Abstract
EmrE, a multidrug antiporter from Escherichia coli, has presented biochemists with unusual surprises. Here we describe the transformation of EmrE, a drug/H+ antiporter to a polyamine importer by a single mutation. Antibiotic resistance in microorganisms may arise by mutations at certain chromosomal loci. To investigate this phenomenon, we used directed evolution of EmrE to assess the rate of development of novel specificities in existing multidrug transporters. Strikingly, when a library of random mutants of EmrE was screened for resistance to two major antibacterial drugs—norfloxacin, a fluoroquinolone, and erythromycin, a macrolide— proteins with single mutations were found capable of conferring resistance. The mutation conferring erythromycin resistance resulted from substitution of a fully conserved and essential tryptophan residue to glycine, and, as expected, this protein lost its ability to recognize and transport the classical EmrE substrates. However, this protein functions now as an electrochemical potential driven importer of a new set of substrates: aliphatic polyamines. This mutant provides a unique paradigm to understand the function and evolution of distinct modes of transport.
Keywords: ion-coupled transporters, membrane proteins, SMR multidrug transporters, putrescine
The emergence of multiple antibiotic resistance poses a serious threat to the treatment of infectious diseases (1). Active removal of antibiotics, both drug-specific and multidrug, are important determinants of resistance (2, 3). Transporters and multidrug transporters (MDTs) are capable of removing noxious compounds away from their target to the medium in the case of microorganisms or to subcellular organelles in fungi and plants. In Escherichia coli, the MDTs are specialized and well organized so that, for example, the AcrAB–TolC complex, considered the major MDT in this organism, removes drugs only from the periplasm, whereas other MDTs such as EmrE and MdfA remove them from the cytoplasm to the periplasm (4). In addition, the multitude of MDTs display overlapping specificities, which introduces a degree of redundancy that can act as a compensatory backup mechanism, should one protein become nonfunctional due to a mutation in a critical residue. Overall, the MDT system provides a very robust survival mechanism. This very survival strategy is what poses a serious threat in the treatment of infectious diseases because it severely limits the use of antibiotics that are also substrates of many of these multidrug transporters. Therefore, we embarked on a study of the flexibility, promiscuity, and evolvability of these transporters to provide tools to understand the mechanisms of emergence of antibiotic resistance.
The Small Multidrug Resistance (SMR) proteins provide an experimental paradigm to study the evolution of the large modern transporters (5). SMRs display a remarkable plasticity regarding topology in the membrane, interaction between subunits, and interaction with substrates (6). The SMR family consists of proton-coupled transporters. The best-studied protein of this family is the E. coli EmrE, which recognizes a wide range of aromatic cations and removes them from the cell in exchange for two protons (5).
We used directed evolution to assess the evolvability of EmrE and to understand the emergence of new specificities in MDTs. A library of EmrE random mutants was screened for resistance to two antibiotics—norfloxacin, a fluoroquinolone, and erythromycin, a macrolide. Macrolides and fluoroquinolones are among the major antibacterial drugs in current clinical use in humans (2). Erythromycin is a large molecule, composed of a 14-membered lactone ring with two sugars, much larger than the common substrates of SMR proteins (2). Strikingly, single mutations confer new specificities to EmrE (Fig. S1). The mutation conferring a robust resistance to norfloxacin resulted from replacement of Leu by Phe in TM2. The new mutant gained a new specificity but also conserved its ability to transport most of the original substrates. On the other hand, the mutation conferring resistance to erythromycin was a replacement of Trp63, a fully conserved and essential amino acid (7). Interestingly, this mutant completely lost the capacity to recognize and remove the classical substrates of EmrE but gained, in addition to its ability to confer resistance to erythromycin, a completely new function—the ability to import polyamines into the cell. Thus, a single mutation in an essential residue transforms a drug/H+ antiporter into a polyamine importer and provides a unique paradigm to understand the coupling mechanism and evolution of distinct modes of transport.
Results
Isolation of Mutants That Confer Resistance to Macrolides and Fluoroquinolones.
To select for mutants that confer novel resistances, a library containing random mutants of EmrE was generated and transformed into E. coli BW25113 ∆emrE ∆mdfA, a strain used in our laboratory to test activity of EmrE mutants (4, 8). About 4 × 105 transformants were inoculated to 10–20 LB–agar plates containing the antibiotic at concentrations not permissive for growth of cells expressing wild-type EmrE. Plasmids were prepared from colonies isolated from such plates in the first screen and used to retransform E. coli BW25113 ∆emrE∆mdfA for phenotype verification. Plasmids from independent colonies were sequenced.
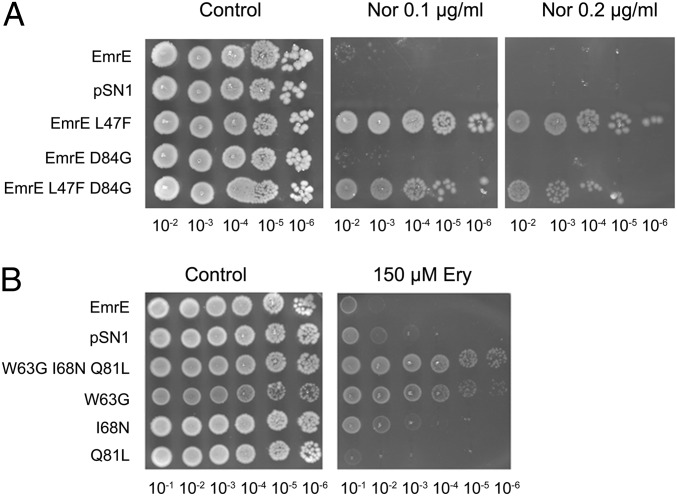
In the norfloxacin screen, an isolate bearing two mutations, L47F and D84G, was found to confer resistance to this antibiotic (Fig. 1A). In this experiment, decimal dilutions of logarithmic cultures were plated and growth was assessed after 24–36 h. To identify which mutation is responsible for the phenotype, mutations were separated, and the growth phenotype of cells bearing plasmids with single mutations revealed that the L47F mutation is sufficient to confer full resistance whereas the EmrE D84G mutant is unable to grow on norfloxacin, similar to what is observed for EmrE or the empty vector (Fig. 1A). The EmrE L47F mutant was also found to confer resistance to another fluoroquinolone, ofloxacin (Fig. S2A). The resistance that the EmrE L47F mutant confers to these antibiotics is very robust and even greater than that conferred by SsmE from Serratia marcescens, the only SMR homolog reported to confer resistance to fluoroquinolones (9). The amino acid at the equivalent position in SsmE is leucine as in wild-type EmrE. This suggests that other determinants in the norfloxacin resistance are likely to exist. Furthermore, it was observed that the specificities of EmrE L47F toward the common substrates of the wild-type protein are altered. The ability of this mutant to confer resistance to acriflavine is moderately reduced, whereas its ability to confer resistance to ethidium is not affected (Fig. S2B). The EmrE L47F mutant, however, loses its ability to confer resistance to methyl viologen (MV). The finding that this position affects the ability of EmrE to confer resistance to MV complements previous studies that located residues in TM2 that influence MV resistance (10).
Fig. 1.
EmrE mutants that confer resistance to norfloxacin and to erythromycin. (A) The L47F mutation is responsible for the resistance to norfloxacin. E. coli BW25113 ∆emrE∆mdfA cells transformed with pSN1 (vector), pSN1-EmrE, or pSN1 with each of the mutants were grown overnight at 37 °C in LB medium containing chloramphenicol. Five microliters of serial dilutions of the culture were spotted onto LB plates containing 30 mM BTP (pH 7.0), 200 μM IPTG, with or without the addition of 0.1 μg/mL or 0.2 μg/mL norfloxacin. Growth was analyzed after an overnight incubation at 37 °C. (B) Replacement of the essential amino acid Trp63 by Gly (W63G) confers resistance to erythromycin. Growth was assayed and analyzed as above except that the plates contained 150 μM erythromycin instead of norfloxacin.
In the erythromycin screen, the mutant isolated carried mutations in three codons that generated the triple mutant W63G, I68N, and Q81L. To identify the residue responsible for the phenotype, genes with the single mutations or combinations were generated, and cells bearing each of them were assayed for erythromycin resistance. The results of such an experiment are shown in Fig. 1B. As can be seen, the originally isolated triple mutant displays weak but distinct growth at all of the dilutions tested. Of all of the three mutants derived from it, only W63G conferred a similar resistance. We conclude that replacement of the bulky Trp residue at position 63 with the smallest amino acid Gly is necessary and sufficient to confer resistance to erythromycin. When the Trp residue was replaced with Ala rather than Gly, an amino acid with only an additional methyl group, the mutant protein did not confer detectable resistance (Fig. S3). The result suggests that the volume of the side chain is critical and that only Gly can support erythromicin resistance. To support the conclusion that the resistance is due to the activity of EmrE, we generated a double mutant that, in addition to the Gly replacement at position 63, carried a mutation in the catalytic Glu14 that was replaced with Cys. Indeed, this double mutant (W63G E14C) does not confer resistance to erythromycin, and the finding shows that Glu14 is essential also for erythromycin resistance. We conclude that functional EmrE is essential for the resistance that is most likely achieved by active removal of the antibiotic from the cytoplasm.
Mutation at Position 63 Inactivates the Previously Known Functions of EmrE.
Trp63 is completely conserved in the SMR family, and we previously showed that it is essential for EmrE function (7). However, because we did not previously use a Gly replacement, we investigated the W63G mutant. A quick estimate of the protein function is based on its ability to allow growth under otherwise nonpermissive conditions. Thus, E. coli cells expressing wild-type EmrE or any of the other mutations obtained in the original mutant (I68N or Q81L) grow in the presence of acriflavine or ethidium (Fig. S4A). However, and as expected from a replacement of an essential residue, cells expressing W63G are unable to grow under these conditions (Fig. S4A). Furthermore, binding of the high-affinity substrate tetraphenyl phosphonium (TPP+) to the purified W63G protein (Fig. S4B) and transport of MV (Fig. S4C) in reconstituted proteoliposomes were completely abolished as well.
We conclude that replacement of the essential bulky Trp with the smallest amino acid completely abolishes the well-characterized function of EmrE and generates a protein with novel specificities.
Deleterious Effect of the W63G Mutation on Growth.
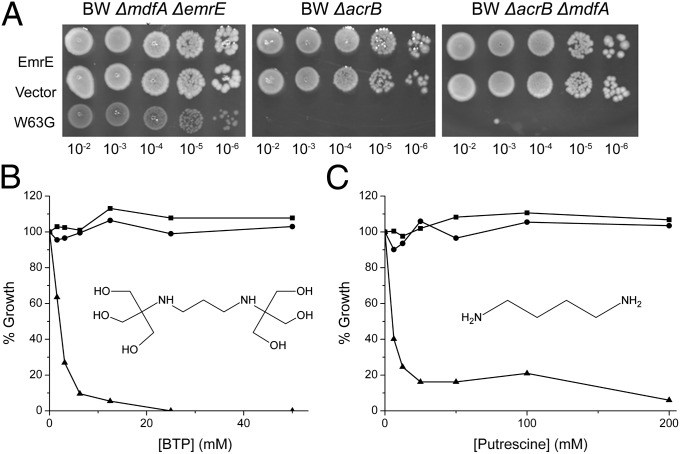
The resistance to erythromycin conferred by the W63G mutant is distinct but weak (Fig. 1B). To provide a more quantitative estimate of the resistance, we determined the IC50 for inhibition of growth in liquid. The IC50s significantly increased from 41 ± 3.5 and 49 ± 2.5 μM for strains bearing EmrE or just the pSN1 plasmid, respectively, to 63 ± 2.5 and 76 ± 2.8 μM for W63G and the triple mutant, respectively. We also screened for function in strains devoid of AcrB, the major multidrug transporter in E. coli that should have lower basal resistance to erythromycin as predicted by previous work (11, 12). Previously, while testing the phenotype of EmrE W63G in BW25113 ∆emrE∆mdfA cells, we observed that these cells grew quite poorly even on the control LB–agar plates (Fig. 1B and Fig. S3, Left). When we tested the resistance to erythromycin in two other BW25113 knockout strains, ΔacrB and ΔacrB ΔemrE, we found that cells bearing the W63G mutation were unable to grow even on the control plates containing no toxins, compared with EmrE and the empty vector (Fig. 2A, Right and Center). These results raised the question of what could cause this growth inhibition of cells expressing the EmrE W63G mutant.
Fig. 2.
Growth of E. coli BW25113 ΔacrB cells expressing the mutant W63G is inhibited by BTP and putrescine. (A) Different strains of E. coli BW25113 cells were transformed with pSN1 (vector), pSN1-EmrE, or pSN1-W63G, and growth was assayed and analyzed as in Fig. 1A. (B) E. coli BW25113 ΔacrB cells transformed with the plasmid pSN1-EmrE (circle), pSN1 (square), or pSN1 with the EmrE W63G mutant (triangle) were grown overnight at 37 °C in LB medium containing chloramphenicol. Cultures were then diluted into LB medium containing 200 μM IPTG and increasing concentrations of BTP (pH 7.0). Growth was analyzed after 6 h incubation at 37 °C. (C) The same experiment conducted for B was repeated, using putrescine instead of BTP. The molecular structure of both BTP and putrescine are shown inside each of their respective graphs. The pKa values of BTP are 6.8 and 9.0 and of putrescine are 9.2 and 10.65.
An early assumption was that this phenomenon could be a result of the overexpression through the induction by isopropyl β-d-1-thiogalactopyranoside (IPTG). This was ruled out because BW25113 ΔacrB cells transformed with the different mutants showed the same growth pattern with and without IPTG (Fig. S5). Interestingly, under all conditions, cells transformed with constructs containing the W63G mutation with different combinations of the other two mutations present in the original screen mutant (I68N and Q81L) show no growth impairment (Fig. S5). This finding suggests that the I68N and Q81L mutations suppress the weak growth of mutants bearing solely the W63G mutation and are, most likely, the reason why the two additional I68N and Q81L mutations were present in the mutant found in the screen: although not involved in the erythromycin resistance, they allowed for a more robust growth than would be achieved with the single W63G mutation.
Because the phenotype of EmrE is sensitive to pH, the solid media used to test growth is buffered with Bis-Tris-Propane (BTP), a buffer that can be used over a broad range of pH values. We previously showed that this buffer is innocuous to many E. coli strains bearing a large variety of plasmids and mutants tested in our laboratory (13). However, because growth was normal in plates with no BTP, we tested growth of BW25113 ΔacrB cells transformed with a plasmid bearing the W63G mutation. Strikingly, growth was practically fully inhibited when tested in liquid media already at 5 mM BTP (Fig. 2B). BTP had no effect even at 50 mM when growth was monitored in cells carrying vector or wild-type EmrE (Fig. 2B). The inhibition is not due to a general effect of buffers because neither the closely related Tris buffer nor 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), another “good buffer,” had a significant effect on growth even at concentrations of up to 50 mM.
In the BTP molecule, a short aliphatic chain connects two tertiary amines (Fig. 2B). Because the growth inhibition does not occur in the presence of Tris, the building block of BTP with a single amine, we looked into similar molecules with two secondary or tertiary amines held together by a short aliphatic chain. Our attention focused on the polyamine putrescine. Concentrations as low as 6 mM putrescine reduced growth to about 40%, and growth was practically fully inhibited at 25 mM (Fig. 2C). Putrescine had no effect on growth of cells expressing EmrE or mock-transformed with the vector (Fig. 2C). This suggests that it is the polyamine chain in the BTP molecule that results in the growth inhibition observed in cells expressing the EmrE W63G mutant.
EmrE W63G Is a Putrescine Transporter.
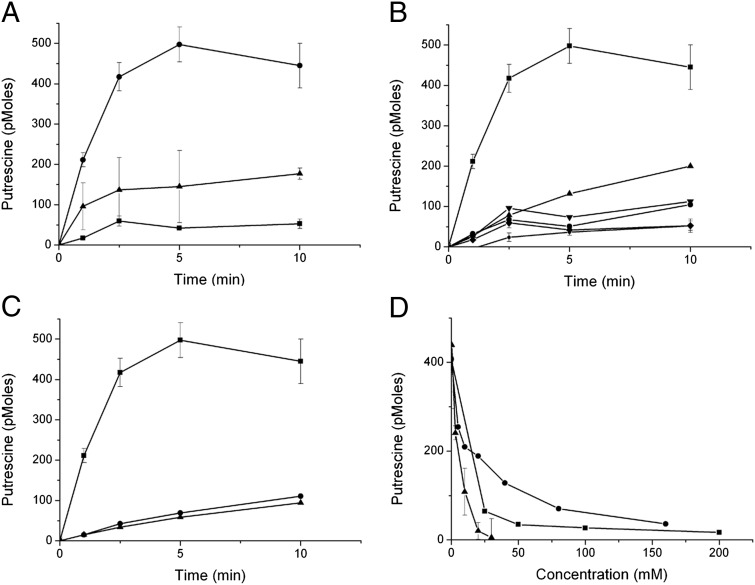
In an attempt to understand how putrescine can cause such an effect on the growth of bacteria expressing EmrE W63G, uptake of radiolabeled putrescine by whole cells was measured. For these experiments, BW25113 ΔpuuP cells were used. In these cells, the chromosomal puuP gene, coding for a putrescine transporter, was inactivated. The cells were grown in minimal medium, and expression was induced before the transport assay by the addition of IPTG. A time-dependent increase in putrescine uptake was observed in cells expressing the EmrE W63G mutant but not wild-type EmrE (Fig. 3A). Uptake was inhibited by preincubation with the uncoupler CCCP. No significant uptake was observed when cells transformed with either wild-type EmrE or with the vector alone were similarly assayed. These findings indicate that the EmrE W63G mutant facilitates energy-dependent accumulation of putrescine in cells, and this accumulation is, most likely, the cause of the observed growth inhibition.
Fig. 3.
[14C]Putrescine is accumulated in cells expressing EmrE W63G. (A–C) E. coli BW25113 ΔpuuP cells transformed with the plasmid pSN1 and the indicated constructs were assayed as described in Materials and Methods. A 30-min incubation with the uncoupler CCCP (50 μM) was used to disrupt the membrane potential. (A) Circles: EmrE-W63G; squares: pSN1; triangles: EmrE-W63G + CCCP. (B) Squares: EmrE-W63G; circles: EmrE; diamonds: pSN1; triangles: EmrE-W63G I68N; inverted triangles: EmrE-W63G Q81L; small circles: EmrE-W63G I68N Q81L. (C) Squares: EmrE-W63G; triangles: EmrE-W63A; circles: EmrE-W63G E14C. (D) Putrescine uptake is inhibited by BTP, spermine, and spermidine. The same putrescine uptake assay as described above was used with BW25113 ΔpuuP cells transformed with pSN1-EmrE W63G. Putrescine uptake was conducted for 2 min using increasing concentrations of BTP (triangles), spermine (circles), and spermidine (squares).
On the other hand and as expected from the finding that it does not confer sensitivity to BTP (Fig. 1B, Left, and Fig. S5), the original triple mutant (W63G I68N Q81L) does not catalyze putrescine accumulation (Fig. 3B). When analyzed separately, we found that the mutation Q81L by itself has only a weak suppressor effect on the putrescine transport activity of the W63G mutant. However, I68N practically completely abolishes transport (Fig. 3B).
A replacement with Ala at position 63 did not confer sensitivity to BTP (Fig. S3), and indeed the W63A mutant does not mediate uptake of putrescine (Fig. 3C). This result suggests that the complete removal of Trp at this position is essential for acquiring the new transport mode.
The W63G E14C mutant is also unable to catalyze putrescine accumulation (Fig. 3C), as expected from its failure to convey sensitivity to BTP (Fig. S3). This result supports the contention that the negative charge at position 14 is critical not only for the exchange activity of EmrE but also for the uptake of the polyamine molecules.
To learn about the specificity of the putrescine transport activity, we tested inhibition by other polyamines. BTP, spermine, and spermidine inhibit putrescine accumulation with relatively similar potency with 50% inhibition in the low millimolar range (Fig. 3D).
To characterize the kinetic properties of this newly evolved transport activity, the apparent Km was determined by measuring initial transport rates at various concentrations of putrescine and found to be 790 ± 46 μM. The Vmax was 940 ± 270 pmoles/min/3 × 106 cells.
EmrE W63G Supports Growth on Putrescine.
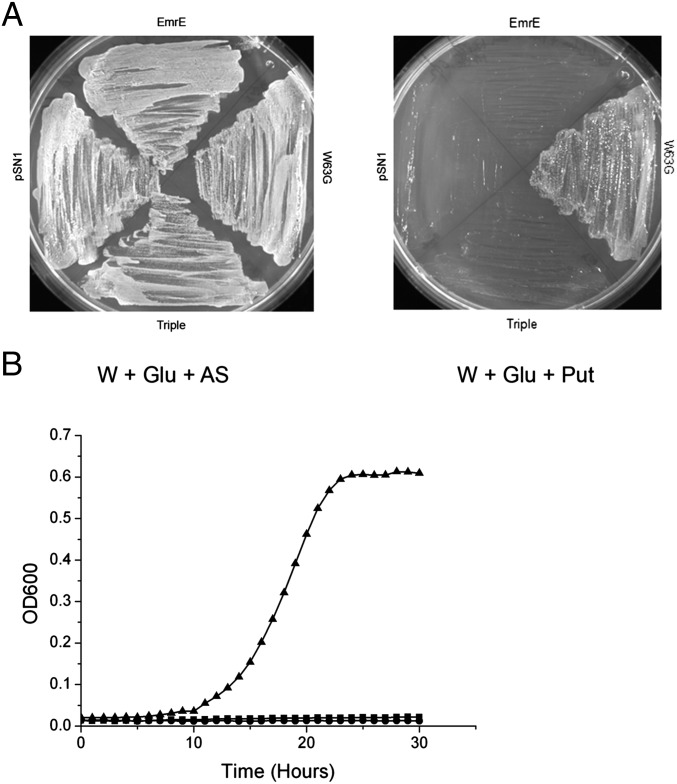
We assume that the inhibition of growth is due to the fact that, in rich medium, the putrescine utilization pathway is repressed (14) and the putrescine transported by the W63G mutant accumulates to high concentrations. However, putrescine can also be used as a carbon and nitrogen source by E. coli under conditions at which the putrescine utilization system is derepressed (such as minimal medium) (15). In addition to the intracellular enzymes required, the metabolism of putrescine needs the product of the PuuP gene, a putative putrescine transporter (15). ΔpuuP cells transformed with either EmrE, W63G, the triple mutant, or with the pSN1 plasmid with no insert were capable of growing in a medium supplied with glucose as carbon and energy source and ammonium as a nitrogen source (Fig. 4A, Left). In contrast, only cells transformed with W63G were able to grow with putrescine as the sole nitrogen source (Fig. 4 A, Right, and B). Transport of putrescine mediated by W63G supports growth under conditions where the putrescine utilization pathway is induced. The same transporter inhibits growth when the metabolism is repressed.
Fig. 4.
EmrE-W63G is capable of supporting growth of BW25113 ΔpuuP when putrescine is the sole nitrogen source. (A) BW25113 ΔpuuP E. coli cells transformed with pSN1, pSN1-EmrE, pSN1-EmrE W63G, or pSN1-EmrE W63G I68N Q81L (triple) were streaked onto minimal medium plates containing W-salts, glucose (as the carbon source), and (NH4)2SO4 (W + Glu + AS) or putrescine (W + Glu + Put) as the sole nitrogen source. Growth was analyzed after a 48-h incubation at 37 °C. (B) Growth was conducted in minimal media with putrescine as the sole nitrogen source, and OD600 readings were taken every hour for 30 h. Triangles: EmrE W63G; circles: EmrE; squares: pSN1 (vector).
Transport Activity of Purified EmrE W63G Reconstituted in Proteoliposomes.
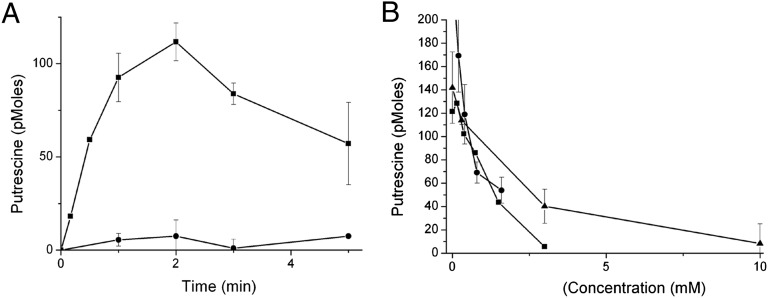
The experiments described above strongly support the notion that the uptake into the cell is energy-dependent and mediated by the W63G mutant. To directly support this contention, the W63G mutant was purified and reconstituted into proteoliposomes. To provide a driving force, we generated a membrane potential by dilution of K+-loaded proteoliposomes into a medium with no potassium in the presence of valinomycin, a K+-specific ionophore. Upon imposition of the potential, W63G proteoliposomes, but not EmrE ones, accumulated putrescine in a time-dependent manner (Fig. 5A). Accumulation was inhibited by agmatine, BTP, and erythromycin (Fig. 5B). In the absence of valinomycin, no significant uptake was observed.
Fig. 5.
EmrE-W63G but not EmrE is capable of membrane potential driven [3H]putrescine uptake when reconstituted into proteoliposomes. Proteoliposomes reconstituted with purified EmrE or EmrE-W63G loaded with 150 mM KCl and 20 mM K–Hepes (pH 7.5) were diluted into a medium containing 150 mM NaCl, 20 mM Na–Hepes (pH 7.5), 100 μM [3H]putrescine, and 10 μM of the potassium ionophore valinomycin (to generate the membrane potential). Transport was then measured over time as described in Materials and Methods. (A) Squares: EmrE-W63G; circles: EmrE B. The same transport reaction was carried out with EmrE-W63G proteoliposomes for 2 min as mentioned above but also including increasing concentrations of agmatine (circles), BTP (squares), or erythromycin (triangles).
Discussion
The original aim of our work was to explore the evolvability of transporters to acquire novel resistances. We used EmrE as an experimental paradigm and subjected it to directed evolution and selection in clinically relevant antibiotics. To our surprise, a single mutation, L47F, was sufficient to confer a very robust resistance to at least two fluoroquinolones, and another single mutation, W63G, conferred a weak but distinct resistance to erythromycin and other macrolides. This high evolvability provides a distinct advantage to the organism but poses a serious challenge to the treatment of infectious diseases by the resistant organism. Drug resistance provided by a transporter is energetically expensive because it requires continuous energy input to remove a drug that will eventually leak back into the cell. However, it provides the first barrier and provides the organism an essential time window needed to accumulate additional less energetically “expensive” mutations.
The norfloxacin resistance conferred by the L47F mutation was even stronger than that reported for SsmE, an SMR protein from S. marcensces that was reported to confer resistance (9). To our knowledge, norfloxacin is the first zwitterionic substrate described for the SMRs. All of the classical well-characterized substrates are cationic, and most of them are aromatic. The ability of this mutant to confer resistance to acriflavine is moderately reduced, whereas its ability to confer resistance to ethidium is not affected. The EmrE L47F mutant, however, loses its ability to confer resistance to MV. The finding that this position, located in TM2, affects the protein’s ability to confer resistance to MV complements previous studies that located residues in TM2 that influence MV resistance (10). The leucine at position 47 is highly conserved in the family, and, when the substitution to phenylalanine was introduced to the homolog BPsmr from Bordetella pertussis, the resulting mutant conferred resistance to norfloxacin, and the ability to recognize the “classical” substrates was not affected by the mutation (Fig. S6). On the other hand, the corresponding mutation in TBsmr, from Mycobacterium tuberculosis, did not confer resistance to norfloxacin and lost its ability to confer resistance to all three other substrates (Fig. S6). These results show that the same mutation in different homologs can result in resistance to a new, chemically different substrate, with varied effects on the resistance to common substrates of the wild-type proteins. In a bioinformatic analysis, several homologs with phenylalanine at this position, for example, a homolog from Bacillus anthracis, were found and further study of these homologs may be valuable.
In the erythromycin screen, a very interesting mutant—EmrE W63G—was found to confer resistance to this antibiotic. This was a very surprising result because tryptophan at position 63 is fully conserved in the family and is essential for the protein’s activity (7). Indeed, it was observed that the EmrE W63G mutant loses its ability to confer resistance to acriflavine and ethidium; it does not bind the high-affinity substrate TPP+ and does not transport MV2+. We speculate that the erythromycin resistance is possible as a result of the large aromatic tryptophan being substituted by the small glycine, thus allowing the movement of the bulky erythromycin molecule, which is much larger than the common EmrE substrates. Moreover, this gain-of-new-function mutation appears to be specific to glycine in this position because alanine with only an additional single methyl in the EmrE W63A mutant eliminates the erythromycin resistance.
An exciting and unexpected feature of the EmrE W63G mutant was revealed by complete serendipity. The original mutant had three mutations, and it grew very well in the host cell. When we separated the mutations, it became evident that E. coli BW25113 ΔemrEΔmdfA cells expressing the single W63G mutant grew quite poorly. This phenomenon is accentuated in BW25113 knockout strains lacking the acrB gene, where EmrE W63G-expressing cells are completely unable to grow on the control plates. This finding suggests that the W63G mutation was found in the screen due to the presence of these two other suppressor mutations. It is not uncommon that overexpression of membrane proteins impairs growth, but in this case we showed, as described in Results, that the presence of the BTP buffer prevents the cells from growing. Because an aliphatic chain in the BTP molecule connects two amines, we tested the effect of similar molecules. We found that putrescine results in a similar dramatic growth inhibition of cells expressing EmrE W63G and showed that this inhibition is due to accumulation of putrescine inside the cells, observed in an assay of putrescine uptake by whole cells and in proteoliposomes reconstituted with purified protein. We also confirmed the transporter function as it complements growth when putrescine is the sole nitrogen source in a strain where a chromosomal putrescine transporter was inactivated and under conditions where the putrescine utilization pathway is derepressed.
The significance of this finding is that a single mutation converts a well-behaved drug-H+ antiporter into a protein that can catalyze a different function: putrescine import. Is this a “broken” transporter that allows movement of some substrates driven by a membrane potential? The W63G mutant is not a nonspecific channel because it is not toxic to the cell unless polyamines are present in the growth medium. Transport of putrescine is saturable and can be competed with by other substrates such as BTP and other polyamines. The specificity is dramatically changed because the W63G mutant cannot perform any of the previously known functions: it cannot confer resistance to ethidium or acriflavine, it does not bind the high-affinity ligand TPP+, and it does not transport MV2+ in proteoliposomes. We speculate that Trp63 serves a dual role as a hydrophobic gate: it provides a binding determinant for aromatic cations by π-cation or stacking interactions (16) and, when a substrate binds, it induces the change in conformation needed for translocating the substrate to the other side of the membrane. When Trp63 is completely removed (replaced with Gly), the gate is removed together with a major binding determinant. We cannot at the moment conclude whether putrescine uptake is coupled to proton movement or simply a uniport mechanism. This is due to the fact that putrescine is a weak base and the protonated form can permeate the membrane passively. Determination of the coupling mode and the stoichiometry requires further investigation, but the finding that the essential Glu14 is neccesary for transport suggests that it may also supply the protons for a symport process.
A mutation phenomenologically similar was previously observed in Bacteriorhodopsin where replacement of a highly conserved and essential aspartate (Asp85) with threonine completely abolished the proton extrusion activity but generated a protein capable of pumping Cl− ions in the opposite direction (17). In other cases, the ability of the Cl−/H+ antiporter was modified so that the coupling to proton movement is abolished but the mutant generated can still translocate Cl− ions; i.e., the specificity was retained (18). The W63G mutation is unique because the transporter can function as an antiporter with erythromycin (as judged from its ability to confer resistance) and as a uniporter/symporter when putrescine or BTP are the substrates. Further study of this phenomenon should provide important clues for understanding the basis of the coupling mechanism and to further document a unique case where the mode of coupling is dictated by the substrate–protein interaction and not only by the native mechanism of the protein.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli BW25113 strain was used throughout this work (Table S1). BW25113 ΔemrEΔmdfA, ΔacrB, and ΔpuuP individual knockouts and combinations were prepared according to Datsenko and Wanner (19) and described by Tal and Schuldiner (4).
The plasmid used for gene expression is pSN1, a derivative of pACYC184 (20). Tagged EmrE is fused at the C terminus to a Myc epitope followed by His6 residues (21). For simplicity, this construct is named EmrE throughout the article.
Growth Conditions.
Cells were grown at 37 °C with shaking in LB or minimal medium A supplemented with 0.5% (wt/vol) glucose (22). The solid medium LB–agar has the same composition but with additional 1.5% (wt/vol) agar. When needed, chloramphenicol (34 μg/mL) was added to the medium.
Resistance to Toxic Compounds on Solid Medium.
For testing resistance on solid medium, E. coli cells were transformed with the indicated plasmid and were grown overnight at 37 °C in LB medium with the corresponding antibiotic. The cultures were diluted to A600 = 0.5, and 5 μL of serial dilutions of the culture was spotted onto LB plates containing 30 mM BTP, pH 7.0, with or without the addition of the indicated concentrations of the antibiotics or toxic compounds. IPTG (200 μM) was added for induction of expression. Growth was visualized after overnight incubation at 37 °C. Ethidium bromide, acriflavine, MV, and erythromycin were obtained from commercial sources. All of the experiments were repeated at least twice.
Resistance to Toxic Compounds in Liquid Medium.
For resistance in liquid medium, overnight cultures were diluted 100-fold into LB medium, grown to early logarithmic phase, and diluted to A600 = 0.01 in LB medium containing 30 mM BTP, pH 7.0 (or another mentioned buffer) and the indicated concentrations of toxic compounds in a 96-well plate (Nunc) at 37 °C with constant shaking. OD600 readings were taken every hour for 48 h using a Synergy 2 Microplate Reader (BioTek). The experiment was carried out in triplicate and repeated twice. For growth in minimal medium, overnight LB cultures were washed with 1× W-salts (23) and resuspended in minimal medium [which contained 1× W-salts, 2% (wt/vol) glucose, 2.5 μg/mL thiamine, 34 μg/mL chloramphenicol, 200 μM IPTG, and 0.5% putrescine] to a final OD of 0.01, and growth was followed as described above.
Expression and Membrane Preparation.
E. coli BW25113 cells carrying the plasmid pSN1 containing the constructs of interest were grown at 37 °C in minimal medium A supplemented with 0.5% (wt/vol) glucose and 34 μg/mL chloramphenicol. When the culture reached an A600 of 1.0, IPTG was added to a final concentration of 2 mM to induce protein production. The cultures were incubated at 37 °C with continuous shaking for 2 h. The cells were then harvested by centrifugation and washed with buffer containing 150 mM NaCl, 15 mM Tris⋅HCl, pH 7.5 (Na buffer), and 250 mM sucrose (Na–sucrose buffer). Cells were then resuspended in Na–sucrose buffer containing 14.7 mM β-mercaptoethanol, 2.5 mM MgSO4, 15 μg/L culture DNase, and 1 mM phenylmethanesulfonyl fluoride and broken using a French press or a LV1 microfluidizer (Microfluidics) at 16,000 psi. Unbroken cells were sedimented by centrifugation, and the membrane fraction was collected by ultracentrifugation at 244,717 × g for 1.5 h at 4 °C and resuspended in Na–sucrose buffer. The membranes were rapidly frozen in liquid nitrogen and stored at −70 °C.
Reconstitution into Proteoliposomes.
Reconstitution was performed by solubilizing 1,200 μL of membranes (containing ∼4.5 μg His-tagged protein) in 2 mL of Na buffer containing 1.5% n-dodecyl β-maltoside (Glycon GmbH), 0.5 mM PMSF, and 15 mM β-mercaptoethanol. After removal of unsolubilized material by centrifugation (208,000 × g for 30 min), imidazole was added to 20 mM, and the His-tagged protein was incubated with Ni2+-nitrilotriacetic acid beads (Qiagen) for 1 h at 4 °C. The beads were washed with at least 2 mL of Na buffer containing 1% n-octyl β-d-glucopyranoside (OG) (Glycon GmbH), 30 mM imidazole, and 15 mM β-mercaptoethanol. The protein was eluted with 500 μL of the same buffer containing 200 mM imidazole and mixed with 375 μL of 10 mg E. coli polar lipid extract (Avanti) in Na buffer and 1.2% OG. Eluted protein and phospholipids were sonicated together in a bath-type sonicator to clarity and diluted in buffer containing 1 mM DTT and either 0.19 M NH4Cl, 15 mM Tris⋅HCl, pH 7.5 (for the generation of a pH gradient in the assay), or 150 mM KCl with 20 mM K–Hepes, pH 7.5 (for the generation of a membrane diffusion potential). After 20 min at room temperature, samples were centrifuged at 208,100 × g for 70 min, and the pellet was resuspended in 100 μL of the corresponding buffer, frozen in liquid air, and stored at −70 °C. Before the transport assay, the proteoliposomes were thawed at room temperature and sonicated lightly to clarity.
Generation of Random Mutagenesis Libraries and Screening.
The GeneMorph II Random Mutagenesis Kit (Stratagene) was used to create a library of mutants. A solution containing Mutazyme II—a blend of two error-prone DNA polymerases—and its appropriate buffer and 200 μM dNTPs was prepared. To obtain mutations in the entire gene, external primers at 20 μM each were used. The product of the PCR was then used as a “megaprimer” to insert the library of mutants into an expression vector. This was carried out as described for the Quick Change mutagenesis protocol (Stratagene).
This library was then transformed into competent DH5α cells and plated, and the transformants were collected and used to prepare plasmid DNA. E. coli BW25113 ∆emrE∆mdfA cells were transformed with the above library, and about 4 × 105 cells were plated on LB plates containing either norfloxacin (0.2 μg/mL) or erythromycin (300 μM), concentrations that are not permissive for cells bearing empty plasmid or wild-type EmrE. A total of 10–20 transformations were carried out in parallel to obtain a sufficient number of mutants, but we usually were able to isolate at least one from each individual plate.
Uptake of [14C]Putrescine in Whole Cells.
E. coli BW25113 ΔpuuP cells were grown overnight at 37 °C in minimal medium A, pH 7.0, supplemented with 0.1% glucose and 34 μg/mL chloramphenicol. Cultures were then diluted to A600 = 0.1 in fresh minimal medium A with the aforementioned supplements and grown at 37 °C until they reached A600 = 0.4. IPTG (2 mM) was then added for induction, and the cultures continued to grow for additional 1.5 h. The cells were then harvested at 3,220 × g for 10 min and resuspended in wash buffer [100 mM potassium phosphate (pH 7.5), 10 mM MgSO4, and 0.5% glucose]. After thorough washing, the cells were resuspended to a final OD of 12 in the wash buffer, and 25 μL resuspended cells were put into a glass reaction tube and kept at 37 °C. The transport reaction was initiated by the addition of 25 μL 206 μM [14C]putrescine (specific activity 4 Ci/mole). This reaction was terminated at the indicated time intervals by the addition of 2 mL of wash buffer followed by immediately passing the cells through a Whatman GF/C filter. The air in the reaction tubes was supplemented with extra oxygen throughout the experiment. Radioactivity in the samples was measured by liquid scintillation counting. Experiments were carried out in duplicates and repeated at least twice.
Uptake of [3H]Putrescine in Proteoliposomes.
Uptake of [3H]putrescine into proteoliposomes was assayed at 25 °C by dilution of 2 μL of potassium chloride-containing proteoliposomes into 198 μL of a potassium-free solution containing 100 μM [3H]putrescine (16.25 Ci/mol; Perkin-Elmer), 150 mM NaCl, and 20 mM Na–Hepes, pH 7.5. A membrane potential was generated by the addition of 10 μM valinomycin, a potassium ionophore. The reaction was stopped at the indicated time intervals by dilution with 2 mL of the aforementioned ice-cold solution and by passing the sample through a GSWP filter (0.22 μm; Millipore). The reaction tubes were then washed with an additional 2 mL of solution. The radioactivity on the filters was estimated by liquid scintillation counting. Experiments were carried out in duplicates and repeated at least twice.
Supplementary Material
Acknowledgments
S.S. is Mathilda Marks-Kennedy Professor of Biochemistry at the Hebrew University of Jerusalem. Work in our laboratory is supported by National Institutes of Health Grant NS16708 and by Israel Science Foundation Grant 11/08.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211831109/-/DCSupplemental.
References
- 1.Nathan C. 2012. Fresh approaches to anti-infective therapies. Sci Transl Med 4:140sr2.
- 2.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Schuldiner S. Undecided membrane proteins insert in random topologies. Up, down and sideways: It does not really matter. Trends Biochem Sci. 2012;37:215–219. doi: 10.1016/j.tibs.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbaz Y, Tayer N, Steinfels E, Steiner-Mordoch S, Schuldiner S. Substrate-induced tryptophan fluorescence changes in EmrE, the smallest ion-coupled multidrug transporter. Biochemistry. 2005;44:7369–7377. doi: 10.1021/bi050356t. [DOI] [PubMed] [Google Scholar]
- 8.Nasie I, Steiner-Mordoch S, Gold A, Schuldiner S. Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters. J Biol Chem. 2010;285:15234–15244. doi: 10.1074/jbc.M110.108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minato Y, Shahcheraghi F, Ogawa W, Kuroda T, Tsuchiya T. Functional gene cloning and characterization of the SsmE multidrug efflux pump from Serratia marcescens. Biol Pharm Bull. 2008;31:516–519. doi: 10.1248/bpb.31.516. [DOI] [PubMed] [Google Scholar]
- 10.Mordoch SS, Granot D, Lebendiker M, Schuldiner S. Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J Biol Chem. 1999;274:19480–19486. doi: 10.1074/jbc.274.27.19480. [DOI] [PubMed] [Google Scholar]
- 11.Ma D, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harel-Bronstein M, et al. MH1, a second-site revertant of an Escherichia coli mutant lacking Na+/H+ antiporters (delta nhaA delta nhaB), regains Na+ resistance and a capacity to excrete Na+ in a delta microH(+)-independent fashion. J Biol Chem. 1995;270:3816–3822. doi: 10.1074/jbc.270.8.3816. [DOI] [PubMed] [Google Scholar]
- 14.Kurihara S, Oda S, Kumagai H, Suzuki H. Gamma-glutamyl-gamma-aminobutyrate hydrolase in the putrescine utilization pathway of Escherichia coli K-12. FEMS Microbiol Lett. 2006;256:318–323. doi: 10.1111/j.1574-6968.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- 15.Kurihara S, et al. The putrescine importer PuuP of Escherichia coli K-12. J Bacteriol. 2009;191:2776–2782. doi: 10.1128/JB.01314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S. The fast release of sticky protons: Kinetics of substrate binding and proton release in a multidrug transporter. Proc Natl Acad Sci USA. 2007;104:17989–17994. doi: 10.1073/pnas.0704425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki J, et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 18.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 19.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yerushalmi H, Lebendiker M, Schuldiner S. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 1996;271:31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 21.Muth TR, Schuldiner S. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 2000;19:234–240. doi: 10.1093/emboj/19.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis BD, Mingioli ES. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurihara S, et al. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J Biol Chem. 2008;283:19981–19990. doi: 10.1074/jbc.M800133200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.