Abstract
The six-layered neocortex is a uniquely mammalian structure with evolutionary origins that remain in dispute. One long-standing hypothesis, based on similarities in neuronal connectivity, proposes that homologs of the layer 4 input and layer 5 output neurons of neocortex are present in the avian forebrain, where they contribute to specific nuclei rather than to layers. We devised a molecular test of this hypothesis based on layer-specific gene expression that is shared across rodent and carnivore neocortex. Our findings establish that the layer 4 input and the layer 5 output cell types are conserved across the amniotes, but are organized into very different architectures, forming nuclei in birds, cortical areas in reptiles, and cortical layers in mammals.
Keywords: development, telencephalon, vertebrate evolution
The evolutionary origins of the mammalian neocortex have been the subject of debate for over a century (1–3). Although the six-layered neocortex is remarkably well conserved in all extant mammals, it is not present in our closest living relatives, the reptiles and birds. Birds in particular are an extremely successful radiation with large brains, but the bird dorsal telencephalon does not include a morphologically identifiable cortex; it is, instead, a collection of nuclei (4).
Studies of the neurochemistry, anatomy, and physiology of the central region of the bird telencephalon, which is called the dorsal ventricular ridge (DVR), have shown conclusively that the DVR is a pallial structure (2). In mammals the pallium (dorsal telencephalon) includes the neocortex and the nuclei of the piriform lobe, specifically the claustrum and parts of the amygdala. A classical hypothesis that has attracted renewed interest from modern neuroembryologists and neuromorphologists proposes a field homology between the nuclei of the bird DVR and the mammalian piriform lobe nuclei, including the amygdala (5, 6). A second, more controversial hypothesis proposes that, despite the gross differences in morphology between bird DVR and mammalian neocortex, these structures share a homology (7–9).
The best evidence for neocortex-DVR homology lies in their circuitries. Both the mammalian neocortex and avian DVR receive ascending sensory input from the thalamus, and both send outputs to brainstem premotor areas. These input and output territories form specific, well-defined populations of neurons in both mammals and birds. In mammals, layer 4 neocortical neurons receive thalamic input, and layer 5 neocortical neurons project to the brainstem. In birds there are separate DVR nuclei that receive projections from the thalamus or send axons to the brainstem. The most developed version of the neocortex-DVR hypothesis proposes homology between these input and output neurons at the cell-type level (7); specifically, that the layer 4 input (L4/I) and layer 5 output (L5/O) neurons of the neocortex share a common ancestry with neurons that populate the input and output nuclei of the DVR.
These two prevailing hypotheses about DVR homology are in direct conflict; one is a field homology argument comparing the DVR with piriform lobe nuclei based on morphology and regional position, and the other is a cell-type homology argument based on similarities in circuitry. Our approach has been to test the cell-type homology hypothesis directly by documenting additional traits of layer 4 and layer 5 neurons and looking for any conservation in neurons of specific DVR nuclei.
Studies over the past decade from a number of laboratories have identified mRNAs that are expressed in subsets of neocortical neurons (10). Some of these genes are enriched in neurons in specific layers, including layers 4 and 5. If the neocortical cell-type hypothesis is correct, a clear prediction is that some of the layer 4 and 5 cell type-specific molecular markers should be expressed in the thalamic input and the brainstem output nuclei of the DVR. For these experiments, we identified mammalian layer 4 and layer 5 markers by comparing mouse and ferret neocortical gene expression patterns. We then studied the expression of these marker genes in the telencephalon of two birds and one reptile (Fig. 1). Our findings fully affirm the cell-type homology hypothesis. Thus, the dissimilarities in pallial morphology seen across mammals, reptiles, and birds reside not in the core input and output cell types, but in the malleability of neuronal cellular morphology and in the structure and organization of neuronal assemblies.
Fig. 1.
Amniote phylogeny highlighting the taxa investigated. Species selected have been extensively studied in systems neuroscience: two mammals, the house mouse (Mus musculus) and the ferret (Mustela putorius furo); one reptile, the red-eared turtle (Trachemys scripta elegans); and two birds, the chicken (Gallus gallus) and the zebra finch (Taeniopygia guttata). Rodents and primates are in the mammalian superorder Euarchontoglires. Carnivores such as the ferret are in the superorder Laurasiatheria. Recent molecular timescale analysis dates the divergence of these superorders at ∼97 Mya (50). Game birds such as the chicken, and passerines such as the zebra finch, are also deeply separated, with the divergence between the Galloanserae clade and the Neoaves clade placed at ∼122 Mya (51). Nonavian reptiles are a paraphyletic group (green disk): there is no common ancestor for all reptiles that is not also an ancestor to birds (yellow node). Turtle phylogeny remains deeply controversial. Illustrated is the classical placement of turtles in a sister group to diapsids. This phylogeny retains some support (52). With this phylogeny there are three radiations of reptiles, each of which presents a dorsal cortex in extant taxa (31). There are prominent alternative phylogenies which conflict. Morphological analyses (53) place turtles within Diapsida as a sister group to squamates (lizards and snakes) and sphenodon (tuatara). Molecular analysis (54) also places turtles in Diapsida, but as a sister group of archosaurs (crocodiles and birds). In either of these alternative arrangements, reptiles remain a paraphyletic group with more than one radiation. Red node, last common ancestor of extant amniotes.
Results
Molecular Evidence for a Neocortical Cell-Type Homology Between Mammals and Birds.
Sensory inputs to the mammalian neocortex are relayed by the dorsal thalamus and terminate densely on neurons in neocortical layer 4 (Fig. 2A). The dorsal thalamus is also the major source of ascending sensory input to the bird DVR, which is targeted to specific nuclei. For example, visual information from the optic tectum is relayed by nucleus rotundus of the avian thalamus to neurons in the entopallium of the DVR (11) (Fig. 2D), and the thalamic auditory nucleus ovoidalis sends projections to neurons in Field L (8). Single-unit physiology has confirmed that visual stimuli activate neurons in the entopallium and that Field L neurons respond to sound (8, 12). In both neocortex and the DVR, there are excitatory interneurons that link the input neurons to output neurons projecting to brainstem. These output neurons are in layer 5 of the neocortex and in the arcopallium nuclear complex of the DVR (13) (Fig. 2 A and D). The claim of the neocortical-DVR homology hypothesis is that the principal input and output neurons of the mammalian neocortex and the avian DVR are homologous to each other at the level of neuronal cell type: the thalamorecipient neurons in neocortical layer 4 are homologous as input cell types to neurons in the avian entopallium and Field L, and brainstem projection neurons in neocortical layer 5 are homologous as output cell types to neurons in the avian arcopallium (9). This cell-type homology is based on one feature of these neurons: their fiber connections. If this proposed homology is correct, then other traits that distinguish layer 4 and layer 5 cell types should identify the proposed homologous cell types in the avian DVR. Here we use gene expression to test for conservation of molecular identity between the mammalian and avian input and output neurons.
Fig. 2.
Molecular test for homology at the level of cell type between mammalian neocortex and the avian DVR. (A) Core circuitry of mammalian neocortex. Input neurons in layer 4 (green) receive sensory information from the dorsal thalamus, and output neurons in layer 5 (red) project to the brainstem. (B) Two-color fluorescence ISH (FISH) demonstrating Eag2 (green) and Er81 (red) gene expression in layers 4 and 5 of mouse somatosensory cortex. (C) Rorb coexpression (yellow) with Eag2 (green) in layer 4 neurons. (D) Core circuitry of the avian DVR illustrated for the visual system. Input neurons in the entopallium (green) receive sensory information from the dorsal thalamus, and output neurons projecting to brainstem lie in the arcopallium (red). (E) Layer 4 marker gene EAG2 is enriched in the entopallium. Coronal cross-section of hatchling chicken brain processed for light microscopic ISH. (F) FISH demonstration of RORB coexpression with EAG2 in entopallial neurons. (G) Layer 4 marker RORB identifies the avian auditory input nucleus Field L. (H and I) Layer 5 markers are expressed in the arcopallium of the chicken (H; PCP4) and zebra finch (I; ER81). 1–6, layers of neocortex; A, arcopallium; Bst, brainstem; dTh, dorsal thalamus; E, entopallium; FL, Field L; M, mesopallium; N, nidopallium; St, striatum.
We identified from the literature, the Allen Brain Atlas (14) and the CORTx browser (15), three robust markers of mouse neocortical layer 4 neurons: Eag2/Kcnh5, a potassium ion channel gene (16, 17); Rorb/Nr1f2, a transcription factor gene (18); and Whrn/Dfnb31 (14), which encodes a cytoplasmic protein (Figs. S1 and S2). Mice, like primates, are in the mammalian superorder Euarchontoglires. To assess these genes as general markers of mammalian layer 4 neurons, we isolated cognate cDNAs from the ferret (19), which lies in the other large mammalian superorder, Laurasiatheria (20) (Fig. 1). In mouse neocortex, Eag2 and Rorb are mainly restricted to layer 4 (Fig. 2 B and Fig. S1). Rorb-expressing neurons form a subset of Eag2-expressing neurons, particularly outside primary sensory areas where Rorb is more weakly expressed (Fig. 2C). Both markers are also detected in far fewer neurons in layers 3 and 5. In the ferret, RORB and EAG2 are also layer 4 markers (19). Whrn is enriched in mouse layer 4, but in ferret neocortex WHRN is expressed prominently in layer 5 (Fig. S1) and was dropped from further analysis.
We tested the cell-type homology hypothesis in two distantly related avian species, the chicken (a game bird) and the zebra finch (a songbird) (21) (Fig. 1). In both species, we found that the orthologous layer 4 marker genes EAG2 and RORB are intensely expressed in most neurons in the entopallium and Field L, as predicted by the cell-type homology hypothesis (Fig. 2 E and G, and Fig. S3). Two-color fluorescence experiments in chicken established that RORB-expressing neurons in the DVR are a subset of the EAG2-expressing neurons, as they are in mouse neocortex (Fig. 2F and Fig. S4). One prominent feature of neocortical layers, which is particularly clear with the layer 4 marker Eag2, is that they form a continuous band across the neocortex (16), linking for example the primary sensory areas with each other. Although the entopallium and Field L are distinct nuclei, the marker EAG2 demonstrated that these cell populations are connected by EAG2-positive cell bridges (Fig. S3C).
We identified seven selective markers of L5/O neocortical neurons that are shared between mouse and ferret (Fig. 2B and Fig. S5): the transcription factor genes Er81/Etv1 and Fezf2/Znf312; the ion channel genes Cacna1h and Kcnn2/Sk2; the sulfatase gene Sulf2; and two genes of uncertain function, Pcp4 and Tmem200a/Kiaa1913 (14, 22–27). We found that the chicken orthologs of six of these layer 5 markers (ER81, FEZF2, CACNA1H, PCP4, SULF2, and TMEM200A) are strongly expressed in the chicken arcopallium, the source in the avian DVR of long-distance output projections (Fig. 2H and Fig. S5 A–F). KCNN2 expression patterns were inconclusive because this mRNA was detected only as small puncta throughout the adult chicken dorsal telencephalon (Fig. S5G). We studied one L5/O marker, ER81, in the zebra finch. The zebra finch arcopallium, including the robust nucleus (RA), a known source of projections to brainstem vocal motor neurons (28), intensely expressed ER81 (Fig. 2I). These findings, that eight confirmed neocortical layer 4 and 5 cell-type markers are enriched in the predicted avian DVR nuclei, provide strong molecular support for cell-type homology between the input and output neurons of the avian DVR and those of the neocortex.
Molecular Organization of the Wulst.
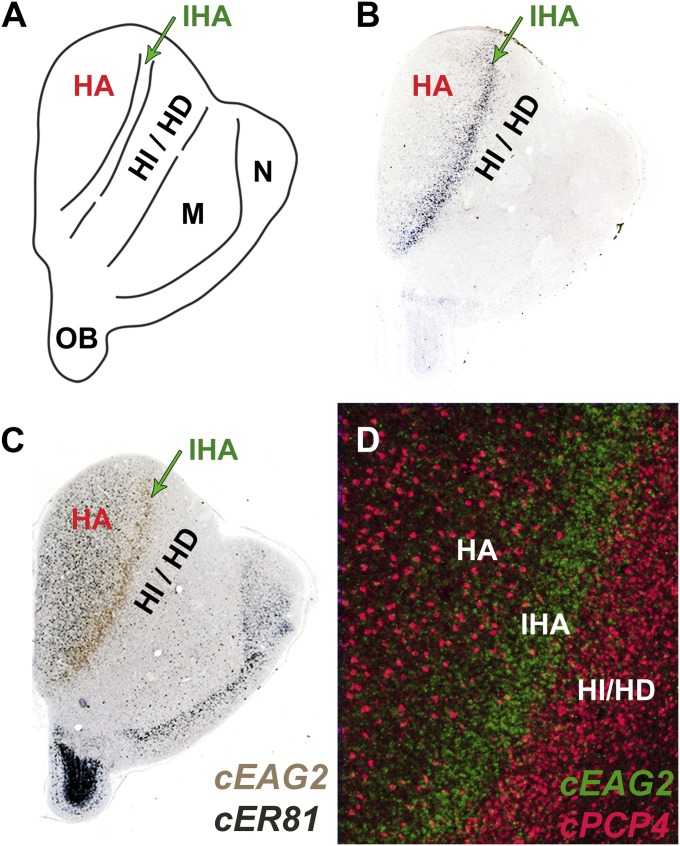
In the bird telencephalon dorsal to the DVR sits a collection of interconnected nuclei called the Wulst. The Wulst, like the DVR, has a transtelencephalic circuitry similar to that of the neocortex (Fig. 3A); it receives lemniscal thalamic input for vision and somatic sensation and is a source of projections to brainstem. As in the DVR, the input and output neurons are segregated into separate nuclei. The thalamic input to the Wulst terminates in a thin, radially oriented nucleus called the interstitial part of the hyperpallium apicale (IHA). Extratelencephalic output from the Wulst originates in the hyperpallium apicale (HA), which flanks the IHA dorsomedially (29, 30). The cell-type homology hypothesis strongly predicts that these Wulst input and output neurons will share the molecular profile of the L4/I and L5/O neurons of the DVR and the neocortex. This is what we found. EAG2 and RORB mRNAs are concentrated in the IHA, and the layer 5 markers ER81, FEZF2, PCP4, and SULF2 are intensely expressed in the HA (Fig. 3 and Fig. S6).
Fig. 3.
The Wulst has L4/I and L5/O cell types. (A) Diagram of a coronal cross-section through rostral chicken telencephalon illustrating the nuclear organization of the Wulst. The IHA (green) is the target of thalamic input and the HA (red) contains Wulst output neurons projecting to brainstem. (B) The L4/I marker EAG2 is expressed as a stripe, filling the IHA (green arrow). (C and D) The neurons of the output nucleus HA are enriched in the L5/O marker genes ER81 (C) and PCP4 (D). (C) Two-color ISH documents that the EAG2-dense IHA (brown) lies immediately lateral to the HA (dark purple). (D) High-power FISH shows that the IHA (EAG2, green) is precisely inserted between the large-celled HA and the more densely packed hyperpallium intercalatum (HI)/hyperpallium densocellulare (HD) territory (PCP4, red). M, mesopallium; N, nidopallium.
Turtle Cerebral Cortex Has a Layer 4 Cell Type.
Reptiles have both a DVR and a cerebral cortex. The reptile cerebral cortex is not a neocortex because it consists of three layers, with pyramidal cells densely packed only in layer 2 (31). The pyramidal neurons are excitatory and are known to receive inputs from the thalamus and to project to subcortical targets (32). These observations led to a completely different hypothesis about neocortical origins (3, 33, 34). This hypothesis holds that the three-layered cortex of reptiles is ancestral; that the reptilian cortical pyramidal neurons are homologous only to deep-layer neocortical projection neurons; and that the superficial layers of the neocortex, including the L4/I neurons, are a mammalian innovation. According to this hypothesis, layer 4-like neurons in avian pallium would represent convergent evolution. The alternative to this hypothesis, that L4/I cell types are conserved between birds and mammals, would require that reptiles have lost L4/I cell types. This “reptile loss of layer-4 cell types” is a challenging position to maintain because (nonavian) reptiles are a paraphyletic group with multiple branches (Fig. 1), thereby entailing multiple independent losses of L4/I cell types.
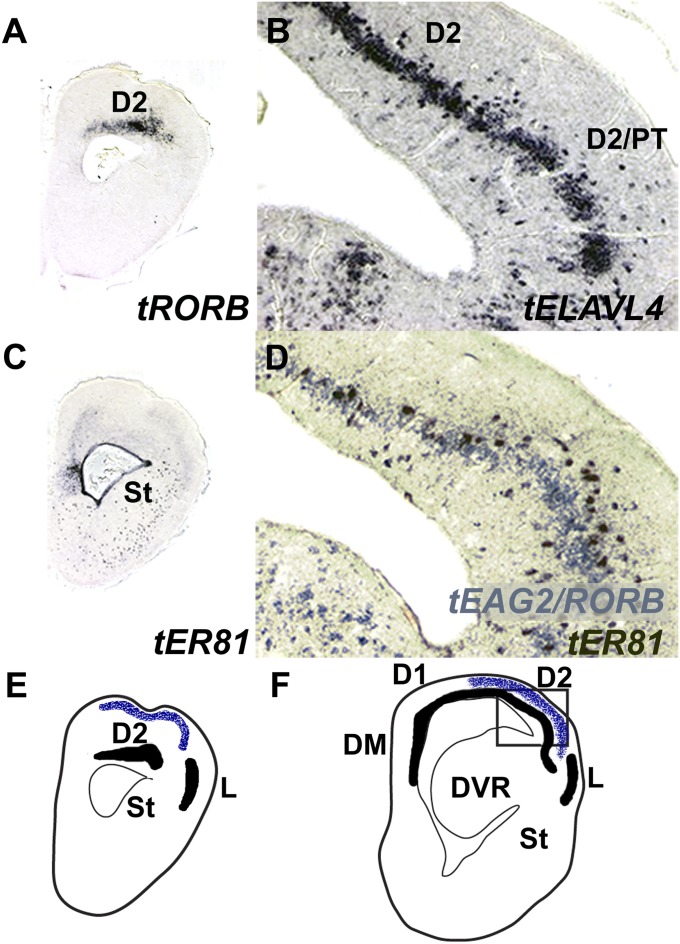
We explored a third possibility, that reptile cortex contains a molecularly distinct population of L4/I neurons. We investigated the expression patterns of the L4/I markers EAG2 and RORB and the L5/O marker ER81 in the well-studied red-eared turtle (32, 33). We found that EAG2 and RORB are expressed in layer 2 of turtle cortex, establishing the presence of the L4/I cell type in reptile cerebral cortex (Figs. 4 A and D and 5B). Both molecular markers are restricted to the anterior half of the cortex, with strong expression in cortical area D2 rostrally. Turtle dorsal cortex is dominated by visual sensory processing (32, 33). Desan (35) identified the cortical targets of ascending visual thalamic input with transneuronal transport of radiolabeled proline from the turtle retina. These projections terminated in outer layer 1 of rostral cortical area D2 (Fig. 4 E and F), closely matching the areal distributions of the EAG2/RORB-expressing cells. These observations establish that molecularly defined L4/I cell types are present in the cerebral cortex of at least one reptile, the turtle, where they populate a cortical territory that receives transthalamic sensory input.
Fig. 4.
L4/I cell-type markers are expressed in turtle dorsal cortex and fall within the target territory of transthalamic retinal input. (A and C) Serially adjoining coronal cross-sections through rostral turtle telencephalon processed for ISH. At this level, dorsal cortical area D2 is enriched in the L4/I marker RORB (A) and almost free of the L5/O marker ER81 (C). (B and D) Serially adjoining cross-sections taken from a more posterior level and photographed at high power (see box in F). (B) ISH for the pan-neuronal marker ELAVL4 illustrates the pyramidal neuron-dense layer 2 of dorsal cortex. (D) Two-color three-probe ISH demonstrates that the L4/I cell types (light blue) and the L5/O cell types (maroon) are distinct populations, but are intermixed within the pyramidal layer at this rostrocaudal level. (E and F) Chartings redrawn from Desan (35) to illustrate visual system input to turtle cerebral cortex at levels corresponding to the ISH cross-sections illustrated (E for A and C; F for B and D). The pyramidal layer is filled in black, and the axon terminals are charted in blue stippling. Transthalamic visual input targets the outer layer of rostral cortical area D2, where neurons expressing the L4/I marker genes are found. D1 and D2, medial and lateral halves of the dorsal cortical area (35); DM, dorsomedial cortical area; DVR, turtle dorsal ventricular ridge; L, lateral cortical area, which is the target of olfactory bulb input (35); PT, pallial thickening, which appears at the lateral edge of the dorsal cortical area and is placed by Desan within area D2 (35); St, striatum.
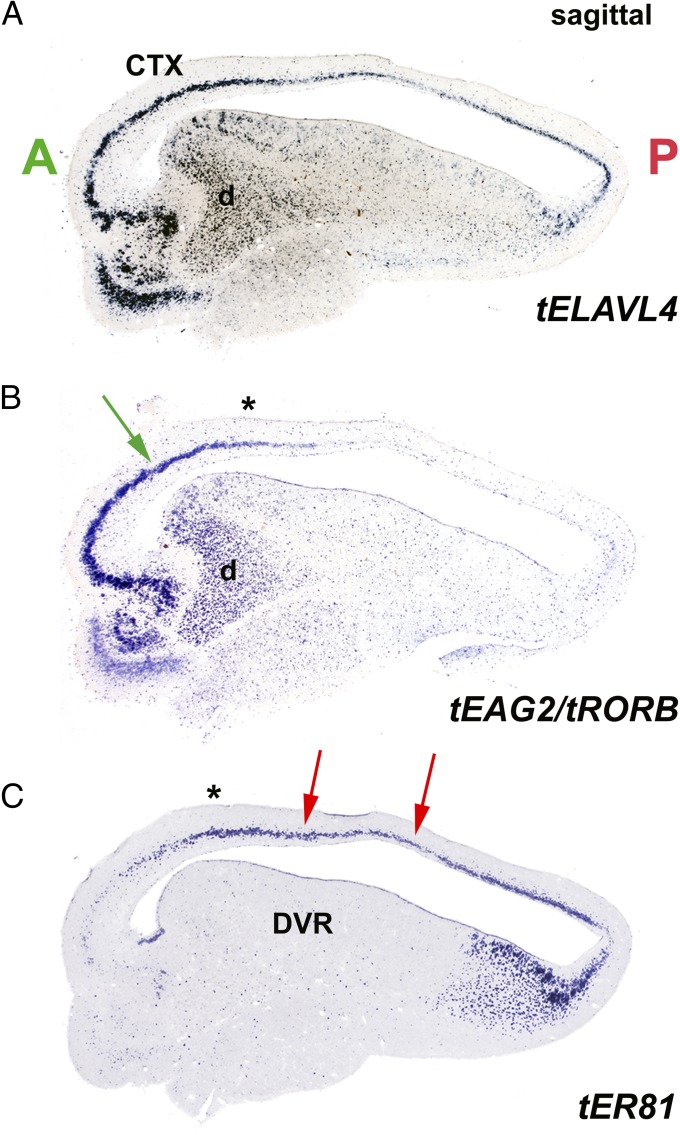
Fig. 5.
L4/I and L5/O cell-type markers identify multiple cerebral cortical fields in the turtle. (A) ELAVL4 ISH demonstrates the pyramidal layer of turtle cortex (CTX) and the neurons of the DVR in a parasagittal cross-section through turtle telencephalon. (B) Two-probe one-color ISH for L4/I markers EAG2 and RORB demonstrates strong expression rostrally in cortical area D2 (green arrow). Labeling in the underlying DVR picks out the thalamorecipient sensory zones of the turtle anterior DVR, including the dorsal area (d) that receives visual input from the nucleus rotundus (4). (C) The L5/O marker ER81 labels intermediate and posterior fields of turtle cortical area D2 (red arrows). (B and C) Asterisks indicate registration between these serially adjoining parasagittal sections, serving to mark the transitional field in area D2 that contains both cell types. A, anterior; P, posterior.
The L5/O marker ER81 is also present in turtle cortex, being densely expressed in the posterior two-thirds of the dorsal cortex, with scattered cells anteriorly (Figs. 4 C and D and 5C). Near the center of the dorsal cortex, there is a broad transitional territory where L4/I and L5/O marker genes are prominently expressed (Figs. 4 B and D and 5). Two-color in situ hybridization (ISH) demonstrated that in the region of overlap, the L4/I and the L5/O molecular markers are expressed by different cells (Fig. 4D). Thus, the turtle dorsal cortex is organized into distinct cortical fields, an anterior field of L4/I neurons that receives ascending thalamic input, a posterior field enriched in L5/O neurons, and a central transitional field with both cell types, which also receives transthalamic visual input (Figs. 4 and 5). An organization of three-layered cerebral cortex into cortical fields is not unusual. An analogous example is provided by the mammalian hippocampus, a three-layered cortex with separate fields, CA1 and CA3, containing pyramidal cells that differ in their connections and their molecular identities and that have between them a transitional field, CA2 (36).
Discussion
Birds are with mammals the animals with the largest brains for their body size (1); they represent a tremendously successful radiation of vertebrates, with a range of impressive behavioral specializations and cognitive abilities (37–39). Our aim in this study was to test a specific hypothesis, first set forth by Karten in 1969 (7), that the core pallial neuronal network responsible for these specializations and abilities is conserved between birds and mammals.
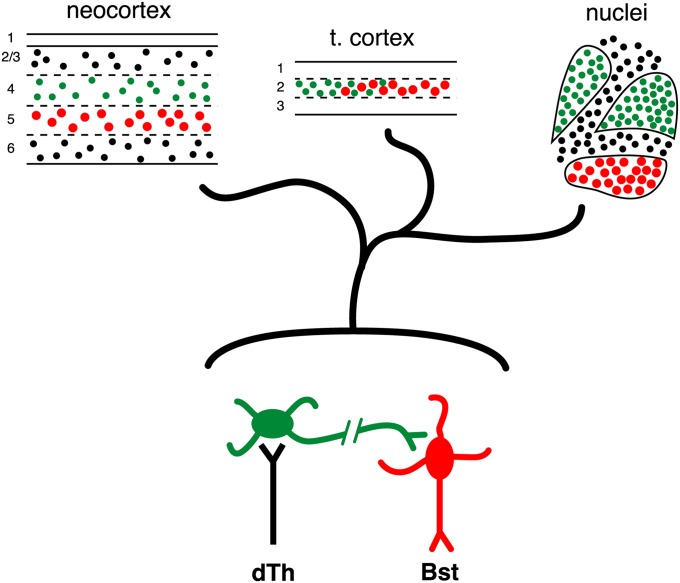
Our results affirm Karten’s hypothesis and establish that the neuronal circuitry of the avian pallium features cell types with the connectional and molecular properties of neocortical input and output neurons. The finding that multiple traits, including the selective expression of transcription factor and ion channel genes, identify input and output cell types in the dorsal telencephalon of two taxa of mammals and three taxa of birds and reptiles provides strong support for L4/I and L5/O cell-type homology across the amniotes. This conservation at the level of cell type stands in sharp contrast to the substantial diversity in the structural arrangement of these tissues, which form cortical layers in mammals, cortical fields in the turtle, and nuclei in the DVR and the avian Wulst (Fig. 6). In the face of these differences, one notable similarity is the tendency of these cell types to form continuous territories, as seen for the layers across the mammalian neocortex, the cell bridges between the input nuclei of the avian DVR, and the continuities between the turtle cortex and DVR at the anterior and posterior ends of the telencephalon (Fig. S3C and Fig. 5). There is no fossil record for internal brain organization, so we cannot know how L4/I and L5/O cells were organized in the most recent common ancestor of the amniote crown group. An attractive speculation is that the ancestral amniote pallium, whether nuclear or cortical in structure, featured L4/I and L5/O cell types that were aggregated into separate tissue compartments and were, as in the turtle, segregated along the anterior/posterior axis.
Fig. 6.
Structural diversity in the organization of conserved input and output cell types in amniote telencephalon. In the mammalian neocortex (Upper Left), input (green) and output (red) neurons are assigned to specific layers. In turtle dorsal cortex (Upper Center), the L4/I and L5/O cell types reside in a single layer but are partially segregated along the anterior-posterior axis. In the DVR (Upper Right) and avian Wulst, the L4/I and the L5/O cell types are segregated into nuclei. Bst, brainstem; dTh, dorsal thalamus; t. cortex, turtle dorsal cortex.
In the canonical neocortical circuit, layer 4 projects to upper layers 2 and 3 (L2/3), which in turn connects with layer 5 (Fig. 2A). A natural question is whether L2/3-type excitatory interneurons are also conserved in avian pallium. Recently, Suzuki et al. (40) proposed that molecularly defined L2/3 neurons are present in chick pallium. Although their specific claims are problematic (two of their three proposed upper-layer markers are expressed in all neocortical layers in the mouse; SI Text and Figs. S7 and S8), this important point warrants further study.
For many vertebrate neural cell types, such as the constituent neurons of the spinal cord and brainstem, the developmental mechanisms underlying cell-fate determination have benefited from investigation in model systems other than the mouse. Indeed, for the neural crest and telencephalic GABAergic neurons, there is evidence that many features of developmental specification are conserved from lampreys to mammals (41, 42). The major cell types to escape this comparative approach are the excitatory neurons of the neocortex. The finding of molecularly defined L4/I and L5/O neurons across amniotes now permits other developmental models, such as the chicken embryo system, to be applied to the study of neocortical cell-type specification. This advance will be particularly important for understanding L4/I development; this cell type is challenging to study in mammals because layer 4 is thin and difficult to dissect (15) and its neurons cannot be retrogradely labeled from distant targets, a strategy that proved crucial in identifying L5/O determinants (26). The L4/I nuclei in the chick are large and readily distinguished from adjoining nuclei even during late embryogenesis, which will allow for RNAseq searches for L4/I determinants.
Because the amniote L4/I and L5/O cell types are homologous at the level of molecular identity, we anticipate substantial conservation in the gene regulatory networks involved in cell-type specification. Because these cell types assemble into such different architectures, we do not expect full conservation of developmental mechanisms, particularly in the spatial and temporal organization of neuronal generation and migration. A clear example of differences in the cellular mechanism is the “inside-out” pattern of neocortical neuronal generation, where deep layer neurons are born first, more superficial neurons are born later, and newly born neurons migrate radially from the ventricular and subventricular zones past earlier-born neurons to settle in the cortical plate (43). Cortex development in the turtle looks very different. First, turtle dorsal telencephalon has a ventricular zone without a discernable subventricular zone that may in mammals contribute to L4/I neurogenesis (44). Second, turtle cortical neurons are produced in a brief temporal window, in a radial “outside-in” pattern with only minor medial-lateral and anterior-posterior gradients (45). Because L4/I and L5/O neurons populate specific cortical fields, it seems likely that L4/I and L5/O neurons in the turtle are generated by spatial rather than temporal mechanisms, with some ventricular zone territories primarily devoted to generating L4/I or L5/O cell types. The generation of the chick pallium differs from that of mammals and reptiles; it has an “outside-in” pattern of neurogenesis, with radial migration away from the ventricular zone, followed by a massive anterior-to-posterior migration during the second half of pallial neurogenesis (46, 47). These turtle and bird observations suggest that the differences in amniote dorsal telencephalon architecture are due to specific differences in the cellular aspects of development, such as the spatial arrangements of ventricular zone progenitor territories and the migration patterns of postmitotic neurons.
Our data establish that the neuronal circuitry of the avian telencephalon features cell types with the connectional and molecular properties of mammalian neocortical input and output neurons. In the bird, these neurons are not pyramidal in shape and do not participate in a neocortical architecture. Instead, the avian layer L4/I and L5/O neurons, like the other neurons of the bird pallium, are arranged into nuclei. These nuclei can, however, form highly structured processing centers, as seen in the sharply circumscribed components of the bird song system and the arrangement of Field L subnuclei into lamina with columnar interconnections (2, 48). A modern evolutionary perspective would expect that the varied neuronal architectures of the amniote pallium—a multilayered neocortex, a cortex with a single cellular layer, or an amalgam of nuclei—are each likely to have specific advantages and specific limitations. A challenge for evolutionary neurobiologists is to understand which circuit properties are enabled and which are precluded in each of these design architectures.
Materials and Methods
Full methods are provided in SI Materials and Methods.
Chickens were harvested at hatching or between 2 and 3 wk of age. The juvenile chicks and adult mice, ferrets, turtles, and male zebra finches were deeply anesthetized with sodium pentobarbital (120 mg/kg) and perfused through the heart with 4% paraformaldehyde in 0.1 M PBS (pH 7.4). Brains were removed and cryoprotected. Sections were cut at 32–36 μm on a sledge microtome and mounted on slides. cDNAs for mouse Er81 and chicken ER81 and RORB were gifts from T. Brown (Pfizer, Groton, CT), T. Jessell (Columbia University, New York), and M. Becker-Andre (Geneva Biomedical Reseach Insitute, Plan-Les-Ouates, Switzerland), respectively. All other cDNAs were isolated by PCR. Section ISH was carried out as previously described (19, 49). All procedures conform to the National Institutes of Health guidelines on animal experimentation and were approved by the University of Chicago Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Jay Goldberg, Elizabeth Grove, Daniel Margoliash, and Amanda Marma for help and discussions, and M. Becker-Andre, T. Brown, and T. Jessell for reagents. This work was supported by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequence data reported in this paper have been deposited in the GenBank database (accession nos. JX648196, JX827243, JX827244, JX827245, JX827246, JX827247, JX827248, JX827249, JX827250, JX827251, and JX845579).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204773109/-/DCSupplemental.
References
- 1.Striedter G. Principles of Brain Evolution. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 2.Reiner A, et al. Avian Brain Nomenclature Forum Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler AB, Hodos W. Comparative Vertebrate Neuroanatomy: Evolution and Adaptation. 2nd Ed. Hoboken, NJ: Wiley; 2005. [Google Scholar]
- 4.Ulinski PS. Dorsal Ventricular Ridge: A Treatise on Forebrain Organization in Reptiles and Birds. New York: Wiley; 1983. [Google Scholar]
- 5.Bruce LL, Neary TJ. The limbic system of tetrapods: A comparative analysis of cortical and amygdalar populations. Brain Behav Evol. 1995;46:224–234. doi: 10.1159/000113276. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez AS, Pieau C, Repérant J, Boncinelli E, Wassef M. Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: Implications for the evolution of telencephalic subdivisions in amniotes. Development. 1998;125:2099–2111. doi: 10.1242/dev.125.11.2099. [DOI] [PubMed] [Google Scholar]
- 7.Karten HJ. The organization of the avian telencephalon and some speculations on the phylogeny of the amniote telencephalon. Ann N Y Acad Sci. 1969;167:164–179. [Google Scholar]
- 8.Wild JM, Karten HJ, Frost BJ. Connections of the auditory forebrain in the pigeon (Columba livia) J Comp Neurol. 1993;337:32–62. doi: 10.1002/cne.903370103. [DOI] [PubMed] [Google Scholar]
- 9.Karten HJ. Evolutionary developmental biology meets the brain: The origins of mammalian cortex. Proc Natl Acad Sci USA. 1997;94:2800–2804. doi: 10.1073/pnas.94.7.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 11.Krützfeldt NO, Wild JM. Definition and novel connections of the entopallium in the pigeon (Columba livia) J Comp Neurol. 2005;490:40–56. doi: 10.1002/cne.20627. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Q, Li DP, Wang SR. Looming-sensitive responses and receptive field organization of telencephalic neurons in the pigeon. Brain Res Bull. 2006;68:322–328. doi: 10.1016/j.brainresbull.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Zeier H, Karten HJ. The archistriatum of the pigeon: Organization of afferent and efferent connections. Brain Res. 1971;31:313–326. doi: 10.1016/0006-8993(71)90185-5. [DOI] [PubMed] [Google Scholar]
- 14.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 15.Belgard TG, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saganich MJ, et al. Cloning of components of a novel subthreshold-activating K(+) channel with a unique pattern of expression in the cerebral cortex. J Neurosci. 1999;19:10789–10802. doi: 10.1523/JNEUROSCI.19-24-10789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig J, et al. Cloning and functional expression of rat eag2, a new member of the ether-à-go-go family of potassium channels and comparison of its distribution with that of eag1. Mol Cell Neurosci. 2000;16:59–70. doi: 10.1006/mcne.2000.0851. [DOI] [PubMed] [Google Scholar]
- 18.Schaeren-Wiemers N, André E, Kapfhammer JP, Becker-André M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 19.Rowell JJ, Mallik AK, Dugas-Ford J, Ragsdale CW. Molecular analysis of neocortical layer structure in the ferret. J Comp Neurol. 2010;518:3272–3289. doi: 10.1002/cne.22399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goloboff PA, et al. Phylogenetic analysis of 73060 taxa corroborates major eukaryotic groups. Cladistics. 2009;25:211–230. doi: 10.1111/j.1096-0031.2009.00255.x. [DOI] [PubMed] [Google Scholar]
- 21.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 22.Hevner RF, et al. Beyond laminar fate: Toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 23.Talley EM, et al. Differential distribution of three members of a gene family encoding low voltage-activated (T-type) calcium channels. J Neurosci. 1999;19:1895–1911. doi: 10.1523/JNEUROSCI.19-06-01895.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sailer CA, et al. Regional differences in distribution and functional expression of small-conductance Ca2+-activated K+ channels in rat brain. J Neurosci. 2002;22:9698–9707. doi: 10.1523/JNEUROSCI.22-22-09698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagamine S, Koike S, Keino-Masu K, Masu M. Expression of a heparan sulfate remodeling enzyme, heparan sulfate 6-O-endosulfatase sulfatase FP2, in the rat nervous system. Brain Res Dev Brain Res. 2005;159:135–143. doi: 10.1016/j.devbrainres.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 27.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wild JM. Descending projections of the songbird nucleus robustus archistriatalis. J Comp Neurol. 1993;338:225–241. doi: 10.1002/cne.903380207. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu T, Cox K, Karten HJ. Intratelencephalic projections of the visual wulst in pigeons (Columba livia) J Comp Neurol. 1995;359:551–572. doi: 10.1002/cne.903590404. [DOI] [PubMed] [Google Scholar]
- 30.Wild JM, Williams MN. Rostral wulst in passerine birds. I. Origin, course, and terminations of an avian pyramidal tract. J Comp Neurol. 2000;416:429–450. [PubMed] [Google Scholar]
- 31.Ulinski PS. The cerebral cortex of reptiles. In: Jones EG, Peters A, editors. Cerebral Cortex. Vol 8A. New York: Plenum; 1990. pp. 139–216. [Google Scholar]
- 32.Connors BW, Kriegstein AR. Cellular physiology of the turtle visual cortex: Distinctive properties of pyramidal and stellate neurons. J Neurosci. 1986;6:164–177. doi: 10.1523/JNEUROSCI.06-01-00164.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall JA, Foster RE, Ebner FF, Hall WC. Visual cortex in a reptile, the turtle (Pseudemys scripta and Chrysemys picta) Brain Res. 1977;130:197–216. doi: 10.1016/0006-8993(77)90270-0. [DOI] [PubMed] [Google Scholar]
- 34.Molnár Z. Evolution of cerebral cortical development. Brain Behav Evol. 2011;78:94–107. doi: 10.1159/000327325. [DOI] [PubMed] [Google Scholar]
- 35.Desan PH. 1988. The organization of the cerebral cortex of the pond turtle, Pseudemys scripta elegans. PhD dissertation (Harvard University, Cambridge, MA)
- 36.Tole S, Christian C, Grove EA. Early specification and autonomous development of cortical fields in the mouse hippocampus. Development. 1997;124:4959–4970. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- 37.Kenward B, Weir AA, Rutz C, Kacelnik A. Behavioural ecology: Tool manufacture by naive juvenile crows. Nature. 2005;433:121. doi: 10.1038/433121a. [DOI] [PubMed] [Google Scholar]
- 38.Raby CR, Alexis DM, Dickinson A, Clayton NS. Planning for the future by western scrub-jays. Nature. 2007;445:919–921. doi: 10.1038/nature05575. [DOI] [PubMed] [Google Scholar]
- 39.Scarf D, Hayne H, Colombo M. Pigeons on par with primates in numerical competence. Science. 2011;334:1664. doi: 10.1126/science.1213357. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki IK, Kawasaki T, Gojobori T, Hirata T. The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev Cell. 2012;22:863–870. doi: 10.1016/j.devcel.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Sauka-Spengler T, Bronner-Fraser M. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol Bull. 2008;214:303–314. doi: 10.2307/25470671. [DOI] [PubMed] [Google Scholar]
- 42.Martínez-de-la-Torre M, Pombal MA, Puelles L. Distal-less-like protein distribution in the larval lamprey forebrain. Neuroscience. 2011;178:270–284. doi: 10.1016/j.neuroscience.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 43.Rakic P. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung AF, Pollen AA, Tavare A, DeProto J, Molnár Z. Comparative aspects of cortical neurogenesis in vertebrates. J Anat. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goffinet AM, Daumerie C, Langerwerf B, Pieau C. Neurogenesis in reptilian cortical structures: 3H-thymidine autoradiographic analysis. J Comp Neurol. 1986;243:106–116. doi: 10.1002/cne.902430109. [DOI] [PubMed] [Google Scholar]
- 46.Striedter GF, Keefer BP. Cell migration and aggregation in the developing telencephalon: Pulse-labeling chick embryos with bromodeoxyuridine. J Neurosci. 2000;20:8021–8030. doi: 10.1523/JNEUROSCI.20-21-08021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones AW, Levi-Montalcini R. Patterns of differentiation of the nerve centers and fiber tracts of the avian cerebral hemispheres. Arch Ital Biol. 1958;96:231–284. [Google Scholar]
- 48.Wang Y, Brzozowska-Prechtl A, Karten HJ. Laminar and columnar auditory cortex in avian brain. Proc Natl Acad Sci USA. 2010;107:12676–12681. doi: 10.1073/pnas.1006645107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 50.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira SL, Baker AJ. A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Biol Evol. 2006;23:1731–1740. doi: 10.1093/molbev/msl038. [DOI] [PubMed] [Google Scholar]
- 52.Lyson TR, Bever GS, Bhullar BA, Joyce WG, Gauthier JA. Transitional fossils and the origin of turtles. Biol Lett. 2010;6:830–833. doi: 10.1098/rsbl.2010.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rieppel O, deBraga M. Turtles as diapsid reptiles. Nature. 1996;384:453–455. [Google Scholar]
- 54.Crawford NG, et al. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 2012;8:783–786. doi: 10.1098/rsbl.2012.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.