Abstract
The circadian clock is an endogenous time-keeping mechanism that enables organisms to adapt to external daily cycles. The clock coordinates biological activities with these cycles, mainly through genome-wide gene expression. However, the exact mechanism underlying regulation of circadian gene expression is poorly understood. Here we demonstrated that an Arabidopsis PSEUDO-RESPONSE REGULATOR 5 (PRR5), which acts in the clock genetic circuit, directly regulates expression timing of key transcription factors involved in clock-output pathways. A transient expression assay and ChIP-quantitative PCR assay using mutated PRR5 indicated that PRR5 associates with target DNA through binding at the CCT motif in vivo. ChIP followed by deep sequencing coupled with genome-wide expression profiling revealed the direct-target genes of PRR5. PRR5 direct-targets include genes encoding transcription factors involved in flowering-time regulation, hypocotyl elongation, and cold-stress responses. PRR5-target gene expression followed a circadian rhythm pattern with low, basal expression from noon until midnight, when PRR9, PRR7, and PRR5 were expressed. ChIP-quantitative PCR assays indicated that PRR7 and PRR9 bind to the direct-targets of PRR5. Genome-wide expression profiling using a prr9 prr7 prr5 triple mutant suggests that PRR5, PRR7, and PRR9 repress these targets. Taken together, our results illustrate a genetic network in which PRR5, PRR7, and PRR9 directly regulate expression timing of key transcription factors to coordinate physiological processes with daily cycles.
Keywords: ChIP-seq, plant
The circadian clock in plants regulates a broad range of biological processes. For example, hypocotyl elongation is observed before dawn and cold-stress responses reach maximal levels in the afternoon in Arabidopsis thaliana (1, 2), all largely because of circadian coordination of these biological processes (clock-output) with daily cycles. The circadian clock mechanism controls the temporal regulation of numerous genes involved in output processes (3–5).
A number of recent studies have described the genetic components of the clock in Arabidopsis. CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) encode morning-expressed MYB transcription factors (TFs) that directly repress TIMING OF CAB EXPRESSION 1 [TOC1, also called PSEUDO-RESPONSE REGULATOR 1 (PRR1)], EARLY FLOWERING 3 (ELF3), ELF4, and LUXARRHYTHMO (LUX) (6–10). ELF3 and LUX associate with upstream region of PRR9, and repress PRR9 expression (11, 12). Expression of PRR9 and PRR7 are activated by CCA1 and LHY (13). CCA1 and LHY are in turn repressed by four PRR proteins, PRR9, PRR7, PRR5, and TOC1 from early daytime through to around midnight (14–16). These TFs form a negative feedback loop for clock function (12, 17, 18). Evidence is accumulating that these TFs directly regulate the expression of genes involved in clock-output pathways. LUX, ELF3, and ELF4 together form the “evening complex” that directly represses expression of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 (19), both of which encode TFs positively regulating hypocotyl elongation (2, 20). CCA1 and LHY bind to the promoter regions of DEHYDRATION-RESPONSIVE ELEMENT BINDING 1 [DREB1, also called C-REPEAT BINDING FACTOR (CBF)] genes encoding TFs involved in cold-stress responses (21). These results suggest that the transcriptional regulators form an interface that allows the clock to regulate output processes.
PRR proteins feature a Pseudo-Receiver (PR) domain at the N terminus and a CONSTANS, CONSTANS-LIKE, and TOC1 (CCT) motif at the C terminus (22, 23). The PR domain resembles the receiver domain of a two-component response regulator, but lacks an aspartate residue that accepts a phosphoryl group from the sensor kinase. The PR domain is involved in protein–protein interactions (24, 25) and TOC1 PR is crucial for transcriptional repression (26). In contrast, PRR9, PRR7, and PRR5 have a repression motif in an intervening region (IR) between the PR and CCT, and act as transcription repressors (14). Previous studies suggest that these three PRRs redundantly regulate expression of clock-output genes (27, 28). However, it is still not known which genes are the direct targets of the three PRRs and how they are regulated by them. Identifying the direct-target genes is critical for illustrating the entire genetic network of clock-output regulation.
To address this issue, we studied domains within PRR5 and found that PRR5 binds to the known target gene CCA1 through the CCT motif in vivo. ChIP followed by deep sequencing (ChIP-seq) coupled with genome-wide expression profiling revealed that a number of genes encoding key TFs for hypocotyl elongation, flowering time, and cold-stress responses were enriched in the population of direct-targets of PRR5. Our results demonstrated that PRR5 functions as a transcriptional repressor that controls various biological processes by directly regulating the timing of expression of its target genes.
Results
PRR5 Associates with CCA1 Through the CCT Motif.
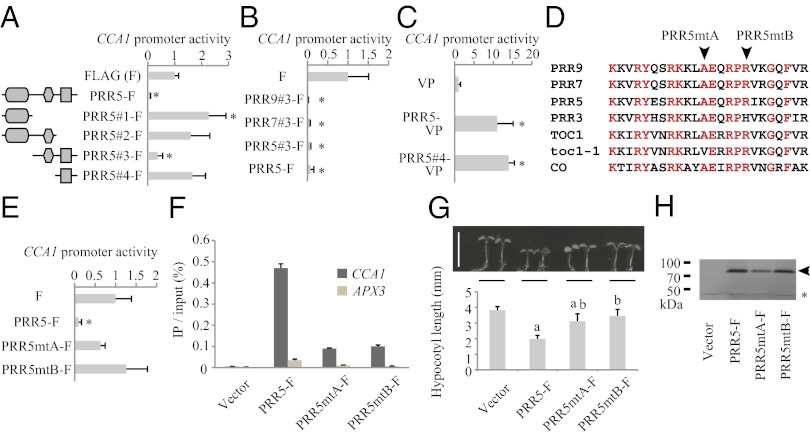
To clarify which specific region of PRR5 represses known target genes, such as CCA1, we performed transient expression assays using a luciferase (LUC) reporter plasmid under the control of the CCA1 promoter (CCA1pro:LUC) with an effector plasmid harboring PRR5 fused to FLAG under the control of the cauliflower mosaic virus 35S promoter (35Spro:PRR5-FLAG), or with a plasmid containing a series of truncated PRR5s (35Spro:PRR5#1-FLAG to 35Spro:PRR5#4-FLAG) (Fig. 1A). Bombardment with 35Spro:PRR5-FLAG (PRR5-FLAG) resulted in a significant reduction of CCA1pro:LUC activity compared with that with the control vector (FLAG), indicating that PRR5 represses the CCA1 promoter in transient assays. Introduction of an effector plasmid harboring IR and CCT of PRR5 (PRR5#3-FLAG) resulted in a significant reduction of CCA1pro:LUC activity, whereas any expression of PR (PRR5#1-FLAG), PR and IR (PRR5#2-FLAG), or CCT (PRR5#4-FLAG) did not, suggesting that both IR and CCT of PRR5 are required for repressing CCA1 promoter activity. The IR and CCT in PRR9 and PRR7 are also sufficient for the repression of CCA1 promoter (Fig. 1B).
Fig. 1.
PRR5 associates with CCA1 through its CCT in vivo. (A) Effect of truncated PRR5 constructs #1 to #4 on CCA1 promoter activity in Arabidopsis seedlings (Right), and a schematic of each construct (Left). Ellipses indicate PR, diamonds indicate a repression motif, and squares indicate a CCT motif. (B) Effect of truncated (full-length IR and CCT) PRR9, PRR7, and PRR5 on CCA1 promoter activity. (C) Effect of PRR5-VP and PRR5#4 (full-length CCT)-VP on CCA1 promoter activity. (D) Amino acid sequence alignment of the C-terminal portion of CCT. Red indicates conserved residues among PRR9, PRR7, PRR5, TOC1, and CONSTANS (CO). Arrows indicate amino acid substitutions in toc1-1 (PRR5mtA) or PRR3 (PRR5mtB). (E) Effect of two CCT mutants of PRR5 on CCA1 promoter activity. (F) ChIP-qPCR for CCA1 and APX3 upstream regions in PRR5-FLAG– or mutated PRR5-FLAG– expressing plants. Percentages of the amplicons coimmunoprecipitated with anti-FLAG antibody relative to input DNA are indicated. (G) Hypocotyl length of PRR5-FLAG– or mutated PRR5-FLAG–expressing plants under 10-h light/14-h dark cycles. Typical seedlings are indicated with a scale bar (Left). (Scale bar, 5 mm.) (H) Expression of PRR5mt-FLAG protein in transgenic plants. The arrow and asterisk indicate FLAG-fused protein and nonspecific bands, respectively. Error bars indicate the SD of biological replicates in A–C, E, and G (n = 15 for G, and 3 for others), and the SD of three technical replicates in F. Asterisks indicate a significant change in CCA1 activity compared with coexpression with FLAG (Student t test; P < 0.05). The “a” and “b” in G indicate one-way ANOVA P < 0.05 compared with Vector and PRR5-F, respectively.
To examine whether a CCT is required for PRR5 association with its target genes, we performed a transient expression assay in which the protein of interest was fused to a tandem construct of the stringent transcriptional activation domain VP16 (VP), such that if the protein of interest associates with the promoter, the VP-fused protein activates promoter activity (29). Expression of PRR5-VP or PRR5#4-VP resulted in significant activation of the CCA1 promoter (Fig. 1C). The TOC1 CCT is sufficient for DNA binding in vitro, and both the toc1-1 and PRR3-type mutations attenuate activity (26). We thus generated two independent mutations [toc1-1 type (mtA: Ala538Val) or PRR3 type (mtB: Arg543His)] within the CCT of PRR5 (Fig. 1D) and assayed for their effect on the CCA1 promoter. Expression of PRR5mtA or PRR5mtB did not result in any significant reduction of CCA1 promoter (Fig. 1E). These results suggest that PRR5 associates with the CCA1 promoter through its CCT.
To further investigate whether a CCT is crucial to the DNA-binding activity of PRR5 in vivo, a ChIP assay was performed for plants overexpressing PRR5-FLAG, PRR5mtA-FLAG, or PRR5mtB-FLAG (Fig. 1F). Plants were grown under 12-h light/12-h dark conditions (LD), and harvested at Zeitgeber time 10 (ZT10 indicates 10 h after lights are turned on), when native PRR5 protein associates with target promoters in vivo. Amplicons located in the upstream region of CCA1 and ASCORBATE PEROXIDASE 3 (APX3) were quantified by quantitative PCR (qPCR). The amplicons located at the CCA1 promoter, but not upstream of APX3, was enriched in ChIP DNA from PRR5-FLAG–expressing plants, indicating that PRR5-FLAG associates with the CCA1 promoter in vivo (Fig. 1F). In contrast, PRR5mtA-FLAG and PRR5mtB-FLAG associated with CCA1 less often. These ChIP-qPCR analyses indicated that a functional CCT is crucial for interactions between PRR5 and the CCA1 promoter in vivo.
To determine the biological importance of the PRR5 CCT, we measured hypocotyl lengths of plants overexpressing PRR5-FLAG, PRR5mtA-FLAG, or PRR5mtB-FLAG (Fig. 1G). The PRR5-FLAG plants grew short hypocotyls, a well-known phenotype of lines that overexpress PRR5 (30). Hypocotyls of PRR5mtA-FLAG plants were significantly shorter than the wild-type (P < 0.05), but the hypocotyls of PRR5mtA-FLAG and PRR5mtB-FLAG were longer than those of PRR5-FLAG, even though exogenous proteins were expressed at levels similar to those of PRR5-FLAG–expressing plants (Fig. 1H).
Identification of Direct-Target Genes of PRR5.
To determine the genes bound by PRR5 on a genomic scale, we conducted ChIP-seq for FLAG-PRR5-GFP protein expressed under the control of the PRR5 promoter in a prr5 mutant background (PRR5pro:FLAG-PRR5-GFP/prr5) (Fig. S1) (14). DNA libraries for deep sequencing were generated from the immunoprecipitated fraction (ChIP DNA) and input DNA fraction (input DNA), and analyzed with an Illumina Genome Analyzer II (Fig. S2). Five-hundred forty-two genomic loci (1,024 genes) were significantly enriched in ChIP DNA compared with input DNA [false-discovery rate (FDR) q < 10−50]. These genes make up the in vivo “PRR5-bound” genes (Fig. S3 and Dataset S1), which potentially contain “PRR5 direct-target” genes, but may also contain some false-positive genes because of inherent problems with ChIP and deep-sequencing procedures (31).
To discover PRR5 direct-target genes another way, we performed a DNA microarray experiment with transgenic Arabidopsis overexpressing PRR5-VP in the wild-type background (35Spro:PRR5-VP). When we compared genome-wide gene expression in PRR5-VP plants and prr9 prr7 prr5 (Fig. S4), significant overlaps (P < 10−16) were found between genes similarly regulated both in PRR5-VP and prr9 prr7 prr5, but not between genes oppositely regulated. LHY expression was up-regulated in PRR5-VP during the daytime (Fig. S4), and hypocotyls were longer for PRR5-VP plants, but not for PRR5-ox, thereby resembling the phenotypes of prr9 prr7 prr5 (28). PRR5-VP plants flowered significantly later than the wild-type (Fig. S4), and thus showed a phenotype similar to prr5. These data suggest that PRR5-VP acts in an inverse manner to PRR5. Because PRR5-VP activates a direct-target of PRR5 (Fig. 1C), genes whose expression is significantly increased in 35Spro:PRR5-VP lines compared with wild-type (FDR q < 0.01) potentially contain PRR5 direct-target genes. This strategy may miss potential activated genes by wild-type PRR5, but 190 genes were obtained as PRR5-VP up-regulated genes (Dataset S2).
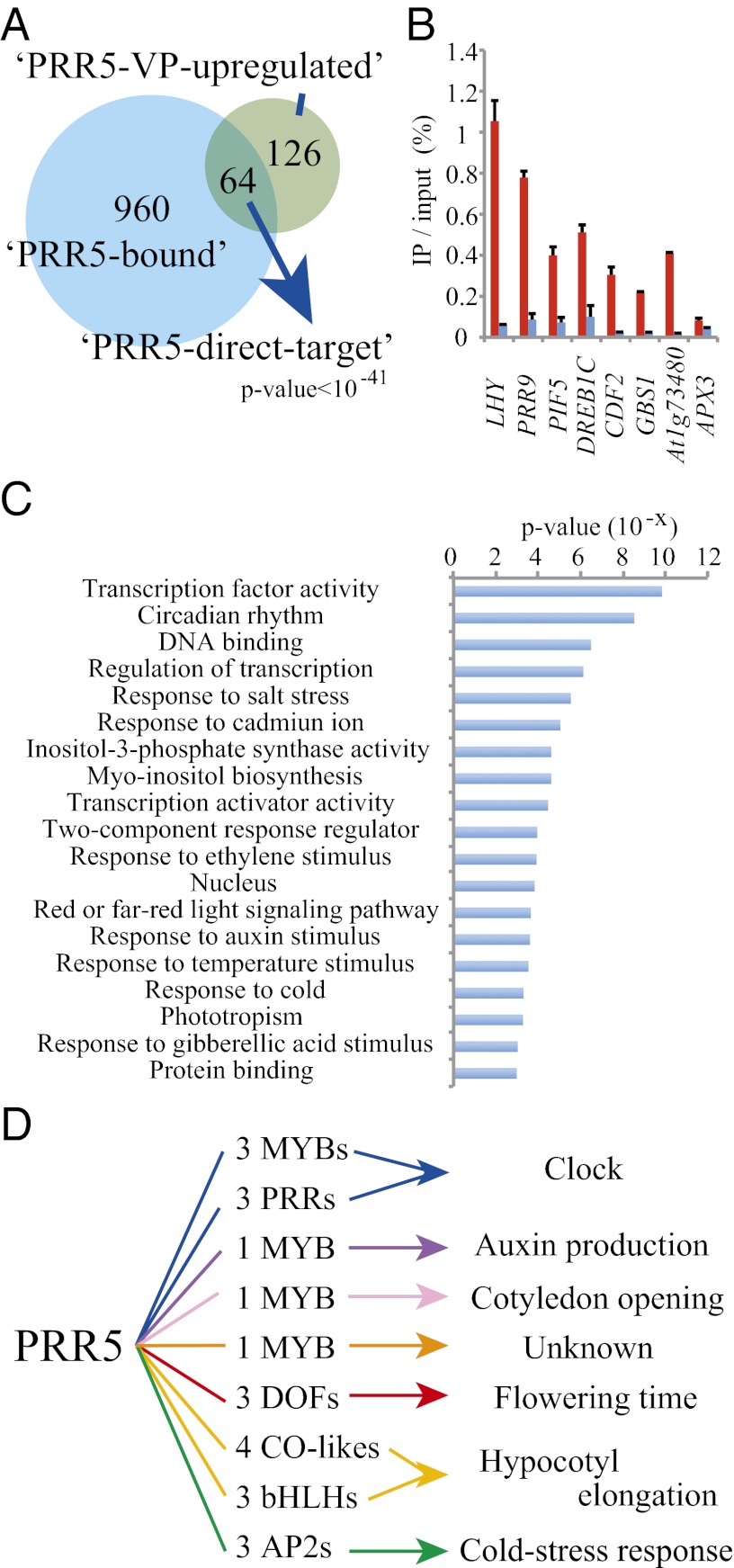
The comparison between PRR5-bound genes and PRR5-VP up-regulated genes delineated 64 direct-target genes of PRR5 (Fig. 2A and Dataset S3), of which two are the known PRR5 direct-target genes, CCA1 and LHY (14). Overlap between the two gene sets was statistically significant (Fig. 2A), supporting the validity of our strategy. On the other hand, the overlap between PRR5-bound genes and down-regulated genes in PRR5-VP was not significant (Fig. S5). ChIP-qPCR experiments (six genes in Fig. 2B, 43 genes in Fig. S6) confirmed PRR5-binding at most of the PRR5-target loci (45 of 49).
Fig. 2.
PRR5 directly controls clock-output pathways by regulating genes encoding transcription factors. (A) Venn diagram of PRR5-bound genes and PRR5-VP up-regulated genes. There is significant overlap between the two gene groups (Fisher’s exact test; P < 10−41). (B) ChIP-qPCR assay for representative PRR5-direct-target genes in PRR5pro:FLAG-PRR5-GFP/prr5. Red and blue bars indicate percentages of amplicons coimmunoprecipitated with anti-GFP antibody relative to input DNA from the sample harvested at ZT10 and ZT22, respectively. Error bars indicate the SD of three replicates. (C) eGO analysis was performed for PRR5 direct-target genes. (D) Schematic indicating that PRR5 targets key transcription factors in clock-output pathways.
TFs are Enriched in PRR5 Direct-Targets.
Significantly enriched Gene Ontology (eGO) analysis was performed to explore the biological functions of the direct targets of PRR5 (Fig. 2C). “Transcription factor activity” was the most enriched category in PRR5 direct-targets (P < 10−9). “Circadian rhythm” was the next enriched group (P < 10−8). “DNA binding,” “regulation of transcription,” “response to salt stress,” and “response to cadmium ion” were also enriched (P < 10−5). We were especially interested in TFs because three categories related to TFs were enriched. The direct-target TF group includes six MYB TFs [CCA1, LHY, EARLY PHYTOCHROME RESPONSIVE 1 (EPR1)/also called as REVEILLE7 (RVE7), RVE1, RVE3, and RVE8], three DOF TFs [CYCLING DOF FACTOR 2 (CDF2), CDF3, and CDF5], four C2C2-CO–like TFs [B-BOX DOMAIN PROTEIN 2 (BBX2), BBX6, BBX24, and BBX29], three bHLH TFs [PIF4, PIF5, and LONG HYPOCOTY IN FAR-RED (HFR1)], three AP2/EREBP TFs (DREB1A, DREB1B, and DREB1C), and three PRRs (PRR9, PRR7, and PRR5). CCA1, LHY, RVE8 (32, 33), and PRRs are known to be involved in clock function, EPR1/RVE7 is in cotyledon opening and flowering-time regulation (34), RVE1 is in auxin production (35), RVE3 is in unknown biological process, DOF TFs are in flowering time regulation (36, 37), C2C2-CO-like (38) and bHLH TFs (2, 20) are in hypocotyl elongation, and AP2/EREBP TFs are in cold-stress responses (39, 40), suggesting that PRR5 controls diverse biological processes by regulating these TFs (Fig. 2D).
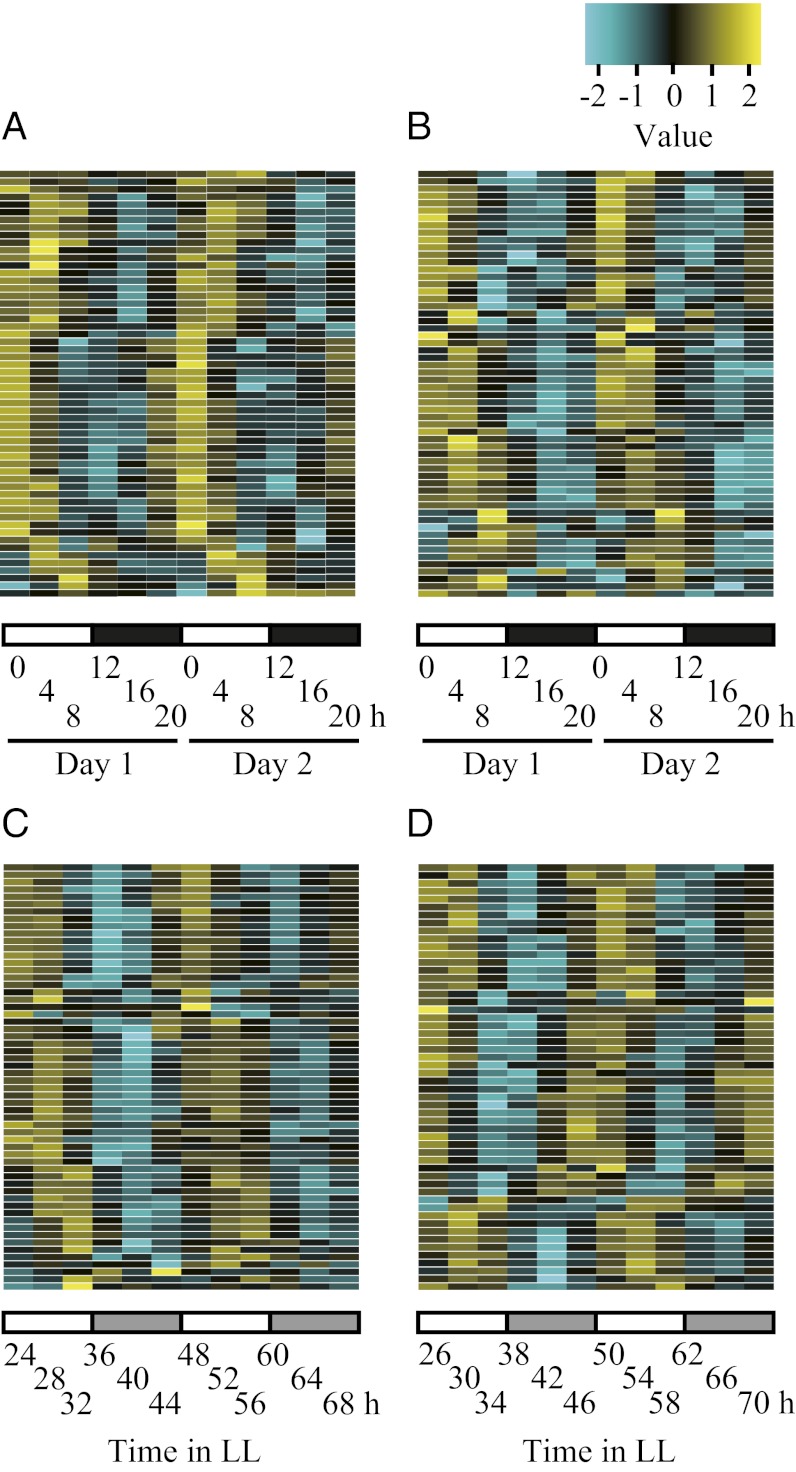
PRR5 Direct-Targets Are Repressed from Noon Until Midnight.
To examine the expression patterns of the target genes under LD, we tested gene expression in a public microarray database [DIURNAL (4, 41)]. Expression of target genes with valid data (see SI Materials and Methods) showed clear diurnal rhythms (Fig. 3 A and B). About 60% of the gene set had an expression peak at ZT0, with the others peaking at ZT4 or ZT8. Even under free-running constant-light conditions, expression of targets with valid data were cyclic, with peaks in subjective dawn to daytime (Fig. 3 C and D). Expression troughs of these genes extended from noon to midnight, when the three PRR (PRR9, PRR7, and PRR5) proteins are expressed (14). We also performed RT-qPCR analysis for “invalid genes in DIURNAL” and found similar expression patterns for genes analyzed in DIURNAL (Fig. S7). The majority of direct-targets are expressed in the morning, and thus showed similar expression patterns to CCA1 and LHY (14), suggesting that these genes and CCA1 and LHY are regulated by PRR5 through the same mechanism.
Fig. 3.
PRR5 direct-targets are repressed during noon until midnight. Expression of direct-targets under LD (A and B), and constant light conditions (C and D). See also SI Materials and Methods for detailed information.
PRR5 Represses Its Direct-Targets.
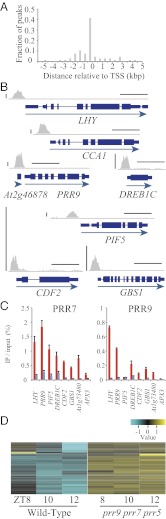
To investigate how PRR5 regulates direct-targets, a PRR5-binding profile in ChIP-seq data were visualized. Apparently, PRR5 preferentially binds to upstream regions of direct-targets, supporting the idea that PRR5 regulates gene expression (Fig. 4 A and B). ChIP-qPCR analyses using PRR5 CCT mutants suggested that PRR5 regulates its representative target genes by associating with their upstream regions through the CCT (Fig. S8).
Fig. 4.
PRR5, PRR7, and PRR9 repress expression of PRR5-targets. (A) Positions of ChIP peaks relative to transcription start sites (TSS) in the PRR5 direct-targets. (B) Examples of peak distributions around direct-targets. Vertical and horizontal bars indicate 0.5-k sequence reads and 1.0 kbp, respectively. (C) ChIP-qPCR assay for representative PRR5 direct-target genes in PRR7pro:FLAG-PRR7-GFP/prr7 and PRR9pro:FLAG-PRR9-GFP/prr9 plants. Red bars indicate percentages of amplicons coimmunoprecipitated with anti-GFP antibody relative to input DNA from the sample harvested when PRR proteins were expressed, and blue bars are from samples when PRRs were not expressed. (D) Expression of the PRR5 direct-target genes in prr9 prr7 prr5 at ZT8, ZT10, and ZT12.
Because PRR5, PRR7, and PRR9 redundantly target CCA1 and LHY (14), we examined whether PRR9 and PRR7 associate with PRR5-target genes using PRR9pro:FLAG-PRR9-GFP/prr9 and PRR7pro:FLAG-PRR7-GFP/prr7 plants (Fig. 4C and Fig. S6). The plants were grown under LD, and samples were collected when PRR proteins are expressed (i.e., ZT4 for PRR9pro:FLAG-PRR9-GFP/prr9, ZT10 for PRR7pro:FLAG-PRR7-GFP/prr7) and not expressed (ZT22 for all plants). Amplicons located in the upstream regions of most of the tested targets were significantly enriched in PRR7pro:FLAG-PRR7-GFP/prr7 when PRR7 is expressed. Similar trends of enrichment were observed for PRR9pro:FLAG-PRR9-GFP/prr9 with some exceptions (e.g., BBX24, CDF3, PIF4, PIF5, and SIGE), indicating that PRR7 and PRR9 share targets with PRR5.
When we surveyed the expression of targets in a microarray dataset for prr9 prr7 prr5 triple mutants (27), most of the targets were up-regulated in prr9 prr7 prr5 compared with the wild-type from ZT8 to ZT12 (Fig. 4D). Taken together, these results suggest that PRR9, PRR7, and PRR5 coordinately act on the upstream region of the PRR5-targets to repress their expression.
Discussion
In this study, a ChIP-qPCR assay using two CCT-motif mutants indicated that PRR5 associates with several target genes through it’s CCT in vivo (Fig. 1F and Fig. S8). We also found that a CCT is crucial for the biological function of PRR5, because mutations in the CCT resulted in attenuation of PRR5 activity leading to hypocotyl shortening (Fig. 1G). It was reported that Gly-to-Trp change in a CCT of a barley homolog of PRR7 is the most likely cause of photoperiod-H1 (42). These results suggest that CCT is essential for target gene recognition, through which PRR5-regulated biological processes are controlled.
An in vitro gel-shift assay showed that recombinant CCTs from PRR9, PRR7, PRR5, and TOC1 bind to the TGTG motif (26). ChIP-seq analysis for TOC1 revealed that G-box (CACGTG)- or evening element (AAAATATCT)-like sequences are enriched in TOC1-bound DNA sequences (16). In our analysis, a G-box motif was found to be enriched around the peak sequences of ChIP DNA (Fig. S9) (43). However, this result should be interpreted with care because ChIP-enriched sequences do not necessarily represent the motif directly bound by PRR5. Because the ChIP procedure involves cross-linking, it is conceivable that bound sequences are a mixture of motifs directly bound and others associated through various protein–protein interactions by PRR5. Previously it was shown that TOC1 occupies the CCA1 promoter region both by direct binding (16, 26) and through interaction with the CHE transcription factor to regulate CCA1 (44). Furthermore, we found that PRR7 and PRR5 associate with BBX24, CDF3, PIF4, PIF5, and SIGE upstream regions, whereas PRR9 associates much more poorly or not at all with these loci (Fig. 4C and Fig. S6). A comparison between TOC1-targets (16) and PRR5-targets reveals that TOC1 binds to 27 genes of 64 PRR5-targets (Dataset S4). Taken together, these results suggest that interactions between PRRs and a certain locus are not solely determined by the CCT binding motif in vivo, and this may cause preferences of target-recognition. Further experiments are required for fully understanding how each PRR is recruited to their target loci in vivo.
Four PRRs directly repress CCA1 and LHY expression from early daytime until midnight (14, 16); however, whether PRR9, PRR7, and PRR5 act as repressors for other target genes was unknown. We proposed that PRR5 represses 64 targets that were found by ChIP-seq coupled with genome-wide expression profiling using PRR5-VP. This strategy might miss genes positively regulated by native PRR5 because native PRR5 may sufficiently activate targets in PRR5-VP plants. To examine the possible PRR5 activation of its targets, 149 down-regulated genes in prr9 prr7 prr5 were compared with PRR5-bound genes. Although the overlap between the down-regulated genes in prr9 prr7 prr5 and PRR5-bound genes was not statistically significant (P > 0.01), 12 genes were found as potential activated targets by PRR5 (Fig. S5). Expression of UBT71B1 and AT4G29700 were slightly but significantly up-regulated in the PRR5-ox line, suggesting that PRR5 potentially activates these genes (Fig. S10). Because PRR5 has a repression motif (14), PRR5 may activate UBT71B1 and AT4G29700 by an unknown mechanism or with some other transcriptional activators.
PRR9, PRR7, and PRR5 seem to regulate about 60% of the PRR5-target genes by the same mechanism by which they control CCA1 and LHY because circadian expression patterns of these genes were similar to those of CCA1 and LHY (genes whose expression peaked at ZT0 in Fig. 3). The expression peak and trough positions of other genes were slightly different from those of CCA1 and LHY, suggesting that these genes are also regulated by other factors. For example, the DREB1A and DREB1B promoters are directly regulated by CCA1 and LHY (21), and PIF4 and PIF5 promoters are regulated by the evening complex (19). Such differences in TF combinations might be one of the bases for shifting the expression timing of target genes.
The most enriched Gene Ontology category for the targets of PRR5 was TF, suggesting that PRR5 functions as a repressor directly regulating key TFs (PIFs, BBXs, CDFs, DREB1s/CBFs) (Fig. 2D). These TFs control a cascade of gene expression involved in output processes. This kind of hierarchical genetic architecture may be effective in orchestrating the expression of genes involved in certain biological process at the appropriate time of day (3, 4). A similar genetic architecture, in which master clock function TFs directly regulate output TFs, was reported in Drosophila, which has a different type of central clock mechanism than plants (45), suggesting that such an architecture is conserved among species.
Materials and Methods
Plant Materials and Growth Conditions.
Transgenic plants and growth conditions are described in SI Materials and Methods.
Transient Expression Assay.
Transient expression assay by particle bombardment was described previously (14). Detailed information is in SI Materials and Methods.
Protein Sequence Alignment.
The alignment for the C-terminal portion of CCT from the proteins was done using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2).
Measurement of Hypocotyl Lengths.
Measurement of hypocotyl lengths under 10-h light/14-h dark conditions was described previously (14).
Protein Gel-Blot Analysis.
Protein gel blotting was performed as previously described (14).
ChIP-qPCR Assay.
The ChIP-qPCR assay was performed as described previously (14). Anti-FLAG antibody (F3165; Sigma-Aldrich) was used for immunoprecipitation of PRR5(mt)-FLAG proteins. Primers used for ChIP-qPCR are listed in Dataset S5.
ChIP-Seq Analysis.
The methods for ChIP-seq are described in SI Materials and Methods. ChIP-seq data were deposited in the National Center for Biotechnology Information GEO (www.ncbi.nlm.nih.gov/gds) under accession no. GSE36361.
Microarrays.
Microarray methods and data analyses are described in SI Materials and Methods. Microarray data for PRR5-VP-expressin plants were deposited with National Center for Biotechnology Information GEO under accession no. GSE36360.
eGO Analysis.
eGO analysis was performed as previously described (46).
Supplementary Material
Acknowledgments
We thank E. Farre for sharing unpublished data; RIKEN Omics Center for reading sequences by Illumina Genome Analyzer II; H. Tsukagoshi, Y. Tada, and M. Nomoto for technical suggestions; and S. Aoki, J. Inaba, S. Oyama, A. Suzuki, S. Suzuki, and T. Umemori for technical support. This work was supported by Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology Grant 20109, and RIKEN Technology Transfer Office Fund (to N.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE36360 (microarray data) and GSE36361 (ChIP-seq data)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205156109/-/DCSupplemental.
References
- 1.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alabadí D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 7.Mizoguchi T, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 8.Li G, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 2011;13:616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 9.Lu SX, et al. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazen SP, et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 14.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokhilko A, et al. Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol. 2010;6:416. doi: 10.1038/msb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 17.Nakamichi N. Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol. 2011;52:1709–1718. doi: 10.1093/pcp/pcr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokhilko A, et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimori T, Yamashino T, Kato T, Mizuno T. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1078–1086. doi: 10.1093/pcp/pch124. [DOI] [PubMed] [Google Scholar]
- 21.Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 23.Makino S, et al. Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- 24.Para A, et al. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamichi N, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 28.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 29.Aoyama T, et al. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell. 1995;7:1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato E, Nakamichi N, Yamashino T, Mizuno T. Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 2002;43:1374–1385. doi: 10.1093/pcp/pcf166. [DOI] [PubMed] [Google Scholar]
- 31.Kaufmann K, et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nat Protoc. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
- 32.Rawat R, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuno N, et al. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell. 2003;15:2476–2488. doi: 10.1105/tpc.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawat R, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 37.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 38.Kumagai T, et al. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2008;72:1539–1549. doi: 10.1271/bbb.80041. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 41.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 42.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abruzzi KC, et al. Drosophila CLOCK target gene characterization: Implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.