Abstract
Malarial dihydrofolate reductase (DHFR) is the target of antifolate antimalarial drugs such as pyrimethamine and cycloguanil, the clinical efficacy of which have been compromised by resistance arising through mutations at various sites on the enzyme. Here, we describe the use of cocrystal structures with inhibitors and substrates, along with efficacy and pharmacokinetic profiling for the design, characterization, and preclinical development of a selective, highly efficacious, and orally available antimalarial drug candidate that potently inhibits both wild-type and clinically relevant mutated forms of Plasmodium falciparum (Pf) DHFR. Important structural characteristics of P218 include pyrimidine side-chain flexibility and a carboxylate group that makes charge-mediated hydrogen bonds with conserved Arg122 (PfDHFR-TS amino acid numbering). An analogous interaction of P218 with human DHFR is disfavored because of three species-dependent amino acid substitutions in the vicinity of the conserved Arg. Thus, P218 binds to the active site of PfDHFR in a substantially different fashion from the human enzyme, which is the basis for its high selectivity. Unlike pyrimethamine, P218 binds both wild-type and mutant PfDHFR in a slow-on/slow-off tight-binding mode, which prolongs the target residence time. P218, when bound to PfDHFR-TS, resides almost entirely within the envelope mapped out by the dihydrofolate substrate, which may make it less susceptible to resistance mutations. The high in vivo efficacy in a SCID mouse model of P. falciparum malaria, good oral bioavailability, favorable enzyme selectivity, and good safety characteristics of P218 make it a potential candidate for further development.
Keywords: drug resistance; drug target; structure-informed drug discovery; slow-binding inhibitors; 2,4-diaminopyrimidines
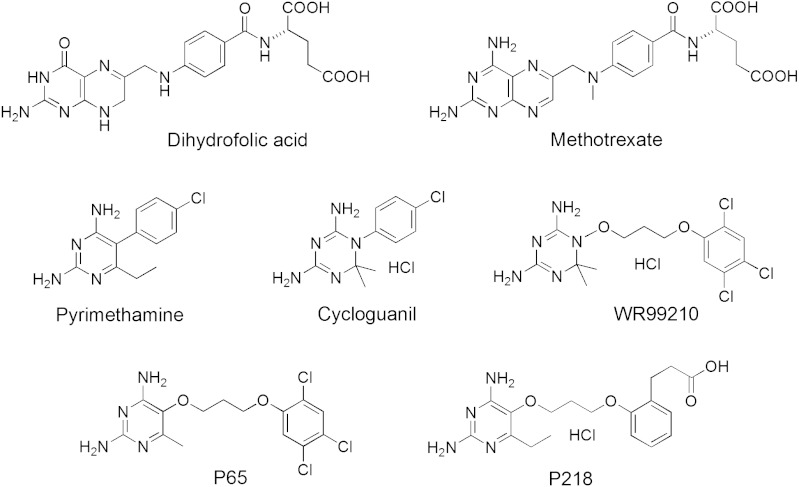
Malaria continues to be a major infectious disease, with a global estimate of 200–500 million cases per year, and annual mortality of some 1.2 million (1). The usefulness of such antimalarials as chloroquine and pyrimethamine (PYR) is now vastly decreased owing to the emergence of Plasmodium strains resistant to these drugs. Although there are now effective drug combinations based on artemisinin (ACTs), signs of emerging resistance have already been reported in Southeast Asia (2). Some apparent successes early in the process of identifying new, effective antimalarials for human use have been achieved with novel small synthetic analogs of the artemisinin family of drugs (3, 4), with whole-organism, high-throughput screens of chemical databases (5, 6), and with chemical genomic profiling assisted by knowledge of the Plasmodium falciparum (Pf) genome (7; for recent reviews, see refs. 8–10). Currently, nevertheless, there still exist very few well-defined clinically validated targets against which antimalarial drug discovery efforts can be directed. The best known such target is P. falciparum dihydrofolate reductase (DHFR), which is inhibited by the antimalarials PYR and cycloguanil (CG) (Fig. 1).
Fig. 1.
Chemical structures of dihydrofolate and DHFR inhibitors. P65 and P218 are new inhibitors introduced in this study.
DHFR and thymidylate synthase (TS) catalyze successive steps in recycling folates for use in synthesis of thymidylate, purines, and methionine. DHFR inhibitors have a long history as anticancer agents and as anti-infective drugs against bacterial and protozoal pathogens. In Plasmodia, DHFR and TS coexist as a single-chain bifunctional enzyme, in contrast to prokaryotes and higher eukaryotes where the two proteins are distinct monofunctional enzymes. In bifunctional DHFR-TS, individual DHFR and TS domains have polypeptide folds closely related structurally to those of their respective monofunctional counterparts (11). Crystal structures for wild-type bifunctional DHFR-TS from P. falciparum and for the highly PYR-resistant quadruple mutant enzyme (QM) have been reported by our group (12). We have also investigated inhibitor binding to both wild-type and mutant forms of PfDHFR-TS, revealing a probable structural basis for reduced binding of PYR and CG to mutation-compromised PfDHFR-TS targets (13). Clinical isolates of the parasite resistant to PYR carry various combinations of mainly four point mutations—at codons 51, 59, 108, and 164 (N51I, C59R, S108N, and I164L)—in the DHFR domain portion of the DHFR-TS gene (14). S108N, recognized as the first mutation, led to reduced binding affinities of inhibitors like PYR, with a rigid p-chlorophenyl substituent at the 5-position (8, 9). The binding affinities are reduced further by additional mutations that result in conformational and other changes preferentially affecting binding of the inhibitors with relatively less effect on the substrates (12, 13). The highly resistant quadruple mutant carries the above mutations at all four codons.
We describe the results of an iterative process in which cocrystal structures of PfDHFRs (with their inhibitors and substrate) along with efficacy and pharmacokinetic profiling have been used in the design, characterization, and preclinical development of a selective, highly efficacious, and orally available antimalarial drug candidate that potently inhibits both wild-type PfDHFR and mutated forms of the enzyme known to have arisen in response to widespread clinical use of PYR. As other validated antimalarial drug targets emerge based on current and future research, and as first-generation drugs against these targets make their way into clinical use, mutation-mediated drug resistance will likely appear, necessitating the search for new drugs capable of overriding the resistance. We foresee the type of integrated, structure-informed discovery approach described here as having the potential to identify new drug candidates useful against other validated but resistance-compromised drug targets.
Results and Discussion
Compound Design: Choice of 2,4-Diaminopyrimidine as the Chemical Scaffold.
The observation that 2,4-diaminoheterocycles antagonize the activity of folic acid (15) led to the development of methotrexate (MTX) (Fig. 1), a powerful antitumor drug still in clinical use today (16). The chemically simple substitution of a 4-oxo group in folate by a 4-amino group in MTX increases binding to, and inhibition of, human DHFR (hDHFR) by at least five orders of magnitude. The molecular basis of this result was deduced from the cocrystal structure of Escherichia coli DHFR containing bound MTX (17). Various diamino heterocycles, including pteridines, quinazolines, pyridopyrimidines, pyrimidines, and triazines, have served as scaffolds for good DHFR inhibitors (15, 16). Certain of these heterocycles have proven superior to others in terms of their usefulness in achieving species-selective DHFR inhibition, which is a required attribute of any nontoxic antimalarial drug targeting this enzyme. Both PYR, a 2,4-diaminopyrimidine, and CG, a 4,6-diamino-1,2-dihydro-1,3,5-triazine (Fig. 1), bind in the expected manner to quadruple mutant PfDHFR, making key polar interactions with the enzyme similar to those for MTX (12).
A 4,6-diamino-1,2-dihydro-1,3,5-triazine WR99210 (Fig. 1) was advanced as an antifolate-based antimalarial with high activity against both wild-type and PYR resistance–associated PfDHFR (18). Yuvaniyama et al. (12) showed crystallographically that WR99210 could avoid steric clash with the side chain of Asn-108 in PYR-resistant quadruple mutant PfDHFR because of its flexible (2,4,5-trichlorophenoxy)propoxy side chain. Further development of WR99210 as an antimalarial was terminated because of its low bioavailability and severe gastrointestinal toxicity although some additional work continues on PS-15, a biguanide precursor of WR99210, which is converted in vivo to the active drug (19, 20).
Recognizing the importance of conformational flexibility in avoiding steric clashes with known mutations in PYR-resistant DHFR (and also to better exploit possibilities for optimizing inhibitor species selectivity), we decided to focus our design efforts on diamino-heterocycle scaffolds with one ring rather than two (i.e., triazines and pyrimidines). In order to choose between the two, we needed to understand why WR99210 has very low bioavailability. Although 4,6-diamino-1,2-dihydro-1,3,5-triazines and 2,4-diaminopyrimidines both satisfy the canonical geometrical and chemical hydrogen-bonding requirements for binding deep in the DHFR active site, these inhibitor scaffolds differ significantly in their basicity. Triazines such as WR99210 and CG are much more basic (pKa 10–11) than pyrimidines like PYR (pKa 6–7) (21). Thus, at the slightly acidic pH of the gastrointestinal track, the triazines are fully protonated whereas 2,4-diaminopyrimidines exist as a mixture of protonated and unprotonated forms. In order to test the hypothesis that protonation of the free molecule could affect bioavailability, we synthesized P65 (Fig. 1), which is the 2,4-diaminopyrimidine analog of WR99210, and determined for both compounds the enzyme inhibition constants, in vitro cell-based antimalarial activities, and in vivo antimalarial efficacies in a mouse malaria model (Table 1, Table S1 and Fig. S1). The oral bioavailability of P65 in rats was found to be 83%, compared to less than 1% for WR99210 (Table 1). Studies in Caco-2 cell monolayers confirmed that the poor oral bioavailability of WR99210 was likely the result of poor intestinal permeability, whereas P65 had high permeability in the same model system. Taken together, these results suggest that WR99210 is poorly absorbed, consistent with the deleterious effect that protonation could have on absorption by passive diffusion. The less basic 2,4-diaminopyrimidine scaffold is better suited for oral bioavailability, and so this structural core was chosen for further chemical elaboration.
Table 1.
Characteristics of flexible triazine (WR99210) and flexible pyrimidine (P65 and P218) inhibitors
| Pyrimethamine | WR99210 * | P65 * | P218 | |
| Biological activity | ||||
| Ki quadruple mutant P. falciparum DHFR (nM) ± SD | 385 ± 163 | 1.9 ± 0.8 | 5.59 ± 0.1 | 0.54 ± 0.12 |
| IC50 wild-type P. falciparum (TM4) (nM) ± SD | 58 ± 33 | 0.57 ± 0.1 | 229 ± 68 | 4.6 ± 1.9 |
| IC50 quadruple mutant P. falciparum (V1/S) (nM) ± SD | > 100,000 | 18 ± 12 | 3,490 ± 1,610 | 56 ± 20 |
| Oral ED90 P. chabaudi (mg/kg) † | 1.1 | 74.22 | 1.53 | 0.75 |
| Permeability and oral bioavailability properties | ||||
| Measured pKa ± SD | 6.8 ‡ | 10.4 ± 0.3 | 6.6 ± 0.2 | 4.9 ± 0.003 (carboxylate) 7.3 ± 0.003 (amine) |
| Caco-2 Papp (cm/s) | not available | < 0.12 × 10-6 § | 86 × 10-6 | 21 × 10-6 |
| Oral bioavailability in rats (%) ± SD | ∼100 | < 1 | 82.6 ± 21 | 46.3 ± 11.4 |
IC50, inhibitory concentration that reduces parasite growth by 50% in vitro; Ki, binding constant.
*WR99210 prepared as hydrochloride; P65 prepared as free base.
†P. chabaudi AS, PYR-sensitive.
‡Calculated value using ACD/Labs software (Advanced Chemistry Development, Toronto), Version 9.0.
§Acceptor concentrations were below the lower limit of quantitation (LLQ) of the assay; value represents the maximum value estimated using the LLQ.
Table 1 shows that P65 and WR99210 are both low nanomolar inhibitors of wild-type and quadruple mutant PfDHFR. In order to study interaction of P65 with quadruple mutant PfDHFR-TS, we solved the cocrystal structure at a resolution of 2.38 Å (Table S2) and compared it with the previously determined structure for WR99210 bound to the same enzyme (12). The diamino-heterocycles of the two inhibitors bind identically at the active site of the quadruple mutant enzyme (Fig. S2A). The better in vitro efficacy of WR99210 compared to P65 may be because of new permeability pathways of organic cations (22). However, in spite of the fact that P65 is 200-fold less potent than WR99210 in an in vitro cell-based infectivity assay with a Plasmodium falciparum strain (V1/S) harboring the quadruple mutant PfDHFR, it has a far greater in vivo activity than WR99210 by the oral route (Table 1) because of its superior oral bioavailability.
The Side Chain: Opportunities for Tighter Binding and DHFR Selectivity.
The goals of the design process were to build unique chemical functionality into an analog of P65 that would increase affinity for wild-type and quadruple mutant PfDHFR while improving selectivity for PfDHFR over its human counterpart in order to reduce the potential for human DHFR–mediated host toxicity. The 2,4-diaminopyrimidine anchor of P65 provides the minimum functionality necessary to achieve good binding deep in the DHFR active site. When combined with a flexible five-atom linker the conformational rigidity that would occur with the 2,4-diaminopteridine, quinazoline, or pyridopyrimidine scaffolds is avoided. Larger conformationally constrained ligands cannot adapt as well to changes in the geometry of the binding site and, as a consequence, are highly susceptible to drug-resistance mutations, often by steric exclusion (23, 24). Increased flexibility alone, however, can result in lower specificity and decreased affinity because of unfavorable entropy changes on binding, effects that can be compensated for by favorable enthalpic interactions (24).
At the opposite end of the DHFR active site from the substrate’s bound pteridine ring is a conserved Arg (Arg122 in PfDHFR, Arg70 in human DHFR), which forms charge-mediated hydrogen bonds with the α-carboxylate of the dihydrofolate substrate. The aliphatic portion of the Arg side chain is tightly packed by protein residues that restrict side-chain mobility, providing a rigid docking site for the substrate’s α-carboxylate group. We reasoned that a molecule anchored at one end by 2,4-diaminopyrimidine deep in the DHFR active site and at the other end by strong charge-mediated hydrogen bonds with Arg122 would be a potent inhibitor of PfDHFR. Joining these chemical groups with a flexible linker could also reduce the possibility of resistance resulting from amino acid mutations. DHFR inhibitors designed to interact with the conserved Arg have previously been reported by other groups (25, 26). Although Arg122 is identically conserved in all chromosomally encoded DHFRs, comparison of structures for Pf and human DHFRs shows species-dependent amino acid differences for several residues in the vicinity of the conserved Arg. Inhibitor designs that take advantage of these structural differences to control access to the Arg docking site provide a mechanism for enhancing species selectivity.
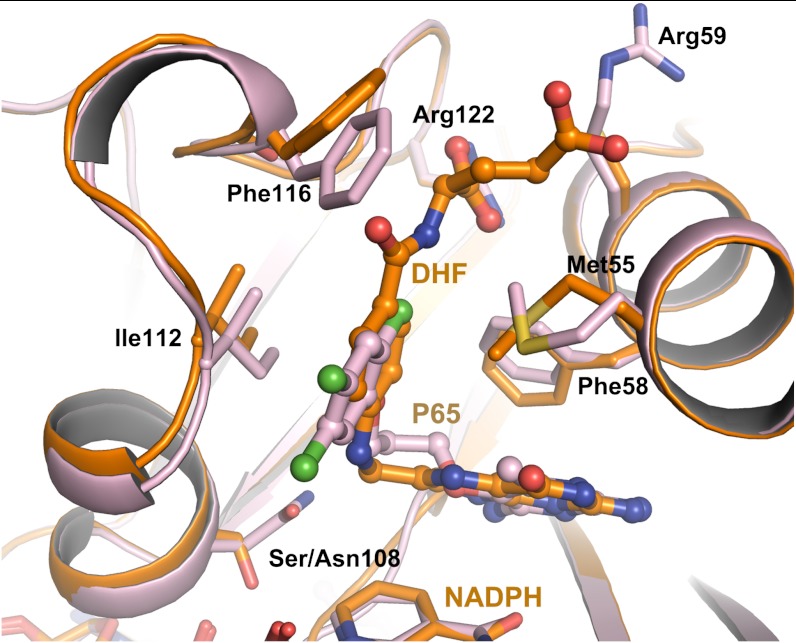
There are three regions of the DHFR active site where nonhomologous single amino acid substitutions in Pf and human DHFR in the vicinity of Arg122/Arg70 could affect how an inhibitor carboxyl group might bind differently to these two enzymes. Met55, Cys/Arg59, and Phe116 in PfDHFR are replaced at structurally equivalent positions in human DHFR by Phe31, Gln35, and Asn64. In addition, proximate to the trichlorophenyl group of P65 bound to quadruple mutant PfDHFR is a stretch of five amino acids (112–116) immediately C-terminal to helix αC that forms a tight turn at Ile112 followed by a single turn of α-helix αD (see Fig. 2). Comparison of structures for dihydrofolate bound to wild-type PfDHFR determined here (Fig. 2, Table S2, and Results and Discussion) and folate bound to human DHFR [Protein Data Bank (PDB) ID code 2W3M] show that the orientation of αD in PfDHFR is tilted slightly inward toward the invariant Arg compared to its human counterpart, resulting in a movement of about 0.5 Å toward Arg122 at Phe116 and away from Arg122 by a similar amount at Ile112. The structure of P65 bound to quadruple mutant PfDHFR shows a further tilt of αD that places Phe116 almost 1.5 Å closer to conserved Arg122 than in the corresponding residue Asn64 in human DHFR.
Fig. 2.
Superposition of active-site regions for P65 bound to quadruple mutant PfDHFR-TS (light purple), and for dihydrofolate bound to wild-type PfDHFR-TS (orange). Note that the 2-chloro atom (green) of P65 points toward the side chain of Arg122.
Examination of the X-ray cocrystal structure of P65 with quadruple mutant PfDHFR suggested that substitution at the 2′-chloro or the 3′-hydrogen of the trichlorophenyl group by an alkyl or alkyloxy group carrying a terminal carboxylate could favorably position the carboxylate group for interaction with Arg122 (Fig. 2). P218 (Fig. 1) is such a molecule with the trichlorophenyl group of P65 replaced by a 2′-carboxyethylphenyl group. All together, a total of 14 molecules were designed, synthesized, and characterized in this series. P218 was identified as the best compound out of this larger series that will be described elsewhere.
P218 Binds Differently to Plasmodium falciparum and Human DHFRs.
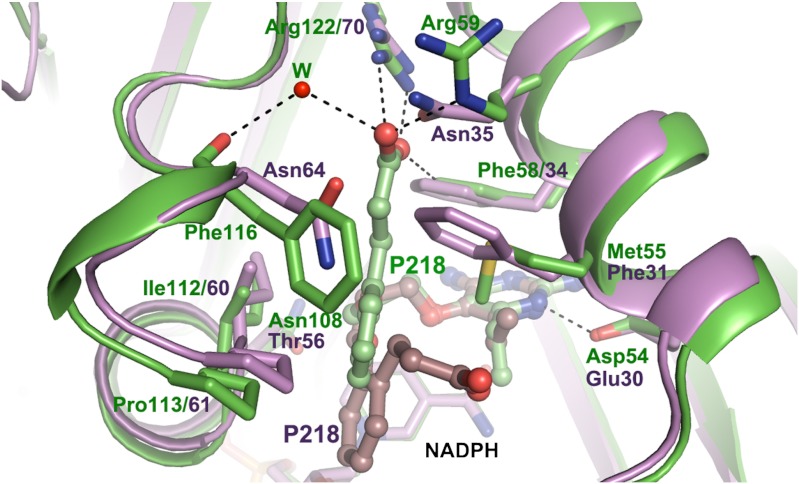
In order to study binding modes of P218 to PfDHFR-TS and to human DHFR, we solved the respective cocrystal structures at resolutions of 2.40 Å and 2.05 Å (Table S2). The carboxylate of P218 bound to quadruple mutant PfDHFR makes two charge-mediated hydrogen bonds with Arg122, but in human DHFR the P218 side chain has no direct interaction with Arg70 (Fig. 3). Rather, it folds back toward the inhibitor’s pyrimidine ring displacing the phenyl side chain of Phe31, which rotates into a new position at the back of the substrate binding site (SI Text, Fig. S2D). These remarkable differences in P218 binding to Pf and human DHFR can be understood in terms of how species-specific binding determinants mentioned above stabilize carboxyethyl interactions with Arg122 in PfDHFR but destabilize interaction with structurally equivalent Arg70 in the human enzyme.
Fig. 3.
Binding of P218 in quadruple mutant PfDHFR-TS (protein, green; inhibitor, light green) and human DHFR (protein, purple; inhibitor, brown), highlighting the different side-chain conformations for bound P218. Dashed lines indicate hydrogen bonds. W marks the ordered water molecule position in the PfDHFR-TS structure linking the P218 carboxylate and the backbone carbonyl of Phe116. Note that the P218 carboxylate interacts with the guanidinium of conserved Arg122 in the complex with the PfDHFR-TS but not in human DHFR. The different binding modes are a result of nonhomologous amino acid substitutions at positions 55, 59, and 116 (Pf numbering).
P218 Is a Slow-Off Tight-Binding Inhibitor of Both Wild-Type and Quadruple Mutant DHFR.
P218 binds to PfDHFR with biphasic kinetics, suggesting a two-step mechanism (27, 28) (Fig. S3):
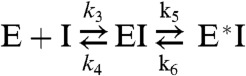 |
The first step is rapid equilibrium binding of inhibitor to enzyme forming the encounter complex (EI), followed by a second step involving slow enzyme isomerization to form a higher-affinity complex (E*I). Kinetic constants for P218 and PYR are shown in Table 2 along with the respective dissociative half-lives t1/2 from the high-affinity final DHFR conformational state. P218 is a subnanomolar inhibitor of both wild-type and quadruple mutant DHFR with binding to the high-affinity DHFR conformational state up to 20 times stronger than that for the initial encounter complex. Although the slow-on/slow-off binding is also shown for PYR with wild-type PfDHFR, it is absent in the binding of PYR with the quadruple mutant, which is weakly bound compared with P218. Both P218 and PYR bind poorly to human DHFR, and show no subsequent slow-on/slow-off phase.
Table 2.
Kinetic characteristics for binding of P218 and PYR to WT and QM PfDHFR and hDHFR
| Inhibitor/enzyme | k5 (s-1) | k6 (s-1) | t1/2 (min) * | Residence time (min) * |
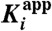 (nM) (nM) |
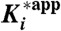 (nM) (nM) |
| PYR/WT | 0.025 ± 0.0004 | 0.0024 ± 0.0006 | 5 ± 1 | 7 ± 2 | 8.20 ± 1.65 | 2.08 ± 0.13 |
| P218/WT | 0.048 ± 0.0036 | 0.0017 ± 0.0002 | 7 ± 1 | 10 ± 1 | 11.61 ± 0.62 | 0.51 ± 0.032 |
| PYR/QM † | - | - | - | - | 2996 ± 1048 | - |
| P218/QM | 0.031 ± 0.0031 | 0.00125 ± 0.00056 | 11 ± 3 | 16 ± 4 | 7.37 ± 1.30 | 0.53 ± 0.13 |
| PYR/hDHFR † | - | - | - | - | 5,406 | - |
| P218/hDHFR † | - | - | - | - | 2,841 ± 319 | - |
Kinetic parameters are defined as in SI Materials and Methods.
*Residence time = 1/koff, and t1/2 = 0.693/koff, where koff is k4k6/(k4 + k5 + k6)). In the two-step mechanism, k4≫k5 and k6, then koff is approximately equal to k6.
†No slow-binding phase observed.
Compounds such as P218 that display slow-off rates have been shown in certain cases to have significant clinical advantages over rapidly reversible inhibitors (29). Once such a compound is bound to its target, receptor activity is shut down for a period of time even after the concentration of compound in systemic circulation has been significantly depleted. In vivo, the extended duration of cellular efficacy for such slow-off inhibitors is often seen to be much longer than predicted simply on the basis of the dissociative half-life. This has been attributed to a proximity effect whereby dissociation of the inhibitor from its target creates a high local concentration of ligand in the vicinity of the binding pocket (30). Following dissociation, immediate rebinding may also be favored by slow relaxation of the high-affinity conformational state of the target protein.
In Vitro/in Vivo Activity and Exploratory Safety Studies of P218.
P218 was found to be highly active against PYR-resistant P. falciparum with quadruple mutant PfDHFR in vitro (Table 1). It was also highly active in vivo both against Plasmodium chabaudi in CD-1 mice (Table 1) and against quadruple mutant P. falciparum in SCID mice (ED50 = 0.3 mg/kg, ED90 = 1 mg/kg). As expected, PYR was not efficacious (up to 50 mg/kg) in this model. P218 shows good drug-like physicochemical properties, is a potent and selective inhibitor of wild-type and resistant malaria, is safe, and has a good pharmacokinetic profile in preliminary studies in rats and mice (Table S1 and Fig. S1). Oral administration of P218 to rats at 30 mg/kg resulted in good oral bioavailability (46%), a reasonable half-life (7.3 h), and minimal distribution into the brain. P218 did not induce gene mutations in the Ames test. Also, it did not significantly reduce the human ether-a-go-go related gene (hERG) currents in stably transfected CHO cells at concentrations up to 30 μM, and did not inhibit cytochrome P450 enzymes at concentrations up to 30 μM. In an exploratory repeat-dose oral-toxicity study in rats conducted over 5 d with a 5-d recovery period, some toxicity signs were observed at 300 mg/kg/d; however, effects were minimal at a dose of 100 mg/kg/d, and the no observed adverse effect level (NOAEL) was considered to be approximately 100 mg/kg/d.
The Binding Mode of P218 May Make It Less Susceptible to Drug Resistance than PYR.
There is growing awareness that drug discovery efforts against evolving infectious diseases should attempt to deal with potential drug-resistance issues early in the drug discovery process, aided by high-resolution structural information. For example, most primary active-site drug-resistant mutations in HIV protease occur where the bound inhibitors protrude beyond the substrate-binding footprint or envelope (31). Similarly, for hepatitis C virus NS3/4A protease, mutations conferring severe resistance occur where the protease extensively contacts the drugs but not the natural viral substrates (32). These results suggest that drugs designed to fit within the substrate envelope will be less susceptible to resistance because mutations blocking inhibitor binding will also interfere with substrate binding (33, 34).
In the case of PfDHFR, similar mechanisms have led to PYR resistance. From bacterial surrogate studies it was found that three evolutionary pathways account for almost 90% of the simulated emergence of PYR resistance, and in all three cases the stepwise acquisition of full PYR resistance begins with the S108N mutation (35). These conclusions are consistent with those from earlier work in which kinetic studies of putative intermediates found as PfDHFR polymorphisms in natural populations indicated similarly favored pathways of drug resistance initiated by the S108N mutation (36). Our group has previously shown that the S108N mutation compromises binding of PYR to PfDHFR by sterically interfering with binding of the inhibitor’s chlorophenyl group (12).
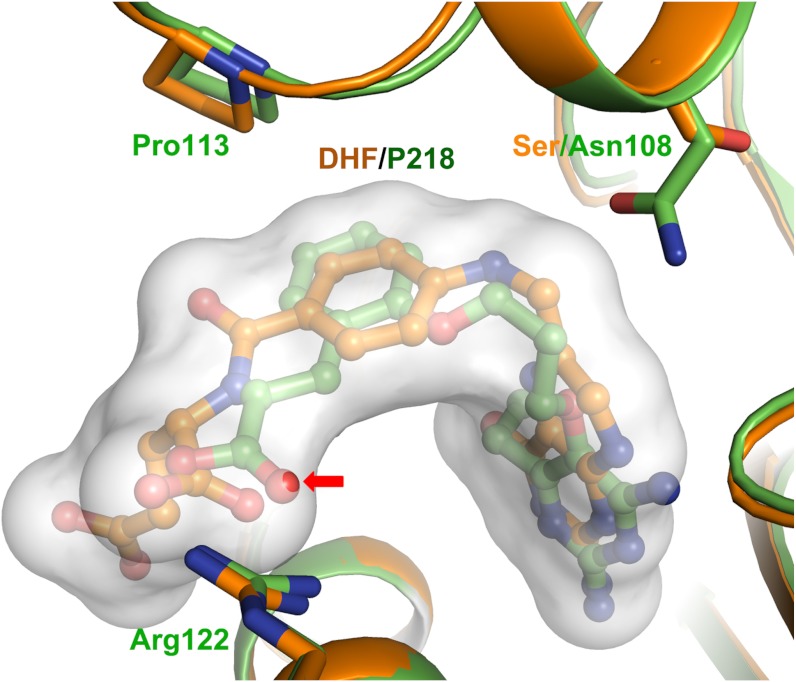
In order to understand protein/substrate interactions for PfDHFR at the molecular level, we determined the cocrystal structure of dihydrofolate bound to PfDHFR-TS at a resolution of 2.5 Å (Table S2). Fig. 4 shows a superposition of the P218 cocrystal structure with PfDHFR-TS onto the dihydrofolate substrate cocrystal structure with the substrate envelope rendered as a transparent surface. For P218 bound to PfDHFR-TS, one carboxylate oxygen slightly penetrates the dihydrofolate substrate envelope (Fig. 4). Phe58 contacts this oxygen in the P218 cocrystal structure with quadruple mutant PfDHFR-TS. Mutation of Phe58 could affect the carboxylate interaction with Arg122 and weaken inhibitor binding. However, this residue makes important stabilizing interactions with the substrate’s pteridine ring and benzoyl group, and is identically conserved in all known DHFR sequences, making mutation of Phe58 highly unlikely because of the expected adverse effect on substrate binding and parasite fitness. The above arguments suggest that, unless other changes not foreseen from our structural analysis occur, clinical resistance to P218 may take longer to emerge than was the case for PYR.
Fig. 4.
Superposition of P218 (green) and the dihydrofolate substrate envelope (orange) bound to quadruple mutant and wild-type PfDHFR-TS, respectively, showing the fit of the P218 in the substrate envelope. Note that one of the carboxyethyl oxygens slightly penetrates the dihydrofolate substrate envelope (arrow).
Conclusions
PYR targets malarial DHFR, but resistance to the drug is now widespread owing to site-specific amino acid mutations in DHFR that reduce drug binding. Extensive efforts to discover a second-generation DHFR-targeted antimalarial drug with efficacy against PYR-resistant strains have been unsuccessful. In this study, we report the results of an iterative drug discovery process in which X-ray structures of Pf and human DHFR cocrystallized with substrates and inhibitors were used along with efficacy and pharmacokinetic profiling in the design and characterization of a selective, highly efficacious antimalarial drug development candidate. P218 is a subnanomolar inhibitor of both wild-type PfDHFR and the clinically relevant highly PYR-resistant quadruple mutant enzyme. P218 is a significantly weaker binder to human DHFR despite the high amino acid sequence homology between the malarial and human enzymes, thus minimizing the potential for human DHFR–associated host toxicity.
In order to reduce the potential for resistance, P218 was designed to incorporate significant intramolecular flexibility, linking polar groups having highly favorable enthalpic interactions with conserved enzyme residues at opposite ends of the substrate-binding site. Selectivity for PfDHFR, compared to the human enzyme, is achieved by designing the linker portion of P218 to allow inhibitor access for binding to a conserved Arg in the case of the protozoal enzyme but not for human DHFR. Cocrystal structures of P218 with Pf and human DHFRs show that the compound binds much differently to the two enzymes. Moreover, P218 binds to PfDHFR within the envelope footprint of the dihydrofolate substrate itself, which could reduce emergence of resistance to P218 owing to the expected adverse effect mutations could have on substrate binding and parasite fitness. We also show that P218 binds to both wild-type and quadruple mutant PfDHFR, but not to human DHFR, according to a two-step mechanism involving an initial encounter complex followed by a slower enzyme isomerization that further tightens binding for P218 to the protozoal enzymes by at least 20-fold. Thus, inhibition of PfDHFR by P218 follows tight-binding/slow-off kinetics, with the inhibitor having a long residence time on the tight-binding conformational state of the enzyme. This study demonstrates that P218 not only has potent in vitro and in vivo antimalarial activity against wild-type and PYR-resistant malarias, but it also possesses suitable pharmacological, metabolic, and safety profiles to warrant further consideration for clinical development. For such purpose, its combination with another antimalarial compound might be warranted to delay further possible future resistance.
Materials and Methods
Synthesis of Inhibitors.
In this study, a modified procedure (37–39) has been employed for the preparation of P65 and P218. The synthetic route involved O-alkylation of 5-hydroxy-6-substituted-2,4-diaminopyrimidine with appropriate alkyl bromides, using lithium hydroxide monohydrate as a base, in N, N-dimethylformamide at room temperature. An alkylation of 5-hydroxy-6-methyl-2,4-diaminopyrimidine with 3-(2,4,5-trichlorophenoxy)propyl bromide (40) gave P65. The analogous reaction between 5-hydroxy-6-ethyl-2,4-diaminopyrimidine and methyl 3-(2-(3-bromopropoxy)phenyl)propanoate, which was prepared from methyl 3-(2-hydroxyphenyl)propanoate (41), provided P218 (SI Text).
X-Ray Structural Studies.
Preparation and crystallization of recombinant PfDHFR-TS and human DHFR enzymes were carried out as previously reported (42–44). X-ray data collection and structure determination are described in Table S2.
Enzyme Kinetic Studies.
Preparation of PfDHFR and hDHFR (43, 45) and preliminary kinetic properties and binding affinity (Ki) in microplate format were as described previously (28) (SI Text).
In Vitro Antimalarial and Cytotoxicity Studies.
P. falciparum, TM4 (wild type), and V1/S strain (N51I+C59R+S108N+I164L) were maintained, and antimalarial activity was determined by [3H]-hypoxanthine incorporation assay as described in detail elsewhere (45–47). Cytotoxicity against Vero cells, a kidney fibroblast line from African green monkey, was determined as previously described (48).
In Vivo Antimalarial Efficacy Studies.
All in vivo tests were performed under the UK Home Office Animals (Scientific Procedures) Act, 1986. Oral efficacy was determined in CD-1 mice infected with P. chabaudi AS (SI Text).
Caco-2 Permeability Studies.
Unidirectional permeability studies were conducted in the apical to basolateral (A–B) direction in the presence of a pH gradient across Caco-2 cell monolayers (SI Text).
In Vivo Pharmacokinetic Studies.
Pharmacokinetic studies were approved by the Monash University Animal Ethics Committee. All studies were conducted in accordance with National Health and Medical Research Council (NHMRC) Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, sixth edition, 1997. Pharmacokinetic studies were conducted in adult male Sprague–Dawley rats or male Swiss outbred mice, and compounds were administered by either i.v. injection (i.v. infusion in rats) or oral gavage. Blood samples were taken in rats via a cannula inserted in the carotid artery, or, in mice, via terminal cardiac puncture under anesthesia, and plasma was assayed for compound by liquid chromatography/MS. Concentration-versus-time data were analyzed using noncompartmental methods (SI Text).
Exploratory Safety Studies.
In vitro Ames and hERG tests were conducted by Harlan Laboratories SA (Barcelona) and bSYS, GmbH (Witterswil, Switzerland), respectively. A repeat-dose oral-toxicology study (Harlan Laboratories SA) was conducted in male Wistar rats with once daily dosing for 5 d followed by a 5-d recovery period. Treatments included a vehicle control, 100 mg/kg P218, and 300 mg/kg P218 (SI Text).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Parichart Naruphontjirakul, Anchalee Tonsomboon, Panuwat Padungros, Nattawut Yotapan, Jittra Kornsakulkarn, SanJay Babu Katiyar, Mark Robinson, Adrian Russell, Francis Chiu, Julia Morizzi, Vivian Cody, and Peter Tonge for their technical support; and Tim Wells and Jeremy Burrows for technical advice. Parasites, TM4, and V1/S were generous gifts from S. Thaithong (Department of Biology, Faculty of Science, Chulalongkorn University, Bangkok, Thailand), and D. Kyle (University of South Florida) [through Malaria Research and Reference Reagent Resource Center (MR4)], respectively. This work was supported by Medicines for Malaria Venture (MMV), the Wellcome Trust, and Thailand Tropical Diseases Research (T2) Programme. S.K. is an international research scholar of Howard Hughes Medical Institute (HHMI, USA).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204556109/-/DCSupplemental.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4DP3, 4DPH, and 4DPD for PfDHFR-TS, and 4DDR for human DHFR).
References
- 1.Murray C, et al. Global malaria mortality between 1980 and 2010: A systematic analysis. Lancet. 2012;379:413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 2.White NJ. Artemisinin resistance: The clock is ticking. Lancet. 2010;376:2051–2052. doi: 10.1016/S0140-6736(10)61963-0. [DOI] [PubMed] [Google Scholar]
- 3.Vennerstrom JL, et al. Identification of an antimalarial synthetic trioxolane drug development candidate. Nature. 2004;430:900–904. doi: 10.1038/nature02779. [DOI] [PubMed] [Google Scholar]
- 4.Charman SA, et al. Synthetic ozonide drug candidate OZ439 offers new hope for a single-dose cure of uncomplicated malaria. Proc Natl Acad Sci USA. 2011;108:4400–4405. doi: 10.1073/pnas.1015762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guiguemde WA, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamo FJ, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows JN, Chibale K, Wells TNC. The state of the art in anti-malarial drug discovery and development. Curr Top Med Chem. 2011;11:1226–1254. doi: 10.2174/156802611795429194. [DOI] [PubMed] [Google Scholar]
- 9.Olliaro P, Wells TNC. The global portfolio of new antimalarial medicines under development. Clin Pharmacol Ther. 2009;85:584–595. doi: 10.1038/clpt.2009.51. [DOI] [PubMed] [Google Scholar]
- 10.Wells TNC, Alonso PL, Gutteridge WE. New medicines to improve control and contribute to the eradication of malaria. Nat Rev Drug Discovery. 2009;8:879–891. doi: 10.1038/nrd2972. [DOI] [PubMed] [Google Scholar]
- 11.Knighton DR, et al. Structure of and kinetic channelling in bifunctional dihydrofolate reductase-thymidylate synthase. Nat Struct Biol. 1994;1:186–194. doi: 10.1038/nsb0394-186. [DOI] [PubMed] [Google Scholar]
- 12.Yuvaniyama J, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10:357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 13.Yuthavong Y, et al. Malarial (Plasmodium falciparum) dihydrofolate reductase-thymidylate synthase: Structural basis for antifolate resistance and development of effective inhibitors. Parasitology. 2005;130:249–259. doi: 10.1017/s003118200400664x. [DOI] [PubMed] [Google Scholar]
- 14.Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth B. Selective inhibitors of bacterial dihydrofolate reductase: Structure activity relationships. In: Hitchings GH, editor. Inhibition of Folate Metabolism in Chemotherapy. Berlin: Springer-Verlag; 1983. pp. 107–127. [Google Scholar]
- 16.Bertino JR. Methotrexate: Historical aspects. In: Cronstein BN, Bertino JR, editors. Methotrexate. Basel: Birkhauser; 2000. pp. 1–8. [Google Scholar]
- 17.Matthews DA, et al. Dihydrofolate reductase: X-ray structure of the binary complex with methotrexate. Science. 1977;197:452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- 18.Rieckmann KH. The in vitro activity of experimental antimalarial compounds against strains of Plasmodium facliparum with varying degrees of sensitivity to pyrimethamine and chloroquine. WHO Tech Rep Ser 529. 1973:58. [Google Scholar]
- 19.Rieckmann KH, Yeo AE, Edstein MD. Activity of PS-15 and its metabolite, WR99210, against Plasmodium falciparum in an in vivo-in vitro model. Trans R Soc Trop Med Hyg. 1996;90:568–571. doi: 10.1016/s0035-9203(96)90326-0. [DOI] [PubMed] [Google Scholar]
- 20.Canfield CJ, et al. PS-15: A potent, orally active antimalarial from a new class of folic acid antagonists. Am J Trop Med Hyg. 1993;49:121–126. doi: 10.4269/ajtmh.1993.49.121. [DOI] [PubMed] [Google Scholar]
- 21.Baker BR, Jordaan JH. Analogs of tetrahydrofolic acid XXVIII. Mode of pyrimidine binding to dihydrofolate reductase pH profile studies. J Pharm Sci. 1965;54:1740–1745. [Google Scholar]
- 22.Staines HM, Rae C, Kirk K. Increased permeability of the malaria-infected erythrocyte to organic cations. Biochim Biophys Acta. 2000;1463:88–98. doi: 10.1016/s0005-2736(99)00187-x. [DOI] [PubMed] [Google Scholar]
- 23.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discovery. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 24.Velazquez-Campoy A, et al. Thermodynamic dissection of the binding energetics of KNI-272, a potent HIV-1 protease inhibitor. Protein Sci. 2000;9:1801–1809. doi: 10.1110/ps.9.9.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosowsky A, Forsch R, Queener S. Inhibition of Pneumocystis carinii, Toxoplasma gondii, and Mycobacterium avium dihydrofolate reductases by 2,4-diamino-5-[2-methoxy-5-(omega-carboxyalkyloxy)benzyl]pyrimidines: Marked improvement in potency relative to trimethoprim and species selectivity relative to piritrexim. J Med Chem. 2002;45:233–241. doi: 10.1021/jm010407u. [DOI] [PubMed] [Google Scholar]
- 26.Kuyper LF, et al. Receptor-based design of dihydrofolate reductase inhibitors: Comparison of crystallographically determined enzyme binding with enzyme affinity in a series of carboxy-substituted trimethoprim analogues. J Med Chem. 1985;28:303–311. doi: 10.1021/jm00381a008. [DOI] [PubMed] [Google Scholar]
- 27.Copeland RA, Harpel MR, Tummino PJ. Targeting enzyme inhibitors in drug discovery. Expert Opin Ther Targets. 2007;11:967–978. doi: 10.1517/14728222.11.7.967. [DOI] [PubMed] [Google Scholar]
- 28.Copeland RA. Methods of Biochemical Analysis. Vol 46. New york: Wiley; 2005. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists; pp. 1–265. [PubMed] [Google Scholar]
- 29.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discovery. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 30.Lewandowicz A, et al. Achieving the ultimate physiological goal in transition state analogue inhibitors for purine nucleoside phosphorylase. J Biol Chem. 2003;278:31465–31468. doi: 10.1074/jbc.C300259200. [DOI] [PubMed] [Google Scholar]
- 31.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: Lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Romano KP, Ali A, Royer WE, Schiffer CA. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc Natl Acad Sci USA. 2010;107:20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali A, et al. Molecular basis for drug resistance in HIV-1 protease. Viruses. 2010;2:2509–2535. doi: 10.3390/v2112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalam MN, Schiffer CA. New approaches to HIV protease inhibitor drug design II: Testing the substrate envelope hypothesis to avoid drug resistance and discover robust inhibitors. Curr Opin HIV AIDS. 2008;3:642–646. doi: 10.1097/COH.0b013e3283136cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozovsky ER, et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci USA. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirawaraporn W, et al. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hull R. Pyrimidines. Part I. The Synthesis of Some 5-Hydroxypyrimidines. J Chem Soc. 1956;60:2033–2035. [Google Scholar]
- 38.Ponsford RJ. 4,179,562. US Patent. 1979
- 39.Hill J, Sharp BW, Warburton D, Walker RB. 4,374,136. US Patent. 1983
- 40.Mamalis P, Outred DJ. 3,723,429. US Patent. 1973
- 41.Meier C, et al. Second-generation cyclosal-d4TMP pronucleotides bearing esterase-cleavable sites-the “trapping” concept. Eur J Org Chem. 2006:197–206. [Google Scholar]
- 42.Chitnumsub P, et al. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductase-thymidylate synthase from Plasmodium falciparum. Acta Crystallogr Sect D. 2004;60:780–783. doi: 10.1107/S0907444904001544. [DOI] [PubMed] [Google Scholar]
- 43.Kamchonwongpaisan S, et al. Stoichiometric selection of tight-binding inhibitors by wild-type and mutant forms of malarial (Plasmodium falciparum) dihydrofolate reductase. Anal Chem. 2005;77:1222–1227. doi: 10.1021/ac0487597. [DOI] [PubMed] [Google Scholar]
- 44.Cody V, Pace J, Lin L, Gangjee A. The Z isomer of 2,4-diaminofuro[2,3-d]pyrimidine antifolate promotes unusual crystal packing in a human dihydrofolate reductase ternary complex. Acta Crystallogr Sect F. 2009;65:762–766. doi: 10.1107/S1744309109025548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarnchompoo B, et al. Development of 2,4-diaminopyrimidines as antimalarials based on inhibition of the S108N and C59R+S108N mutants of dihydrofolate reductase from pyrimethamine-resistant Plasmodium falciparum. J Med Chem. 2002;45:1244–1252. doi: 10.1021/jm010131q. [DOI] [PubMed] [Google Scholar]
- 46.Kamchonwongpaisan S, et al. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J Med Chem. 2004;47:673–680. doi: 10.1021/jm030165t. [DOI] [PubMed] [Google Scholar]
- 47.Yuthavong Y, et al. Development of a lead inhibitor for the A16V+S108T mutant of dihydrofolate reductase from the cycloguanil-resistant strain (T9/94) of Plasmodium falciparum. J Med Chem. 2000;43:2738–2744. doi: 10.1021/jm0009181. [DOI] [PubMed] [Google Scholar]
- 48.Skehan P, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.