Abstract
Most membrane-enveloped viruses depend on host proteins of the endosomal sorting complex required for transport (ESCRT) machinery for their release. HIV-1 is the prototypic ESCRT-dependent virus. The direct interactions between HIV-1 and the early ESCRT factors TSG101 and ALIX have been mapped in detail. However, the full pathway of ESCRT recruitment to HIV-1 budding sites, which culminates with the assembly of the late-acting CHMP4, CHMP3, CHMP2, and CHMP1 subunits, is less completely understood. Here, we report the biochemical reconstitution of ESCRT recruitment to viral assembly sites, using purified proteins and giant unilamellar vesicles. The myristylated full-length Gag protein of HIV-1 was purified to monodispersity. Myr-Gag forms clusters on giant unilamellar vesicle membranes containing the plasma membrane lipid PI(4,5)P2. These Gag clusters package a fluorescent oligonucleotide, and recruit early ESCRT complexes ESCRT-I or ALIX with the appropriate dependence on the Gag PTAP and LYP(X)nL motifs. ALIX directly recruits the key ESCRT-III subunit CHMP4. ESCRT-I can only recruit CHMP4 when ESCRT-II and CHMP6 are present as intermediary factors. Downstream of CHMP4, CHMP3 and CHMP2 assemble synergistically, with the presence of both subunits required for efficient recruitment. The very late-acting factor CHMP1 is not recruited unless the pathway is completed through CHMP3 and CHMP2. These findings define the minimal sets of components needed to complete ESCRT assembly at HIV-1 budding sites, and provide a starting point for in vitro structural and biophysical dissection of the system.
Keywords: confocal microscopy, host-pathogen interaction, membrane traffic, virus budding
Most membrane-enveloped viruses utilize host proteins of the endosomal sorting complex required for transport (ESCRT) machinery for their release (1–3). This dependence was first described for HIV-1, where mutation of an ESCRT-binding motif (late domain) in the viral protein Gag was shown to stall virus bud release at a late stage in virus assembly (4, 5). Late domains have subsequently been found in other retroviruses and numerous other virus families including, e.g., flavi-, filo- and rhabdoviruses (6). Indeed, the only well-characterized case of ESCRT-independent release of a membrane-enveloped virus is influenza virus (7). In normal cell physiology, ESCRTs function in cytokinesis, formation of intralumenal vesicles in multivesicular endosomes, and vesicle release from the plasma membrane (8–10). These seemingly disparate processes all involve membrane budding away from the cytoplasm, and ESCRTs are the only described protein machinery to perform vesicle formation with this topology, which is analogous to virus release.
The initial events in HIV-1 ESCRT recruitment are well characterized. The structural protein of HIV-1, Gag, binds early-acting ESCRT factors through two late-domain motifs in its C-terminal p6 domain: a PTAP sequence that interacts with the TSG101 subunit of ESCRT-I (11–16) and a LYP(X)nL motif that interacts with the ESCRT-associated protein ALIX (17–22). The ultimate effect of both ESCRT-I and ALIX binding is the assembly of the ESCRT-III complex and the AAA ATPase VPS4 at the virus bud neck, leading to membrane scission and virion release. ALIX does this by directly recruiting the major ESCRT-III subunit CHMP4 (19, 20).
However, it is the interaction of HIV-1 with the ESCRT-I complex that is the major pathway for HIV-1 release under most conditions. It is not clear how ESCRT-I directs ESCRT-III assembly because there is little evidence that ESCRT-I can directly bind to ESCRT-III in solution or in large-scale interaction screens (18). In the yeast ESCRT system, ESCRT-I recruits the ESCRT-II complex, which in turn binds to the upstream ESCRT-III subunit Vps20 (CHMP6), initiating ESCRT-III assembly (23–25). However, RNA knockdown experiments have been interpreted as indicating that ESCRT-II and CHMP6 might be dispensable for HIV-I release (26). It has been suggested that ALIX could serve as an alternative to ESCRT-II in bridging ESCRT-I and ESCRT-III, given that ALIX directly binds to both ESCRT-I and ESCRT-III. It also seems possible that in the context of the HIV-1 Gag assembly on membranes, local concentration effects or membrane-induced conformational changes could alter the interaction properties. The connectivity between ESCRT-I and ESCRT-III is thus still a major unanswered question in the HIV-1 release field.
In yeast, the ordered assembly of the ESCRT-III complex has been deduced from genetics (27) and recapitulated in vitro (28, 29). The core yeast subunits assemble in the order Vps20, Snf7, Vps24, and Vps2 (27). These are all absolutely required for function in yeast (24) and for sustained function in vitro (29), and correspond to the human ESCRT-III subunits CHMP6, CHMP4A/B/C, CHMP3, and CHMP2A/B. The late-acting ancillary ESCRT-III proteins Did2, Ist1, and Vps60, which are important but not strictly essential for normal function in yeast, correspond to human CHMP1A/B, IST1, and CHMP5. Detailed models for the ESCRT scission reaction in normal endosomal function have been deduced (30, 31) that involve all of these components. Consistent with the analogy to yeast ESCRTs, simultaneous knockdown of CHMP4A/B/C or CHMP2A/B reduces HIV-1 budding to the late-domain independent baseline (32). Less consistent with the yeast-based mechanism, CHMP6, CHMP3, and CHMP1A/B knockdowns have little or no HIV-1 release phenotype (26, 32). These differences are hard to reconcile with live-cell imaging detecting CHMP1B at bud sites (33) and with the finding that CHMP2A and CHMP3 coassemble in vitro into tubules but do not do so on their own (34). The differences in the results with yeast ESCRT-III proteins and the knockdown, imaging, and tubule formation data for human proteins called out for further investigation.
Despite the many cellular studies of ESCRTs in vesicle formation and virus release, biochemical tools have been lacking for analyzing the assembly of the ESCRT machinery at viral budding sites on a membrane. Here, we have reconstituted the recruitment of ESCRT proteins to HIV-1 Gag assembly sites in a completely defined in vitro system. As compared to previous solution phase studies of ESCRT interactions with the Gag p6 domain or peptides derived from p6, here the system is based on full-length Gag that is oligomerized on a membrane. Because ESCRT proteins interact with membranes and form oligomers, these properties might be expected to profoundly impact the nature of the interactions. We purified the myristylated full-length Gag protein of HIV-1. Myr-Gag, intially in a monodisperse state, assembles into clusters on the surface of synthetic giant unilamellar vesicles (GUVs). The Gag clusters recruit nucleic acid and ESCRT proteins, recapitulating the known properties of Gag in cells. We find a strict dependence on ALIX or ESCRT-II for the assembly of the ESCRT-III complex, and an interdependence of late ESCRT-III subunits CHMP3 and CHMP2A. The model system developed here sheds light on the mechanism of ordered assembly of ESCRT complexes at HIV-1 bud sites.
Results
HIV-1 Gag Clusters on PI(4,5)P2-Containing GUVs.
We sought to reconstruct fully defined HIV-1 Gag assembly sites on membranes from purified and synthetic components. Previous biochemical studies of HIV-1 Gag assembly, as well as that of the closely related Rous sarcoma virus Gag, utilized constructs that lacked the N-terminal myristylation and were truncated in the N-terminal MA domain and the C-terminal, ESCRT-binding, p6 domain (35–37). These measures were taken to avoid problems with Gag protein aggregation. However, these constructs are not suitable for the purposes of this study, because they bind neither to membranes nor to ESCRTs. To circumvent aggregation problems in the context of intact HIV-1 Gag, we expressed full-length Gag in Escherichia coli with maltose-binding protein (MBP) fused to its C terminus. This increased the solubility, protected the flexible C-terminal p6 domain from proteolytic degradation, and left the N terminus accessible for myristylation. N-terminal myristylation was achieved by coexpressing N-myristyl transferase from Saccharomyces cerevisiae by analogy to methods used to generate Myr-MA domain (38–40), and confirmed by mass spectrometry (Fig. S1). The C-terminal MBP tag was removed by Tobacco etch virus protease digestion at low concentration after initial purification steps, which produced a myristylated Gag protein that eluted as a monodisperse peak from a gel filtration column (Fig. S2).
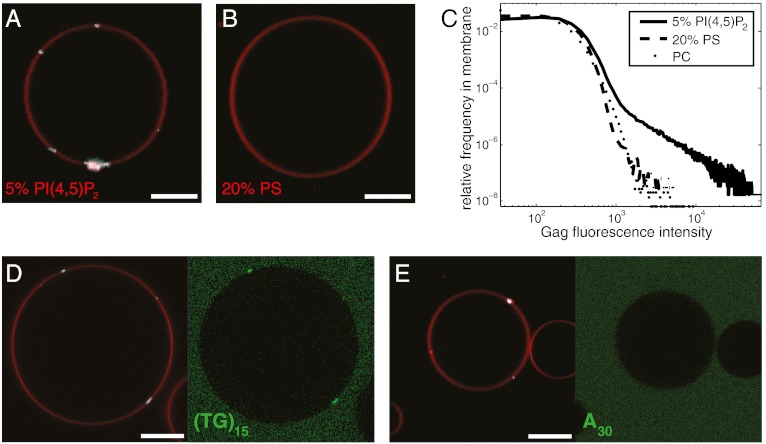
The matrix domain of Gag has a binding site for the plasma membrane lipid PI(4,5)P2, and this binding site is crucial for Gag targeting to the plasma membrane (40, 41). When added at 100 nM to GUVs containing 5% PI(4,5)P2, Gag, labeled with the fluorophore Atto 594, formed bright clusters of variable size, ranging from diffraction-limited puncta to sheets of a few micrometers, on the GUV membrane (Fig. 1A). No strong binding of Gag was observed on GUVs containing the same negative charge density in the form of 20% phosphatidylserine (PS) (Fig. 1B), nor on GUVs with no net charge in the membrane (Fig. 1C). The Gag puncta recruited the fluorescently labeled DNA oligonucleotide (TG)15, (Fig. 1D) but not A30, (Fig. 1E), consistent with earlier results obtained with truncated Gag constructs containing an intact NC domain (35). To quantify the recruitment of labeled oligonucleotides to Gag puncta, we recorded 10 z stacks of GUV images at random positions, typically totaling approximately 50 GUVs and approximately 50–100 Gag puncta, and calculated the percentage of Gag puncta that had an oligonucleotide fluorescence intensity more than 1.5 times that of the surrounding membrane. Three independent repeats of this quantitation yielded 100 ± 0% of Gag puncta positive for (TG)15 recruitment, and 15 ± 23% of Gag puncta positive for A30 recruitment, confirming the visual impression that only (TG)15 is efficiently packaged by Gag puncta. Taken together, these data show that the reconstituted Gag puncta recapitulate the lipid dependence and nucleic acid packaging properties of Gag assembly in cells and in solution.
Fig. 1.
HIV-1 Gag puncta assemble on PI(4,5)P2-containing GUVs. (A and B) Confocal microscopy images of GUVs containing 5% PI(4,5)P2 or 20% PS. When added to GUV exterior at 100 nM, Gag binds the outer leaflet of the PI(4,5)P2-containing GUV, forming clusters (A). Gag does not bind to GUVs with similar charge density in the form of 20% PS (B). (C) Brightness histograms of Gag fluorescence calculated in GUV membrane for GUVs containing POPC and 25% cholesterol (PC), along with 5% PI(4,5)P2 (5% PIP2), or 20% POPS (PS). Each curve is calculated from a total of approximately 50 GUVs in 10 z stacks recorded on random positions. (D) Fluorescent DNA oligo (TG)15 at 1 μM is packaged by Gag puncta. (E) Fluorescent DNA oligo A30 at 1 μM is not packaged by Gag puncta. Membrane is red, Gag is white, and fluorescent oligonucleotide is green. Scale bar, 10 μm.
Late-Domain–Dependent Recruitment of ESCRT-I and ALIX.
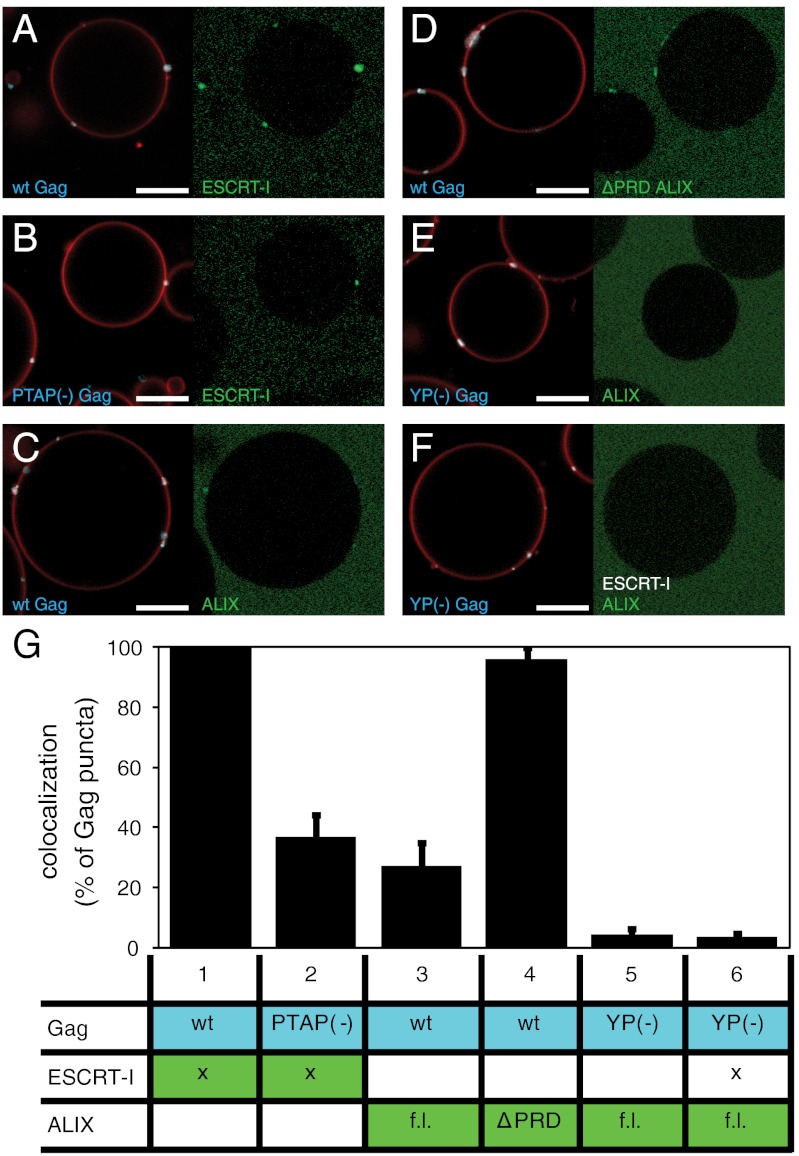
When 30 nM fluorescently labeled ESCRT-I was added together with 100 nM Gag to PI(4,5)P2-containing GUVs, it was recruited to the Gag puncta (Fig. 2A). A Gag protein containing the late-domain–inactivating PTAP → LIRL mutation displayed reduced ESCRT-I recruitment (Fig. 2B), indicating that the ESCRT-I recruitment is largely PTAP-dependent in our reconstituted system. We hypothesize that the remaining PTAP-independent recruitment of ESCRT-I is due to its lipid binding MABP domain (42). We quantitated ESCRT-I recrutiment to Gag puncta as described above for oligonucleotides, in three independent repeats each typically containing approximately 200–400 Gag puncta. This quantitation confirmed the visual impression that ESCRT-I binding is largely PTAP-dependent (Fig. 2G, columns 1 and 2).
Fig. 2.
Recruitment of early ESCRT proteins to Gag puncta. (A–F) 30 nM ESCRT-I or 1 μM ALIX [full-length (f.l.) or ΔPRD], as indicated, were added to GUVs together with 100 nM Gag [wild-type (wt) or late-domain mutant as indicated]. Representative images are shown from experiments detailed in G, with Gag and membrane fluorescence in Left and ESCRT fluorescence in Right. ESCRT-I is strongly recruited to wt Gag puncta (column 1) and moderately to puncta formed by PTAP(-) Gag (column 2). Full-length ALIX is moderately recruited to Gag puncta (column 3), whereas the ΔPRD-ALIX is strongly recruited (column 4). Full-length ALIX is not colocalizing to YP(-) Gag puncta, in the presence or absence of ESCRT-I (columns 5 and 6). (G) Quantification of early ESCRT recruitment to Gag puncta. Each experiment is represented by one row in the table, showing what proteins were added (marked with x if wild type in all experiments). Flourescently labeled proteins have a colored background in the table. For each experiment, 10 z stacks were recorded at random positions. Gag puncta on GUV membranes were identified by an automated MATLAB script, and the percentage of Gag puncta having an ESCRT-I/ALIX fluorescence > 1.5 times the surrounding membrane was calculated and is shown in the bar graphs above each column. All six experiments were carried out on the same day with the same batch of GUVs for comparability, and the error bars represent the standard deviation of three independent repeats of the experiments. Membrane is red, Gag is white, and fluorphore-labeled ESCRT-I/ALIX is green. Scale bar, 10 μm.
Intact ALIX at 1 μM colocalized to Gag puncta, but less strikingly than ESCRT-I (Fig. 2C). It has been proposed that the C-terminal proline-rich domain (PRD) of ALIX maintains it in an autoinhibited conformation (43, 44). When the C-terminal PRD of ALIX was deleted, recruitment was strongly enhanced (Fig. 2D and column 4 of Fig. 2G) and was comparable in efficiency to ESCRT-I recruitment. This indicates that membrane localization is not sufficient to release ALIX autoinhibition.The inactivating mutation YP → SR in the ALIX-binding LYP(X)nL late domain in Gag completely abrogated ALIX recruitment (Fig. 2E and column 5 of Fig. 2G), indicating that the reported interaction between the NC domain of Gag and Bro1 domain of ALIX (45, 46) was not sufficient to recruit detectable levels of ALIX in this system. The ALIX PRD contains a P(S/T)AP motif that is capable of binding to the TSG101 subunit of ESCRT-I (18), although the motif is not essential for ALIX function in HIV-1 budding (19, 20). We tested whether the addition of unlabeled ESCRT-I could rescue the recruitment of ALIX to the LYP(X)nL mutant Gag. No significant rescue of ALIX recruitment to YP → SR Gag puncta by ESCRT-I was found (Fig. 2F and column 6 of Fig. 2G). These data show that the reconstituted Gag assemblies recruit the most upstream ESCRT factors with the same late-domain dependence seen for HIV-1 budding from infected cells.
Recruitment of CHMP4.
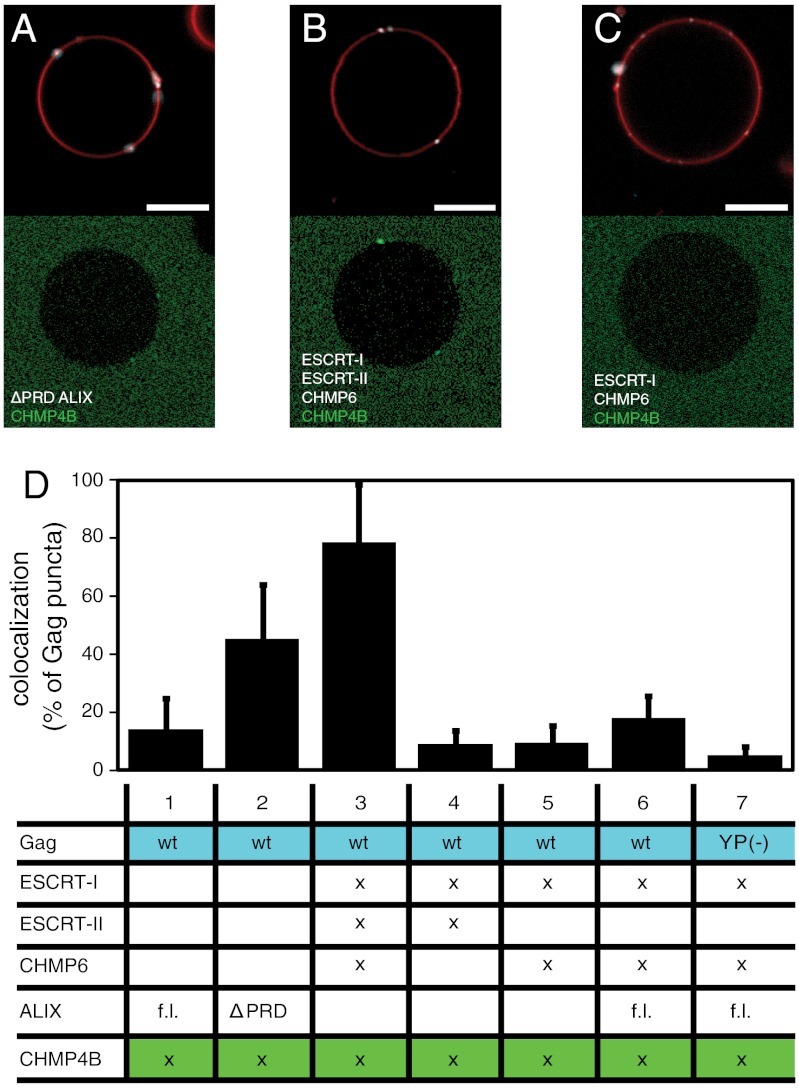
CHMP4 is a critical component of the ESCRT-III complex, and is one of the few components essential for all known functions of the ESCRT pathway. The different pathways for the upstream initiation of the ESCRT cascade converge at the stage where CHMP4 is recruited. Fluorescently labeled Gag and CHMP4B were added to GUVs at 100 and 300 nM, respectively, along with various combinations of unlabeled upstream ESCRT factors. This CHMP4B concentration is higher than what was required for bud neck localization and membrane scission in the in vitro reconstitution of yeast ESCRT vesicle formation (28). The degree of recruitment of CHMP4B to Gag puncta was quantitated as above for ESCRT-I and ALIX. Concentrations of upstream ESCRTs were chosen such as to give a robust recrutiment of CHMP4B in the positive experiments (Fig. 3A and B). This yielded an ALIX concentration of 4 μM, which is close to the solution Kd value for its interaction with the Gag late-domain motifs (19, 21). This suggests that the presence of the membrane does not enhance the ALIX–Gag interaction, which would be consistent with the absence of known lipid-binding domains in ALIX.
Fig. 3.
Recruitment of CHMP4B by upstream ESCRT proteins. (A) Atto 488–labeled CHMP4B at 300 nM is recruited to Gag puncta by ΔPRD-ALIX at 4 μM. (B) Atto 488–labeled CHMP4B at 300 nM is recruited to Gag puncta by ESCRT-I (100 nM), ESCRT-II (200 nM), and CHMP6 (400 nM). (C) Same protein combination as in B except that ESCRT-II is omitted. (D) Quantification of CHMP4B recruitment to Gag puncta. Statistics of CHMP4B recrutiment was performed as for Fig. 2G, and the table shows added proteins analogously to Fig. 2G. Membrane is red, Gag is white, and fluorphore-labeled CHMP4B is green. Scale bar, 10 μm.
However, the 100 nM ESCRT-I concentration needed was much lower than the solution Kd of approximately 27 μM (11), suggesting that the colocalization on a membrane potently increases the Gag–ESCRT-I affinity. This would be consistent with a possible role for the acidic lipid-binding MABP domain of MVB12 (42). These results also parallel the 3–4 order of magnitude increase in affinity between ESCRTs and ubiquitin when the latter is tethered to a membrane (47). Indeed, the effective approximately 3 order of magnitude increase in affinity seen for the ESCRT-I:Gag interaction represents one of the most dramatic differences between the more realistic context of this study and the solution analysis of binary interactions between small fragments.
Full-length ALIX weakly recruited CHMP4B to Gag puncta (Fig. 3D, column 1), consistent with the relatively weak colocalization of full-length ALIX with Gag. The activated C-terminal PRD truncation construct of ALIX recruited CHMP4B more potently at the same concentration (Fig. 3 A and D, columns1 and 2). The strongest recruitment was caused by the addition of 100 nM ESCRT-I, 200 nM ESCRT-II, and 400 nM CHMP6 (Fig. 3B and column 3 of Fig. 3D). This precisely recapitulates the pattern expected by analogy to yeast proteins in vivo (23, 24, 48) and in vitro (28). To investigate the roles of ESCRT-II and CHMP6 in CHMP4 recruitment, we repeated this experiment with either ESCRT-II or CHMP6 omitted. Either of these omissions completely abrogated recruitment of CHMP4B (Fig. 3C and columns 4 and 5 of Fig. 3D). Adding ALIX led to a modest increase in CHMP4 recruitment that was below the threshhold of statistical significance (Fig. 3D, column 6). Even this modest enhancement, however, was dependent on the LYP(X)nL motif in Gag (Fig. 3D, column 7). Thus, ALIX is not acting to replace ESCRT-II and CHMP6 downstream of ESCRT-I, but instead bypasses the entire PTAP and ESCRT-I–dependent pathway. These findings demonstrate that human ESCRT proteins studied in vitro in a realistic model system for HIV-1 budding recruit CHMP4 through an ordered mechanism that is conserved from yeast to humans.
Assembly of Downstream ESCRT-III Subunits.
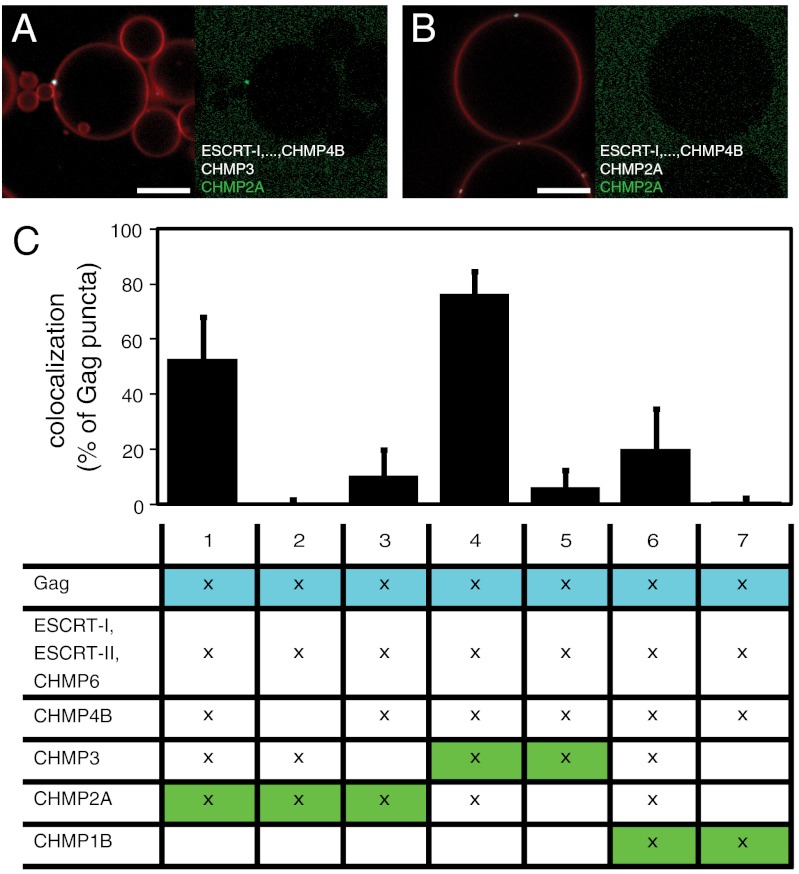
After analyzing the CHMP4 recruitment pathway, we proceeded to analyze the interdepedence of the assembly of the most downstream subunits of the ESCRT-III complex, CHMP3, CHMP2, and CHMP1. Because the combination of ESCRT-I, ESCRT-II, and CHMP6 provided the strongest recruitment of CHMP4B (Fig. 3D), these proteins were added to GUVs together with Gag and unlabeled CHMP4B, all at the same concentrations as used for the CHMP4B recrutiment assay above. The exact stoichiometry of the assembled human ESCRT-III complex is not known, but because the later CHMPs by analogy to the yeast system are thought to be less abundant than CHMP4, we added CHMP3, CHMP2A, and CHMP1B at a lower solution concentration of 100 nM. When CHMP3 and fluorescently labeled CHMP2A were added, CHMP2A was strongly recruited to the Gag puncta (Fig. 4A and column 1 of Fig. 4C). Omitting CHMP4B caused a complete loss of CHMP2A recruitment (Fig. 4C, column 2). The replacement of the unlabeled CHMP3 by the same amount of unlabeled CHMP2A caused a nearly complete loss of recruitment of fluorescent CHMP2A (Fig. 4B and column 3 of Fig. 4C). When the fluorescent label was instead placed on CHMP3, a strong recruitment was found in the presence of CHMP2A (Fig. 4C, column 4), but virtually no recruitment when CHMP2A was replaced by unlabeled CHMP3 (Fig. 4C, column 5). Taken together, these results show that the recruitment of CHMP3 and CHMP2A to Gag puncta is highly synergistic and is completely dependent on CHMP4B recruitment. When fluorophore-labeled CHMP1B was added together with unlabeled CHMP3 and CHMP2A, a moderate recruitment to some Gag puncta was detected (Fig. 4C, column 6). When CHMP3 and CHMP2A were replaced by the same amount of unlabeled CHMP1B, no recruitment of fluorescent CHMP1B was detected (Fig. 4C, column 7). These findings are summarized and placed in context in the model shown in Fig. 5.
Fig. 4.
Interaction between ESCRT-III subunits. (A) Atto 488–labeled CHMP2A at 100 nM is recruited to a Gag punctum in the presence of ESCRT-I (100 nM), ESCRT-II (200 nM), CHMP6 (400 nM), CHMP4B (300 nM), and CHMP3 (100 nM). (B) Atto 488–labeled CHMP2A at 100 nM is not recruited to a Gag punctum in the presence of ESCRT-I (100 nM), ESCRT-II (200 nM), CHMP6 (400 nM), CHMP4B (300 nM), CHMP3 (100 nM), and unlabeled CHMP2A (100 nM). (C) Quantification of late CHMP recruitment to Gag puncta, analogous to Fig. 2G. The fluorescently labeled CHMP is shown on a green background. Labeled or unlabeled CHMP3, CHMP2A, and CHMP1B are present at 100 nM where marked with an x. In experiments 3, 5, and 7, omission of CHMP3, CHMP2A, and CHMP3 + CHMP2A were compensated by adding the same amount of unlabeled CHMP2A, CHMP3, or CHMP1B, respectively. Membrane is red, Gag is white, and fluorphore-labeled CHMPs are green. Scale bar, 10 μm.
Fig. 5.
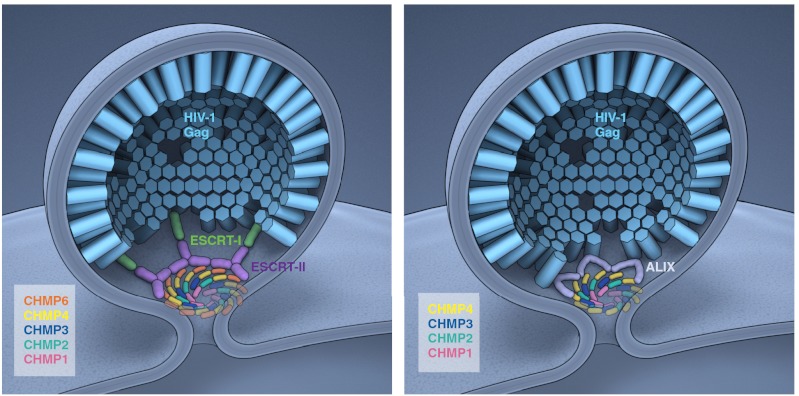
Ordered assembly of ESCRTs at HIV-1 budding sites. Schematic depiction of ESCRT-I–dependent (Left) and ALIX-dependent (Right) assembly cascades. The Gag–ESCRT and ESCRT–ESCRT interactions in this model are based on the present study, whereas their three-dimensional arrangement is extrapolated from other available data. The ESCRT-III subunits are thought to drive membrane scission by forming a dome at the membrane neck (30). In the HIV-1 setting, it is not known if the putative dome forms from the inside (as depicted here) or the outside (3). The detailed arrangment of the subunits in the dome is not known. They are depicted here as forming whorls, which provides for all subunits of a given type to maintain equivalent interactions with other subunit types. Concentric circles, each consisting of a unique subunit composition, would also meet this criterion, but a single spiral would not. Because HIV-1 assembly sites in cells will, on average, contain approximately 2,400 Gag molecules at the time of release (52), even the small subset of Gag molecules which expose their p6 domains at the rim of the Gag lattice could provide enough opportunity for the ESCRT-I–mediated and the ALIX-mediated pathways to both contribute to ESCRT-III assembly at the same assembly site.
Discussion
The major finding in this study is that the ESCRT assembly cascade involved in HIV-1 virion release can be reconstituted from a minimal set of purified components. HIV-1 Gag clustering on membranes requires only the intact Gag protein itself and PI(4,5)P2. This is consistent with the ability of truncated Gag constructs to self-assemble in the absence of lipids (35, 37) and the ability of purified Gag MA domain to bind to PI(4,5)P2 (40, 41). These membrane-bound Gag clusters are capable of selectively packing nucleic acid with sequences that favor NC binding, but this is not required for their assembly. The ESCRT assembly pathway is more complex. Previous analyses of Gag–ESCRT interactions, and ESCRT–ESCRT interactions in HIV-1 budding, have focussed on deletional and imaging experiments in cells, and on binary protein interaction studies in solution or in yeast two-hybrid screens. Reconstitution is complementary to these other approaches because it asks whether a particular minimal set of proteins is sufficient. In this case, we have found two different but overlapping sets of proteins and complexes that are minimally sufficient to reconstruct ordered assembly: ESCRT-I, ESCRT-II, CHMP6, CHMP4, CHMP3, CHMP2, and CHMP1; and ALIX, CHMP4, CHMP3, CHMP2, and CHMP1.
These results agree well with imaging studies of HIV-1 budding, to the extent that these proteins have been examined. ALIX, CHMP4B, CHMP4C, and CHMP1B have been directly observed in real time at budding sites during HIV-1 Gag budding (33). These results are also congruent with much of the knockdown data. HIV-1 budding is highly sensitive to knockdown of subunits of ESCRT-I (2), CHMP4A/B/C (32), and CHMP2A/B (32). Smaller decreases in HIV-1 budding efficiency are seen when CHMP3 and CHMP1A/B mRNAs are knocked down (32). The importance of ALIX in budding has been demonstrated in the context of its ability to rescue the budding PTAP-defective Gag when overexpressed (19, 20).
The main discrepancy between our reconstitution and the published knockdown data is that in the latter case, essentially no budding defect is observed when ESCRT-II subunits or the most upstream ESCRT-III subunit CHMP6 are knocked down (26). In the knockdown study, protein bands for ESCRT-II EAP20/VPS25 and CHMP6 were still visible on Western blots following knockdown, and it cannot be ruled out that the residual levels of these proteins are above the threshhold needed for function. Alternatively, it is possible that in cells, some other factor takes that place of ESCRT-II and CHMP6. ALIX is the most obvious factor that could do so, because it binds to both ESCRT-I and CHMP4. Yet we find that in the context of YPXL-deleted Gag, ALIX does not rescue the assembly pathway. This observation is consistent with the presence of a single P(S/T)AP binding site in the TSG101 UEV domain, which cannot bind simultaneously to both HIV-1 Gag and ALIX. We are left with two possiblities: that ESCRT assembly cascade is robust enough to proceed even with residual ESCRT-II and CHMP6 concentrations that are a fraction of normal levels, or that there exists an unidentified ESCRT-I–CHMP4 bridging factor other than ALIX.
CHMP2A and CHMP3 coassemble into tubes in vitro that may represent structures involved in membrane neck scission (34). The yeast cognates of CHMP2 and CHMP3 are Vps2 and Vps24, which seem to preferentially heterodimerize with one another (23). Thus, it seems fitting that there is a strong codependence of CHMP2A and CHMP3 recruitment in the reconstituted system. The main contradictory observation comes from knockdown studies, which find a > 100-fold drop in viral titer following CHMP2A/B knockdown, but only a 2-fold drop when CHMP3 is knocked down (32). The authors of that knockdown study propose that CHMP4B binds directly to CHMP2A, bypassing the need for CHMP3. Our data suggest that the most efficient recruitment occurs when a CHMP3:CHMP2A copolymer is jointly recruited by CHMP4. As discussed above for the ESCRT-II:CHMP6 pair, it seems that either copolymerization is robust enough to occur at sharply reduced CHMP3 levels, or some unknown factor can copolymerize with CHMP2A in cells.
The long-term aim of this project is to reconstitute the entire ESCRT-mediated HIV-1 assembly and release process, including the scission of the membrane necks of virus-like particles (VLPs). This final step is thought to be mediated by ESCRT-III, possibly in conjuction with the ATPase VPS4 (33, 49). In the conception of these experiments, we had anticipated that inefficient release of Gag puncta into the GUV interior might be seen even in the absence of ESCRTs, because high levels of Gag overexpression have been shown to drive budding (50). If ESCRT-III assembly were sufficient for scission, as seen for the yeast proteins, we would have expected to see a large increase in VLP release upon completion of ESCRT assembly. Contrary to expectations, we see essentially no release of VLPs in either the presence or absence of ESCRTs. With respect to the ESCRTs, an obvious possiblility is that VPS4 activity might be involved in release. However, the lack of baseline ESCRT-independent scission suggests to us the main scission defect is more likely in the nature of the Gag assembly, or in a missing RNA or lipid factor.
Increasingly detailed structural and biophysical models of ESCRT-dependent membrane budding and scission in normal endosomal function are becoming available. In essence, these models invoke a collar consisting of upstream ESCRTs (31, 51) that templates the activation and nucleation of an ESCRT-III dome (30). The latter dome pulls membrane with it as it grows to a tip, bringing the width of the neck down to the critical proximity needed for spontaneous membrane scission (30). This study is a step on the path to developing an equally detailed and insightful model of ESCRT mechanism in HIV-1 release. Such a model cannot be contemplated until a complete parts list for the reaction is known. Now that the list of the minimal set of components needed for assembly is at hand, the in vitro system should be a useful addition to the toolkit for deconstructing the release mechanism and ultimately interfering with it therapeutically. Further progress in advancing mechanistic insight will require superresolution optical imaging and improved electron tomographic analysis of budding events in cells, together with increasingly sophisticated reconstitutions.
Materials and Methods
Formation of GUVs.
GUVs containing palmitoyl-oleoyl-phosphatidylcholine (POPC) (70 mol%), cholesterol (25 mol%), brain PI(4,5)P2 (5 mol%), and the fluorophore Atto 647–DOPE (0.1 mol%) were prepared in 600 mM sucrose essentially as described previously (29). To prevent PI(4,5)P2 from segregating from other lipids, the lipid mix and indium–tin oxide-coated glass slides were preheated to 60 °C prior to spreading the lipids on the slides, and the electroswelling was performed for 1 h at 60 °C. In Fig. 1 B and C, GUVs designated “20% PS” contained 55% POPC, 25% cholesterol, 20% palmitoyl-oleoyl-phosphatidylserine (POPS), and 0.1% Atto 647–DOPE; and GUVs designated “PC” had the POPS replaced by POPC. All lipids were obtained from Avanti Polar Lipids, except Atto 647–PE, which was from ATTO-TEC.
Reconstitution Reactions and Confocal Microscopy.
In a Lab-Tek II chambered coverglass (Fisher Scientific), 150 μL of GUVs were mixed with 150 μL of isoosmotic buffer (20 mM Tris, pH = 7.4, 300 mM NaCl) containing proteins at concentrations stated in Results. The mix was gently stirred and incubated 10 min at room temperature before imaging. In the case of late ESCRT-III recruitment, CHMP3, CHMP2A, and CHMP1B were added after 10 min incubation, and imaged after another 5 min. Images were acquired using a Zeiss LSM780 confocal microscope equipped with a GaAsP detector and a Plan-Apochromat 63 × N.A. = 1.40 objective. The Atto 647–DOPE membrane marker, Atto 594–labeled Gag, and the Atto 488–labeled ESCRTs were excited with 633-, 561-, and 488-nm lasers, respectively. z stacks of GUVs were acquired at positions selected solely on the basis of containing GUVs, without observing the fluorescence channels. Per reconstitution experiment, 10 z stacks of 100 × 100 μm were acquired, each stack consisting of 10 images, spaced at 1 μm. All experiments shown in the same figure were done on the same day with the same GUV batch for comparability. Each experiment series was repeated on at least three separate occasions with different batches of GUVs.
Image Analysis.
Colocalization analysis was performed by custom made scripts for MATLAB (MathWorks, Inc). The membrane fluorescence channel was used to create a binary mask. Within this mask, corresponding to the GUV membrane, Gag puncta were identified and counted as areas with Gag fluorescence above 104/pixel with the given microscope settings. A given Gag punctum was considered positive for colocalization if its average 488-nm (oligonucleotide or ESCRT) fluorescence was > 1.5 times the average in the GUV membrane outside Gag puncta.
Supplementary Material
ACKNOWLEDGMENTS.
We thank E. Freed for critically reading the manuscript, E. Tyler for Fig. 5, E. Boura for providing ESCRT-II, and Hurley lab members for constructive discussions. L.-A. C. was supported by a European Molecular Biology Organization long-term fellowship and a Human Frontier Science Program long-term fellowship. This work was supported by the Intramural Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, and the Intramural AIDS Targeted Anti-viral Program of the Office of the Director, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211759109/-/DCSupplemental.
References
- 1.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin-Serrano J, Neil SJD. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 3.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang MJ, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald B, Martin-Serrano J. No strings attached: The ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 13.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 16.Im YJ, et al. Crystallographic and functional analysis of the ESCRT-1/HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strack B, et al. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 18.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 19.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munshi UM, et al. An Alix fragment potently inhibits HIV-1 budding—Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 22.Zhai Q, et al. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 23.Babst M, et al. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 24.Babst M, et al. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 26.Langelier C, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–873. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabrikant G, et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boura E, et al. Solution structure of the ESCRT-I and -II supercomplex: Implications for membrane budding and scission. Structure. 2012;20:874–886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jouvenet NJN, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell S, et al. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci USA. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu F, et al. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J Virol. 2001;75:2753–2764. doi: 10.1128/JVI.75.6.2753-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang C, et al. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saad JS, et al. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saad JS, et al. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono A, et al. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boura E, Hurley JH. Structural basis for membrane targeting by the MVB12-associated β-prism domain of the human ESCRT-I MVB12 subunit. Proc Natl Acad Sci USA. 2012;109:1901–1906. doi: 10.1073/pnas.1117597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, et al. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai QT, et al. Activation of the retroviral budding factor ALIX. J Virol. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus Type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dussupt V, et al. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boura E, et al. Endosomal sorting complex required for transport (ESCRT) complexes induce phase-separated microdomains in supported lipid bilayers. J Biol Chem. 2012;287:28144–28151. doi: 10.1074/jbc.M112.378646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Baumgartel VBV, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 50.Fang Y, et al. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:1267–1283. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT complexes: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlson LA, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.