Abstract
Ribosome protection proteins (RPPs) confer tetracycline resistance by binding to the ribosome and chasing the drug from its binding site. The current model for the mechanism of action of RPPs proposes that drug release is indirect and achieved via conformational changes within the drug-binding site induced upon binding of the RPP to the ribosome. Here we report a cryo-EM structure of the RPP TetM in complex with the 70S ribosome at 7.2-Å resolution. The structure reveals the contacts of TetM with the ribosome, including interaction between the conserved and functionally critical C-terminal extension of TetM and the decoding center of the small subunit. Moreover, we observe direct interaction between domain IV of TetM and the tetracycline binding site and identify residues critical for conferring tetracycline resistance. A model is presented whereby TetM directly dislodges tetracycline to confer resistance.
Keywords: antibiotic, protein synthesis, RNA, tigecycline, translation
The translational apparatus represents one of the major targets within the bacterial cell for antibiotic treatment (1). One well-characterized class of antibiotics in clinical use is the tetracyclines. Tetracyclines are broad-spectrum antibiotic agents that bind to elongating ribosomes and inhibit delivery of the ternary complex EF-Tu, GTP and aminoacylated-tRNA (EF-Tu•GTP•aa-tRNA) to the A-site (1). Consistently, crystal structures of the small (30S) ribosomal subunit in complex with tetracycline reveal the primary binding site to be located in helix 34 (h34) of the 16S rRNA, in a position overlapping with the anticodon stem-loop of A-site tRNA (A-tRNA) (2, 3). The widespread use of tetracyclines during the past 60 years has led to an increase in acquired tetracycline resistance determinants among clinically important pathogenic bacteria, limiting the utility of many members of this class (4). Of the variety of tetracycline-specific resistance mechanisms, efflux and ribosome-protection are the most common (5). The third generation of tetracycline derivatives, such as tigecycline (Tgc), display enhanced antimicrobial activity, overcoming efflux and ribosome protection mechanisms (6–8).
Ribosome protection is mediated by so-called ribosome protection proteins (RPPs), with the most prevalent and best characterized being TetO and TetM (5, 9). TetO and TetM exhibit ∼75% sequence similarity with each other (10) and ∼45% similarity with elongation factor G (EF-G) (SI Appendix, Fig. S1). Based on the presence of conserved nucleotide binding motifs (i.e., G1–G5), RPPs are grouped together within the translation factor superfamily of GTPases (11). Accordingly, TetO and TetM catalyze the release of tetracycline from the ribosome in a GTP-dependent manner (12, 13). Although GTPase activity is required for multiturnover of RPPs, GTP hydrolysis is however not strictly required to dislodge tetracycline because drug release also occurs in the presence of nonhydrolyzable GTP analogues (12, 13).
A cryo-EM reconstruction of TetO•GDPγS•70S complex at 16-Å resolution revealed density for TetO within the intersubunit space with an overall position similar to EF-G (14). In contrast to EF-G, the tip of domain IV of TetO does not significantly overlap with the A-tRNA, but rather interacts with h34 adjacent to the tetracycline binding site (14). Binding of TetO to the ribosome leads to protection of C1214 and to a lesser extent C1054 within h34 from chemical modification, whereas the reactivity of A1408 in h44 is enhanced (15). As TetO is not observed to directly interact with C1054 or A1408 (14), TetO was suggested to chase tetracycline from the ribosome indirectly via inducing local disturbances within h34 (9, 14, 15). Moreover, the conformational changes were proposed to persist after TetO has dissociated from the ribosome, preventing rebinding of tetracycline as well as stimulating delivery of the ternary complex (9, 16).
To gain further structural insights into the interaction of RPPs with the ribosome and the mechanism of RPP-mediated tetracycline release, we have determined a cryo-EM structure of the RPP TetM bound to the 70S ribosome at 7.2-Å resolution. The improved quality of the map allows us to present the first molecular model for TetM as well as a detailed account of TetM interactions with the ribosome. Surprisingly, the higher resolution enables us to observe density for a loop in domain IV of TetM that interacts directly with the tetracycline binding site, indicating that RPP action uses a direct mechanism of action to dislodge and release tetracycline from the ribosome.
Results and Discussion
Cryo-EM Structure of a TetM•70S Complex.
Escherichia coli 70S ribosomes (0.4 μM) were mixed together with Tgc (10 μM), and subsequently incubated at 37 °C for 20 min in the presence of purified recombinant Enterococcus faecalis TetM protein (4 μM) and the nonhydrolysable GTP analogue GDPNP (500 μM). Pelleting assays confirmed that binding of TetM to the 70S ribosome was observed in the presence of Tgc, suggesting that analysis of this complex by cryo-EM may enable TetM and Tgc to be visualized simultaneously on the same ribosome. Thus, to analyze the interplay between TetM and Tgc, the TetM•70S complex formed in the presence of Tgc was selected for analysis by cryo-EM and single particle reconstruction. From a total of 406,687 particles, in silico sorting yielded three main subpopulations of 70S ribosomal particles (SI Appendix, Fig. S2A): rotated (40%) and nonrotated (33%) 70S ribosomes without density for TetM and a TetM-bound 70S ribosome (27%) exhibiting a nonrotated conformation. Further refinement of the TetM•70S complex resulted in a final density map (Fig. 1A) with a resolution of 7.2 Å (EMD-2183), as determined by using the Fourier shell correlation cutoff value of 0.5 (SI Appendix, Fig. S2B). The absence of a subpopulation of TetM bound to rotated 70S ribosomes suggests that TetM binds preferentially to the nonrotated state, i.e., analogous to the expected (posttranslocational) substrate resulting from tetracycline inhibition.
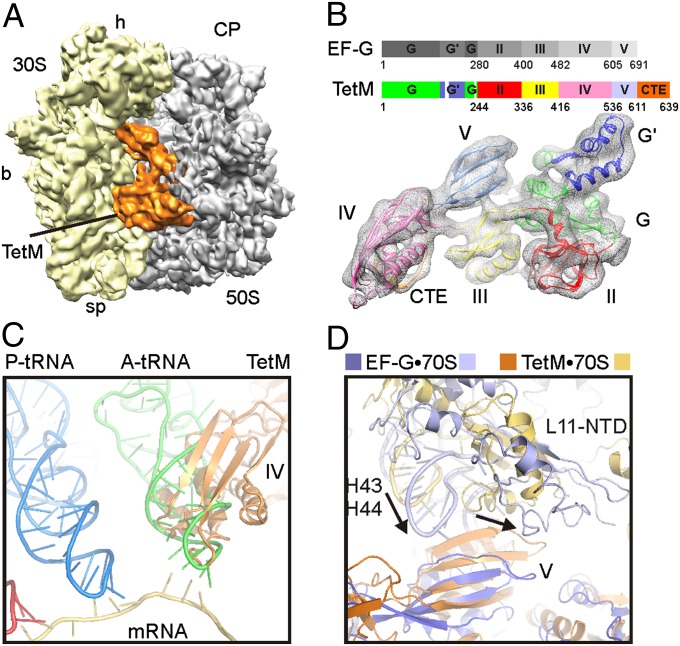
Fig. 1.
Cryo-EM reconstruction of a TetM•70S complex. (A) Final map of the TetM•70S complex with TetM (orange), 30S (yellow), and 50S (gray). (b, body; CP, central protuberance; h, head; sp, spur). (B) Schematic color code of the domain structure of EF-G and TetM (domain I, G′ subdomain, II, III, IV, V, and CTE are shown in green, blue, red, yellow, pink, pale blue, and orange, respectively), with fit of the homology model for TetM into the extracted cryo-EM density (gray mesh). (C) Relative binding position of domain IV of TetM (orange) compared with mRNA (tan), A-tRNA (green), and P-tRNA (blue). (D) Relative positions of domain V of TetM (orange) and EF-G (22) (blue) with their respective stalk base regions (H43/H44 and L11-NTD) colored pale blue and yellow, respectively. Arrows in D indicate the shift in the position of the stalk base between TetM and EF-G.
Molecular Model for the TetM•70S Complex.
The quality and resolution of the density map for the TetM•70S complex allowed an unambiguous fit of molecular models for the E. coli 30S and 50S subunits (17–19) (SI Appendix, Fig. S3). A large additional density within the subunit interface was attributed to TetM (Fig. 1A), as expected from the similarity in location of TetO (14) and EF-G (20–22) on the ribosome. In the absence of a crystal structure of any RPP, a homology model for TetM was built on the basis of the high sequence similarity between TetM and EF-G (10). Like EF-G, E. faecalis TetM is comprised of five domains, labeled I through V (Fig. 1B). Major differences include two truncations within domain I (G domain) of TetM, namely 31 residues lacking between β1G and αAG within the G′ subdomain and a second truncation of 18 residues between αE1 and β12 in domain I (SI Appendix, Fig. S1). An additional difference is the presence of a conserved C-terminal extension (CTE) in TetM (and all other RPPs), which has no counterpart in EF-G (Fig. 1B) and therefore could not be generated on the basis of homology modeling. Distinct secondary structure features, particularly the rod-like cylinders of α-helices, were observed clearly at higher thresholds (SI Appendix, Fig. S4), enabling the individual domains I to V of the TetM homology model (PDB 3J25) to be unambiguously fitted as rigid bodies to the extracted electron density for TetM (Fig. 1B).
Localization of TetM on the Ribosome.
The overall orientation of TetM on the ribosome is similar to that observed previously for TetO (14), although no direct comparison can be made because the TetO map was not deposited in a public database. TetM significantly overlaps with the anticodon stem-loop of the A-tRNA (Fig. 1C), as observed for EF-G bound to a posttranslocation state ribosome (22). However, the binding position of TetM does not overlap in position with the mRNA, and, unlike EF-G, TetM does not appear to encroach on the P-site (SI Appendix, Fig. S5). Moreover, whereas the overall orientation of TetM on the ribosome is similar to that of EF-G (22), EF-G is shifted in position relative to TetM, being located closer to the 30S subunit and further away from the stalk base of the 50S subunit (Fig. 1D and SI Appendix, Fig. S6). The overall orientation of TetM appears rather to structurally mimic more closely the position of EF-Tu•tRNA bound to the 70S ribosome (SI Appendix, Fig. S6). Based on the fit of the molecular model of TetM and the 70S ribosome to the cryo-EM density, a list of interactions was compiled (SI Appendix, Table S1 and Fig. S7). At 7.2-Å resolution, the interactions can only be approximated, and thus the closest residues between TetM and the ribosome at the site of density fusion are listed. In general, the contacts are similar to those reported previously for other translational GTPases, such as EF-G (21, 22), LepA (23), and, at the domain level, TetO (14), and are discussed in more detail in the SI Appendix.
CTE of TetM Interacts with the Ribosomal Decoding Site.
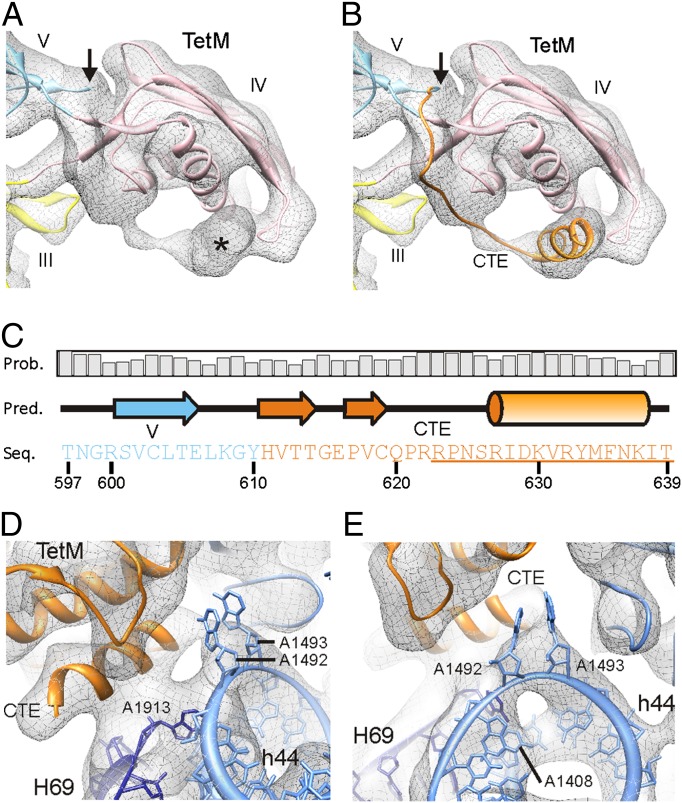
The homology model for TetM based on the EF-G template encompasses residues 1 to 610, leaving 29 C-terminal residues that are not included in the initial TetM model. However, fitting of the initial TetM homology model to the TetM•70S map revealed additional unassigned density that initiates at the C terminus of domain V, traverses domain IV, and eventually forms a large rod-like density adjacent to the tip of domain IV (Fig. 2A). This rod-like density remains present at high thresholds and exhibits the characteristics of an α-helix (SI Appendix, Fig. S4). Therefore, the CTE of TetM was modeled as a short 11-aa α-helix (residues 627–637) connected to domain V by a flexible unstructured loop (Fig. 2B), in accordance with secondary structure predictions (Fig. 2C).
Fig. 2.
Localization and interaction of the CTE of TetM. (A) Density (gray mesh) of domain IV of TetM fitted with homology model (colors are as in Fig. 1B). The arrow indicates the site where the homology with EF-G ends, yet additional density is observed extending from domain V toward domain IV (asterisk). (B) Same as A but with extended CTE (orange), modeled based on (C) PSIPRED secondary structure prediction, with sequence (Seq.), prediction (Pred.), and probability (Prob.) as indicated. Underlined residues were deleted to create TetM-ΔCTE. (D and E) Interaction of the CTE of TetM (orange) with H69 (blue) of the 23S rRNA and h44 of the 16S rRNA (pale blue), modeled with A1492 and A1493 flipped out of h44 [Protein Data Bank ID 2XQD (25)].
The CTE interacts with a loop region at the tip of domain IV of TetM, but also with H69 of the 23S rRNA (Fig. 2D). Additionally, density is seen connecting the distal end of helix-αA of the TetM-CTE with helix 44 of the 16S rRNA, in close proximity to A1492 and A1493 (Fig. 2E). As these bases are known to be flexible and can flip out of helix 44 during mRNA–tRNA decoding (24), the flipped-out conformation of these bases was also fitted (25) (Fig. 2 D and E). We believe this flipped-out conformation not only correlates with the fused electron density between h44 and the CTE (Fig. 2 D and E), but also explains the hole in the density of helix 44 caused by the absence of A1492 and A1493 stacking within the helix (Fig. 2E). Binding of TetO to the ribosome leads to an enhancement in the chemical reactivity of A1408 of the 16S rRNA to DMS modification (15). Consistently, the stacked conformation of A1493 would protect A1408 from modification (SI Appendix, Fig. S7 C and D), whereas the flipped-out conformation would expose A1408, allowing easier access for DMS modification (Fig. 2E). Collectively, these results suggest that binding of both TetM (and TetO) to the ribosome leads to the flipping out of A1492 and A1493—a conformation that is stabilized via interaction with the CTE of TetM. The enhancement of A1408 is also observed when TetO is bound with GTP rather than GDPNP (15), suggesting that the flipped-out conformation of A1492 and A1493 remains after the RPP has left the ribosome.
Domain IV of TetM Directly Encroaches Upon the Tetracycline Binding Site.
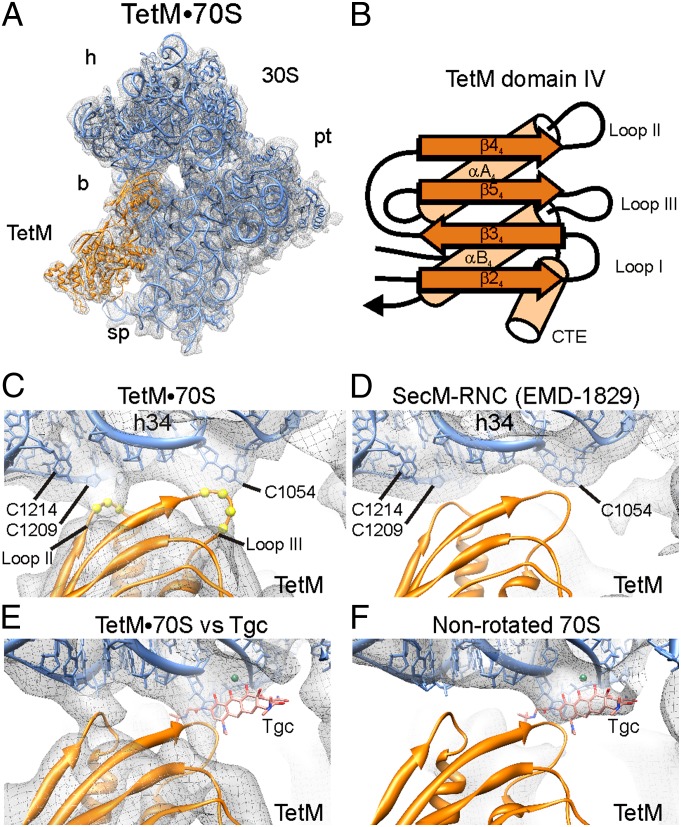
Domain IV of TetM interacts with the cleft between the head and body of the small subunit (Fig. 3A). Sequence alignments (SI Appendix, Fig. S1) and homology modeling suggest that domain IV of TetM is structurally analogous to EF-G, containing a four-stranded β-sheet and two α-helices, with an overall βββαβα topology (Fig. 3B). Three loops protrude from one end of domain IV of TetM, hereafter referred to as loops I, II, and III (Fig. 3B). The proline-rich loop I, located between β24 and β34, is bent significantly to allow interaction with the RPP-specific C-terminal helix-αACTE (SI Appendix, Fig. S8A), whereas, in contrast, loop I of EF-G is longer and adopts an extended conformation on the ribosome that establishes interaction with the P-tRNA (22) (SI Appendix, Fig. S8B). Loop II between β44 and αA4 interacts with the proximal end of h34 of the 16S rRNA, with residues Ser465–Leu466–Gly467 (465SLG467) coming into close proximity with the backbone of C1209 and the nucleobase of C1214 (Fig. 3C). Consistently, binding of TetO to the ribosome protects C1214 from DMS modification (15, 16).
Fig. 3.
Interaction of domain IV of TetM at the tetracycline binding site. (A) Overview of cryo-EM density (gray mesh) with the 30S subunit (blue) and TetM model (orange). (b, body; h, head; pl, platform; sp, spur.) (B) Schematic representation of secondary structure elements for TetM-domain IV, with α-helices, β-sheets, and loops I to III indicated as well as the CTE. (C) Cryo-EM density (gray mesh) reveals interaction of loop II (spheres for Cα of Ser465–Leu466–Gly467) with the backbone of h34 (C1208–C1209), and loop III (spheres for Cα of Tyr507–Ser508–Pro509–Val510) with C1054 of the 16S rRNA. (D) Same view as in C for SecM-stalled ribosome nascent chain complex (SecM-RNC) with an empty A-site (EMD-1829) (26). (E and F) Electron density map (gray mesh) of (E) TetM•70S showing lack of density for Tgc in comparison with (F) the nonrotated 70S map without TetM from sorting (SI Appendix, Fig. S2) that reveals density for Tgc.
Loop III of TetM linking β54 to helix αB4 is less well-resolved than loops I and II, yet additional electron density is clearly observable within the TetM•70S map (Fig. 3C), which, in contrast, is absent in other cryo-EM maps lacking A-site ligands, such as the SecM-stalled ribosome nascent chain complex (26) (Fig. 3D). This additional density in the TetM•70S map fuses directly with C1054 of the 16S rRNA, a component of the primary tetracycline binding site (2, 3), and is consistent with the protection of C1054 from DMS modification observed upon TetO binding to the ribosome (15). Although domain IV of TetO was separated by 6 Å from the tetracycline binding site in the previous cryo-EM reconstruction of the TetO•70S complex (14), we believe that this arises from the limited resolution of the TetO•70S complex: Filtering of the TetM•70S cryo-EM map to a similar resolution as the TetO•70S complex also leads to loss of density for loop III of domain IV of TetM (SI Appendix, Fig. S9).
Interplay of TetM and Tgc on the Ribosome.
Although the TetM•70S complex was formed in the presence of 10 μM Tgc, no density for the drug is observable in the TetM•70S map (Fig. 3E). As we observe the presence of a small population of 70S ribosome containing TetM (27%; SI Appendix, Fig. S2A), we conclude that TetM can bind to the Tgc•70S complex and recycle Tgc; however, it does so too inefficiently (compared with tetracycline) as to allow translation levels necessary for viability. Density for Tgc is, however, clearly present in the nonrotated (Fig. 3F) and rotated (SI Appendix, Fig. S10 A–C) 70S ribosomes where TetM is not bound. Because there is no structure reported for Tgc•70S, the observed binding position of Tgc was modeled into the primary tetracycline binding site (2, 3) based on similarity of the chemical structures of the two drugs (SI Appendix, Fig. S10 D–H). In contrast to tetracycline, which has multiple binding sites on the ribosome (2, 3), a careful examination of the TetM•70S map reveals no additional density for Tgc within any of the secondary tetracycline binding sites (SI Appendix, Fig. S10 I–M). Given that the TetM•70S complex was formed with 10 μM Tgc, which is ∼100 times its IC50 for in vitro translation (8), and that Tgc has a ∼10-fold higher affinity for the ribosome than tetracycline (7, 8, 27), the absence of secondary binding sites illustrates the increased specificity of Tgc compared with tetracycline.
Interplay of TetM and Tetracycline on the Ribosome.
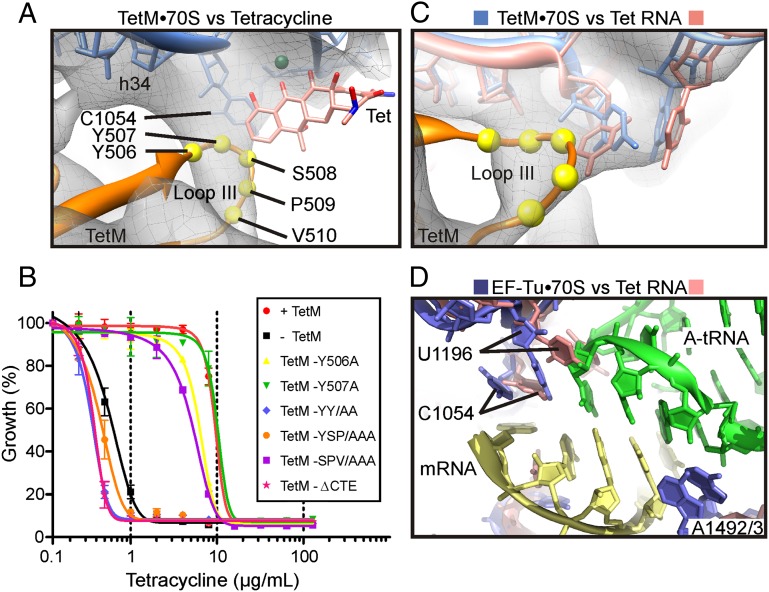
Compared with Tgc, tetracycline that lacks the C9-glycyl side chain (SI Appendix, Fig. S10 G and H) exhibits significantly less overlap with the TetM density and would still permit interaction between the side chains within loop III and C1054 in h34 of the 16S rRNA (Fig. 4A). In contrast, the attached C9-glycyl side chain of Tgc would prevent access of the residues of loop III of TetM (Fig. 3 E and F), leading us to suggest that this steric hindrance contributes, together with the increased affinity of Tgc (7, 8, 27), to explaining how Tgc overcomes TetM-mediated resistance whereas tetracycline cannot (8). Curiously, loop III of EF-G contains a highly conserved histidine residue (H583; SI Appendix, Fig. S8B), which has been shown to be critical for the translocation activity of EF-G (28). The equivalent residue to H583 of E. coli EF-G in TetM is tyrosine 507 (Y507; SI Appendix, Fig. S1), which, in the TetM•70S model, comes into close proximity of C1054 (Fig. 4A). Moreover, loop III contains a number of residues that are highly conserved in RPPs, in particular the 508SPVS511 motif that directly follows Y507 (SI Appendix, Fig. S1). Thus, to investigate whether residues located within loop III of TetM are important for conferring tetracycline resistance, alanine-scanning mutagenesis was used to generate TetM variants with single-residue mutations Y506A, Y507A, S508A, P509A, V510A, and S511A. In addition, we introduced a premature stop codon at residue 623, thus truncating the last 17 aa (Fig. 2C, underlined) and generating a TetM variant lacking the CTE α-helix (ΔCTE).
Fig. 4.
The role of loop III residues in TetM in tetracycline resistance. (A) Cryo-EM density of TetM•70S map (gray mesh) reveals relative proximity of loop III residues Tyr506–Tyr507–Ser508–Pro509–Val510 (spheres for Cα) to C1054 of the 16S rRNA and the primary tetracycline binding site (Tet) (2). (B) Growth curves of WT E. coli strain BL21 (−TetM, black) in the presence of increasing concentrations of tetracycline (0–128 μg/mL) compared with the WT strain harboring a plasmid encoding WT E. faecalis TetM (+TetM, red), C-terminally truncated TetM (TetM-ΔCTE, pink), TetM-YSP/AAA, (orange), TetM-SPV/AAA (purple), TetM-YY/AA (blue), TetM-Y506A (yellow), and TetM-Y507A (green). (C) Cryo-EM density of TetM•70S map (gray mesh) with comparison of relative positions of C1054 and U1196 of 16S rRNA from TetM•70S model [based on EF-Tu•70S structure (25), blue] and tetracycline-30S structure (pale red) (2). (D) Conformation of C1054 and U1196 of 16S rRNA from EF-Tu•70S structure (blue) (25), with A-tRNA (green) and mRNA (yellow) compared with tetracycline-30S structure (pale red) (2).
The growth of WT E. coli strain BL21 (−TetM) in the presence of increasing concentrations of tetracycline (0–128 μg/mL) was compared with the same strain bearing a plasmid overexpressing E. faecalis TetM (+TetM) or one of the TetM variants (Fig. 4B). In the absence of TetM protein, the WT E. coli strain (Fig. 4B, black squares) is sensitive to tetracycline with minimal inhibition concentration (MIC50) of ∼0.6 μg/mL, whereas, as expected, overexpression of E. faecalis TetM (Fig. 4B, red circles) raises the MIC50 by 14-fold to ∼10 μg/mL (Fig. 4B). Surprisingly, no single alanine substitution within loop III of TetM exhibited a significant effect on the ability of TetM to confer tetracycline resistance (shown for Y506A and Y507A in Fig. 4B), whereas, in contrast, deletion of the CTE of TetM extensively inhibited resistance activity, decreasing the MIC50 value (∼0.4 μg/mL; Fig. 4B, blue triangles) to that observed in the absence of TetM overexpression. Subsequent analysis of double and triple mutations within loop III of TetM led to the identification of the Y506A–Y507A (YY/AA) and Y507A–S508A–P509A (YSP/AAA) TetM variants as inactive, whereas the S508A–P509A–V510A (SPV/AAA) triple mutant retained activity (Fig. 4B).
Conclusion
The cryo-EM structure of the TetM•GDPNP•70S complex reveals that loop III of domain IV directly interacts with the tetracycline binding site in proximity to C1054, but does not appear to overlap dramatically with the binding position of tetracycline. This suggests that TetM dislodges tetracycline from its binding site on the ribosome by disrupting the reported stacking interaction between the aromatic ring D of tetracycline and the nucleobase of C1054 (2), in agreement with a previous proposal (15). In the model of the TetM•70S complex, the best fit for EM density in terms of C1054 and the neighboring U1196 was using the 30S subunit from the EF-Tu•70S structure (25) (Fig. 4C, blue). In contrast, the position of C1054 and U1196 when tetracycline is bound does not fit the density as well (Fig. 4C, pink), supporting the suggestion that TetM alters the conformation of these nucleotides to chase tetracycline from the ribosome (15). Additionally, the mutagenesis data (Fig. 4B) suggest that residues especially Y506 and Y507 within loop III of TetM are likely to be important for inducing this conformational change.
Resistance to tetracycline by TetM, however, not only requires that TetM chase the drug from its binding site, but that it also prevent immediate rebinding of tetracycline to the ribosome (9). This has been proposed to be brought about via TetM-induced conformational changes within the ribosome that prevent tetracycline rebinding, and yet promote binding of the ternary complex EF-Tu•GTP•aa-tRNA (9, 15, 16). Indeed, our findings provide structural support for this model, as the conformation of C1054 and U1196 in the TetM•70S structure appears to be most compatible with binding of EF-Tu•GTP•aa-tRNA (Fig. 4D) and would disfavor rebinding of tetracycline. Moreover, binding of TetM to the ribosome induces A1492 and A1493 to adopt a flipped-out conformation (Fig. 2 C and D), analogous to that observed during decoding of the mRNA by EF-Tu•GTP•aa-tRNA (25) (Fig. 4D). Footprinting experiments suggest that the flipped-out conformation of A1492 and A1493 persists upon dissociation of TetM from the ribosome (16), which promote binding of the ternary complex. Finally, we observe that TetM also induces a conformation in the stalk base, analogous to that observed in the EF-Tu•70S structure (25). Collectively, we believe these structural features imparted by TetM on the ribosome would contribute to the synergistic effect that TetM has been proposed to have on EF-Tu binding to the ribosome (16).
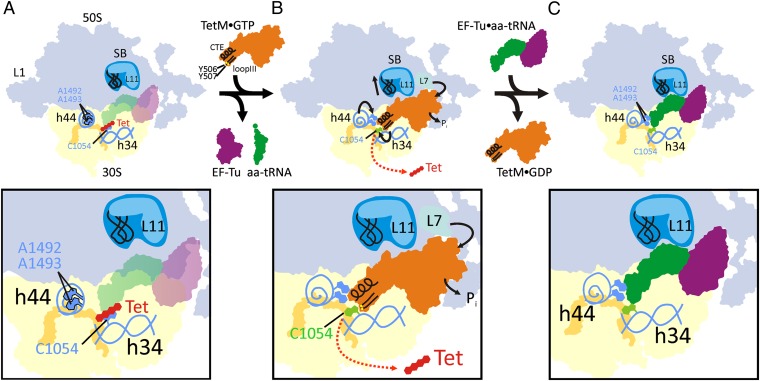
In conclusion, our structure suggests that TetM confers resistance to tetracycline using a direct mechanism (Fig. 5). (i) TetM employs residues including Y506 and Y507 within loop III of domain IV to directly interact and alter the conformation of nucleotide C1054 within h34 of the 16S rRNA that comprises part of the tetracycline binding site (Fig. 5A). (ii) The altered conformation of C1054 perturbs stacking interaction with tetracycline, leading to its dissociation from the ribosome and prevents rebinding (Fig. 5B). (iii) The altered conformation of C1054, together with the flipped-out conformation of A1492 and A1493 induced by interaction with the CTE of TetM, as well as the closed conformation of the stalk base, promote rapid binding of the ternary complex EF-Tu•GTP•aa-tRNA (Fig. 5C).
Fig. 5.
A model for the mechanism of TetM-mediated tetracycline resistance. (A) Binding of tetracycline (Tet, red) and interaction with C1054 (light blue) within h34 of the 30S subunit prevents delivery of the ternary complex consisting of EF-Tu•GTP (purple) and aa-tRNA (green) to the ribosomal A-site. (B) The tetracycline-bound ribosome is recognized and bound by TetM•GTP (orange; CTE and loop III are shown in black with the important residues Y506 and Y507 as yellow spheres). TetM binding induces nucleotides A1492 and A1493 to flip out of h44 and interact with the CTE of TetM. Loop III of TetM dislodges tetracycline from the ribosome (red arrow) and prevents rebinding by changing the conformation of C1054 (changed conformation in green). The stalk base (SB) adopts a position similar to when EF-Tu is bound, and, additionally L7 interacts with the G′ subdomain of TetM to catalyze Pi release and TetM dissociation. (C) Despite the dissociation of TetM•GDP, the conformational changes in the ribosome remain, preventing rebinding of tetracycline and promoting binding of ternary complex EF-Tu•GTP•aa-tRNA so translation can continue.
Materials and Methods
E. faecalis TetM and E. coli ribosomes were purified as described previously (29), and ribosome binding was verified by using pelleting assays as described for EF-G (30). Cryo-EM data collection on a Titan Krios transmission electron microscope (FEI Company) and processing using the SPIDER software package (31) was as described previously (32). The protein homology model of E. faecalis TetM was generated by using HHPred (33) and Modeler (34). The TetM homology model and ribosome crystal structures (18, 19, 35, 36) were fitted as rigid bodies to the cryo-EM density by using Coot (37) and Chimera (38). The QuikChange mutagenesis kit (Qiagen) was used to introduce site-specific mutations into the tetM gene according to the manufacturer’s instructions, and minimal inhibitory concentrations were determined as described previously (8). Detailed materials and methods can be found in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Thomas Becker and Agata Starosta for helpful comments. This work was supported by Deutsche Forschungsgemeinschaft FOR1805 (Grant WI3285/2-1 to D.N.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3J25). The cryo-EM map of the TetM•70S complex has been deposited in the EMDatabank (accession no. EMD-2183).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208037109/-/DCSupplemental.
References
- 1.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103(7):1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 3.Pioletti M, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20(8):1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra I. New developments in tetracycline antibiotics: Glycylcyclines and tetracycline efflux pump inhibitors. Drug Resist Updat. 2002;5(3-4):119–125. doi: 10.1016/s1368-7646(02)00051-1. [DOI] [PubMed] [Google Scholar]
- 7.Bergeron J, et al. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob Agents Chemother. 1996;40(9):2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman TH, et al. Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob Agents Chemother. 2012;56(5):2559–2564. doi: 10.1128/AAC.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell SR, Tracz DM, Nierhaus KH, Taylor DE. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother. 2003;47(12):3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor DE, Chau A. Tetracycline resistance mediated by ribosomal protection. Antimicrob Agents Chemother. 1996;40(1):1–5. doi: 10.1128/aac.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 12.Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol. 1996;178(11):3246–3251. doi: 10.1128/jb.178.11.3246-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trieber CA, Burkhardt N, Nierhaus KH, Taylor DE. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol Chem. 1998;379(7):847–855. doi: 10.1515/bchm.1998.379.7.847. [DOI] [PubMed] [Google Scholar]
- 14.Spahn CM, et al. Localization of the ribosomal protection protein Tet(O) on the ribosome and the mechanism of tetracycline resistance. Mol Cell. 2001;7(5):1037–1045. doi: 10.1016/s1097-2765(01)00238-6. [DOI] [PubMed] [Google Scholar]
- 15.Connell SR, et al. The tetracycline resistance protein Tet(o) perturbs the conformation of the ribosomal decoding centre. Mol Microbiol. 2002;45(6):1463–1472. doi: 10.1046/j.1365-2958.2002.03115.x. [DOI] [PubMed] [Google Scholar]
- 16.Connell SR, et al. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 2003;22(4):945–953. doi: 10.1093/emboj/cdg093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5-Å resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 18.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103(43):15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seidelt B, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326(5958):1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: The mechanism of translocation. Proc Natl Acad Sci USA. 1998;95(11):6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25(5):751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326(5953):694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15(9):910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 24.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28(5):259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 25.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330(6005):835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhushan S, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyltransferase center. PLoS Biol. 2011;19:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson MW, et al. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob Agents Chemother. 2006;50(6):2156–2166. doi: 10.1128/AAC.01499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000;300(4):951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 29.Mikolajka A, et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol. 2011;18(5):589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma MR, et al. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G) J Biol Chem. 2010;285(6):4006–4014. doi: 10.1074/jbc.M109.062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 32.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482(7386):501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 35.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: The structure of the L11-RNA complex. Cell. 1999;97(4):491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 36.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17(9):1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.