Abstract
Microbial communities and their associated metabolic activity in marine sediments have a profound impact on global biogeochemical cycles. Their composition and structure are attributed to geochemical and physical factors, but finding direct correlations has remained a challenge. Here we show a significant statistical relationship between variation in geochemical composition and prokaryotic community structure within deep-sea sediments. We obtained comprehensive geochemical data from two gravity cores near the hydrothermal vent field Loki’s Castle at the Arctic Mid-Ocean Ridge, in the Norwegian-Greenland Sea. Geochemical properties in the rift valley sediments exhibited strong centimeter-scale stratigraphic variability. Microbial populations were profiled by pyrosequencing from 15 sediment horizons (59,364 16S rRNA gene tags), quantitatively assessed by qPCR, and phylogenetically analyzed. Although the same taxa were generally present in all samples, their relative abundances varied substantially among horizons and fluctuated between Bacteria- and Archaea-dominated communities. By independently summarizing covariance structures of the relative abundance data and geochemical data, using principal components analysis, we found a significant correlation between changes in geochemical composition and changes in community structure. Differences in organic carbon and mineralogy shaped the relative abundance of microbial taxa. We used correlations to build hypotheses about energy metabolisms, particularly of the Deep Sea Archaeal Group, specific Deltaproteobacteria, and sediment lineages of potentially anaerobic Marine Group I Archaea. We demonstrate that total prokaryotic community structure can be directly correlated to geochemistry within these sediments, thus enhancing our understanding of biogeochemical cycling and our ability to predict metabolisms of uncultured microbes in deep-sea sediments.
Keywords: taxonomic profiling, ultraslow-spreading ridge, amplicon sequencing
Marine sediments host the largest reservoir of organic carbon in the world and outnumber any other environment with respect to microbial cell abundance (1, 2). The microbial activity in this habitat has a profound impact on the global carbon cycle through the remineralization of sedimentary organic carbon, thus ultimately regulating the oxygen level of the atmosphere (3). Extensive sampling and drilling efforts during the past decade have changed our perception of microbial life in the inaccessible deep seafloor fundamentally (4–17). This pioneering work has led to novel insights into the composition and abundance of potentially active groups of Archaea and Bacteria in deep-sea sediments.
The density of prokaryotic cells in coastal and continental margin sediments typically is 108–109 cells/cm3 in the top sediment layers and declines with increasing depth in a logarithmic fashion (2). Even though cell abundance in organic-poor open-ocean sites can be several orders of magnitude lower, it may still exceed 105 cells/cm3 at depths close to 1,000 m below the seafloor (mbsf) (12). This omnipresence of prokaryotic cells was demonstrated recently by Roussel et al. (16) who detected viable cells at a sediment depth of 1,626 mbsf, supporting the idea that temperature is the ultimate limit for microbial survival at depth (18). In general, marine sediment communities appear to be dominated by a restricted number of bacterial and archaeal phyla, including Chloroflexi, Planctomycetes, Japanese Sea division 1 (JS-1), a diverse spectrum of Proteobacteria, the Deep Sea Archaeal Group (DSAG), Marine Group I (MG-I), the Miscellaneous Crenarchaeotic Group (MCG), and the South African Goldmine Euryarchaeotal Group (SAGMEG) (reviewed in refs.13, 19, and 20). These groups have been found in a variety of different marine sediments, including coastal and open-ocean sites, based on 16S rDNA clone libraries (6, 9, 21, 22), 16S rRNA clone libraries (15), and metagenomic shotgun sequencing (23, 24). The vast majority of these communities seem to be alive and active, although the fractions of dead and dormant cells vary substantially among locations (15, 25, 26).
Geochemical pore water profiles in marine sediments show distinct redox zones (27), suggesting that each of these zones are shaped by organisms with specific metabolic traits. In agreement with this notion, several studies have demonstrated down-core stratification of microbial community composition (28–31). Furthermore, a few specific microbial groups, such as anaerobic methane oxidizers (ANME) (32, 33) and anaerobic ammonium oxidizers (anammox) (34) have been associated consistently with specific redox zones. In addition, samples obtained from markedly different sedimentary settings (e.g., high or low temperature, presence or absence of methane, high or low carbon load) have been found to show distinct microbial community compositions (7, 35, 36). Nonetheless, an explicit, quantitative correlation between stratigraphic variability in geochemical properties and concomitant changes in the structure of the total microbial community or the relative abundance of individual taxa has remained elusive. This lack of correlation could be explained in part by limited datasets, low spatial resolution, and insufficient depth of the taxonomic profiling. Here we address these challenges and quantitatively explore the relationship between microbial community structure and multivariate geochemistry in highly stratified deep-sea sediments.
We investigated two 3-m-long sediment cores from the ultraslow-spreading Arctic Mid-Ocean Ridge system, one of the few places where substantial amounts of sediment accumulate in the mid-ocean rift valley. The cores were retrieved from two sites, one 15 km southwest (SW) and one 15 km northeast (NE) of the Loki’s Castle Vent Field (37). In addition to the hemipelagic and glaciomarine sediments derived from the Bear Island Fan system, the rift valley also receives episodic input of volcanogenic and metallogenic hydrothermal material. The resulting stratification of the cored sediments allowed us to study the influence of changes in sediment geochemistry on the microbial subsurface communities on a compact depth scale. We used deep sequencing of 16S rRNA gene amplicon tags, covering both the archaeal and bacterial domains, to obtain a detailed qualitative and quantitative taxonomic inventory from selected sediment horizons in both cores. We linked estimates of the relative abundance distribution of the entire prokaryotic community to copy numbers of marker genes from both domains as well as to 16 different geochemical and geophysical parameters of sedimentary solids and solutes. We found that stratigraphic variation in the structure of the microbial community as well as in the relative abundance of individual taxa can be correlated directly to stratigraphic variation in geochemical properties across both cores.
Results
Core Descriptions.
Core GC6, retrieved SW of the vent field, consisted of highly stratified hemipelagic-glaciomarine sediments with layers of different colors varying on a centimeter scale from light to darker brown and gray (Fig. 1A). Distinct coarser layers of detrital pyrite, likely of hydrothermal origin [71 and 95 cm below sea floor (cmbsf)], and of altered volcanoclastic material (65 and 232 cmbsf) generally were reflected by Fe enrichments in X-ray fluorescence (XRF) core scanner profiles (Fig. 1A) and by elevated Fe2O3 and S concentrations in quantitative geochemical sediment analyses (Table 1). The XRF profiles also revealed five distinct Mn-enriched layers, the uppermost of which (22 cmbsf) was interpreted as the lower boundary of the oxic layer (Table 1). Overall, the reduction potential (Eh) profile in GC6 (Table 1) covaries with the color changes, with higher values in the brown sections and lower but still positive values in the gray sections.
Fig. 1.
Characteristics of gravity cores GC6 (A) and GC12 (B) including geochemical data and relative abundances of the four most dominant bacterial and archaeal taxa/phyla. (Left to Right) Photograph of the archive half core; XRF core scanner maps of normalized iron and manganese content; pore water concentrations of ammonium, nitrate, manganese, iron and sulfate; organic carbon content in the sediment (weight %); total number of 16S rRNA gene copies/g sediment (wet weight) as measured by qPCR; percent of total SSU reads obtained from the given taxa in the amplicon library in each horizon. Note that different scales on the x-axis are color coded to indicate the different respiration processes, based on pore water geochemistry: blue, aerobic oxidation; red, nitrate reduction; purple, manganese reduction; brown, iron reduction; green, sulfate reduction. Delta, Deltaproteobacteria; Epsilon, Epsilonproteobacteria; P.mycetes, Planctomycetes; Thermoplas, Thermoplasmata.
Table 1.
Context data
| Solid phase |
Interstitial phase |
Gene copies/g sediment |
|||||||||||||||||
| Depth (cmbsf) | Fe2O3 (W %) | MnO2 (W %) | Nitrate (ppm) | S (W‰) | Eh (mV) | TIC (%C) | TOC (%C) | Fe/Ti (counts) | Mn/Ti (counts) | AT | pH | *NO3− (μM) | NH4+ (μM) | SO42- (mM) | Mn2+ (μM) | Fe2+ (μM) | Archaeal (SSU) | Bacterial (SSU) | Archaeal (amoA) |
| GC6 | |||||||||||||||||||
| 16 | 3.2 | 0.6 | 50.4 | 0.6 | 320 | — | 0.5 | 46.0 | 1.3 | 3.0 | 7.6 | 4.9 | 6 | 29.0 | 0.0 | 0.0 | 7.5 x 106 ± 2.5 | 1.3 x 106 ± 0.3 | 2.0 x 106 ± 0.5 |
| 29 | 7.1 | 0.1 | 75.6 | 0.3 | 90 | — | 1.2 | 41.4 | 0.4 | — | — | 0 | — | — | — | — | 8.3 x 107 ± 0.9 | 1.0 x 107 ± 0.0 | 2.4 x 106 ± 0.5 |
| 65 | 6.6 | 0.1 | 39.0 | 0.6 | 45 | 0.3 | 0.4 | 58.6 | 0.8 | 3.2 | 7.8 | 0 | 23 | 29.0 | 123.1 | 0.0 | BD | 3.4 x 103 ± 0.1 | BD |
| 88 | 11.4 | 0.2 | — | — | 40 | 1.51 | 0.7 | 63.8 | 0.8 | 3.0 | 8.3 | 0 | 8 | 28.9 | 166.4 | 34.0 | 2.0 x 108 ± 0.5 | 1.0 x 108 ± 1.0 | 1.6 x 106 ± 0.5 |
| 95 | 12.4 | 0.2 | 21.3 | 3.7 | — | 2.53 | 0.1 | 62.8 | 1.0 | 3.1 | 7.9 | 0 | 43 | 29.8 | 143.2 | 9.0 | 1.9 x 104 ± 1.3 | 4.3 x 104 ± 1.0 | 2.5 x 103 ± 1.0 |
| 156 | 6.1 | 0.1 | 42.4 | 0.5 | 155 | 0.4 | 0.4 | 43.2 | 0.6 | 3.2 | 7.7 | 0 | 49 | 28.0 | 201.5 | 0.0 | 1.1 x 106 ± 0.0 | 1.1 x 106 ± 0.3 | 2.4 x 104 ± 0.0 |
| 174 | 3.0 | 0.1 | 44.5 | 1.1 | 215 | 0.6 | 0.4 | 46.8 | 0.8 | 3.1 | 7.7 | 0 | 48 | 29.5 | 184.9 | 0.0 | 1.7 x 107 ± 0.8 | 5.5 x 106 ± 0.2 | 3.9 x 104 ± 0.0 |
| 232 | 20.2 | 0.2 | 25.1 | 0.6 | 120 | 0.05 | 0.2 | 99.3 | 1.1 | 3.1 | 7.6 | 0 | 1 | 28.2 | 129.8 | 0.0 | 3.0 x 104 ± 0.4 | 1.8 x 105 ± 0.2 | 1.5 x 103 ± 0.9 |
| 296 | 5.9 | 0.1 | 33.7 | 0.7 | −7 | 0.5 | 0.3 | 38.7 | 0.3 | 3.0 | 7.9 | 0 | 23 | 29.6 | 54.4 | 22.4 | 5.1 x 104 ± 1.3 | 1.8 x 104 ± 0.5 | 4.7 x 103 ± 4.0 |
| GC12 | |||||||||||||||||||
| 19 | 6.4 | 0.2 | 42.3 | 0.7 | 286 | 0.7 | 0.6 | 46.0 | 1.3 | 2.4 | 7.6 | 21.4 | 9 | 28.4 | 0.0 | 0.0 | 3.2 x 105 ± 0.5 | 1.9 x 105 ± 0.0 | 3.0 x 104 ± 1.1 |
| 35 | 4.1 | 0.9 | 16.9 | 0.4 | 350 | 0.02 | 0.2 | 41.4 | 0.4 | 2.9 | 7.5 | 9.4 | 9 | 28.8 | 0.0 | 0.0 | 6.4 x 105 ± 0.9 | 1.0 x 106 ± 0.1 | 2.9 x 106 ± 1.9 |
| 81 | 6.7 | 0.1 | 32.6 | 0.5 | −118 | 0.3 | 0.4 | 58.6 | 0.8 | 3.5 | 7.3 | 0 | 45 | 31.3 | 66.4 | 136.1 | 2.2 x 106 ± 0.2 | 3.0 x 106 ± 0.7 | 1.5 x 104 ± 1.4 |
| 126 | 5.0 | 0.1 | 23.7 | 0.5 | −165 | 0.1 | 0.6 | 63.8 | 0.8 | 3.1 | 7.3 | 0 | 65 | 27.8 | 80.0 | 94.9 | 1.7 x 106 ± 0.5 | 4.0 x 105 ± 0.9 | 2.8 x 105 ± 0.3 |
| 207 | 7.0 | 0.1 | 36.3 | 1.7 | −110 | 0.3 | 1.3 | 99.3 | 1.1 | 3.4 | 7.3 | 0 | 94 | 27.1 | 94.8 | 195.2 | 1.4 x 106 ± 0.2 | 5.2 x 105 ± 0.6 | 1.7 x 104 ± 0.3 |
| 310 | 5.4 | 0.1 | 25.2 | 0.7 | −155 | 0.3 | 1.0 | 38.7 | 0.3 | 3.5 | 7.3 | 0 | 146 | 23.4 | 64.1 | 188.0 | 5.1 x 105 ± 1.5 | 1.4 x 106 ± 0.0 | 5.3 x 104 ± 0.0 |
Context data for gravity core GC6 and GC12. See Materials and Methods for details on each measured parameter. All gene copy numbers are given per gram of wet weight sediment. Values marked with an asterisk were measured in gravity cores retrieved at the same position in year 2010. –, missing value; BD, below detection;; W%, percent by weight; %C, percent of carbon.
Core GC12, retrieved NE of the vent field, consisted of hemipelagic-glaciomarine sediments in the upper half and glacigenic debris flows in the lower half. The oxic/anoxic transition zone was marked by a single Mn-enriched layer at 38 cmbsf, and the Eh profile again covaried with the sediment color (Table 1).
Total organic carbon (TOC) content varied between 0.1–1.2% of weight in GC6 (Table 1, Fig. 1A) and 0.2–1.3% of weight in GC12 (Table 1 and Fig. 1B). These values were slightly higher than commonly reported for open-ocean sediments and likely reflect input from the Bear Island Fan and the high productivity of the Arctic Ocean. However, these sediments still are considered a low organic carbon system (<1%) compared with continental margin sites.
We recognized four to five chemical redox zones as defined by Canfield and Thamdrup (38) based on pore water depth profiles of dissolved nitrate, ammonium, manganese, iron, and sulfate (Table 1 and Fig. 1). These profiles include an oxic, nitrogenous, manganous, ferruginous, and sulfidic zone (Fig. 1, Left), the last deduced from sulfate depletion in GC12. Despite their proximity, the two cores showed clear differences: Although dissolved Mn2+ followed a typical diagenetic profile in both cores, the concentration in GC6 was higher than usually reported for deep-sea sites (up to 200 μM at 156 cmbsf) even in metal-rich deep-sea sediments (39). An increase in NH4+ coincided with the detection of dissolved Mn2+ in both cores, likely as a result of oxygen depletion. However, the down-core increase was less pronounced for GC6 and even decreased in the two volcanoclastic horizons. In GC6 dissolved Fe2+ increased in the deepest horizon as Mn2+ decreased, probably defining the shift from the manganous to the ferruginous zone. In GC12, on the other hand, Fe2+ already was elevated in the manganous zone and increased with depth in an irregular manner. A decrease in SO42− to 23 mM in the lower part of GC12 indicates microbial sulfate reduction. In contrast, no indication of sulfate reduction was observed in GC6. However, traces of H2S were detected at 88 cmbsf, between the two detrital pyrite layers. This horizon also showed traces of dissolved Fe2+ and a significant pH increase and NH4+ decrease.
NO3− was depleted from the interstitial phase below the oxic layers. Interestingly, however, NO3− could be extracted from the solid phase of all horizons in both cores (Table 1), suggesting that it was adsorbed to mineral phases, as observed in soils (40).
Composition and Diversity of the Microbial Community.
For each of the 15 sampled sediment horizons, we generated a 16S rRNA gene amplicon library, with one primer set covering the V5–V8 region of both bacterial and archaeal taxa (Materials and Methods). Pyrosequencing yielded a total of 59,364 high-quality sequence reads after extensive filtering. In total, 4,790 reads were unique, with an average length of 231 bp.
From the nine horizons in core GC6, 1,668 different operational taxonomic units (OTUs) (cutoff, 97% sequence identity) were divided into 50 classes (39 bacterial and 11 archaeal) representing 33 different phyla. For the six horizons in core GC12, 1,135 OTUs were identified within 48 different classes (36 bacterial and 12 archaeal) distributed among 38 phyla (for a complete list of represented taxa and their abundance on phylum and class level, see Table S1). A comma-separated text file listing the taxonomic affiliations of all OTUs and their distribution across datasets can be downloaded from http://services.cbu.uib.no/supplementary/jorgensen2012. At the class rank, more than 85% of all observed taxa were present in both cores, but their abundance varied greatly between cores and among horizons within each core (Fig. 1 and Table S1). Approximately 50% of the OTUs present in core GC12 were shared with GC6. It is noteworthy that none of the 2,277 unique OTUs was represented in all 15 horizons.
Determination of Bacterial and Archaeal Abundance.
Quantitative PCR (qPCR) was used to estimate total numbers of bacterial and archaeal small subunit (SSU) rRNA genes separately (Table 1). The total numbers in GC12 (Archaea plus Bacteria) varied between 5.1 × 105 and 5.2 × 106 16S rRNA gene copies/g sediment (wet weight), comparable to the abundance in other open-ocean sediments (41, 42). GC6, on the other hand, exhibited stronger variability between horizons, with both Bacteria and Archaea up to 100-fold more abundant in the horizon where H2S and Fe2+ were detected in the pore water (3 × 108 16S rRNA gene copies/g sediment in total at 88 cmbsf). In contrast, abundances were barely above the detection limit in the volcanoclastic layers.
The archaeal 16S rRNA gene copy numbers made up 3–89% of the total number of SSU rRNA gene copies in core GC6 and 31–75% in core GC12 (Fig. S1). Thus, the microbial population fluctuates between a bacterial- and an archaeal-dominated community even within these relatively short cores. The ratio between bacterial and archaeal SSU rRNA gene copies as determined by qPCR supported the relative amount of Bacteria versus Archaea as estimated from the amplicon library (Fig. S1), indicating that the primers used for the amplification for deep sequencing allowed a high rate of coverage and amplified with relatively little bias under the applied conditions.
Principal Components and Correlation Analyses.
Principal component analysis (PCA) was performed independently on the relative abundance data (Table S1) and on the geochemical data excluding all gene copy numbers, but including depth (Table 1). Sample scores on the first principal component (PC1) of the two datasets showed a significant rank-order correlation at the class level (ρ = 0.671, P = 0.02) (Fig. S2A), and the strength of this relationship increased substantially when oxic layers were removed (ρ = −0.883, P = 0.003) (Fig. S2B). This result demonstrates that changes in the geochemical structure of the sediments, especially below the oxic zone (PC1 explained 41% of the variance), covary with changes in the overall community structure (PC1 explained 33% of the variance at class level).
Next, we plotted the original data for each individual geochemical parameter against PC1 scores of the relative abundance data and found the strongest correlations with TOC (ρ = 0.621, P = 0.016) (Fig. 2A), dissolved SO42− in the pore water (ρ = 0.691, P = 0.006) (Fig. 2B), and the relative content of iron and manganese in the solid phase [ρ = −0.821, P < 0.000 (Fig. 2C) and ρ = 0.582, P = 0.025 (Fig. 2D), respectively].
Fig. 2.
Significant correlations (α = 0.05) between variation in microbial community structure and context data. Microbial community variation is measured by PC1 scores on relative abundance data at the class level. (A) Organic carbon content (% C). (B) Pore water sulfate concentration (mM). (C) Relative iron content in solid-phase Iron values as measured by counts by XRF and normalized to Ti counts. (D) Relative content of manganese measured in the solid phase. Manganese values are measured as counts by XRF and normalized to titanium (Ti) counts. Correlations are given as Spearman’s rank-order correlation (ρ). Blue circles indicate values from gravity core GC6; red triangles indicate values from gravity core GC12. Color shading indicates depth in sediment (light, shallow; dark, deep).
Finally, pairwise comparisons uncovered several strong linkages between the relative abundance of individual taxonomic groups and specific geochemical parameters (Fig. S3) as well as striking patterns of co-occurrence among microbial groups at different taxonomic levels (Fig. S2 C–F). The most important results are discussed in more detail below.
Distribution of the Most Abundant Bacterial Phyla.
Expressing the relative abundance of taxa as the proportion of total SSU rRNA gene tags allows the depth variation of the most abundant bacterial phyla to be compared with down-core variation in selected context data (Fig. 1).
Planctomycetes exhibited a high diversity in SSU rRNA gene sequences, represented by 461 different OTUs in GC6 and 311 in GC12 (97% cutoff). The great majority of these sequences could be assigned to the family of Planctomycetaceae, with little similarity to characterized relatives. We found a significant positive correlation between the abundance of this group and the SO42− concentration in pore water in core GC12 (r = 0.861, P = 0.028) (Fig. S3H). On the other hand, their relative abundance correlated positively with total inorganic carbon content in GC6 (r = 0.815, P = 0.026) (Fig. S3A). Although the former result suggests a link to the sulfur cycle, the latter is difficult to interpret. We detected low abundances of the Candidatus Scalindua group in the uppermost two horizons of GC12 (0.2 and 0.9% of the total SSU rRNA gene pool). This group is represented by members able to oxidize ammonium under anaerobic conditions (anammox) using nitrite as the electron acceptor (43). Interestingly, their appearance, albeit at very low numbers, in deeper layers of GC6 (156 and 174 cmbsf) coincided with relatively high numbers of reads affiliated with Nitrospina, Nitrosococcus, and MG-1 (all potentially involved in the nitrogen cycle). Chloroflexi were among the most dominant bacterial groups in both cores, with the majority of reads affiliating within the class of Dehalococcoides, but representatives from SAR202, Ktedonobacteria, Caldilineae, and Anaerolineae were present also. None of the 56 different Chloroflexi OTUs (97% cutoff) from the two cores showed close sequence similarity to any of the few cultured species (maximum similarity was 90% to Dehalococcoides ethanogenes). Members of the candidate division JS-1 often co-occur with Chloroflexi in anoxic sediment zones (13). This co-occurence also was observed in our cores. No inferences can be drawn about potential physiologies of either group, but, notably, the relative abundance of JS-1 in GC6 had a significant positive correlation with that of the family Desulfobacteraceae (r = 0.912, P = 0.011) (Fig. S2C) within the class of Deltaproteobacteria. Epsilonproteobacteria represented up to 39.5% of total reads in GC12 horizons and exhibited a unique depth profile. All reads that could be assigned to genus level affiliated with phylotypes involved in the sulfur cycle, including genera within the Helicobacteraceae (Sulfurimonas and Sulforovum) and to a lesser extent within Campylobacteraceae (Arcobacter and Sulforospirillum). A high proportion of reads could be resolved only to the family of Helicobacteraceae but displayed a depth distribution similar to that of the above-mentioned groups. Interestingly, the same OTUs are present in high numbers in both the uppermost (oxic) and lowermost (anoxic) horizons, indicating organisms with a facultative mode of energy metabolism (either reducing or oxidizing sulfur compounds), as has been shown for some cultured representatives (44, 45). In GC12 a significant negative correlation was found between the abundance of this group and SO42− concentration in the pore water (r = −0.896, P = 0.015) (Fig. S3G), indicating involvement in the sulfur cycle. In GC6, on the other hand, Epsilonproteobacteria were virtually absent, indicating minor importance of the sulfur cycle, in agreement with the geochemical data.
The presence of Epsilonproteobacteria in the deeper layers of GC12 could indicate elevated temperatures at depth. However, high abundances of the same bacterial OTUs in the uppermost and lowermost layers and the lack of any overall depth trends suggest a moderate temperature gradient, if any. Deltaproteobacteria was the most abundant class of proteobacteria in GC6, with a strong positive correlation with Mn2+ concentration in the pore water (r = 0.895, P = 0.003) (Fig. S3B). This correlation could be attributed to the abundance of Nitrospina and Sh765_TZT_29, an uncultured group within the Deltaproteobacteria (Silva taxonomy), pointing to their likely involvement in the manganese cycle.
Distribution of the Most Abundant Archaeal Phyla.
As observed for the Bacteria, most archaeal taxa were present (at the class level) in both cores, but the relative abundance varied greatly between and within cores (Fig. 1).
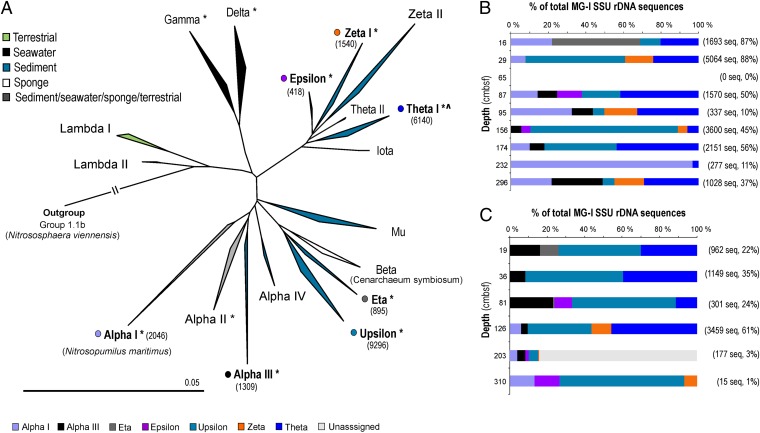
MG-I, also named “Marine Group I.1a,” now assigned to the Thaumarchaeota phylum (46, 47), was the most abundant in six of the nine horizons in GC6. This group contributed as much as ≥87% of the total number of SSU rRNA gene-sequence reads in the two top layers (16 and 29 cmbsf). They also dominated in GC12 except for the two deepest horizons (203 and 310 cmbsf). These Archaea not only constitute one of the most abundant microbial planktonic groups in the oceans (48, 49) but also are a highly abundant component in marine sediments (29, 50, 51). Although only aerobic growth has been reported for Thaumarchaeota thus far (52-55), we found MG-I–related organisms in anoxic horizons in our study. Their presence was particularly evident at 126 cmbsf in GC12 (61% of total reads) and at 88, 156, and 174 cmbsf in GC6 (45–56% of total reads), giving an absolute estimate of 4.2 × 105 to 1.5 × 108 MG-I–affiliated 16S rRNA gene copies/g sediment (wet weight) in these horizons. Phylogenetic analysis (Fig. 3) of all published full-length sequences of MG-I for which the habitat origin was assigned unambiguously in the database entry (Materials and Methods) demonstrated a separation of planktonic (gamma, delta), sponge-associated (beta), and terrestrial (lambda I, II) taxa from those found predominantly in the sediment (upsilon, eta, iota, theta, epsilon, zeta, and mu clusters; no sequences related to cluster kappa fulfilled the requirements mentioned in Materials and Methods) (Fig. 3A). With the exception of sequences affiliated with cluster alpha 1 (9%), all MG-I–related sequences in GC6 and GC12 were affiliated with the lineages that are found predominately in sediments (Fig. 3 B and C). This finding augments the earlier observations by Durbin and Teske (50), who stated that novel clusters distinct from planktonic MG-I organisms can be discovered in marine sediments. A similar habitat-specific clustering has been shown for the archaeal ammonia monooxygenase (amoA) (56, 57). To explore the potential of sedimentary MG-I archaea to oxidize ammonia, we quantified the archaeal amoA gene, which is used as a genetic marker for this metabolism. Copy numbers ranged from 1.5 × 103 to 2.4 × 106 and from 1.5 × 104 to 2.9 × 106/g sediment (wet weight) for GC6 and GC12, respectively (Table 1). We found a significant correlation of archaeal amoA gene copy numbers with the abundance of group MG-I (r = 0.929, P < 0.000) (Fig. 4C and Fig. S3E) and total numbers of archaeal 16S rRNA genes (r = 0.861, P = 0.003) (Figs. 4B and Fig. S3F) in GC6. Both marker genes (amoA and 16S rDNA) were found in the same order of magnitude in all samples. More intriguingly, the MG-I abundances also correlated with total NO3− concentration (r = 0.827, P = 0.011) (Fig. 4A and Fig. S3D) and, to a lesser but still significant extent, with TOC content (r = 0.692, P = 0.039) (Fig. S3C).
Fig. 3.
Phylogenetic analysis and depth distribution of MG-I. (A) Phylogeny based on SSU rRNA gene information from all published sequences available in the Silva database (release 104). The nomenclature follows that used by Durbin and Teske (50), but the additional group names lambda I, lambda II, and mu are given. The tree is reconstructed by NJ using the Felsenstein correction. Topology and clusters are supported by RaxML and PhyML reconstructions on the same dataset. Clusters marked with an asterisk contain sequences retrieved from marine hydrothermal environments. The sediment cluster marked with ^ contains a subcluster of freshwater/terrestrial sequences. Numbers in parentheses indicate the total number of reads from our study that affiliated with that particular group. (B and C) Depth distribution of MG-1 16S rRNA gene sequences affiliating with each cluster obtained in this study from core GC6 (B) and core GC12 (C). Numbers in parentheses indicate the number of reads from that horizon assigned to MG-1 and the percentage of the total. Color codes correspond to the groups in A.
Fig. 4.
Covariance between relative abundance of MG-I and context data. The depth distribution of the relative abundance of MG-I in core GC6 strongly covaries with (A) nitrate concentration (ppm) extracted from the solid phase (Pearson’s r = 0.827, P = 0.011); (B) archaeal SSU rDNA (r = 0.827, P = 0.011); and (C) archaeal amoA gene copies (r = 0.929, P < 0.000). Gene copy numbers are estimated by qPCR and given per gram of sediment (wet weight). See also Fig. S3.
The DSAG, also referred to as “Marine benthic group B,” is the most abundant single group of organisms in deep-sea sediments, along with the MCG (20). They were a dominant archaeal constituent in both cores. The relative abundance of DSAG in GC12 (∼25% of total reads) correlated significantly with both TOC and Fe2O3 concentration (r = 0.869, P = 0.025, and r = 0.819, P = 0.046, respectively) (Fig. 5 A and B and Fig. S3 I and J). This observation is consistent with a heterotrophic lifestyle (14) possibly coupled to the reduction of iron oxides rather than to sulfur compounds, as previously proposed (7).
Fig. 5.
Covariance between relative abundance of DSAG and context data. The depth distribution of the relative abundance of DSAG in core GC12 strongly covaries with (A) TOC (Pearson’s r = 0.869, P = 0.025) and (B) Fe2O3 concentrations in the sediment (Pearson’s r = 0.819, P = 0.046). See also Fig. S3.
SSU rDNA signatures of the MCG often are found in high numbers in clone libraries from deep-sea sediments [reviewed by Teske and Sørensen (19)]. In our samples they constitute up to ∼10% of all prokaryotic SSU rRNA genes in certain layers. This group, like the DSAG, has been proposed to be anaerobic heterotrophs (14) and has no cultured representatives. Unlike the DSAG, however, they populate a wide variety of habitats and exhibit larger diversity at the 16S rDNA level. No correlation for this group with any of the measured geochemical parameters was found within our dataset.
Methanomicrobia SSU rDNA signatures were among the four most dominant of the archaeal classes in GC12. The obtained sequences affiliated with ANME groups 1, 2a, 2b, and 2c, all of which are thought to be involved in anaerobic oxidation of methane (AOM) (58). Both their relative and total abundance increased with depth, reaching a maximum of 3.6% of all reads at the deepest horizon (310 cmbsf).
Notably, we found a tight correlation of ANME-1 phylotypes with both Methylococcales and Campylobacterales (Fig. S2 D–F), suggesting a common metabolism or a syntrophic partnership.
Discussion
The proximity of the two investigated sediment cores to hydrothermal active sites combined with sediment input from the Bear Island Fan has resulted in a compact redox zonation profile and high concentrations of dissolved metals in both cores. These properties make the sediments interesting model sites to study the variation of subsurface microorganisms in the context of geochemical changes. There were substantial differences between the two cores. For example, the GC12 core is distinguished from GC6 by a clear signature of shallow sulfate reduction and much higher concentrations of dissolved iron (Table 1). Despite such differences, the PCA analysis (Fig. S2 A and B) shows that the samples from both cores share a major axis of variation that links changes in the overall composition of the microbial community to changes in the overall geochemical composition. More specifically, the structure of the prokaryotic communities is coupled to variation in the iron and manganese content of the minerals, TOC, and pore water SO42− concentration (Fig. 2). Our results provide quantitative evidence for the common assumption that organic carbon is one of the fundamental factors shaping microbial communities, supporting the argument that heterotrophic organisms play an important role in deep-sea sediments (14, 20). The content of iron and manganese within the sediments is strongly related to microbial community structure, and, although the causal relationships are highly complex, our results suggest that mineralogy is a key determinant. Finally, we attribute the correlation between abundance and sulfate concentration to the effect of microbial sulfur metabolisms on pore water chemistry, rather than vice versa.
By directly correlating the abundance of each individual taxonomic group to the individual context data, we were able to make predictions about the metabolism of the most dominating organisms in the deep-sea sediments, in particular MG-I, DSAG, and Epsilon- and Deltaproteobacteria. In addition, strong correlations in the relative abundances of different taxa (Fig. S2 C–F) might help elucidate syntrophic partnerships and/or common metabolic preferences.
The archaeal class MG-I belongs to the newly defined phylum Thaumarchaeota, represented by two pure cultures and a few enrichments (59) that were shown to gain energy from aerobic oxidation of NH4+ to NO2−. Their main carbon source is CO2, but coassimilation of organic carbon has been reported for this group (52). The majority of MG-I sequences retrieved in our study (21,644 reads) clustered in phylogenetic groups mainly associated with marine sediments, suggesting the presence of a specialized sediment population (Fig. 3).
None of these groups has any characterized members, but at least four observations support their ability to oxidize ammonia to nitrite in the sediments from GC6: (i) the tight correlation between the abundance of this group and amoA gene copy numbers (Fig. 4C and Fig. S3E); (ii) a high positive correlation of the MG-I with nitrate concentration (Fig. 4A and Fig. S3D); (iii) their co-occurrence with several other phylotypes normally linked to the nitrogen cycle, such as members related to anammox and nitrite-oxidizing Nitrospina; and (iv) the fact that the increase in ammonium concentration below the oxic zone is less pronounced than would be expected otherwise (Fig. 1). The presence of an active group of ammonia-oxidizing archaea is surprising, because the sediments are considered to be anoxic below the two uppermost sampling depths. However, MG-I–related sequences have been reported previously from supposedly anoxic environments, although the metabolic implications of these findings have not been addressed (7, 29, 39, 60). Their presence in anoxic horizons can have several explanations: (i) sediment-specific phylogenetic groups of MG-I may have the ability to oxidize ammonium with an alternative electron acceptor; (ii) oxygen could be produced intracellularly, as recently described for the methane-oxidizing group NC10 (50); or (iii) amo genes present in the organisms could have some other function, as suggested by Mussmann and colleagues (61). In summary, our data strongly support ammonia oxidation, but the electron acceptor remains unknown. In this context it is noteworthy that members of Methylococcales, another group assumed to be comprised exclusively of aerobes, also occur in highly reduced horizons in this study.
The DSAG is proposed to represent heterotrophic organisms based on indirect evidence from stable carbon isotopes in archaeal lipids (14). A strong correlation between the relative abundance of DSAG and organic carbon content in our study (Fig. 5A and Fig. S3I) supports this suggestion. Their energy-yielding metabolism is a subject of debate, and several studies have proposed direct or indirect coupling to methane oxidation (7, 14, 62) as well as possible sulfate reduction (7). Here we argue that labile iron oxides serve as the terminal electron acceptor either in the direct oxidation of organic carbon or as an energy-yielding metabolism coupled to another electron donor, such as CH4, NH4+, or sulfur compounds. We base this proposal on the significant correlation between iron oxide and the relative abundance of DSAG in core GC12 (Fig. S3J). The correlation between DSAG and dissolved Fe2+ in the pore water was positive but not significant (Fig. S3K). (GC6 showed a similar relationship but was omitted from statistical analysis because of the low number of observations.)
It is well known that the iron and sulfur cycles are tightly linked; hence Fe2+ in the pore water also could result from sulfate reduction to sulfide and the subsequent reaction with iron oxide (63). However, our sulfate profiles do not suggest any significant sulfate reduction except at the deepest horizon in GC12. Thus, iron oxide is the most likely electron acceptor, and the results discussed above, as well as theoretical energy yield, point to organic carbon as the most likely donor. Alternatively, this pathway could involve AOM coupled to iron oxide reduction, a process that potentially yields more energy than AOM coupled to sulfate reduction. Such a metabolism has been demonstrated recently based on geochemical data by Beal and colleagues (64).
In summary, these highly stratified sediments offered a unique possibility to correlate stratigraphic variation in geochemical properties directly to stratigraphic variation in the structure of the microbial community as well as to the relative abundance of individual taxa. We combined deep sequencing of bacterial and archaeal SSU rRNA genes, using a single primer set with broad target coverage, with an extensive set of environmental context data, including nitrate from the solid phase. We used PCA to distill the covariance structures of the prokaryotic community and the geochemical data separately, thereby revealing a significant correlation between overall changes in geochemistry and overall changes in community composition. Four geochemical components were linked closely to the taxonomic distribution of microorganisms: the total organic carbon, iron, and manganese content in the minerals and the sulfate concentration in pore water. Our findings yield testable predictions about the metabolisms of the most typical and abundant microbial lineages found in the deep subsurface, including the DSAG and sediment lineages of potentially anaerobic MG-1.
Materials and Methods
Site Location and Shipboard Sampling.
After retrieval, the cores immediately were cut in sections and split into archive and working halves. Sampling of pore water and sediment for geochemical and microbial analyses from the working halves was conducted onboard the ship as quickly as possible. The two gravity cores, GC6 ∼15 km SE of the vent field (73°21.39′N, 7°33.90′E, 3,280 mbsl), and GC12 ∼15 km NE of the vent field (73°45.80′N, 8°27.83′E, 3,250 mbsl), were selected for microbiological studies. The in situ bottom water temperature was −0.2 °C when cores were retrieved. Samples were collected at depths of 16, 29, 65, 87, 95, 156, 174, 232, and 296 cm in GC6 and 19, 36, 81, 126, 203 and 301 cm in GC12. All samples were collected with sterile 10-mL syringes and were processed immediately or snap-frozen in liquid nitrogen before storage at −80 °C. Pore water was extracted with Rhizon samplers from approximately the same depths as the microbiological samples and was analyzed immediately onboard for pH, alkalinity, sulfide, and ammonium or was stored at 4 °C until later onshore analysis of dissolved ions. Eh measurements of the sediment were carried out onboard. After sampling was completed, the sediment cores were stored in plastic boxes at 4–6 °C on the ship and later at the University of Bergen core repository.
DNA Extraction.
DNA was extracted from ∼0.5 g of sediment in each sample using a FastDNA spin for soil kit in conjunction with the FastPrep-24 instrument (MP Biomedicals) following the manufacturer’s protocol applying the poly(A) modification described by Hugenholtz et al. (65).
DNA Amplification for the 16S rRNA Gene Library.
Different primer combinations were evaluated in silico before final selection using the Ribosomal Database Project (RDP) (66), to select the lowest possible degeneracy while maintaining high overall prokaryotic target coverage. In addition, the 454 GS (454 Life Sciences, Roche) FLX technology requires that the length of the amplified product be within 300–800 bp. The optimal primer combination was found to be Uni787F (5′-ATTAGATACCCNGGTAG-3′) (67) and Uni1391R (5′-ACGGGCGGTGWGTRC-3′), modified from ref. 68. These primers target the V5–V8 region on the 16S rRNA gene and cover 87% and 94%, respectively, of all prokaryotes in the RDP without mismatch (as of May, 2011). The stringency of the performed PCR was kept at a minimum to obtain as much taxonomic diversity as possible, with one mismatch increasing the coverage of both primers to 98%. Furthermore, the chosen primer combination had the least bias toward any one specific taxonomic group (“universal” prokaryotic primers often are problematic, in that they tend to have mismatches against specific taxonomic groups such as Verrucomicrobia, Planctomycetes, and Chloroflexi). DNA from each horizon was PCR amplified in triplicate using the above-mentioned primer combination under the following thermal conditions: 95 °C for15 min, then 25–30 cycles of 94 °C for 45 s, 53 °C for 45 s, 72 °C for 1 min followed by 72 °C for 7 min before cooling at 4 °C. Each reaction (25 μL) contained 1× HotStar Taq Master Mix (Qiagen), template DNA, and 1.2 μM of each primer. To ensure correct amplicon length, the PCR product was evaluated by gel electrophoresis. The triplicate PCR products then were pooled to minimize PCR drifting and were purified using GenElute PCR Clean-Up kit (Sigma). A new round of PCR was performed using the same specific primers and thermal conditions but linked to the 454 Life Sciences A and B pyrosequencing adaptor sequence. In addition, the forward primer was labeled with a unique barcode (one for each analyzed sediment horizon) as described by Hamady et al. (69). To minimize PCR bias, the number of cycles was held to a minimum (25–30 in the first PCR and five in the second). Amplicons were purified again, and the concentration and quality were controlled by gel inspection, spectrometry (Cary-300 Bio UV-vis; Varian), and BIO-analyzer (Agilent Technologies). All amplicons (a total of 15 unique samples) then were pooled in a 1:1 ratio based on DNA concentration (∼20 ng/μL from each sample) and sequenced using multiplex GS FLX pyrosequencing (without titanium chemistry) at the Norwegian High-Throughput Sequencing Centre in Oslo, Norway.
Pyrosequencing flowgrams (SSF files) have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accession number SRP009131.
Filtering and Removal of Noise from Amplicon Sequence Data.
The dataset (84,580 reads) was filtered and cleaned from noise by using AmpliconNoise (70) software. In short, this method includes four steps: filtering, flowgram clustering, sequence clustering, and chimera removal. In the filtering step, reads were truncated at 600 flows, and those with fewer than 360 flows or a noisy signal (flow intensity 0.5–0.7, equivalent to a degenerate base) before this position were removed. In addition, all reads not matching the barcode and primer sequences were removed. In the sequence-clustering step, reads were truncated at 240 bp. AmpliconNoise generated a set of de-noised and chimera-filtered sequences, each with a set of reads most likely to be derived from it. Barcode and primer sequences were removed before further analysis such as taxonomic classification and linkage clustering.
Taxonomic Evaluation.
To assign the filtered, de-noised, and chimera-filtered sequences to taxa, we modified and updated the Silva SSUref database release 100 (71) with respect to taxonomy, based on the most recent literature. This database (available at http://services.cbu.uib.no/supplementary/community-profiling/) is described in Lanzén et al. (72).
The sequences were aligned to this database using Blastn (NCBI), and the results were analyzed and assigned to specific taxa using the software MEGAN version 3.7 (73) by applying a lowest common ancestor algorithm [for details see Lanzén et al. (72)].
OTU Clustering.
All unique de-noised sequences were clustered into OTUs using maximum linkage clustering of pairwise distances. The distance matrix was generated using the Needleman–Wunsch algorithm as implemented in NDist, and clustering was carried out using FDist (both programs are distributed with AmpliconNoise) (70). A 3% distance cutoff was used to define OTUs.
Phylogenetic Analysis of MG-1.
The phylogenetic relationship within the MG-I was evaluated to find habitat-specific groups and to calculate the affiliation of MG-I sequences from this study. The tree was calculated based on all published full-length (>900 bp) sequences available in the Silva database release 104 (71), applying the archaeal positional filter and removing highly variable positions (if a sequence was described as “unpublished” in the ARB entry field “journal,” it was excluded from the calculation). Length was truncated to ARB position 1773-31131 leaving 784 valid columns for calculation. After calculation, sequences were removed if the ARB entry field “Isolation source” contained one of the following words: mangrove, marsh, or estuary (to avoid uncertainties in the habitat type), leaving 659 sequences. Clusters containing one or more sequences obtained from hydrothermal environments are marked with an asterisk. We do not label these clusters as hydrothermal in Fig. 3, because sequences obtained from marine hydrothermal settings are inherently difficult to assign unambiguously to a specific habitat (e.g., seawater, sediment, chimney, microbial mat). The phylogenetic tree (Fig. 3A) was calculated by Neighbor Joining (NJ) using the Felsenstein correction (74). To verify the tree topology, RaxML (75) and PhyML (76) algorithms likewise were applied on the same dataset; both supported the displayed clustering and grouping. The nomenclature follows that reviewed by Durbin and Teske (50). In addition, we have identified and named three clusters: lambda I and II (exclusively from terrestrial habitats) and mu (exclusively from marine sediments).
All unique sequences from each sediment horizon that could be assigned to MG-I from our dataset (123 sequences from a total of 21,644) were added to the tree using the parsimony tool in the ARB package. Adding this many short sequences distorts the branch length; hence they are not included in the displayed tree. To confirm the affiliation of our sequences, an NJ tree was calculated based on the above-mentioned database sequences and our sequences but with all truncated to a length of 230 bp. All affiliations were confirmed, with a few exceptions for which the sequences could not be phylogenetically resolved. The depth distributions of our sequences and cluster to which they affiliate are displayed for both gravity core GC6 (Fig. 3B) and gravity core GC12 (Fig. 3C).
Real-Time qPCR.
Archaeal 16S rRNA genes were quantified using the prokaryotic primer Uni519F (5′-CAGCMGCCGCGGTAA-3′) (77) and the archaeal-specific primer Arc908R (5′-CCCGCCAATTCCTTTAAGTT-3′) [modified from Jurgens et al. (78)]. Each reaction (25 μL) contained 1× QuantiTech Sybr Green PCR master mixture (Qiagen), 0.8 μM of each primer, and 1 μL template DNA. The thermal cycling program was 15 min at 95 °C, then 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 45 s. The quantification standard consisted of a dilution series of a known amount of linearized fosmid 54d9 (79) and a copy number of archaeal 16S rRNA genes between 10 and 107 copies/μL [calculated as described by Leininger et al. (80)]. Genomic DNA from Escherichia coli was used as a negative control. The R2 value for the standard curve was 0.99, and the slope value was −3.26, giving an estimated amplification efficiency of 102%.
Bacterial 16S rRNA genes were quantified using the bacterial-specific primer bac341f (5′-CCTACGGGWGGCWGCA-3′) [modified from Ishii and Fukui (81)] and the prokaryotic 519r (5′-TTACCGCGGCKGCTG-3′) (77). The quantification standard consisted of a dilution series (between 1 × 102 and 1 × 107 copies/μL) of a known amount of purified PCR product obtained from genomic E. coli DNA by using the bacterial 16S rRNA gene-specific primers 8F/1392R (68, 82). Sulfolobus solfataricus genomic DNA was used as negative control. The R2 value for the standard curve was 0.99, and the slope value was −3.06, giving an estimated amplification efficiency of 112%. The thermal cycle program was 15 min at 95 °C, then 35 cycles of 95 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s.
Archaeal amoA genes were quantified using the archaeal amoA-specific primers CrenamoA23f (5′-ATGGTCTGGCTWAGACG-3′) (80) and CrenamoA616r (5′-GCCATCCABCKRTANGTCCA-3′) (83). Each reaction (25 μL) contained 1× QuantiTech Sybr Green PCR master mixture (Qiagen), 1.2 μM of each primer, and 1 μL template DNA. The thermal cycling program was 15 min at 95 °C, then 40 cycles of 95 °C for 30 s, 50 °C for 45 s, and 72 °C for 45 s. The quantification standard was the same as used in archaeal and crenarchaeal 16S rRNA gene quantification. Genomic DNA from E. coli was used as a negative control. The R2 value for the standard curve was 0.99, and the slope value −3.51, giving an estimated amplification efficiency of 93%. All qPCR experiments were performed with the Step-OnePlus real-time PCR system (Applied Biosystems) using SYBRGreenI as the fluorescent dye. To confirm product specificity, melting curve analyses were performed after each run for all experiments, and each qPCR setup contained samples, standard series, negative controls, and blanks, all in triplicate.
XRF Core Scanning.
The archived core halves were scanned using the nondestructive ITRAX XRF core scanner system at Bergen Geoanalytical Facility, University of Bergen. Samples were irradiated with 3 kW Mo. Step size was 0.5 mm for XRF analysis with a count time of 10 s. Manganese and iron were normalized to titanium counts.
Organic and Inorganic Carbon Measurements.
Coulometric titration (CM5012 CO2 Coulometer; IUC, Inc.) was used to determinate the total inorganic carbon (TIC) and the total carbon (TC) contents in 12 of the 15 samples. Analytical precision determined by analysis of replicate standards for TIC and TC was ±0.02% C and ±0.03% C, respectively. The TOC contents were calculated by subtracting TIC contents from the TC contents, leading to an error of ±0.05% C for TOC contents. The remaining three samples were measured applying the method described for nitrogen in the Solid-Phase Geochemistry section below.
Pore Water Chemistry.
Pore water was extracted using Rhizon samplers, and aliquots were analyzed onboard for pH by using a mobile pH meter, for alkalinity by an autotitrator, and for sulfide and ammonium by spectrophotometric methods (84). Onshore analysis of sulfate was performed by ion chromatography, and dissolved iron and manganese were analyzed by inductively coupled plasma optical emission spectrometry.
Solid-Phase Geochemistry.
Eh was measured in all layers directly by electrodes (SP 50×; Consort) inserted into undisturbed sediment as soon as cores were split.
Nitrates were measured in an aqueous solution of (NH4)2SO4 (2 M) and sediment (5:1 ratio) through an ion-selective electrode (nitrate ion combination epoxy electrode no. 31503; Phoenix). Nitrogen and sulfur were determined using an elemental analyzer for CHNS-O based on the principle of dynamic flush combustion coupled with gas chromatography. Nitrogen was measured using a GC packed column for CHNS polytetrafluoroethylene, 2 m (Eurovector), and sulfur was measured using a GC packed column for sulfur, 1 m, 6 × 5 mm (Eurovector). Each sample was dried and powdered before analysis. Iron oxide and manganese oxide contents were determined by atomic absorption spectrometry after digestion of samples by microwave, using H2O2+HCl, HNO3, and HF.
Statistical Analyses.
PCA was used to summarize the community structure (relative abundance data) and the geochemical structure (context data) of the sediments quantitatively. Each sample was standardized to zero mean and unit SD. The relative abundance data also were subjected to a centered log-ratio transformation with multiplicative zero replacement (85, 86) to remove any forced correlations imposed by the constant-sum constraint. PCA uses the covariance structure to rotate the original data onto a new set of orthogonal (independent) axes oriented in the direction of maximal remaining variance. By projecting the samples onto the first axis (PC1), we effectively reduced the community (or geochemical) structure to a single variable accounting for the greatest proportion of variance. If the overall community structure were causally related to the overall geochemical structure of the sediment, then we would expect a significant, monotonic (if not strictly linear), arbitrarily signed relationship between the PC1 scores of the relative abundance and geochemical datasets. We tested this hypothesis using the Spearman rank-order correlation (ρ).
The community–geochemistry relationship was dissected further by correlating the relative abundance PC1 scores against the original geochemical measurements, again using Spearman's ρ. Finally, we mined the relative abundance data directly to explore linkages among individual taxa and linkages between individual taxa and geochemical variables, using Pearson's product–moment correlation.
Supplementary Material
Acknowledgments
We thank S. Monsen and all the onboard scientific members and crew members of the research vessel G.O. Sars during the 2008 H2Deep expedition. We thank H. Haflidasson for help with the core scanning and T. Urich for fruitful discussions. The Norwegian High-throughput sequencing Centre at the University of Oslo performed the sequencing (http://www.sequencing.uio.no). The project was funded by the H2Deep project through the European Science Foundation program (EuroMARC). Additional funding was provided by Norwegian Research Council through the Centre for Geobiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: 16S rRNA gene sequences are deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP009131). A comma-separated text file listing the taxonomic affiliations of all operational taxonomic units and their distribution across datasets can be downloaded from http://services.cbu.uib.no/supplementary/jorgensen2012. Data have been deposited in the Pangaea database (doi:10.1594/PANGAEA.786687).
See Commentary on page 16756.
See Author Summary on page 16764 (volume 109, number 42).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207574109/-/DCSupplemental.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes RJ, Cragg BA, Wellsbury P. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol J. 2000;8(1):11–28. [Google Scholar]
- 3.Berner RA. Burial of organic carbon and pyrite sulfur in the modern ocean; its geochemical and environmental significance. Am J Sci. 1982;282(4):451–473. [Google Scholar]
- 4.Rochelle PA, Fry JC, Parkes RJ, Weightman AJ. DNA extraction for 16S ribosomal-RNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett. 1992;100(1–3):59–65. doi: 10.1111/j.1574-6968.1992.tb14019.x. [DOI] [PubMed] [Google Scholar]
- 5.Marchesi JR, Weightman AJ, Cragg BA, Parkes RJ, Fry JC. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol Ecol. 2001;34(3):221–228. doi: 10.1111/j.1574-6941.2001.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 6.Reed DW, et al. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl Environ Microbiol. 2002;68(8):3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inagaki F, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA. 2006;103:2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inagaki F, et al. Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of okhotsk. Appl Environ Microbiol. 2003;69:7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormas KA, Smith DC, Edgcomb V, Teske A. Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176) FEMS Microbiol Ecol. 2003;45:115–125. doi: 10.1016/S0168-6496(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 10.Newberry CJ, et al. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ Microbiol. 2004;6:274–287. doi: 10.1111/j.1462-2920.2004.00568.x. [DOI] [PubMed] [Google Scholar]
- 11.Parkes RJ, et al. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature. 2005;436:390–394. doi: 10.1038/nature03796. [DOI] [PubMed] [Google Scholar]
- 12.Parkes RJ, et al. Deep bacterial biosphere in Pacific Ocean sediments. Nature. 1994;371:410–413. [Google Scholar]
- 13.Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol. 2008;66:181–196. doi: 10.1111/j.1574-6941.2008.00566.x. [DOI] [PubMed] [Google Scholar]
- 14.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sørensen KB, Teske A. Stratified communities of active Archaea in deep marine subsurface sediments. Appl Environ Microbiol. 2006;72:4596–4603. doi: 10.1128/AEM.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roussel EG, et al. Extending the sub-sea-floor biosphere. Science. 2008;320:1046. doi: 10.1126/science.1154545. [DOI] [PubMed] [Google Scholar]
- 17.D’Hondt S, et al. Subseafloor sedimentary life in the South Pacific Gyre. Proc Natl Acad Sci USA. 2009;106:11651–11656. doi: 10.1073/pnas.0811793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen BB, Boetius A. Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol. 2007;5:770–781. doi: 10.1038/nrmicro1745. [DOI] [PubMed] [Google Scholar]
- 19.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 20.Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev. 2011;75:361–422. doi: 10.1128/MMBR.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, et al. Community structure of archaea from deep-sea sediments of the South China Sea. Microb Ecol. 2010;60:796–806. doi: 10.1007/s00248-010-9746-y. [DOI] [PubMed] [Google Scholar]
- 22.Webster G, et al. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:65–85. doi: 10.1111/j.1574-6941.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- 23.Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA. 2008;105:10583–10588. doi: 10.1073/pnas.0709942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biddle JF, White JR, Teske AP, House CH. Metagenomics of the subsurface Brazos-Trinity Basin (IODP site 1320): Comparison with other sediment and pyrosequenced metagenomes. ISME J. 2011;5:1038–1047. doi: 10.1038/ismej.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schippers A, et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 26.D’Hondt S, Rutherford S, Spivack AJ. Metabolic activity of subsurface life in deep-sea sediments. Science. 2002;295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 27.Froelich PN, et al. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: Suboxic diagenesis. Geochim Cosmochim Acta. 1979;43(7):1075–1090. [Google Scholar]
- 28.Durbin AM, Teske A. Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ Microbiol. 2011;13:3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x. [DOI] [PubMed] [Google Scholar]
- 29.Roussel EG, et al. Archaeal communities associated with shallow to deep subseafloor sediments of the New Caledonia Basin. Environ Microbiol. 2009;11:2446–2462. doi: 10.1111/j.1462-2920.2009.01976.x. [DOI] [PubMed] [Google Scholar]
- 30.Fry JC, Webster G, Cragg BA, Weightman AJ, Parkes RJ. Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:86–98. doi: 10.1111/j.1574-6941.2006.00144.x. [DOI] [PubMed] [Google Scholar]
- 31.Webster G, et al. Distribution of candidate division JS1 and other Bacteria in tidal sediments of the German Wadden Sea using targeted 16S rRNA gene PCR-DGGE. FEMS Microbiol Ecol. 2007;62:78–89. doi: 10.1111/j.1574-6941.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- 32.Hoehler TM, Alperin MJ, Albert DB, Martens CS. Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogen-sulfate reducer consortium. Global Biogeochem Cycles. 1994;8(4):451–463. [Google Scholar]
- 33.Boetius A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 34.Strous M, et al. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 35.Nunoura T, et al. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the Southern Okinawa trough. Appl Environ Microbiol. 2010;76:1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durbin AM, Teske A. 2012. Archaea in organic-lean and organic-rich marine subsurface sediments: An environmental gradient reflected in distinct phylogenetic lineages. Front Microbiol, 10.1038/ncomms1124.
- 37.Pedersen RB, et al. Discovery of a black smoker vent field and vent fauna at the Arctic Mid-Ocean Ridge. Nat Commun. 2010;1:126. doi: 10.1038/ncomms1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canfield DE, Thamdrup B. Towards a consistent classification scheme for geochemical environments, or, why we wish the term ‘suboxic’ would go away. Geobiology. 2009;7:385–392. doi: 10.1111/j.1472-4669.2009.00214.x. [DOI] [PubMed] [Google Scholar]
- 39.Sørensen KB, Lauer A, Teske A. Archaeal phylotypes in a metal-rich and low-activity deep subsurface sediment of the Peru Basin, ODP Leg 201, Site 1231. Geobiology. 2004;2(3):151–161. [Google Scholar]
- 40.Harmand J-M, Ávila H, Oliver R, Saint-André L, Dambrine E. The impact of kaolinite and oxi-hydroxides on nitrate adsorption in deep layers of a Costarican Acrisol under coffee cultivation. Geoderma. 2010;158(3–4):216–224. [Google Scholar]
- 41.Schippers A, Neretin LN. Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ Microbiol. 2006;8:1251–1260. doi: 10.1111/j.1462-2920.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- 42.D'hondt SL, Jorgenen BB, Miller DJ, et al. 2003 [Google Scholar]
- 43.Schmid M, et al. Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol. 2003;26:529–538. doi: 10.1078/072320203770865837. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto M, Nakagawa S, Shimamura S, Takai K, Horikoshi K. Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ Microbiol. 2010;12:1144–1153. doi: 10.1111/j.1462-2920.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- 45.Sievert SM, et al. USF Genomics Class Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 47.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 48.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol. 2004;70:781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durbin AM, Teske A. Sediment-associated microdiversity within the Marine Group I Crenarchaeota. Environmental Microbiology Reports. 2010;2(5):693–703. doi: 10.1111/j.1758-2229.2010.00163.x. [DOI] [PubMed] [Google Scholar]
- 51.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 56.Dang H, et al. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J Appl Microbiol. 2009;106:1482–1493. doi: 10.1111/j.1365-2672.2008.04109.x. [DOI] [PubMed] [Google Scholar]
- 57.Pester M, et al. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2012;14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knittel K, Boetius A. Anaerobic oxidation of methane: Progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 59.Pester M, Schleper C, Wagner M. The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H, et al. Dominance of putative marine benthic Archaea in Qinghai Lake, north-western China. Environ Microbiol. 2008;10:2355–2367. doi: 10.1111/j.1462-2920.2008.01661.x. [DOI] [PubMed] [Google Scholar]
- 61.Mussmann M, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teske AP. Microbial communities of deep marine subsurface sediments: Molecular and cultivation surveys. Geomicrobiol J. 2006;23(6):357–368. [Google Scholar]
- 63.Canfield DE. Reactive iron in marine sediments. Geochim Cosmochim Acta. 1989;53:619–632. doi: 10.1016/0016-7037(89)90005-7. [DOI] [PubMed] [Google Scholar]
- 64.Beal EJ, House CH, Orphan VJ. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 65.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roesch LFW, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lane DJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lanzén A, et al. Exploring the composition and diversity of microbial communities at the Jan Mayen hydrothermal vent field using RNA and DNA. FEMS Microbiol Ecol. 2011;77:577–589. doi: 10.1111/j.1574-6941.2011.01138.x. [DOI] [PubMed] [Google Scholar]
- 73.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- 75.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 76.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 77.Ovreås L, Forney L, Daae FL, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Treusch AH, et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 80.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 81.Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 84.Grasshoff K, Kremling K, Ehrhardt M. Methods of Seawater Analysis. 2nd Ed. Weinheim, NY: Wiley-VCH; 1999. [Google Scholar]
- 85.Aitchison J. The Statistical Analysis of Compositional Data. Monographs on Statistics and Applied Probability. London: Chapman and Hall; 1986. p. 416. [Google Scholar]
- 86.Martín-Fernández J, Barceló-Vidal C, Pawlowsky-Glahn V. Dealing with Zeros and Missing Values in Compositional Data Sets Using Nonparametric Imputation. Math Geol. 2003;35(3):253–278. [Google Scholar]