by fine-tuning three functional behaviors of a bistable switch built with dual rbSSRs, and illustrate the need for tuning by showing that the genomic context of a host strain can have profound effects on the switch’s behavior. We also show that rbSSR sequences are stable over more than 200 generations, but that destabilization of the repeats in a mutator strain focuses mutations to the spacer region, which could be used to tune and select for optimized gene networks in vivo. These results are broadly applicable to rapidly engineering functional gene circuits and scaling up circuit complexity by enabling the creation of expression libraries that thoroughly and predictably sample the parameter space of a gene network.
Results and Discussion
Explorability and Predictability of rbSSRs.
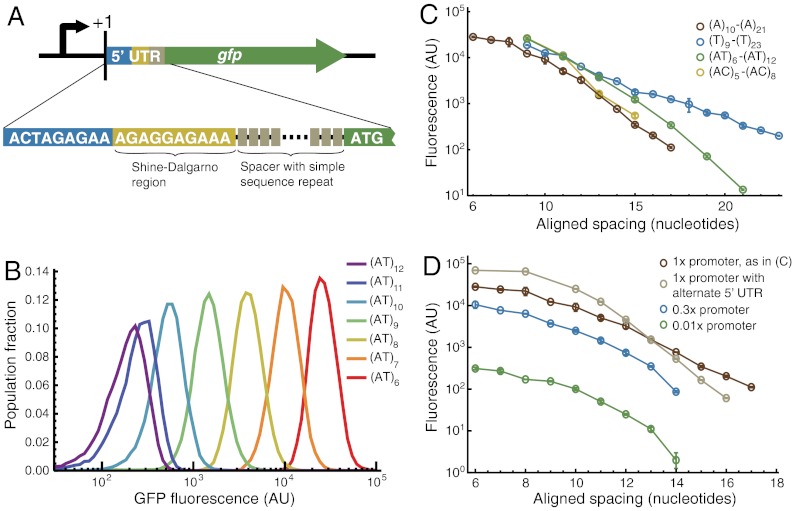
To understand the resolution and limits of translational control with rbSSRs, we experimentally examined four rbSSR spacer motifs: (A)n, (T)n, (AT)n, and (AC)n; that is, n repeats of either a single or a pair of nucleotides. For each motif, we constructed a parent plasmid with a constitutive promoter, a strong Shine–Dalgarno region, and an initial rbSSR spacer, driving the expression of a gfp gene (Fig. 1A). Taking advantage of the inherent instability of repeats during replication, especially in PCR (31), we generated plasmid libraries by amplifying a region of each parent plasmid flanking the rbSSR sequence and re-inserting the mutated amplicons into pre-cut plasmid backbones (Fig. S1A). The resulting plasmid libraries were transformed into E. coli and screened visually and via cytometry for unique fluorescence levels to produce a strain library (rbSSR-GFP) of repeat lengths for the four spacer motifs.
Fig. 1.
The rbSSR construct and rbSSR-GFP library characterization. (A) The rbSSR construct. A simple sequence repeat is embedded in the spacer region of the ribosome binding site between the Shine–Dalgarno sequence and the coding sequence of the target gene, in this case gfp. (B) Fluorescence distributions of the (AT)6–(AT)12 rbSSR-GFP library expressed from constitutive promoter J23100 and measured by flow cytometry. Varying the spacer length evenly samples a large expression range over three orders of magnitude with uniform noise properties. (C) Mean GFP expression levels for rbSSR-GFP libraries generated from parent plasmids (A)20, (T)20, (AT)12, and (AC)8 (Table S4). Increasing the spacer length generally lowers gene expression, though the trend of the decline is sensitive to the nucleotide composition of the repeats. Each library uses identical promoter (J23100) and 5′ UTR sequences. Error bars represent standard error from three colonies. (D) Mean GFP expression levels for multiple (A)n rbSSR-GFP libraries, as in C. Tuning translational and transcriptional efficiencies through regulatory sequences in the 5′ UTR or promoter regions (Table S5), respectively, can be used in conjunction with rbSSR libraries to more broadly sample the expression space.
We measured the fluorescence output of rbSSR-GFP library strains via flow cytometry of exponentially growing cells. The mean intensity decreased linearly in log fluorescence over a 100- to 1,000-fold range as the number of rbSSR repeats increased (Fig. 1B). The cell-cell variation in GFP levels for each strain is considerably more uniform than what is observed for more noise-prone tuning approaches, such as the dose-response of an inducible promoter (32) (Fig. S2). The rate at which fluorescence intensity decreases depends on the nucleotide composition of the spacer, with the steepest decline for (A)n and the most gradual decline for (T)n (Fig. 1C). The overall trend of the decline roughly corresponds to computational predictions (18), but with increasing disparity as the nucleotide composition of the spacer deviates from poly-(A) residues (Fig. S3).
Creating rbSSR libraries is compatible with existing combinatorial or compuational tuning approaches for transcription (12, 17) and translation (18, 19) rates, as well as for RNA stability (21), and results in efficient sampling of the expression space with a predictable mapping from sequence to expression. By coupling rbSSRs with promoters of different strengths or by altering the 5′ untranslated region (UTR) of the transcript, different regions of the expression space spanning nearly five orders of magnitude can be sampled (Fig. 1D). Through PCR mutagenesis of the rbSSR, we generated no fewer than nine bases of repeat sequence, which—depending on the nucleotide composition of the RBS—results in spacing near the optimum of five bases (29) (Fig. S3A).
Scaling Up Complexity: Fine-Tuning a Bistable Switch.
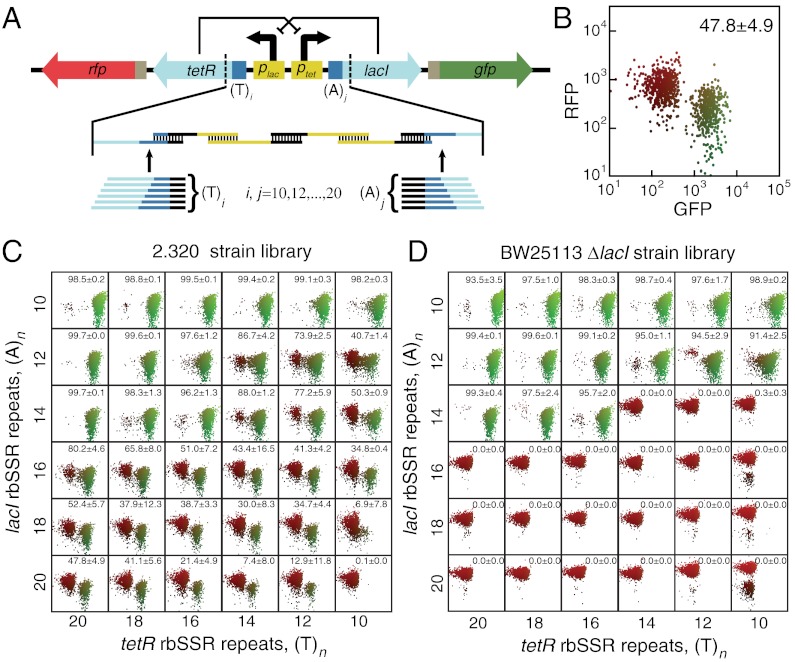
To demonstrate that rbSSRs can be used to fine-tune functional gene networks, we built an rbSSR-enhanced bistable switch (Fig. 2A) using the same architecture as the mutually inhibitory switch described by Gardner, Cantor, and Collins (33). For our switch, rbSSRs separately drive the expression of the repressor proteins LacI and TetR, which are expressed bicistronically with GFP and RFP, respectively.
Fig. 2.
Construction of rbSSR-BSS library and its characterization in two lacI- strains. (A) The mutually inhibitory bistable switch architecture with (T)i rbSSR upstream from tetR and (A)j rbSSR upstream from lacI repressor genes. A combinatorial library was built using oligos that encode different length rbSSR sequences. (B) Scatter plot showing fluorescence distribution of (T)20/(A)20 rbSSR-BSS variant in strain 2.320. The horizontal axis is the GFP fluorescence level, and the vertical axis is the RFP fluorescence level. The color indicates whether the cell is in the red, green, or mixed subpopulation, and the number represents the mean and standard error of the percent of cells in the green state, measured from three colonies. Each scatter plot displays 1,000 points selected at random from the associated cytometry data. (C) Grid of scatter plots for the 2.320 strain library plotted as in B. The comparative strengths of rbSSR pairs determine the distribution of cells in the LacI- and TetR-dominant states. (D) Grid of scatter plots for the BW25113 ΔlacI strain library showing the fluorescence distributions of the majority colony type. The host strain affects the behavior of the switch library primarily by sharpening the boundary between strains in which one or the other state dominates and results in fewer bimodal constructs.
The dominant state and spontaneous switching rate between states for this circuit depend on the initial state of the system, the expression strength of each repressor gene, the stability of the associated proteins and mRNAs, the rate of leaky transcription for each repressor/promoter combination (see Fig. S4), plasmid copy number, and circuit-host interactions that affect global expression dynamics and growth rate. As a result, it is difficult to predict a priori if one state will dominate or if each state will be equally likely when the switch is expressed in a given strain. In fact, our first assays with this switch architecture, untuned and expressed in E. coli strain DH5α, showed that the switch was only stable in one state: Despite an initial bias to the LacI-dominant state, cells that spontaneously switched to the TetR-dominant state eventually swept the population due to a growth rate advantage (Fig. S5).
Depending on the values of the network parameters, the mutual inhibition circuit architecture can produce a range of useful behaviors. If both states are highly stable, cells can act as molecular detectors, responding to environmental signals and retaining the detected state over tens of generations in the absence of the signal (33). If one state is sufficiently less stable than the other, cells can act as programmable timers when initialized in the less stable state (12). If the two switch states are well-balanced, cells could perform coin-flipping behaviors as a noisy switch. A synthetic bistable switch encoding coin-flipper behaviors could be a useful foundational circuit for implementing synthetic multicellular systems that emerge from individual cells, such as bet-hedging, pattern formation, and division of labor (34).
To make our switch easily tunable, we incorporated poly-(T) and poly-(A) rbSSRs between identical, unoptimized 5′ UTR sequences and the tetR and lacI genes, respectively. From this design we built a 36-strain combinatorial library (rbSSR-BSS) using oligo assembly (35) (Fig. S1C) of six single-stranded DNA fragments that encode the regulatory sequences for the two operons, including rbSSR spacers of length 10 to 20 repeats in steps of two (Fig. 2A). This library represents a coarse sampling (30%) of the reachable rbSSR expression space from 10 to 20 repeats. We transformed sublibrary assemblies that contained all six poly-(A) variants for each poly-(T) rbSSR into lacI- expression strain 2.320 for screening via flow cytometry and sequence verification.
We initially characterized the behaviors of the rbSSR-BSS strain library by observing the natural bias of each switch variant in fresh, uninduced 2.320 transformants. We used flow cytometry to capture a scatter plot of red and green fluorescence for the cells during exponential growth (Fig. 2B). We combined these results to produce a grid of scatter plots for the entire library (Fig. 2C) and observed a suprisingly broad range of switch distributions. As expected, a strong rbSSR for one repressor coupled with a weak rbSSR for the opposing repressor results in a natural bias toward the strong rbSSR state. However, the distribution of switch states is bimodal for the majority of the library variants, and the fraction of cells in the green state, while consistent for a single strain, shifts predictably as rbSSR strength is varied.
Context Matters.
The context in which a gene network such as the bistable switch is expressed can dramatically affect the performance of the network (4, 36), but its effect is, in general, poorly understood and not easily incorporated into modeling gene network performance. To illustrate this point, we transformed each variant of the rbSSR-BSS library into a second lacI- strain of E. coli—BW25113 ΔlacI—and performed the same initial characterization assay described above. While the genotypes of these two strains differ (see SI Text), one might expect the two to be functionally identical with regard to the circuit’s behavior because neither strain expresses LacI or TetR from the genome. In fact, the behaviors of the second strain, shown in Fig. 2D, are profoundly different. The same rbSSR pairs that generate bimodality in the original strain generally produce unimodal behaviors in the new strain. Also, while we observed little colony-to-colony variation in the distributions of switch states for the first strain library, nearly half of the strains in the second library exhibited two distinct colony phenotypes—one biased toward LacI-dominant cells, and the other biased toward TetR-dominant cells. For these strains, the distribution of colony types shifted predictably with rbSSR strength (Fig. S6).
To further characterize the behavior of the switch library, we performed induction and microscopy assays on subsets of the strain libraries. First, to observe the long-term stability of each switch, we forced strains to the TetR- and LacI-dominant states using the chemical inducers IPTG and aTc, respectively. After a period of induced growth, we washed the inducers away and monitored fluorescence distributions over 96 h of continuous growth (Fig. S7). We found that construct (T)12/(A)12 in both strain backgrounds exhibits robust bistable behaviors. In strain 2.320, the bistability is maintained for the duration of the experiment. For the weaker rbSSR pairing (T)16/(A)14, the LacI-dominant state is less robust, leading to mixed or TetR-dominant populations after 96 h. On the other hand, strains we found to have low growth rates when in the naturally dominant state (Table S1) behaved like monostable timer circuits, completely switching state after initialization to the slow-growth state. Finally, to investigate the switching dynamics of the uninduced switch, we performed microcolony growth assays from single cells for a subset of the 2.320 strain library (Table S2). Along the diagonal for roughly equal rbSSR lengths, we observed a few rbSSR pairings with a significant fraction of cells in transition between states, which resulted in mixed-state microcolonies (Fig. S8 and Movies S1–S3).
The difference between Fig. 2 C and D and the range of functional behaviors observed when varying rbSSR pairings both underscore the importance of tuning and highlight limitations of tuning a single network parameter. In the initial characterization assay, we estimate roughly 40 generations pass from the initial plating of transformants to the cytometry assay. Over that period, switch variants in strain 2.320 appear to approach steady-state distributions relatively quickly, evidenced by consistent fluorescence distributions across colonies. By contrast, the same constructs in BW25113 lacI- appear to maintain some initial state, resulting in two distinct colony phenotypes. For this circuit, it is likely that adjusting an additional network parameter, such as circuit copy number (37) or promoter leak (17) could compensate for the contextual differences between the two strains. Although it is unlikely that these differences could be predicted for two arbitrary strains, it is reasonable to expect that two arbitrary strains could be tuned to behave the same way—suggesting that building tuning into a system is an extremely important design consideration.
Stability and Evolvability of rbSSRs.
Although we are not aware of simple sequence repeats embedded within RBS spacers in any natural context, these repeat sequences are known to accelerate evolutionary adaptation when situated within coding sequences and other regulatory regions (30). SSRs undergo insertion/deletion mutations at rates four to five orders of magnitude higher than arbitrary sequences of the same length (38, 39). Some bacteria utilize SSR variability to strictly control protein expression via frameshift mutations in coding sequences or to alter transcription rates via insertions or deletions to promoter spacers (40). Repeats embedded in promoter and gene coding sequences are also found to be responsible for environmental adaptations in many higher organisms (30). When used in synthetic gene circuits, the instability of SSRs could be detrimental if it caused the performance of a circuit to degrade or detune over time. On the other hand, repeat variation could be a powerful tool to optimize circuit performance through directed evolution by focusing mutations to regions of the circuit that have strong and predictable effects on gene expression.
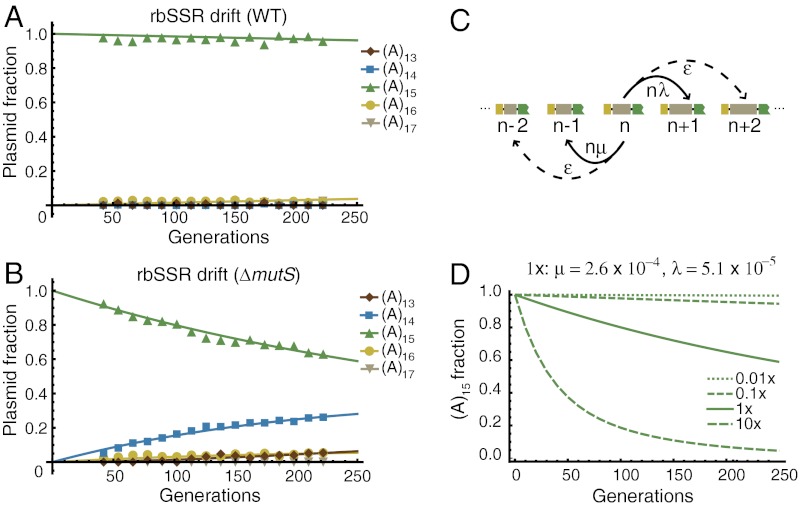
To examine the long-term stability of rbSSR sequences, we measured the sequence drift of the rbSSR-GFP gene circuit using DNA sequence trace analysis of serially-passaged wild-type and mutator strains over approximately 220 generations. Specifically, we transformed the (A)15 rbSSR-GFP plasmid into wild-type strain BW25113 and mismatch-repair deficient strain BW25113 ΔmutS, and cultured each strain in triplicate over 16 serial passages. We extracted plasmid DNA from each overnight culture for sequence trace analysis (see Fig. S9). For the wild-type strain, we observed no mutations in the gfp gene, including the regulatory and rbSSR sequences (Fig. 3A), which demonstrates that rbSSR sequences can remain stable for very long periods. However, when propagated in the mutator strain, we observed a strong bias for SSR deletions as the fraction of (A)14 plasmids increased steadily over time (Fig. 3B).
Fig. 3.
The stability of rbSSR sequences in vivo. (A) Distributions of (A)13 to (A)17 rbSSR repeats in the plasmid population as a function of estimated cell generation for strain BW25113. The distribution of rbSSR repeats was essentially unchanged after 220 generations. Curves are least-absolute-deviations fits to the model in C. (B) Distributions as in A for strain BW25113 ΔmutS. The fraction of plasmids with the original construct steadily decreased as the fraction of plasmids with a single-unit deletion increased. The fraction of plasmids with a single-unit insertion also increased, though at a slower rate. (C) Birth–death model of SSR variation. The probability of repeat unit insertion is nλ and repeat unit deletion is nμ per repeat unit per generation, given a replication template of repeat unit length n. The probability ϵ that two deletions or insertions occur in one generation is assumed to be zero. (D) Predictions for the fraction of (A)15 plasmids over time for multiple mutation rates, with 1× corresponding to the rates obtained from a fit of the model in C to the data in B. Mutation rates of 0.05× or below fit the data in A for the wild-type strain, suggesting that the mutator strain is at least 20 times more likely than wild-type to insert or delete a repeat.
By fitting a model of insertion/deletion mutations based on a birth-death process to the data (Fig. 3 C and D and SI Text), we inferred the mutation rates to be 2.6 × 10-4 deletions and 5.1 × 10-5 insertions per base pair of repeat sequence per generation, which are within the range of reported rates (38). We also inferred the SSR mutation rate in the mutator strain to be at least 20 times greater than in the wild-type strain, which is consistent with previous work (39). While we observed no mutations in the promoter or the gfp coding sequence for the mutator strain, we did find that rbSSR instability varied among replicates (Fig. S10) with one replicate resulting in a final distribution of plasmids with repeats (A)13 through (A)17. Note that these experiments were performed without any intentional selection, and the results are likely primarily due to drift.
Conclusion
The experiments described in this report suggest that a complex gene network may require substantial tuning to function as desired. Our approach to tuning synthetic bacterial gene networks uses a very simple construct: A sequence repeat in the spacer region of the RBS. Sequence repeats seem ideal for this purpose for a variety of reasons. First, the relationship between the length of the repeat and the strength of the resulting RBS is clear. Second, the range of expression obtained by coupling rbSSR libraries with other regulatory sequences, such as promoters, is large. Third, genetic instability can be focused on the RBS spacer, allowing rapid exploration of the expression space via PCR or combinatorial assembly methods. Additional experiments must be performed to demonstrate the potential of these repeats as tools for tuning gene networks in vivo via selection.
Our study also demonstrates that the range of behaviors of a gene network library, in our case of a bistable switch, can be substantial and highly sensitive to the host context. We have shown that cells expressing the switch can act as a molecular detector or a timer, with possible applications in medical diagnostics or industrial bioprocessing. However, growth inhibition strategies for the timer that rely on protein overexpression are not mutationally robust, so other strategies may be required to increase circuit stability. We have also shown that the same switch architecture, tuned properly, produces noisy coin-flipping behaviors that could be used as a core circuit element to initiate cell differentiation for synthetic multicellular systems.
We believe the same approach we have used to engineer highly tunable elements with simple sequence repeats can be extended to other network parameters in bacteria (40, 41) and to higher organisms (42) by tuning the spacing between known regulatory motifs such as those responsible for transcription initiation (12) or intron splicing efficiency (43). To continue scaling up functionally complex behaviors in synthetic gene networks, these approaches will likely need to be combined with tuning methods that control parameters that are untunable by our methods, including network connectivity, protein-protein interactions, and enzyme catalytic efficiencies. The present tuning approach will likely be a part of comprehensive strategies for fine-tuning gene circuits to perform optimally in a given context.
Materials and Methods
Strains and Media.
GFP library construction and assays were carried out in MG1655. Bistable switch library constructs were screened in 2.320 [Coli Genetic Stock Center (CGSC) accession number 6440] and assayed in strains 2.320 and BW25113 ΔlacI. Serial passage experiments for in vivo mutation rate analysis were carried out in wild-type BW25113 (CGSC accession number 7636) and mutator BW25113 ΔmutS strains. Strains BW25113 ΔlacI and BW25113 ΔmutS were created from JW0336-1 (CGSC accesstion number 8528) and JW2703-2 (CGSC accession number 10126), respectively, using plasmid pFLP2 (44) to excise the FRT-flanked kanamycin cassettes, followed by sucrose counterselection to cure the pFLP2 plasmid (45). M9 minimal media (M8000, Teknova) supplemented with 50 μg/mL kanamycin was used for growth and fluorescence assays. Serial passages were performed in LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) supplemented with 50 μg/mL kanamycin.
GFP Library Generation.
The backbone for the rbSSR-GFP libraries was generated from a parent plasmid with a p15A replication origin containing (A)20, (T)20, (AC)8, or (AT)12 by digestion with endonucleases XbaI and NdeI to excise a small region including the rbSSR, followed by gel extraction and purification. Spacer variation for the rbSSR-GFP libraries was generated by PCR with Phusion Flash master mix (F548, Finnzymes) using the parent plasmids as templates. PCR amplicons were purified by gel extraction. The backbone and rbSSR amplicon libraries were joined using Gibson assembly (46), and transformed directly into expression strain MG1655 for screening. Repeat lengths with fewer than nine base pairs were not observed from PCR reactions; plasmids for repeats (A)6–(A)9 (see Fig. S3A) were thus constructed by ordering oligos encoding each spacer, followed by PCR amplification and Gibson assembly to a PCR-amplified vector backbone. All constructs were verified by sequencing. A more detailed description of the method is found in the SI Text.
Bistable Switch Library Generation.
The backbone for rbSSR-BSS library construction was prepared from a parent plasmid with a p15A replication origin by digestion with endonucleases AclI and SnaBI, followed by gel extraction and purification. A set of six primers—O1R, O2F, O3R, O4F, O5R, and O6F (see Table S3)—was ordered (IDT) to encode the regulatory region and introduce rbSSR sequences. Six variant oligos encoding rbSSRs (T)10,12,…,20 and (A)10,12,…,20 were ordered for primers O1R and O6F, respectively. A short PCR amplicon from tetR was used to join the digested backbone to the oligos. Combinatorial libraries of all poly-(A) repeats for each poly-(T) repeat were generated using Gibson assembly with the backbone fragment, the PCR amplicon, and the proper mix of oligos, and transformed directly into expression strain 2.320 for screening. The regulatory regions between digestion sites for each library clone were verified by sequencing. A more detailed description of the method is found in the SI Text.
Cell Growth and Plate Reader Measurements.
For GFP assays, freshly streaked colonies were transferred in triplicate to 200 μL M9 minimal media in 96-well plates (Costar 3795) and grown to saturation overnight at 37 °C in a shaker. The cultures were then diluted 1∶100 in 200 μL prewarmed fresh broth in 96-well plates (Costar 3904), grown at 37 °C to OD600 0.15 to 0.2 in a plate reader (Biotek) with shaking. Optical densities (600 nm) and GFP measurements (485-nm excitation, 510-nm emission) were taken every 10 min. When grown to target density, 10 μL of each culture was transferred to 100 μL 1× PBS (Gibco) with 34 μg/mL chloramphenicol chilled at 4 °C. Bistable switch assays were performed similarly, with the exception that freshly transformed colonies were selected at random from plates after incubation for 12–14 h at 37 °C, and fluorescence measurements were made in the plate reader for RFP (590-nm excitation, 632-nm emission) in addition to optical density and GFP.
Flow Cytometry Measurements.
Diluted cultures from the plate reader measurements were transferred to a flow cytometer (C6 with CSampler, Accuri). To prevent well–well contamination, blank wells containing PBS were read after each sample well. GFP measurements (488-nm excitation, 533-nm emission) and RFP measurements (488-nm excitation, 610-nm emission) were recorded for 50,000 events per sample. Cells were gated using a rectangular gate in forward scatter and side scatter. For the GFP assays, background fluorescence levels from cells containing an empty vector without gfp were subtracted from the geometric mean of GFP expression for each sample culture. Fluorescence levels in both the GFP and RFP channels were used to generate the scatter plots for the bistable switch assays. Cytometry measurements were analyzed using custom Matlab scripts (see SI Text). For color compensations, 4.5% of the red channel fluorescence was subtracted from the green channel fluorescence, and 7.8% of the green channel fluorescence was subtracted from the red channel fluorescence. These percentages were determined using experimental data from strains with plasmids expressing only RFP or GFP, respectively. Cutoffs from color-compensated levels of 500 and 750 were used for classifying cells as “red” or “green,” respectively (see SI Text).
rbSSR Variation in Vivo.
Freshly transformed colonies of wild-type and mutator strains containing the (A)15 rbSSR-GFP plasmid were transferred in triplicate to 6 mL of LB supplemented with kanamycin and grown to saturation overnight at 37 °C in a shaker. Overnight cultures were mixed and 1 μL transferred into 6 mL fresh broth, which was grown to saturation. Plasmid minipreps from overnight cultures were prepared, sequenced, and the sequencing traces were analyzed as described below. This process was followed through 16 passages.
Chromatogram Processing of in Vivo rbSSR Passages.
Chromatogram trace data from sequencing reactions were converted to csv files with the abiparser.py Python script (http://www.bioinformatics.org/groups/?group_id=497) and imported into Mathematica for analysis. Trace data were separated by nucleotide identity into individual channels and four isolated bases—A30, T99, T115, and G143—were selected for calculating repeat distributions. Each isolated nucleotide was located at least five bases from the nearest nucleotide of the same type (Fig. S9). For each isolated nucleotide, peak heights over a 7-peak window were normalized to calculate a distribution of repeats (A)13 to (A)17. The mean of these four distributions was used to populate the serial passage dataset.
Note that we consistently observed an apparent 5–8% contribution from a single-unit deletion for the (A)15 sequence traces. This noise, possibly an artifact of polymerase slippage in the sequencing reaction, was observed in all rbSSR (A)15 sequencing traces, including the initial sequence verification of the (A)15 rbSSR-GFP construct and all passages of the wild-type strain across three replicates, with a mean contribution of 6.6% (n = 50). To compensate for this noise we adjusted the peak height Pn for repeat length n to  for each target nucleotide when computing distributions.
for each target nucleotide when computing distributions.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank Georg Seelig for helpful discussions and advice on experiments and the manuscript, and David Thorsley for discussions on modeling mutation rates. This work was supported by the National Science Foundation (NSF) through the Molecular Programming Project (Grant 0832773) and an NSF Graduate Research Fellowship (R.G.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 16758.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205693109/-/DCSupplemental.
References
- 1.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 2.Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 3.Arkin AP, Fletcher DA. Fast, cheap and somewhat in control. Genome Biol. 2006;7:114. doi: 10.1186/gb-2006-7-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danchin A. Scaling up synthetic biology: Do not forget the chassis. FEBS Lett. 2012;586:2129–2137. doi: 10.1016/j.febslet.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purnick PE, Weiss R. The second wave of synthetic biology: From modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach M, Voigt CA. Prokaryotic gene clusters: A rich toolbox for synthetic biology. Biotechnol J. 2010;5:1277–1296. doi: 10.1002/biot.201000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy K, Adams R, Wang X, Balazsi G, Collins J. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callura J, Dwyer D, Isaacs F, Cantor C, Collins J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol Syst Biol. 2011;7:471. doi: 10.1038/msb.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring cells: Synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox RS, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutalik VK, Qi L, Guimaraes JC, Lucks JB, Arkin AP. Rationally designed families of orthogonal RNA regulators of translation. Nat Chem Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- 20.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 21.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 22.Andersen JB, et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poelwijk FJ, de Vos MGJ, Tans SJ. Tradeoffs and optimality in the evolution of gene regulation. Cell. 2011;146:462–470. doi: 10.1016/j.cell.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng XJ, et al. Optimizing genetic circuits by global sensitivity analysis. Biophys J. 2004;87:2195–2202. doi: 10.1529/biophysj.104.044131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong TS, Zhurina D, Schwaneberg U. The diversity challenge in directed protein evolution. Comb Chem High Throughput Screening. 2006;9:271–288. doi: 10.2174/138620706776843192. [DOI] [PubMed] [Google Scholar]
- 27.Haseltine EL, Arnold FH. Synthetic gene circuits: Design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 28.Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 31.Shinde D, Lai YL, Sun FZ, Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 2003;31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci USA. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 34.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson DG, Smith HO, Hutchison CA, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 36.Bagh S, et al. Plasmid-borne prokaryotic gene expression: Sources of variability and quantitative system characterization. Phys Rev E. 2008;77:021919. doi: 10.1103/PhysRevE.77.021919. [DOI] [PubMed] [Google Scholar]
- 37.Loinger A, Biham O. Analysis of genetic toggle switch systems encoded on plasmids. Phys Rev Lett. 2009;103:068104. doi: 10.1103/PhysRevLett.103.068104. [DOI] [PubMed] [Google Scholar]
- 38.Torres-Cruz J, van der Woude MW. Slipped-strand mispairing can function as a phase variation mechanism in Escherichia coli. J Bacteriol. 2003;185:6990–6994. doi: 10.1128/JB.185.23.6990-6994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987;15:5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Bacterial contingency loci: The role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 41.Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 42.Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 45.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.