Abstract
We report an action of the protein kinase WNK3 on the neuronal mRNA splicing factor Fox-1. Fox-1 splices mRNAs encoding proteins important in synaptic transmission and membrane excitation. WNK3, implicated in the control of neuronal excitability through actions on ion transport, binds Fox-1 and inhibits its splicing activity in a kinase activity-dependent manner. Phosphorylation of Fox-1 by WNK3 does not change its RNA binding capacity; instead, WNK3 increases the cytoplasmic localization of Fox-1, thereby suppressing Fox-1–dependent splicing. These findings demonstrate a role of WNK3 in RNA processing. Considering the implication of WNK3 and Fox-1 in disorders of neuronal development such as autism, WNK3 may offer a target for treatment of Fox-1–induced disease.
Keywords: cytoplasmic retention, FMNL3
Mammalian Fox family members recognize a (U)GCAUG element through a conserved RNA recognition motif (RRM) required for their mRNA splicing activity (1, 2). Depending on the position of the (U)GCAUG stretch relative to the target exon, Fox proteins promote either exon inclusion or skipping (3). Fox-1 splices pre-mRNAs in a tissue-specific manner. In brain it has been implicated in autism and other neuronal developmental disorders (2–7). Comparative profiling of splicing in autism spectrum disorder (ASD) and normal brain demonstrated differential Fox-1–mediated splicing of target exons yielding proteins involved in synaptic function in autistic brain (8). Analysis of Fox-1−/− brains revealed splicing changes affecting synaptic transmission and membrane excitation (9).
With No Lysine (K) (WNK) protein kinases are large enzymes, notable for a uniquely positioned catalytic lysine, that regulate ion balance and blood pressure (10, 11). In contrast to WNK1, which is ubiquitous, WNK3 is highly expressed in brain, kidney, and some other tissues (12, 13). WNK3 modulates neuronal excitability through control of ion cotransporters (11). Duplication or deletion of the genomic location encompassing the WNK3 gene has been found in individuals with neurodevelopmental disorders including ASD and schizophrenia (14–16). Nonsynonymous mutations in WNK3 identified in ASD patients may be disease-relevant (17). WNK proteins are pleiotropic, suggesting versatile signal transducing roles of WNK3 in addition to its actions on ion transport. In the present study, we found that WNK3 is associated with RNA binding proteins, including Fox-1, and we show that WNK3 modulates Fox-mediated mRNA splicing by causing its subcellular relocalization.
Results
WNK3 Binds mRNA Processing Factors.
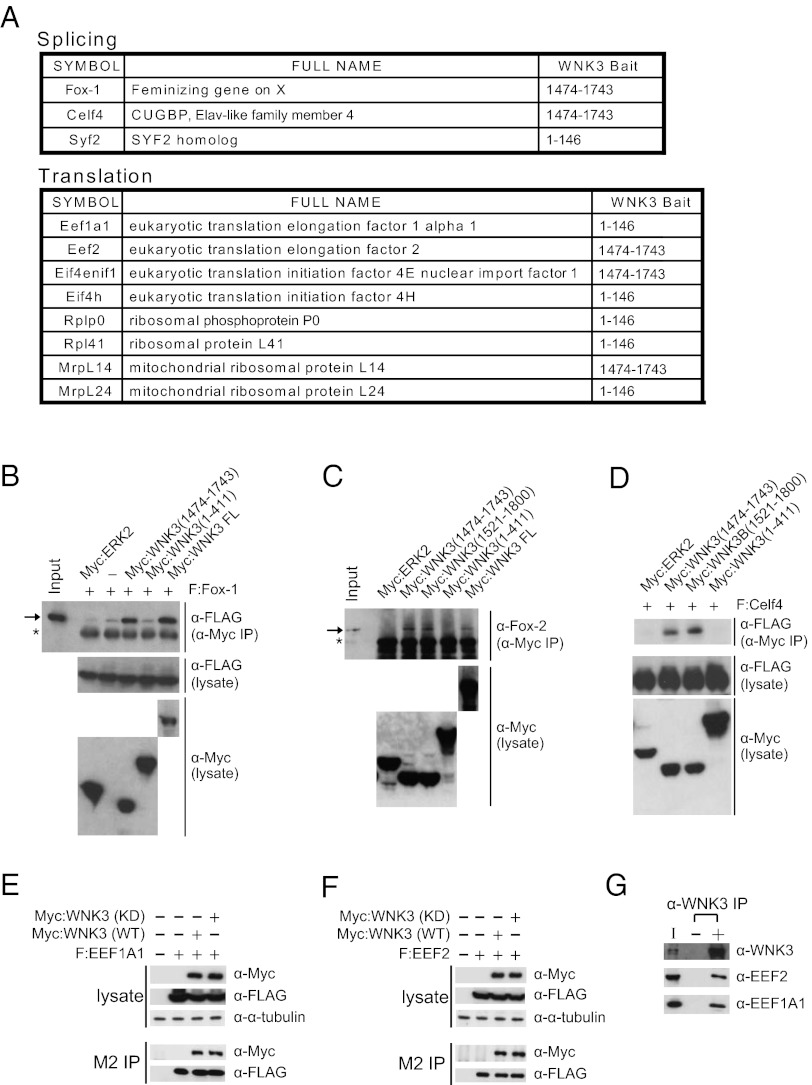
To explore unique biological roles of WNK3, yeast two-hybrid screens were performed with three WNK3 baits and a neonatal mouse brain cDNA library. More than 100 putative WNK3 partners were identified; several are involved in mRNA splicing and translation (Fig. 1A), suggesting a function of WNK3 in these processes. Several of these interactions were confirmed by coimmunoprecipitation (Fig. 1 B–G). Consistent with yeast two-hybrid results, the WNK3 C terminus bound three splicing factors, Fox-1, Fox-2, and Celf4 (Fig. 1 B–D). WNK3 also bound the general translation elongation factor, EEF1A1, located not only at the ribosome but also in RNA processing complexes (18), and another translation elongation factor, EEF2 (Fig. 1 E–G).
Fig. 1.
WNK3 binds to mRNA processing factors. (A) List of mRNA processing factors identified as WNK3-binding partners from yeast two-hybrid screens. (B) FLAG-tagged Fox-1 (F:Fox-1) was expressed in HEK293 cells with or without Myc-tagged proteins as indicated. The protein complex was coimmunoprecipitated with anti-Myc antibody and analyzed by immunoblotting. (C) Myc-tagged proteins were expressed in HEK293 cells and anti-Myc antibody was used for coimmunoprecipitation to detect bound endogenous Fox-2. In B and C, the arrow marks the position of F:Fox-1 or endogenous Fox-2, respectively. In both, the asterisk represents the position of IgG. (D) Transfection and coimmunoprecipitation were as described in B and proteins were immunoblotted. (E and F) Proteins were expressed in HEK293 cells as indicated and then immunoprecipitated with M2 (anti-FLAG antibody) resin followed by immunoblotting. Alpha-tubulin was used as a loading marker. (G) HEK293 cells were lysed and immunoprecipitated with anti-WNK3 antibody. Associated proteins were detected by immunoblotting with the indicated antibodies.
Mapping Interacting Regions on WNK3 and Fox-1.
To evaluate the significance of these interactions we focused on the convergence of tissue specificity and physiological and pathological involvement of WNK3 and Fox-1 and analyzed their interaction in more detail. We found that the coiled-coil domain in the C terminus of WNK3 was important to its interaction with Fox-1; from coimmunoprecipitation, its binding was equivalent to the full-length protein (Fig. 2A and Fig. S1A and D). Because that coiled coil is conserved among WNKs, it seemed likely that Fox-1 would bind other WNKs. Two-hybrid tests indicated no interaction of Fox-1 with the comparable region of WNK2 or WNK4 (Fig. S1B). Models of the surface-charge distribution of the coiled coils in WNK1 and WNK3 showed regions that could confer specificity (Fig. S1C). We tested binding of Fox-1 to WNK1 directly and found no coimmunoprecipitation, suggesting that Fox-1 binds WNK3 with selectivity (Fig. S1A).
Fig. 2.
Mapping the interacting regions on WNK3 and Fox-1. (A) Schematic representation of protein domains in human WNK3 (Left). Shaded regions indicate protein domains including Kinase, AI (auto inhibitory), and CC (coiled-coil) domains. Outside these regions, WNKs are poorly conserved. Numbers are the amino acid residues of full-length protein or the boundaries of small fragments linked to GST. GST pull-down assays with bacterially purified F:Fox-1 are presented (Right). (B) Schematic view of mouse Fox-1. RRM = RNA recognition motif. Fragments were generated for WNK3 interaction assays and binding results are summarized on the right. (C) Coimmunoprecipitation of Myc-WNK3 (1474–1743) with FLAG-tagged Fox-1 fragments from HEK293 cells.
WNK3 isoforms include one nonneuronal form (isoform 1, Fig. 2A) and three brain-specific forms (isoforms 2–4) produced by alternative splicing of exons 18b and 22; these exons are absent from the more broadly expressed WNK3 isoform (13, 19). Fragments representing all possible spliced products were generated as GST fusion proteins for in vitro pull-down assays. Fox-1 bound not only fragments containing the WNK3 two-hybrid bait, but also a brain-specific fragment, residues 991–1412, indicating the importance of exon 18b residues (Fig. 2A). This suggests that Fox-1 differentially binds neuronal relative to nonneuronal forms of WNK3.
Based on a series of overlapping Fox-1 fragments, Fox-1 residues 291–326 accounted for its ability to bind WNK3 (Fig. 2 B and C). Brain and muscle forms of Fox-1 are well conserved except in two regions (Fig. S2A), one at the N terminus and a second in the segment that binds WNK3. The RRM and the WNK3-binding region, residues 291–326, are most similar between Fox-1 and Fox-2; thus, this region of Fox-2 is also likely recognized by WNK3 (Fig. S2B).
WNK3 Regulates Splicing by Fox-1 and Fox-2.
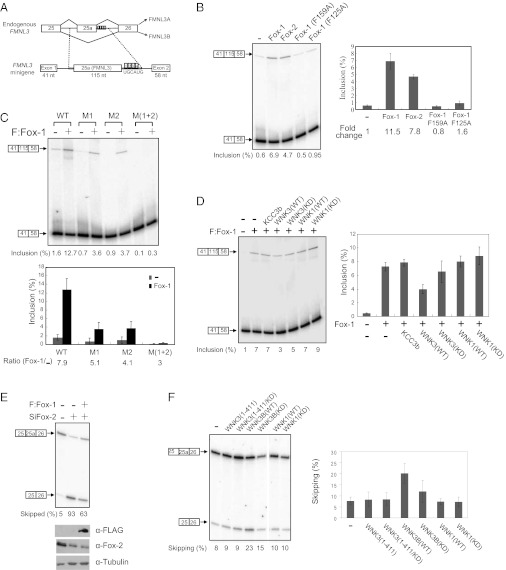
To test the functional relationship between WNK3 and Fox-1, we first established an assay for alternative splicing mediated by Fox-1. The Fox-1 target gene formin-like protein 3 (FMNL3) contains four UGCAUG elements downstream of exon 25a (20). Inclusion of the exon introduces an early stop codon that produces a shorter isoform, FMNL3A (Fig. 3A, Upper) (20, 21). An FMNL3 minigene splicing template was generated by inserting the FMNL3 sequence surrounding exon 25a in between beta-globin constitutive exons 1 and 2 (Fig. 3A). When exogenously expressed with the minigene, Fox-1 and Fox-2 increased the inclusion ratio of exon 25a up to 12-fold (Fig. 3B). F125 and F159 in the Fox-1 RRM domain are crucial for RNA sequence recognition (1, 22, 23). As expected, Fox-1(F125A) and (F159A) mutants failed to induce exon inclusion, supporting the conclusion that increased splicing was due to expression of wild-type Fox proteins (Fig. 3B). To verify that splicing depended on UGCAUG elements in the minigene, mutations were introduced in two of the four contiguous elements that the Fox-1 RRM recognizes (M1 and M2 of four), individually or in combination (Fig. 3A). The expression of single-element mutants (M1 or M2) reduced the inclusion ratio compared with the wild-type template, and activity was further reduced if two elements were mutated [M(1+2)], consistent with the importance of UGCAUG element recognition in Fox-mediated alternative splicing (Fig. 3C).
Fig. 3.
WNK3 regulates splicing activity of Fox-1 and Fox-2. (A) Genomic organization and alternative splicing patterns of an endogenous FMNL3 region including exons 25, 25a, and 26 and introns (Upper). FMNL3 minigene content (Lower). The region marked with dotted lines was inserted between β-globin exons 1 and 2 in the pcAT7-Glo1 plasmid. (B) FMNL3 minigene and FLAG-tagged Fox proteins were expressed in HEK293 cells as indicated and then subjected to the splicing assay described in Experimental Procedures. Alternative splicing products are depicted on the left of the gel. Error bars indicate SD (Right). (C) Minigene splicing assay with wild-type or UGCAUG element-mutated templates in the absence or presence of F:Fox-1 expression (mean ± SD). (D) Minigene splicing assay with F:Fox-1 and Myc-tagged proteins as indicated. Error bars show the SD in three experiments. (E) Endogenous FMNL3 splicing assay. Fox-2 down-regulation by siRNA against Fox-2 (SiFox-2) and F:Fox-1 expression were monitored by immunoblotting. (F) Effect of Myc-tagged WNK proteins on endogenous Fox-2–mediated FMNL3 splicing (mean ± SD).
The minigene was used to test effects of WNK3 on Fox splicing activity. As shown in Fig. 3D, Fox-1–mediated exon inclusion was significantly down-regulated by coexpression with wild-type WNK3, whereas expression of the K-Cl cotransporter (KCC3b), a probable target of WNK3, had no effect. WNK3 kinase activity was required for exon skipping, because inactive WNK3 showed only a marginal effect. Likewise, WNK1 did not alter Fox-induced exon inclusion. We conclude that WNK3 specifically inhibits Fox-1–induced exon inclusion in a kinase activity-dependent manner.
The role of endogenous WNK3 in a neuronal system was tested in stable cell lines expressing control shRNA or shRNA against WNK3 (Fig. S3A). Under conditions promoting neuronal differentiation, knockdown of WNK3 down-regulated the skipping ratio of the endogenous FMLN3 25a exon, further supporting the idea that WNK3 inhibits Fox-mediated exon inclusion (Fig. S3B).
To explore WNK3 regulation of splicing mediated by endogenous Fox proteins, the assay was validated in HEK293 cells, which express a detectable amount of Fox-2 endogenously. As shown in Fig. 3E, about 95% of endogenous FMNL3 exists in the exon 25a-included form in HEK293 cells. Upon Fox-2 knockdown by RNAi, the exon skipping ratio was increased to 93%, indicating the strong dependence on Fox-2 of FMNL3 exon 25a splicing. Up-regulated exon skipping by siFox-2 was partially rescued by Fox-1 overexpression, suggesting that Fox proteins have an overlapping target collection owing to their identical RRM domains. Expression of a WNK3 brain-specific isoform (isoform 4, Fig. 2A) in HEK293 cells dramatically increased the exon 25a skipping ratio, consistent with the minigene results (Fig. 3F). A catalytically inactive WNK3 mutant was less effective than wild-type WNK3.
Because results from the minigene system and analysis of endogenous Fox-2 function revealed the importance of WNK3 kinase activity in regulating splicing, we asked whether the WNK3 kinase domain was sufficient to regulate splicing. Neither wild-type nor inactive WNK3 kinase domains had any effect on Fox-mediated splicing, suggesting that not only its kinase activity but also the region of WNK3 that binds Fox proteins is necessary for proper splicing regulation (Fig. 3F). Confirming the WNK selectivity, WNK1 did not change the exon skipping ratio.
WNK3 Influences the Subcellular Localization of Fox-1 in a Kinase Activity-Dependent Manner.
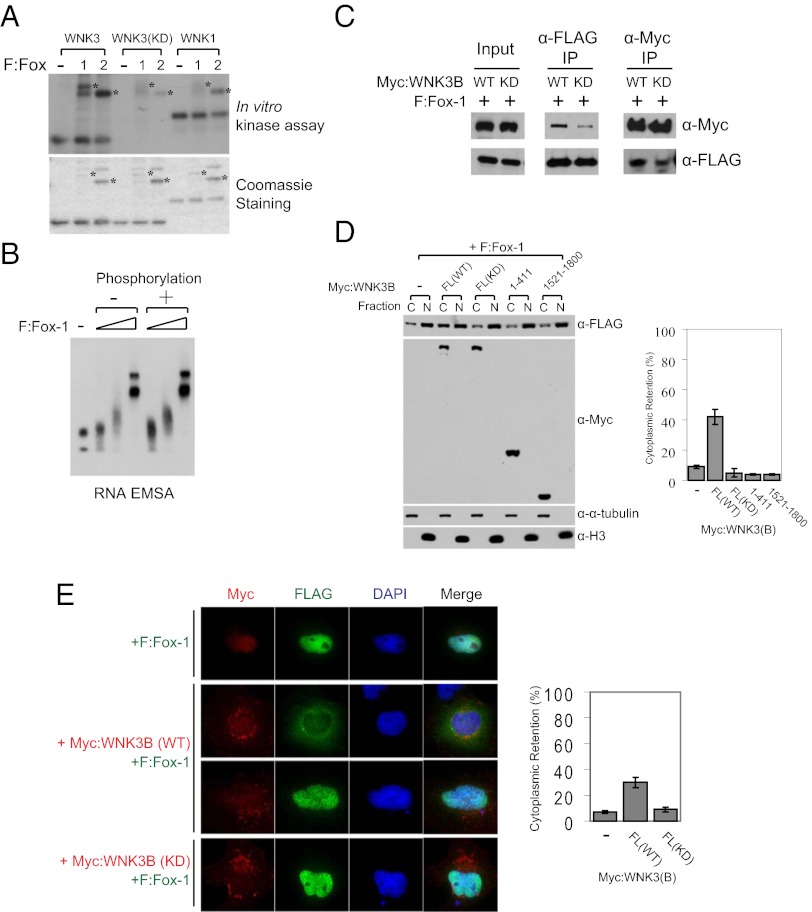
Because the kinase activity of WNK3 is important for the regulation of Fox-mediated splicing, we tested WNK3 kinase activity toward Fox proteins. The WNK3 kinase domain phosphorylated both Fox-1 and Fox-2, whereas neither kinase-dead WNK3 nor wild-type WNK1 did so (Fig. 4A and Fig. S4A). Full-length WNK3 phosphorylated Fox proteins in vitro to an extent comparable to the kinase domain, consistent with the suggestion that the differential effect of WNK3 (1–411) shown in Fig. 3F is due to lack of WNK3 binding (Fig. S4B). Considering the dependence on UGCAUG element binding of Fox proteins for their splicing activity and the requirement of WNK3 kinase activity for Fox negative regulation, we asked whether WNK3 phosphorylation of Fox-1 reduced its RNA recognition. Bacterially purified Fox-1 wild-type and RNA binding mutants (F125A, F159A) were tested in RNA electrophoretic mobility-shift assays (Fig. S5A). The assay was validated by confirming that only wild-type Fox-1 shifted in mobility as a function of increasing protein concentration (Fig. S5B). However, WNK3-mediated phosphorylation of Fox-1 did not inhibit its intrinsic RNA binding activity; phosphorylated Fox-1 recognized the UGCAUG template with a marginal increase in binding (Fig. 4B). Instead, wild-type WNK3 bound to Fox-1 more efficiently than the kinase-dead mutant, even though both proteins contain the C-terminal Fox-1 binding site (Figs. 1B and 4C). More phosphorylated Fox-1 was pulled down with wild-type WNK3 (Fig. S4C). This could be due to an effect of phosphorylation on the interaction or an effect of WNK3 kinase activity on the accessibility of its C-terminal region for interaction with Fox proteins.
Fig. 4.
WNK3 modulates the subcellular localization of Fox-1 in a kinase activity-dependent manner. (A) In vitro kinase assay with purified WNK kinase domains and F:Fox-1 and F:Fox-2 (Upper). Following these reactions, Fox proteins contained ∼2 mol phosphate/mol protein. Coomassie blue staining of proteins used for the assay (Lower). Asterisks indicate positions of Fox proteins. (B) RNA electrophoretic mobility-shift assay with the RNA template including a UGCAUG element and nonphosphorylated or WNK3-phosphorylated F:Fox-1. Triangles represent increasing amounts (0.1, 0.3, and 1 μg) of proteins used. (C) Myc-tagged WNK3 proteins and F:Fox-1 were expressed in HEK293T cells for coimmunoprecipitation. (D) F:Fox-1 was coexpressed with Myc-tagged WNK3 proteins in HEK293T cells and then subjected to subcellular fractionation as described in Experimental Procedures. C and N represent cytoplasmic and nuclear fractions, respectively. Alpha-tubulin and histone 3 (H3) were detected as markers of cytoplasmic and nuclear fractions, respectively. To calculate the percentage of cytoplasmic retention, the cytoplasmic F:Fox-1 intensity was divided by the total intensity of both fractions and multiplied by 100 (mean ± SD). (E) Immunostaining of F:Fox-1 (green), Myc:WNK3 (red), and DAPI (blue). HeLa cells were transfected for 60 h as indicated. Cytoplasmic and nuclear intensities were quantitated individually through ImageJ and calculated as described in D.
As previously suggested, Fox-1 carries a hydrophobic PY nuclear localization consensus sequence recognized by Karyopherin beta 2 (Kapβ2) (4, 24). The sequence lies near the C terminus, which diverges between cytoplasmic and nuclear Fox-1 isoforms (2). Indeed, nuclear Fox-1 directly bound Kapβ2 and the interaction was greatly reduced by addition of GTP-Ran, displaying the appropriate specificity (see Fig. S8A). Because the nuclear localization of Fox proteins is required for their splicing activity (2), we hypothesized that WNK3 changes the localization of Fox-1, thereby affecting its splicing output. Subcellular fractionation and immunofluorescence showed that expression of wild-type, but not kinase-dead, WNK3 increased the amount of Fox-1 in the cytoplasm (Fig. 4 D and E). The kinase domain (1–411) in the absence of C-terminal sequences was insufficient to promote cytoplasmic retention of Fox-1. This result parallels the splicing results (Fig. 3F) and suggests that inhibition of Fox-mediated splicing is due to the increased cytoplasmic and thus decreased nuclear localization of Fox-1 caused by wild-type WNK3. Consistent with the fractionation assay, exogenously expressed Fox-1 resided primarily in the nucleus (Fig. 4E). Expression of wild-type WNK3 resulted in a partial or complete relocalization of Fox-1 to the cytoplasm (Fig. S6). Kinase-dead mutant of WNK3 did not induce cytoplasmic Fox-1, confirming the requirement of its kinase activity. We conclude that WNK3 alters the subcellular localization of Fox-1, thereby regulating Fox-mediated splicing output.
Discussion
The identification of mRNA processing factors as WNK3 binding partners implies an unexpected role of WNK3 in mRNA splicing and translation. Besides its distinct involvement in each event, WNK3 may regulate the complex consisting of the factors implicated in these stages of mRNA processing; this seems more plausible as the coupling of different gene expression events has become evident. This notion has been supported by the recent report showing that Fox-2 and EEF1A, both WNK3 binding proteins, are found in a pre-mRNA 3′ processing complex (18). Perhaps WNK3 affects formation of the complex or coordinates a separate complex including other proteins found as binding partners.
Among the newly identified WNK3-binding partners, knowledge of Fox-1 and WNK3 has revealed several intersections. These factors both have brain-specific isoforms, both have strong mechanistic links to control of neuronal excitability, and both have been implicated in neurological diseases such as ASD and schizophrenia. Current ideas concerning a connection between WNK3 and ASD are limited to the finding of a few genetic alterations in the vicinity of the WNK3 gene in autistic brain and its modulation of neuronal excitation by controlling chloride transporters (11). However, Fox-1 has been shown to affect transcripts encoding many genes implicated in ASD and is responsible for generating proper alternative splicing variants required for normal neuronal excitability and synaptic transmission (8, 9). Our finding that WNK3 regulates Fox-mediated alternative splicing provides an additional tier of evidence that implicates WNK3 in neurological diseases through reprogramming splicing patterns.
Fox regulation by WNK3 is dependent on its kinase activity, which also affects the interaction between the two factors in the cellular environment. It is likely that both of the physical and kinetic relationships are required for proper regulation. There are at least three possibilities about how this physio-kinetic interaction might occur. The first and most expected is that phosphorylation may induce conformational changes in Fox-1 favorable for the interaction with WNK3. This possibility was investigated by mapping WNK3 phosphorylation sites on Fox-2. The sites, S174 and T337, conserved in Fox-1 and Fox-2, were tested through point mutations and kinase assays (Figs. S2B and S7A). However, the mutants functioned equivalently to and were localized like wild-type Fox-2 (Fig. S7 B–D). It is conceivable that additional Fox phosphorylation sites that were not detected could mediate this effect. WNK3 may also phosphorylate other factors that enhance the interaction between WNK3 and Fox-1. Finally, it is also possible that WNK3 catalytic activity induces conformational changes in itself that enable the interaction with Fox-1. Given the major function of WNK proteins as scaffolds, this possibility must be seriously considered.
We report here Kapβ2 as a putative importin for Fox-1. Consistent with its sequence information, Fox-1 is recognized by Kapβ2, suggesting its nuclear import step can be regulated to modulate its splicing output. Indeed, WNK3 induces cytoplasmic retention of Fox-1 and its kinase activity is crucial for this phenomenon. One hypothesis we considered, that WNK3 might inhibit the interaction between Kapβ2 and Fox-1, was not supported experimentally (Fig. S8 B and C). WNK3 may regulate steps of nuclear import other than the recognition by Kapβ2, or Kapβ2 may not be a major importin for Fox-1.
Related to the involvement of WNK3 in neuronal activity, most research has focused on the regulation of proteins at a posttranslational level. Our findings provide a unique mechanism for WNK3 regulation of factors important for neuronal excitability through control of the spliced forms that are expressed. This type of regulation, like control of membrane proteins, can have a potent impact on the pathogenesis of neurodevelopmental diseases.
Experimental Procedures
GST Pull-Down Assays.
GST-tagged proteins were prepared as described (25). One microgram of GST-tagged proteins was immobilized to 20 μL of a 50% (vol/vol) glutathione-Agarose slurry (Pierce) at 4 °C, overnight. After three washes, 100 ng of recombinant FLAG-Fox-1 (F:Fox-1) was incubated with immobilized GST-proteins at 4 °C for 1 h. Reactions were washed three times and analyzed by immunoblotting.
RNA Preparation and RT-PCR for Splicing Assays.
Total RNA was prepared from cells with RNA-Bee (Tel-Test). First-strand cDNA was synthesized from RNA with SuperScript II reverse transcriptase (Invitrogen) using gene-specific reverse primers. cDNA was amplified by PCR under conditions previously described (26). Primer sequences are as follows: for endogenous genes (FMNL3-ex25-F GAACACCGGCCTGTTTATGAG, FMNL3-ex26-R AAGTGCTTCTGCCTCCGAGAG) and for minigenes (DUP-ex1-F AAGGTGAACGTGGATGAAGTTGGT, DUP-ex3-R ACAGATCCCCAAAGGACTCAAAGAAC). To quantify the relative ratio of specific exon included or excluded products, 32P end-labeled primers were added to PCR reactions with few cycles. PCR products were resolved on denaturing gels and exposed using PhosphorImager screens. Radioactivity in bands was quantified using Multi Gauge software (Fuji Film). The percentage of exon inclusion was determined as ([cpm of exon included product/(cpm of exon included product and exon skipped product)] × 100%) and the skipping ratio was determined as ([cpm of exon skipped product/(cpm of exon included product and exon skipped product)] × 100%).
In Vitro Kinase Assays.
FLAG-tagged Fox proteins (F:Fox) were expressed and immunoprecipitated with M2-agarose beads from HEK293 cells for kinase assays. The beads were washed three times with detergent buffer [0.25 M Tris (pH 7.4), 1 M NaCl, 0.1% Triton X-100, 0.1% sodium deoxycholate] and once with 10 mM Hepes (pH 7.6), and then further incubated with indicated proteins in 50 μL of 20 mM Hepes (pH 7.6), 5 μM ATP (5 μCi of [γ-32P]ATP), 10 mM MgCl2, 10 mM glycerol phosphate at 30 °C for 1 h. Reactions were stopped by adding 15 μL of 5× sample buffer followed by boiling for 2 min. The reactions were analyzed by polyacrylamide gel electrophoresis in SDS and autoradiography.
RNA Electrophorectic Mobility-Shift Assay.
For RNA EMSA, phosphorylated and nonphosphorylated F:Fox-1 were prepared as follows. M2-immobilized F:Fox-1 was incubated in the kinase reaction solution as above with or without purified WNK3 kinase domain at 30 °C for 2 h. After washing, proteins were released from beads with elution buffer [20 mM Tris⋅Cl (pH 7.9), 20% glycerol, 0.2 mM EDTA, 100 mM KCl, 0.03% Nonidet P-40, and 0.2 mg/mL FLAG peptide] and the indicated amount of protein was used for the assay.
The RNA sequence template (AAACCAGCAUGAACGAUUUACCAAG) was selected as reported (1) and a biotinylated RNA was obtained from Dharmacon. Biotinylated RNA (20 nM) was incubated with binding buffer [10 mM Hepes (pH 7.3), 20 mM KCl, 1 mM MgCl2, 1 mM DTT, 2 μg tRNA] in the absence or presence of the proteins as indicated. After 30 min at room temperature, the reactions were analyzed in 6% polyacrylamide gels and then transferred to nylon membrane in 0.5× Tris⋅borate-EDTA at 400 mA for 30 min. The membrane was crosslinked with a hand-held UV lamp with a 254-nm bulb for 3 min. Biotinylated RNA on the membrane was visualized using Chemiluminescent Nucleic Acid Detection Module (Pierce) according to the manufacturer’s protocol.
Subcellular Fractionation.
Subcellular fractionation was performed as described (27) with some modifications. Briefly, transfected HEK293 cells were lysed with 0.3 mL of cytoplasmic lysis buffer and centrifuged at 3,500 × g, 4 °C, for 15 min. The pellet was washed with 1 mL of cytoplasmic lysis buffer without Nonidet P-40 and lysed with 0.3 mL of nuclear lysis buffer and centrifuged at 15,000 × g, 4 °C, for 20 min to separate a nucleoplasmic fraction. Insoluble material was further incubated with 0.3 mL of nuclease incubation buffer with 25 U/μL Benzonase at room temperature for 30 min. The reaction was centrifuged at 20,000 × g, 4 °C, for 20 min. The supernatant was collected and mixed with the nucleoplasmic fraction as the nuclear fraction and each fraction was analyzed by immunoblotting.
Other procedures are described in SI Text.
Supplementary Material
Acknowledgments
We thank Kristen Lynch and Lynch laboratory members Jason Jackson, Laura Motta-Mena, and Alan Tong for the gift of the pcAT-Glo1 construct, human genomic DNA, and their help setting up splicing assays. This work was supported by National Institutes of Health Grant GM53032 and Robert A. Welch Foundation Grant I1243.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215406109/-/DCSupplemental.
References
- 1.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhalla K, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 6.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 7.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu B, et al. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 11.McCormick JA, Ellison DH. The WNKs: Atypical protein kinases with pleiotropic actions. Physiol Rev. 2011;91:177–219. doi: 10.1152/physrev.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veríssimo F, Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 13.Holden S, Cox J, Raymond FL. Cloning, genomic organization, alternative splicing and expression analysis of the human gene WNK3 (PRKWNK3) Gene. 2004;335:109–119. doi: 10.1016/j.gene.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Chung BH, et al. Phenotypic spectrum associated with duplication of Xp11.22-p11.23 includes Autism Spectrum Disorder. Eur J Med Genet. 2011;54:e516–e520. doi: 10.1016/j.ejmg.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Edens AC, Lyons MJ, Duron RM, Dupont BR, Holden KR. Autism in two females with duplications involving Xp11.22-p11.23. Dev Med Child Neurol. 2011;53:463–466. doi: 10.1111/j.1469-8749.2010.03909.x. [DOI] [PubMed] [Google Scholar]
- 16.Qiao Y, et al. Autism-associated familial microdeletion of Xp11.22. Clin Genet. 2008;74:134–144. doi: 10.1111/j.1399-0004.2008.01028.x. [DOI] [PubMed] [Google Scholar]
- 17.Piton A, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz-Rangel S, et al. Similar effects of all WNK3 variants on SLC12 cotransporters. Am J Physiol Cell Physiol. 2011;301:C601–C608. doi: 10.1152/ajpcell.00070.2011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh M, Katoh M. Identification and characterization of human FMNL1, FMNL2 and FMNL3 genes in silico. Int J Oncol. 2003;22:1161–1168. [PubMed] [Google Scholar]
- 22.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 24.Lee BJ, et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu SY, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch KW, Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anindya R, Aygün O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.