Abstract
The type II p21-activated kinases (PAKs) are key effectors of RHO-family GTPases involved in cell motility, survival, and proliferation. Using a structure-guided approach, we discovered that type II PAKs are regulated by an N-terminal autoinhibitory pseudosubstrate motif centered on a critical proline residue, and that this regulation occurs independently of activation loop phosphorylation. We determined six X-ray crystal structures of either full-length PAK4 or its catalytic domain, that demonstrate the molecular basis for pseudosubstrate binding to the active state with phosphorylated activation loop. We show that full-length PAK4 is constitutively autoinhibited, but mutation of the pseudosubstrate releases this inhibition and causes increased phosphorylation of the apoptotic regulation protein Bcl-2/Bcl-XL antagonist causing cell death and cellular morphological changes. We also find that PAK6 is regulated by the pseudosubstrate region, indicating a common type II PAK autoregulatory mechanism. Finally, we find Src SH3, but not β-PIX SH3, can activate PAK4. We provide a unique understanding for type II PAK regulation.
Keywords: autoregulation, protein kinase, RHO GTPase effector, signaling
The RHO-family small GTPases RAC1 and CDC42 control many cellular functions, including cytoskeletal organization, morphological changes, cell motility, and cell-cycle progression (1). These enzymes are regulated by cycling between GDP-bound and GTP-bound states, whereby the GTP-bound state binds to and activates multiple effector molecules, triggering distinct downstream events. Pathological mutations in these proteins can alter cellular outcomes, and recurrent activating mutations suffer high mutational burdens in cancer (2). There is consequently significant interest in understanding the intrinsic details of these molecules and their complex signaling pathways.
An important group of proteins that directly interact with RAC1 and CDC42 are the p21-activated kinases (PAKs) (3). These Ste20 family serine-threonine kinases are regulators of the actin cytoskeleton, cell survival, cell adhesion, cytokine signaling, and transcription (3, 4). There are two subgroups of PAK kinase, denoted type I (PAK1, PAK2, and PAK3) and type II (PAK4, PAK5, and PAK6). PAK4 is the best-studied type II PAK family member, is widely expressed (5), and is essential for viability in mice (6). Downstream substrates of PAK4 include the RHO GTPase guanine nucleotide exchange factor H1 (7), the cytoskeletal regulator LIM domain kinase 1 (LIMK1) (8), the adhesion receptor integrin β5 (9), the focal adhesion scaffolding protein paxillin (10), and the apoptotic regulation protein Bcl-2/Bcl-XL antagonist causing cell death (BAD) (11). This array of substrates are thought to mediate effects of PAK4 activation in cells, including loss of focal adhesions (12), cell rounding (12, 13), cytoskeleton changes (13), and protection from apoptosis (11). Together, these observations indicate that PAK4 plays roles both as a regulator of the actin cytoskeleton and as a promoter of cell survival (11). Although the expression profiles of the type II PAKs vary, the signaling pathways of PAK5 (also known as PAK7) and PAK6, which are highly similar to PAK4 at the amino acid level, may be somewhat redundant with PAK4 (3). Indeed, some substrates are common, including BAD (for PAK4 and PAK5) (14) and LIMK1 (for all type II PAKs) (15). Collectively, these findings suggest that the type II PAKs may be regulated by common mechanisms.
The signaling pathways in which the type II PAKs function are often associated with cancer. Indeed, PAK4 is required for oncogenic or metastatic phenotypes of many cancer cell lines (16–18). PAK4 can promote tumorigenesis (19) and breast cancer cell migration (20), and it is the only PAK that is transforming when overexpressed (17). Consistent with their roles in BAD signaling, both PAK4 and PAK5 can protect cancer cells from apoptosis (21, 22). Although acquired somatic missense mutations in type II PAKs have not yet been shown to be transforming, PAK5 has been identified to be among the most frequently mutated kinase genes in human cancer (23). PAK6 is also overexpressed in cancer (17, 24) and could potentially play a role in MAP kinase signaling pathways associated with cancer cell migration (4). Furthermore, a specific PAK4 inhibitor (PF-3758309) has shown efficacy in mouse models of cancer (25), although it failed in clinical trials most likely due to undesirable pharmacology (http://clinicaltrials.gov/ct2/show/NCT00932126). Overall, these data suggest the potential for beneficial effects of improved type II PAK inhibitors in selected tumors. The type II PAKs may therefore represent targets for treatment of specific tumors.
The domain architecture of both the type I and type II PAKs includes an N-terminal GBD [GTPase binding domain, also known as a CDC42/RAC1 interactive binding (CRIB) domain] and a C-terminal protein kinase catalytic domain (Fig. 1A). Additionally, the type I PAKs contain an autoinhibitory domain (AID) overlapping the GBD, which is not observed in the type II PAKs. It is well established that the type I PAKs are activated by binding RHO-family small GTPases through their GBD:AID region. This interaction results in a conformational change that relieves autoinhibition and allows further steps, including autophosphorylation by activation segment exchange (reviewed in ref. 26). The type II PAKs, however, do not contain the conserved AID seen in type I PAKs, and are not regulated in a similar manner. Indeed, although type II PAKs bind small GTPases, in particular CDC42 and less to RAC1 (3), the interaction is generally thought to be important for localization, not direct activation (5, 17, 27–29), which raises the question of whether alternate mechanisms of autoinhibition could be functioning for the type II PAKs. A previous study suggested that PAK5 may be regulated by an autoinhibitory region, although this sequence is not conserved across all type II PAKs (27). A recent study suggested that autoinhibition of type II PAKs is released by CDC42 (30). Furthermore, truncated type II PAKs lacking N-terminal regions have elevated catalytic activity (5, 12). Structural studies of the kinase domains of type II PAKs have suggested that the conformational plasticity of the glycine-rich loop acts as a molecular sensor for ATP binding, governing structural rearrangements important for maintenance of a competent active state (31). How these conformational rearrangements are controlled at the molecular level, however, remains to be elucidated. Moreover, the type II PAKs are generally described as being constitutively catalytically active, and their function in signaling pathways is presumed to be regulated, at least in part, by relocalization via RHO-family GTPases (32). Overall, no clear mechanism has yet been established to provide the switch-like on/off regulation mechanism for the type II PAKs that is so frequently seen for protein kinases. Here we demonstrate that type II PAKs are autoregulated by a conserved pseudosubstrate sequence within the N-terminal region.
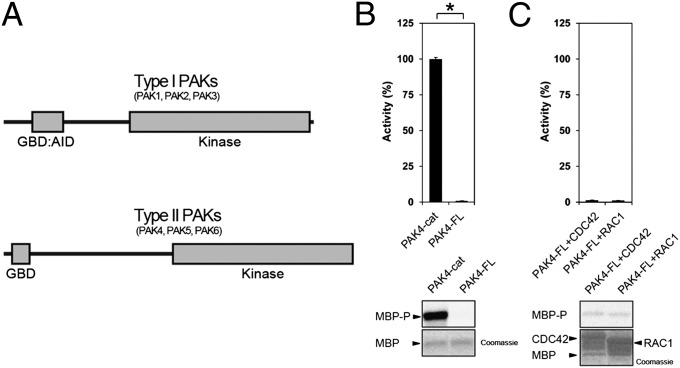
Fig. 1.
PAK schematic and kinase assays for type II PAKs. (A) Schematic diagram for type I and type II PAK family members. The type I PAKs contain an N-terminal GBD that overlaps with an AID (GBD:AID), and a C-terminal kinase domain. The type II PAKs contain an N-terminal GBD and a C-terminal kinase domain. All PAKs contain proline-rich patches between the N-terminal GBD and C-terminal kinase domains. (B) Kinase assay for PAK4-cat and PAK4-FL. Purified PAK4 catalytic domain PAK4-cat135–426 (PAK4-cat and PAK4-FL were assayed for activity toward MBP with [γ-33P]ATP. Reactions were subjected to SDS/PAGE, exposed to phosphor storage screen, scanned, and MBP phosphorylation quantified by optical densitometry (Lower, MBP-P). MBP loading is shown (Lower, MBP, Coomassie). Activities are shown as a percentage of PAK4-cat activity (100%). PAK4-FL displays ∼100-fold less activity than PAK4-cat. *P < 0.01, t test. (C) GTPases do not increase PAK4-FL catalytic activity. PAK4-FL kinase activity toward MBP was assayed in the presence of either purified CDC42 or RAC1 with GMP-PNP and Mg2+. MBP and GTPase loading are shown. Small GTPases do not impact kinase activity of PAK4-FL toward MBP.
Results
PAK4 Catalytic Activity Is Inhibited by Its Amino-Terminal Region.
To investigate the regulation mechanisms of PAK4 we conducted kinase assays using highly purified enzyme expressed in bacteria and myelin basic protein (MBP) as a substrate. We found that the activity of full-length PAK4 (PAK4-FL) was severely reduced in comparison with the isolated PAK4 catalytic domain (PAK4-cat; Fig. 1). Interestingly, PAK4-FL essentially shows a complete lack of detectable kinase activity. We then used this system to address the controversy regarding activation of type II PAKs, in which interaction with small GTPases either does not increase (3, 5, 33) or greatly increases kinase activity (30). We measured the activity of PAK4-FL in the presence of CDC42 or RAC1, GMP-PNP (5′-guanylyl imidodiphosphate), and Mg2+. No change in PAK4-FL catalytic activity was observed on incubation with these GTPases (Fig. 1). Our results are most consistent with a lack of direct regulation of PAK4 by small GTPases. We note that previous studies used kinase immunoprecipitates from cultured cells, which may also harbor proteins that could influence type II PAK activity.
Crystal Structures Reveal a Bound Peptide Only for Full-Length PAK4.
We therefore wondered whether a structure-directed approach would allow us to understand how catalytic activity of PAK4-FL is significantly inhibited compared with the catalytic domain alone. We crystallized PAK4-FL and PAK4-cat in the same crystal form. These crystals diffract X-rays to 3.1-Å and 3.0-Å resolutions, respectively (Table S1 and Fig. 2A). Both PAK4-cat and PAK4-FL crystallize in the active, DFG-in, state with an extended activation loop and S474 phosphorylated. The crystals are in space group P3, distinct from previously solved structures of the PAK4 catalytic domain. Our crystals contain two molecules per asymmetric unit and have solvent channels that comprise an unusually high proportion (∼80%) of the crystal (compared with 50% solvent content for a typical protein crystal) (34). These solvent channels have a diameter of ∼91 Å (Fig. S1A), allowing the N terminus of PAK4-FL to be predominantly unstructured without adversely impacting crystal formation. We confirmed that the PAK4-FL crystals were not proteolytically degraded by SDS/PAGE (Fig. S1B). In this crystal form the catalytic domain structure is experimentally identical between the PAK4-FL and PAK4-cat structures (rmsd ∼0.4 Å).
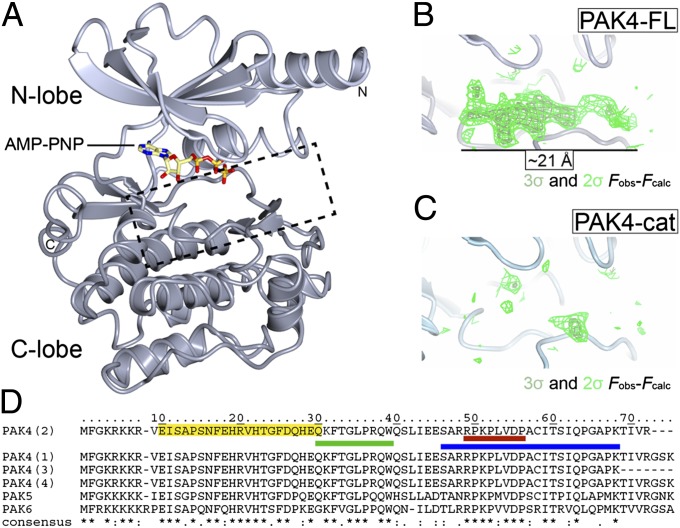
Fig. 2.
Structures of PAK4-cat and PAK4-FL in the P3 crystal form. (A) Overall structure of the PAK4 kinase domain. PAK4 kinase domain shown in ribbon format with N-lobe (light gray) and C-lobe (dark gray) indicated. Bound AMP-PNP shown in stick format. N and C termini are indicated. Region shown in B and C is indicted by a dashed box. (B and C) The PAK4-FL P3 crystal contains a significant region of positive difference electron density in the peptide substrate binding site; this is not observed in the PAK4-cat structure. Two contour levels (2σ and 3σ in dark and light green, respectively) are shown for the unbiased Fobs-Fcalc map. (D) Sequence alignment for the N terminus of human type II PAKs (UniProt accession nos: PAK4, O96013; PAK5, Q9P286; PAK6, Q9NQU5). The four PAK4 isoforms are shown, which are identical between the N terminus and residue K68. (*) indicates identical; (:), highly conserved; (.), semiconserved. The PAK4 GBD (CRIB) domain as defined by Abo et al. (5) is shaded yellow. QKF peptide, green; RPK peptide, red; and 23-mer peptide, blue.
On inspection of the PAK4-FL crystal structure we observed a region of contiguous positive difference (Fobs-Fcalc) electron density in the kinase peptide substrate-binding site of ∼21 Å in length for both copies in the asymmetric unit (Fig. 2B). Positive difference density is not visible in the equivalent region of the PAK4-cat crystals (Fig. 2C). We therefore asked two questions. First, is this region part of the N terminus of PAK4? Second, does this electron density reflect a pseudosubstrate autoinhibition mechanism that results in the observed reduced activity of PAK4-FL compared with PAK4-cat (Fig. 1B)?
To address these questions we first generated hypotheses for the identity of the positive difference density observed in the PAK4-FL crystal. We built a polyalanine chain into the positive difference density for both copies in the asymmetric unit, using the previously determined structure of PAK4 in complex with a peptide substrate as a guide (PDB ID code 2Q0N). We found that the peptide substrate backbone fitted the density well, and following crystallographic refinement we noted that this density was very similar in both copies of PAK4 in the asymmetric unit. We therefore averaged the electron density maps using the graphics program Coot (35) and discovered that in the resulting averaged maps, some residues clearly indicated no or a small side-chain, so were probably Ala, Pro, or Gly, but other locations indicated hydrophobic residues, or residues with a longer side-chain (Fig. S2). We therefore investigated whether the N terminus of PAK4 contained a sequence that matched our hypothesis for arrangement of small and large side-chains. We found two regions that we thought matched with reasonable similarity, F32TGLPR and R49PKPLV (Fig. S2). We and others (30, 33) note the almost complete conservation for the R49PKPLV sequence across all type II PAKs (Fig. 2D).
PAK4 N-Terminal Region R49PKP Inhibits Catalytic Activity.
To investigate whether these regions of PAK4-impacted catalytic activity, we conducted kinase assays using the purified recombinant catalytic domain of PAK4, PAK4-cat135–426, in the presence of N-terminal regions of PAK4 (Fig. 3A). We first synthesized two peptides corresponding to the hypothesized regions (Fig. S2), which we termed the QKF peptide (QKF, Q30KFTGLPRQW) or RPK peptide (RPK, R49PKPLVDP). In these assays the RPK peptide was a moderate inhibitor of PAK4-cat135–426, whereas the QKF peptide was a much weaker inhibitor (Fig. 3B). We then investigated the effect of a series of recombinant N-terminal constructs on kinase activity of PAK4-cat135–426. We expressed and purified protein constructs that included either the GTPase binding domain (GBD) alone (PAK41–30); the GBD and the QKF sequence (PAK41–45); the GBD and both the QKF and RPK sequences (PAK41–70); and two longer N-terminal constructs containing all three regions (PAK41–90 and PAK41–130). We also generated constructs that did not include the GBD, but did include the QKF and RPK regions (PAK431–130), or that excluded the GBD and either the QKF or RPK sequences (PAK480–130 and PAK4110–130; Fig. 3A). We conducted kinase assays using PAK4-cat135–426 in the presence of each of these purified, recombinant proteins (Fig. 3C). Though inclusion of the GBD did not impact kinase activity, inclusion of the R49PKP sequence resulted in reduced phosphorylation of the substrate, MBP. Addition of a construct that included the whole N-terminal region of PAK41–130 resulted in the lowest phosphorylation of MBP, but constructs including residues 46–70 could transinhibit kinase activity with similar potency.
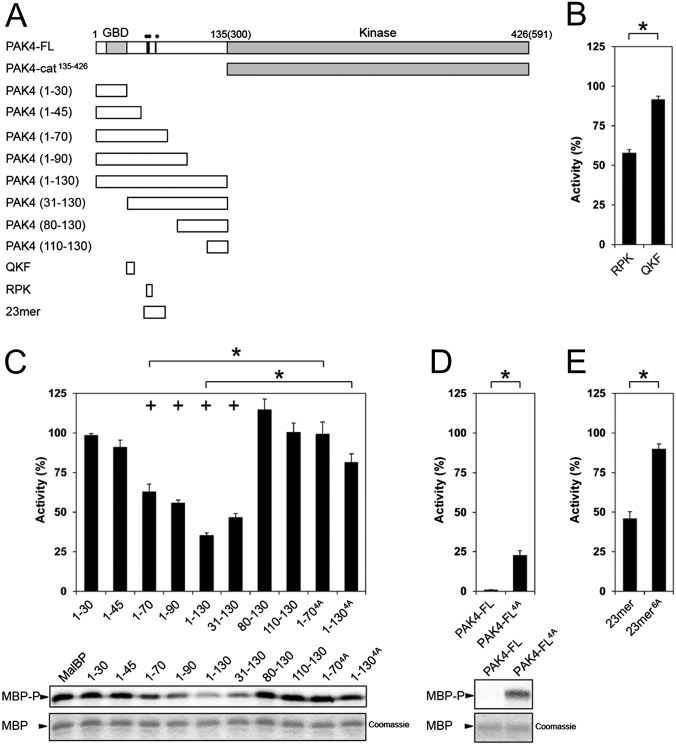
Fig. 3.
PAK4 kinase assays. (A) Schematic of constructs used for PAK4 kinase activity assays. Residues mutated are indicated on PAK4-FL as black lines with two filled circles (••) indicating R49PKP to A49AAA or a single filled circle (•) indicating I59T to A59A. (B) Purified PAK4 catalytic domain PAK4-cat135–426 (PAK4-cat) was assayed for activity toward MBP in the presence of RPK or QKF peptides. Radiolabel incorporation into MBP was determined by optical densitometry following SDS/PAGE and exposure to a phosphor storage screen. Activities are shown as a percentage of the activity of PAK4-cat135–426. For all, kinase assays show SEM for more than three experiments. RPK peptide significantly inhibits kinase activity. *P < 0.01, t test. (C) PAK4-cat135–426 activity in the presence of N-terminal constructs of PAK4. Purified MalBP fusion constructs PAK41–30, PAK41–45, PAK41–70, PAK41–90, PAK41–130, PAK431–130, PAK480–130, and PAK4110–130 were added and compared with addition of MalBP alone (MalBP). PAK41–70, PAK41–90, PAK41–130, and PAK431–130 inhibit kinase activity of PAK4-cat135–426. MBP phosphorylation (Lower, MBP-P) and loading are shown (Lower, MBP, Coomassie). Mutation of R49PKP to A49AAA in either PAK41–70 or PAK41–130 results in a loss of inhibition of kinase activity. Activities are shown as a percentage of the activity of PAK4-cat135–426 with negative control MalBP added. *P < 0.01, t test; +P < 0.01 by t test compared with PAK4-cat135–426 with MalBP alone. (D) Mutation of PAK4-FL restores kinase activity. Purified PAK4-FL containing point mutations R49PKP to A49AAA (PAK4-FL4A) show a significant increase kinase activity with respect to MBP. Activities are shown as a percentage of PAK4-cat135–426 activity. (E) Effect of 23-mer peptide on PAK4-cat135–426 kinase activity. The 23-mer peptide inhibits kinase activity better than the RPK peptide (B). Mutation of R49PKP to A49AAA and I59T to A59A in this peptide (23-mer6A) results in a loss of kinase inhibition. Activities are shown as a percentage of the activity of PAK4-cat135–426.
To validate whether inclusion the R49PKP sequence impacted kinase activity, we introduced quadruple alanine point mutations into the PAK41–70 and PAK41–130 constructs by mutating R49PKP to A49AAA (termed PAK41–70-4A and PAK41–130-4A). We found that PAK41–70-4A and PAK41–130-4A display severely reduced ability to inhibit kinase activity (Fig. 3C), which indicates that a significant portion of transinhibition is lost on introduction of these mutations. We therefore wondered whether introduction of a quadruple alanine mutation into PAK4-FL would impact catalytic activity and potentially relieve the autoinhibition we had initially observed (Fig. 1B). We found that quadruple alanine mutation of R49PKP to A49AAA in the context of PAK4-FL (PAK4-FL4A) resulted in a significant increase in kinase activity (Fig. 3D). For PAK4-FL4A, phosphorylation of MBP was ∼20% of that observed for PAK4-cat135–426 alone, in contrast to ∼1% activity for wild-type PAK4-FL. Finally, we evaluated a longer synthetic peptide (23-mer, S46ARRPKPLVDPACITSIQPGAPK) and a similar peptide with six completely conserved type II PAK residues substituted with alanine (23-mer6A, S46ARAAAALVDPACAASIQPGAPK) as inhibitors of PAK4-cat135–426 activity. We found that though the wild-type 23-mer peptide inhibited catalytic activity better than the shorter RPK peptide (∼50% inhibition at 75 μM), the 23-mer6A peptide showed weak inhibition of PAK4-cat135–426 activity (Fig. 3E). We note that the 23-mer inhibits PAK4 approximately fivefold less potently than the entire N-terminal region (∼50% inhibition at 75 μM and 15 μM, respectively). This could mean that the N terminus harbors additional regions of contact with the catalytic domain, or alternatively that the pseudosubstrate sequence binds with higher affinity when presented in the context of the intact N terminus. We also note that although in trans the pseudosubstrate sequence is a weak inhibitor, in cis the proximity effect is likely to allow for potent autoinhibition. Together these results indicate that the region of PAK4 including and surrounding R49PKP inhibits catalytic activity in vitro.
N-Terminal Region R49PKPLV Binds PAK4 Kinase Domain in a Pseudosubstrate Conformation.
We next wondered whether the N-terminal sequence of PAK4 that inhibits catalytic activity could bind PAK4 kinase domain “in crystal,” and whether this binding would resemble the density observed for the PAK4-FL crystals (Fig. 2B). Therefore, we conducted cocrystallography of PAK4-cat with the RPK peptide, and as a negative control, PAK4-cat with the QKF peptide (Table S1). These cocrystals were obtained in space group P41212 and diffracted to higher resolution than the P3 crystals; they are also found in the active state with S474 phosphorylated. Though cocrystallization of PAK4-cat with the QKF peptide resulted in no unusual difference density in the peptide substrate binding site (Fig. S3D), we found that cocrystallization of PAK4-cat with the RPK peptide resulted in a significant extended region of positive difference density (Fig. S3A) resembling that observed for PAK4-FL (Fig. 2B). Because this new crystal structure was determined to 2.0-Å resolution, we were able to build the RPK peptide into the electron density (Fig. S3B). We then used this density as a model to complete refinement for the PAK4-FL crystal structure (Fig. S3C) and found that the built peptide satisfied the observed electron density well. We next cocrystallized PAK4-cat with the RPK peptide in the P3 crystal form (containing the ∼91-Å diameter solvent channels) and observed strong difference density at the same location (Fig. S3E). We built this structure to 2.6-Å resolution (Fig. S3F). In all three structures that include the RPK sequence bound in the peptide substrate-binding site, the mode of binding is similar (Fig. S3). Each of these structures shows that the RPK sequence binds as a pseudosubstrate with P52 located at the site of the phosphate acceptor residue (Fig. 4 A and B and Fig. S3J). We therefore conclude that the region in the PAK4-FL crystal structure that results in the observed contiguous positive electron density (Fig. 2B) is R49PKPLVD. A detailed description is provided in SI Text.
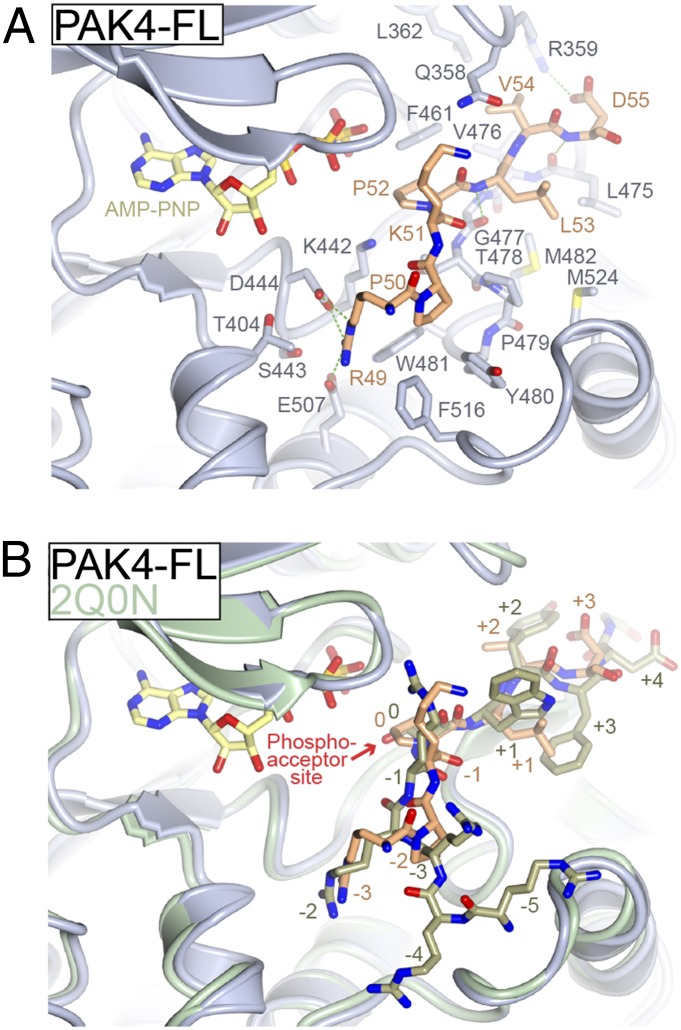
Fig. 4.
Mode of binding for the type II PAK autoinhibitory region. (A) Structural details of pseudosubstrate binding to PAK4 catalytic domain. Structure of PAK4-FL is shown with residues discussed in the text shown in stick format and labeled. Pseudosubstrate is colored orange. Hydrogen bonds are colored green. (B) Comparison of pseudosubstrate binding to a consensus substrate peptide. Crystal structure of PAK4 catalytic domain bound to a consensus substrate sequence (PDB ID code 2Q0N) is shown in green. Labels for substrate and pseudosubstrate indicate number of residues distal from the phosphoacceptor site (labeled and denoted 0).
The mode of binding for the R49PKPLV region to the substrate-binding site is unusual compared with a bound substrate (Fig. 4B). First, P52, the residue at the phosphate acceptor site, does not extend deeply toward the γ-phosphate of ATP. Compared with the previously determined crystal structure of PAK4 in complex with a consensus substrate peptide (PDB ID code 2Q0N), the γ-hydroxyl group of S0 extends up to 2.9 Å deeper into the catalytic cleft. Second, the guanidino head group of R49, located three residues upstream of P52, occupies the same acidic binding site for an important Arg residue located at a different position in the substrate (two residues upstream of the phosphoacceptor site). Third, the path of the pseudosubstrate inhibitor peptide backbone is between 1 Å and 3 Å distal from the path of the bound consensus peptide, and this seems to be facilitated by the presence of P50 bending the peptide backbone away from the bound peptide’s canonical path. Indeed, the Cα residue of R49 is located ∼3 Å from that of the previously observed −2 arginine, allowing R49 to be accommodated in the acidic −2 peptide binding pocket.
Autoinhibitory Pseudosubstrate Inhibits PAK4 Catalytic Activity in Cultured Cells.
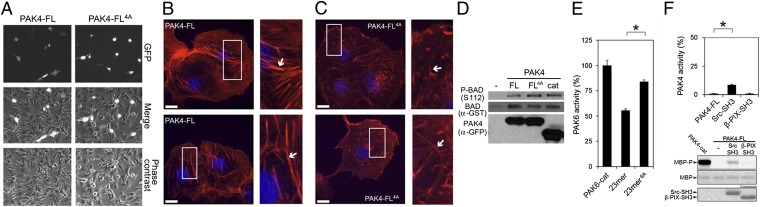
The mechanism for PAK4 autoregulation defined by our crystal structures and biochemical assays also appears to restrain PAK4 activity in cultured cells. Transient expression of a constitutively activate PAK4 mutant was previously shown to cause cell rounding in fibroblasts (13). Consistent with an increase in PAK4-FL4A catalytic activity in cells compared with wild-type PAK4-FL, we observe a similar cell-rounding effect upon expression in NIH 3T3 fibroblasts (Fig. 5A) and reduced actin filament containing stress fibers in COS-7 cells (Fig. 5 B and C and Fig. S4). We next wondered whether alteration of the pseudosubstrate inhibitory region would impact downstream signaling from PAK4. We therefore investigated phosphorylation of a validated PAK4 target, BAD (11). Coexpression with either PAK4-cat or PAK4-FL4A led to consistently higher levels of BAD phosphorylation than expression with PAK-FL (Fig. 5D).
Fig. 5.
Cellular effects of loss of PAK4 pseudosubstrate inhibition and PAK6 pseudosubstrate inhibition. (A) Morphological changes upon transient expression of PAK4-FL or PAK4-FL4A. NIH 3T3 cells transiently transfected with GFP-PAK4-FL4A (PAK4-FL4A) exhibit cell rounding that is not observed with GFP-PAK4-FL (PAK4-FL). (B and C) Reduced actin containing stress fibers in COS-7 cells expressing activated PAK4. Transiently expressed GFP-tagged GFP-PAK4-FL (B) or GFP-PAK4-FL4A (C) were fixed with paraformaldehyde, permeabilized, then stained with rhodamine-conjugated phalloidin and imaged with a spinning-disk confocal-based inverted Olympus microscope. The overall size and extent of stress fiber organization (Insets and arrows) was decreased in cells expressing the PAK4-FL4A mutant compared with cells expressing PAK4-FL. Insets and arrows show reduced stress fibers organization and increase in actin containing membrane ruffles in cells expressing the PAK4-FL4A mutant. Transfection of cells with GFP-PAK4 constructs is validated in Fig. S4. (Scale bars: 10 μm.) (D) Phosphorylation of the PAK4 substrate BAD. Transient overexpression of GFP-PAK4-FL (WT), GFP-PAK4-FL4A (4A), or GFP-PAK4-cat135–426 shows that mutation of the PAK4 pseudosubstrate elevates BAD Ser112 phosphorylation to a level equivalent to that induced by the kinase domain alone (cat). (E) Purified PAK6 catalytic domain (PAK6-cat) was assayed for activity toward MBP in the presence of the 23-mer and 23-mer6A peptides. MBP was loaded onto SDS/PAGE, exposed to phosphor storage screen, scanned, and quantified by optical densitometry. Activities are shown as a percentage of the activity of PAK6-cat. For all kinase assays, SEM is shown, and values are calculated from more than three experiments. The 23-mer peptide significantly inhibits kinase activity. *P < 0.01, t test. (F) Src SH3 domain, but not β-PIX SH3 domain, increases PAK4-FL catalytic activity. PAK4-FL kinase activity toward MBP was assayed in the presence of either purified Src or β-PIX SH3 domain. MBP and SH3 domain loading are shown.
General Mechanism for Type II PAK Regulation.
The region that we found to be autoinhibitory for PAK4 is very well conserved between the type II PAK family (Fig. 2D). Therefore, we wondered whether the mechanism of regulation is general to the all type II PAKs. To test this we generated recombinant PAK6 kinase domain (PAK6-cat) and conducted kinase assays in the presence of the wild-type and 23-mer6A peptides. We found that PAK6 is inhibited to ∼55% activity by the wild-type 23-mer peptide, but that the 23-mer6A peptide shows reduced inhibition (Fig. 5E). Because the majority of the residues in this peptide are conserved between PAK4 and PAK6 (Fig. 2D), we therefore infer that PAK6, and the type II PAKs as a whole, are regulated by a pseudosubstrate autoinhibitory mechanism.
Full-Length PAK4 Can Be Activated By Src SH3 Domain but Not β-PIX SH3 Domain.
To investigate whether PAK4 could be activated by SH3 domain interactions, we conducted kinase activity assays in the presence of either β-PIX or Src SH3 domains. We found that β-PIX SH3 domain did not activate PAK4-FL, but that the SH3 domain of Src could activate PAK4-FL (Fig. 5F).
Discussion
Type II PAKs are important regulators of cell adhesion, migration, proliferation, and survival; however, since their discovery over a decade ago, general cellular mechanisms of type II PAK regulation have remained elusive. Early on, type II PAKs were found to be more active when expressed as a kinase domain alone compared with the full-length protein (5), and PAK5 was found to be regulated by a region encoding residues 60–180 (27). This region, however, is not conserved in the other type II PAKs, and has therefore led to an assumption that these kinases are regulated in alternate manners. Another study investigated the structural biology of type II PAK kinase domains. This study found that glycine-rich loop conformational flexibility could function as a molecular sensor for ATP binding that governs structural rearrangements, which was suggested to be important for maintenance of a competent active state (31)—an effect that though important, may not provide the clear functional switch usually observed for protein kinases. Additionally, if PAK4 exists at some points in an activation-loop unphosphorylated state, it may rapidly activate by a dimerization model similar to that proposed for PAK1 (36). In contrast, a recent study using kinases immunoprecipitated from cultured cells suggested that CDC42 activates the type II PAKs directly by releasing an autoinhibitory conformation (30). In accordance with the previous literature (5, 17, 27–29), however, we do not find evidence for this in our in vitro assays using purified proteins. Because they are constitutively phosphorylated on the activation loop (30, 37), the paradigm has been that the type II PAKs are always “on,” and that the RHO-family GTPases function to transport a proverbial lit fire starter to the fireplace. Our data now provide a distinct “off” conformation for the type II PAKs, occurring even when the activation loop is phosphorylated. This finding fits well with the previous structural descriptions, provides a pan-type II PAK regulation mechanism, explains the differences in activities of the full-length and catalytic domain constructs, and presumably allows coordination of type II PAK activation with delivery to specific subcellular compartments by RHO GTPases.
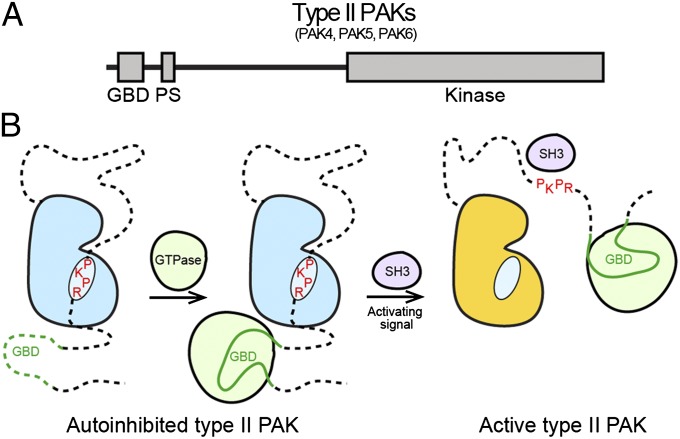
The autoinhibitory pseudosubstrate region (Fig. 6A) that we have found for type II PAKs is in a proline-rich region, suggesting the potential for interactions with SH3 domains. In the type I PAKs, an atypical PxxP motif (PxxxPR) interacts with a nonclassical SH3 domain in the guanine nucleotide exchange factor PIX (38). We tested whether β-PIX or Src SH3 domains could activate PAK4 and found that β-PIX SH3 could not activate PAK4, but surprisingly that Src SH3 domain could (Fig. 5F). There is therefore scope for activation for type II PAKs by pseudosubstrate autoinhibition release through binding to a SH3 domain containing proteins, potentially Src; this mechanism clearly could be dependent on subcellular localization. This two-step activation process is attractive because it would allow first a small GTPase to bind the type II PAK GBD and to relocalize the kinase to the desired position in the cell, and second, an activation signal (e.g., SH3 domain) to release the autoinhibition and activate the kinase (Fig. 6B). Interestingly, this mechanism is distinct from that observed for type I PAKs where an autoinhibitory dimer is disrupted by small GTPase binding (39).
Fig. 6.
Regulation of type II PAKs. (A) Updated schematic diagram for type II PAK family members. The type II PAKs contain a GBD, an autoinhibitory pseudosubstrate (PS), and a C-terminal kinase domain. (B) Schematic showing regulation of type II PAKs. Activation loop-phosphorylated type II PAKs are autoinhibited by the pseudosubstrate sequence RPKP. GTPase binding to the GBD does not activate type II PAKs, but is important for subcellular localization. A second signal (e.g., an SH3 domain) releases pseudosubstrate autoinhibition to allow kinase activity, possibly through direct interaction with the pseudosubstrate sequence or another portion of the N terminus.
There has been much recent interest in PAKs as an emerging target for small-molecule inhibitors to treat human cancer (4). This interest has been driven by both the frequent overexpression and increased activation observed for PAK family members in cancer, although a failed clinical trial of a relatively potent small-molecule inhibitor of the type II PAKs with poor pharmacokinetic properties, PF-3758309, has somewhat dampened enthusiasm for PAK inhibitors. We wondered, however, whether mutations in the type II PAKs had previously been observed in cancer. On analysis of the COSMIC database of cancer mutations we discovered that the autoinhibitory pseudosubstrate region of PAK5 and PAK6 has been found to be mutated in human cancer. PAK5 was found to be mutated at P50 (P50L) in lung cancer (40), and PAK6 in two independent melanomas on P52 (P52L) (2, 41), suggesting dysregulation of this mechanism of type II PAK autoregulation can be associated with human cancer. The discovery of recurrent point mutations in the type II PAKs that deregulate kinase activity may help provide rationale for new innovative therapeutic interventions.
Materials and Methods
Expression and Purification.
We used PAK4 isoform 2, which encodes a 426-aa protein (UniProt accession no. O96013-2). For full-length PAK4, we subcloned residues 5–426 (PAK4-FL) into a modified pET-28 vector with an N-terminal hexahistidine (6xHis) tag removable by tobacco etch virus protease. For catalytic domain PAK4, we subcloned residues 109–426 (PAK4-cat) into the same expression vector. Because residues 121–426 of isoform 2 correspond exactly to residues 286–591 in isoform 1, for clarity we discuss kinase domain residues using the isoform 1 numbering throughout. We use isoform 2 numbering for residues in the N terminus (residues 1–119 are identical between isoform 1 and isoform 2). PAK4-FL elutes from size exclusion chromatography as a monomeric protein, not a dimer, as was previously observed for type I PAKs. PAK6 (UniProt accession no. Q9NQU5) residues 383–674 was generated.
Structure Determination.
For cocrystallizations, we preincubated PAK4 with peptide. PAK4 kinase domain (PDB ID 2CDZ) (31) was used as a molecular replacement search model.
Kinase Assay.
Recombinant purified PAK4-cat135–426 was considered optimally active. Kinase assays were performed using radioactive ATP, and measured activities were controlled for addition of MalBP and compared with PAK4-cat135–426. CDC42, RAC1, Src SH3, and β-PIX SH3 were expressed and purified as N-terminal 6xHis-tagged proteins. PAK4 binding to GTPases was confirmed by pull-down (Fig. S5).
Bcl-2/Bcl-XL Antagonist Causing Cell Death Assay.
HEK 293 cells were cotransfected with pEBG-Bcl-2/Bcl-XL antagonist causing cell death (BAD), and empty pEGFP, pEGFP-PAK4, pEGFP-PAK44A, or pEGFP-PAK4-cat.
Cell Morphology and Confocal Imaging.
NIH 3T3 cells were transfected and examined by fluorescence microscopy. COS-7 cells were transiently transfected and examined with a multicolor spinning-disk confocal system.
Supplementary Material
Acknowledgments
The authors thank the National Synchrotron Light Source beamlines X6A and X29; the Northeastern Collaborative Access Team at the Advanced Photon Source; and K. Draheim, D. Calderwood, and A. Koleske. Support for this work was provided by National Institutes of Health Grants GM102262 (to B.E.T. and T.J.B.) and GM079498 (to B.E.T.); Yale SPORE in Skin Cancer Grant CA121974 (to R.H.), Gilead Sciences Grant YG-001-11 (to J.S. and T.J.B.); China Scholarship Council Grant 2011666003 (to J.G.); and Guangxi University Joint PhD Training Program Fund L300279 (to J.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FIE–4FIJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214447109/-/DCSupplemental.
References
- 1.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 2.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 4.Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–473. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- 5.Abo A, et al. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu J, et al. PAK4 kinase is essential for embryonic viability and for proper neuronal development. Mol Cell Biol. 2003;23:7122–7133. doi: 10.1128/MCB.23.20.7122-7133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T. PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci. 2005;118:1861–1872. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- 8.Dan C, Kelly A, Bernard O, Minden A. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, et al. p21-activated kinase 4 phosphorylation of integrin beta5 Ser-759 and Ser-762 regulates cell migration. J Biol Chem. 2010;285:23699–23710. doi: 10.1074/jbc.M110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells CM, Whale AD, Parsons M, Masters JR, Jones GE. PAK4: A pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–1673. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnesutta N, Qu J, Minden A. The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J Biol Chem. 2001;276:14414–14419. doi: 10.1074/jbc.M011046200. [DOI] [PubMed] [Google Scholar]
- 12.Wells CM, Abo A, Ridley AJ. PAK4 is activated via PI3K in HGF-stimulated epithelial cells. J Cell Sci. 2002;115:3947–3956. doi: 10.1242/jcs.00080. [DOI] [PubMed] [Google Scholar]
- 13.Qu J, et al. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol Cell Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nekrasova T, Jobes ML, Ting JH, Wagner GC, Minden A. Targeted disruption of the Pak5 and Pak6 genes in mice leads to deficits in learning and locomotion. Dev Biol. 2008;322:95–108. doi: 10.1016/j.ydbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Siu MK, et al. p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc Natl Acad Sci USA. 2010;107:18622–18627. doi: 10.1073/pnas.0907481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callow MG, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed T, Shea K, Masters JR, Jones GE, Wells CM. A PAK4-LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell Signal. 2008;20:1320–1328. doi: 10.1016/j.cellsig.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, et al. The pak4 protein kinase plays a key role in cell survival and tumorigenesis in athymic mice. Mol Cancer Res. 2008;6:1215–1224. doi: 10.1158/1541-7786.MCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Li Z, Viklund EK, Strömblad S. P21-activated kinase 4 interacts with integrin alpha v beta 5 and regulates alpha v beta 5-mediated cell migration. J Cell Biol. 2002;158:1287–1297. doi: 10.1083/jcb.200207008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Minden A. PAK4 functions in tumor necrosis factor (TNF) alpha-induced survival pathways by facilitating TRADD binding to the TNF receptor. J Biol Chem. 2005;280:41192–41200. doi: 10.1074/jbc.M506884200. [DOI] [PubMed] [Google Scholar]
- 22.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006;20:1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 23.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur R, Yuan X, Lu ML, Balk SP. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate. 2008;68:1510–1516. doi: 10.1002/pros.20787. [DOI] [PubMed] [Google Scholar]
- 25.Murray BW, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci USA. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eswaran J, Soundararajan M, Kumar R, Knapp S. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008;33:394–403. doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Ching YP, Leong VY, Wong CM, Kung HF. Identification of an autoinhibitory domain of p21-activated protein kinase 5. J Biol Chem. 2003;278:33621–33624. doi: 10.1074/jbc.C300234200. [DOI] [PubMed] [Google Scholar]
- 28.Baldassa S, Calogero AM, Colombo G, Zippel R, Gnesutta N. N-terminal interaction domain implicates PAK4 in translational regulation and reveals novel cellular localization signals. J Cell Physiol. 2010;224:722–733. doi: 10.1002/jcp.22172. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, et al. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 30.Baskaran Y, Ng YW, Selamat W, Ling FT, Manser E. Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 2012;13:653–659. doi: 10.1038/embor.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eswaran J, et al. Crystal structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure. 2007;15:201–213. doi: 10.1016/j.str.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey A, et al. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene. 2002;21:3939–3948. doi: 10.1038/sj.onc.1205478. [DOI] [PubMed] [Google Scholar]
- 34.Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Wu JW, Wang ZX. Structural insights into the autoactivation mechanism of p21-activated protein kinase. Structure. 2011;19:1752–1761. doi: 10.1016/j.str.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Schrantz N, et al. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem. 2004;279:1922–1931. doi: 10.1074/jbc.M311145200. [DOI] [PubMed] [Google Scholar]
- 38.Manser E, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 39.Lei M, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 40.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 41.Wei X, et al. NISC Comparative Sequencing Program Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.