Abstract
The conserved Ndc80 complex is an essential microtubule-binding component of the kinetochore. Recent findings suggest that the Ndc80 complex influences microtubule dynamics at kinetochores in vivo. However, it was unclear if the Ndc80 complex mediates these effects directly, or by affecting other factors localized at the kinetochore. Using a reconstituted system in vitro, we show that the human Ndc80 complex directly stabilizes the tips of disassembling microtubules and promotes rescue (the transition from microtubule shortening to growth). In vivo, an N-terminal domain in the Ndc80 complex is phosphorylated by the Aurora B kinase. Mutations that mimic phosphorylation of the Ndc80 complex prevent stable kinetochore-microtubule attachment, and mutations that block phosphorylation damp kinetochore oscillations. We find that the Ndc80 complex with Aurora B phosphomimetic mutations is defective at promoting microtubule rescue, even when robustly coupled to disassembling microtubule tips. This impaired ability to affect dynamics is not simply because of weakened microtubule binding, as an N-terminally truncated complex with similar binding affinity is able to promote rescue. Taken together, these results suggest that in addition to regulating attachment stability, Aurora B controls microtubule dynamics through phosphorylation of the Ndc80 complex.
Keywords: mitosis, Hec1, single molecule, optical trap, total internal reflection fluorescence microscopy
During mitosis, replicated chromosomes are segregated by the mitotic spindle, a bipolar array of dynamic microtubules. Each chromatid is linked to a bundle of microtubules (a “K-fiber”) by a kinetochore. To ensure accurate chromosome segregation, regulatory mechanisms detect and correct errors in attachments between kinetochores and spindle microtubules. The conserved Aurora B kinase plays a crucial role in the resolution of aberrant kinetochore-microtubule attachments (1). Aurora B has many identified targets at the kinetochore, and it is generally thought that phosphorylation of these targets triggers the release of incorrect attachments (2–7). However, emerging evidence suggests that Aurora B activity does not always result in kinetochore-microtubule detachment. For example, early in mitosis when merotelic attachments are more prevalent, phosphorylation of the Ndc80 complex (the key microtubule-binding component of the kinetochore) is relatively high, yet kinetochores do not appear to release from their K-fibers (8, 9). Similarly, syntelic attachments formed in the presence of a reversible Aurora B inhibitor are not immediately released when the kinase is reactivated (10). Instead, the K-fiber microtubules disassemble, carrying the kinetochores back to the centrosome, where the attachments are corrected by an unknown mechanism. These results suggest that Aurora B additionally acts to regulate microtubule dynamics as a part of its mechanism of error correction.
Additional findings suggest that Aurora B modulates microtubule dynamics through regulation of the Ndc80 complex. A component of the Ndc80 complex, the Hec1 protein, has a disordered N-terminal tail that is targeted by Aurora B in vivo (11, 12). In PtK cells, preventing phosphorylation of these target sites not only results in hyperstable kinetochore-microtubule attachments, but also damped kinetochore oscillations (8). The abnormal oscillations could be explained by direct or indirect contributions from the Ndc80 complex. The Ndc80 complex could itself directly control microtubule dynamics in response to Aurora B activity. An alternative (but not mutually exclusive) explanation is that phosphorylation of Hec1 alters the localization of other factors that modulate dynamics. These factors may include the microtubule stabilizer EB1 and the microtubule depolymerase MCAK, both of which are also targets of Aurora B (13–19).
Here, we show that the human Ndc80 complex directly stabilizes the tips of disassembling microtubules, slows the rate of disassembly, and promotes microtubule rescue (the transition from microtubule shortening to growth) in vitro. In contrast, the Ndc80 complex with mutations mimicking Aurora B phosphorylation was impaired in its ability to influence microtubule dynamics, even when tracking with the tips of disassembling microtubules. This diminished ability of the phosphomimetic complex to affect dynamics is not solely a result of weakened microtubule binding, as an N-terminally truncated complex with similar affinity was still able to promote rescue. These results suggest that Aurora B modulates microtubule dynamics through regulation of the Ndc80 complex, and this mechanism could be separable from effects on attachment stability.
Results
Characterization of Full-Length Human Ndc80 Complex.
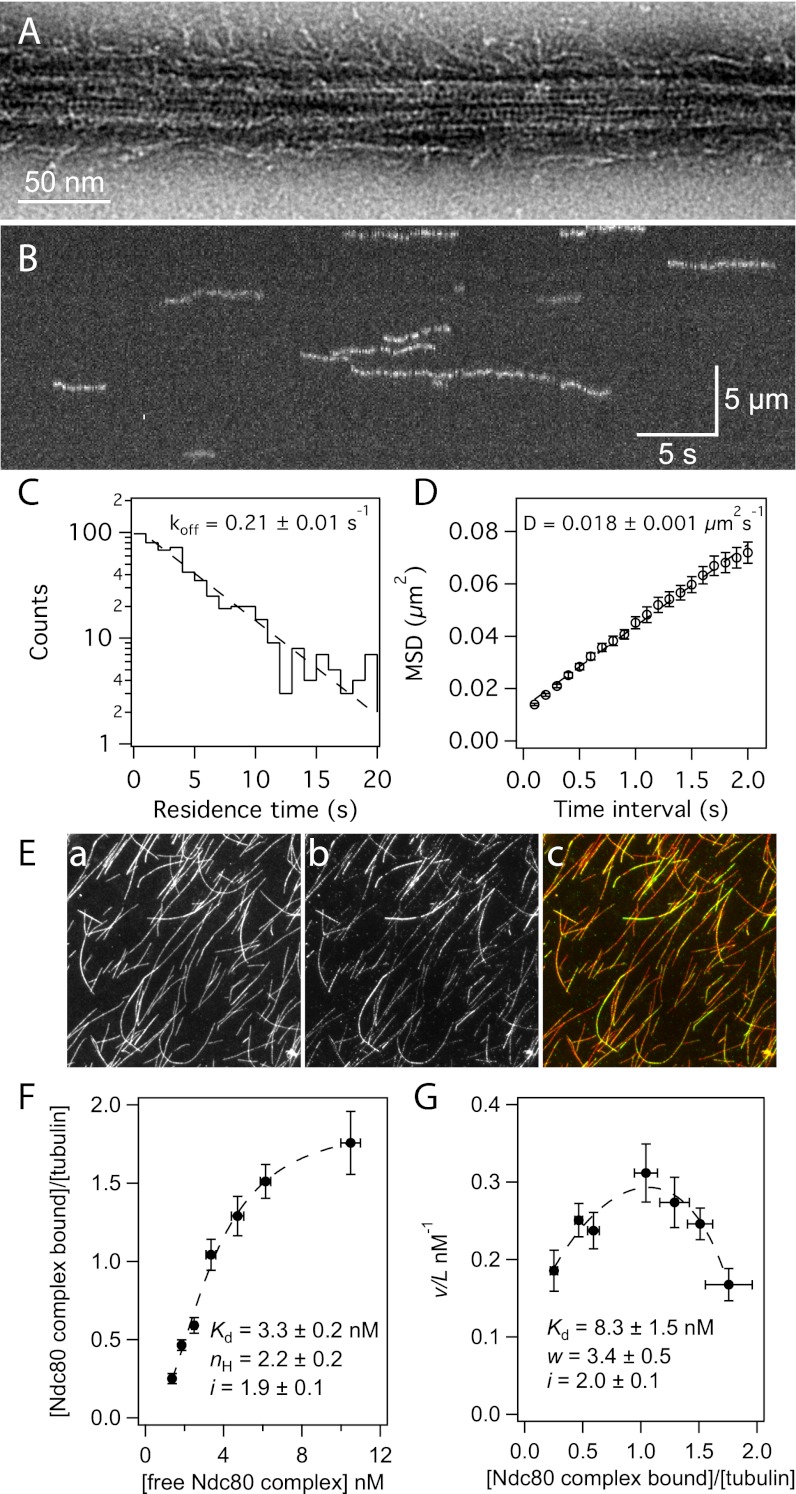
The conserved Ndc80 complex is an essential microtubule-binding component of the kinetochore (20). Although the Ndc80 complex from yeast and worms has been extensively studied in vitro (21–26), most work on the human complex has been limited to the use of truncated forms (26–31). We expressed and purified full-length human Ndc80 complex from Escherichia coli for in vitro characterization (Fig. S1). As seen by negative-stain EM, this recombinant Ndc80 complex bound to taxol-stabilized microtubules (Fig. 1A). Using total internal reflection fluorescence (TIRF) microscopy, we visualized single molecules of GFP-tagged Ndc80 complex on taxol-stabilized microtubules (Fig. 1B and Fig. S2) and measured their dissociation and diffusion rate constants (koff = 0.21 ± 0.01 s−1, D = 0.018 ± 0.001 μm2⋅s−1) (Fig. 1 C and D). The affinity and cooperativity of microtubule binding were measured using a bulk microtubule binding assay that measures the amount of GFP-tagged complex bound to microtubules over varying concentrations of complex (Fig. 1E) (32, 33). Based on a standard Hill model fit (34), the Ndc80 complex binds microtubules with a strong apparent affinity (Kd = 3.3 ± 0.2 nM) and has a Hill coefficient of 2.2 ± 0.2 (Fig. 1F). The Hill model describes cooperativity arising from allosteric changes that enhance ligand binding to a protein. In our binding assay, cooperativity is likely based on interactions between Ndc80 complexes that occur when they are bound to microtubules. Therefore, we used a model previously developed by McGhee and von Hippel that describes cooperativity between ligands binding to a polymer lattice (35). Fitting the binding data with this model (Fig. 1G) also showed a strong apparent affinity (Kd = 8.3 ± 1.5 nM) and cooperativity between Ndc80 complexes on the microtubule lattice (w = 3.4 ± 0.5). Compared with the Hill model fit, the McGhee and von Hippel model fit yielded a weaker apparent Kd for a single complex. Thus, interactions between complexes bound to the microtubule contribute to the Kd predicted by the Hill model. This finding is supported by the observation that at high concentrations, truncated Ndc80 complex binds microtubules in clusters (28). Fits to both models revealed a lattice occupancy of approximately two Ndc80 complexes per tubulin dimer, consistent with cryo-EM reconstructions that showed a 4-nm spacing of the truncated complex on microtubules (28).
Fig. 1.
The human Ndc80 complex binds to and diffuses along the microtubule lattice. (A) Negative-stain electron micrograph of the Ndc80 complex on a taxol-stabilized microtubule. (B) A representative kymograph showing the binding and diffusion of Ndc80 complex (5 pM complex in solution) on taxol-stabilized microtubules. Position along the microtubule is depicted on the vertical axis over time on the horizontal axis. (C) Residence time distributions of GFP-tagged Ndc80 complex on microtubules fit with a single exponential (dashed line) to calculate the off-rate constant, koff. (D) Mean-squared displacement (MSD) ± SEM vs. time lag. A linear fit to the data (dashed line) was used to determine the diffusion constant, D. (C and D) n = 584. (E) Representative image from the bulk microtubule binding assay with GFP-tagged Ndc80 complex (2 nM) on taxol-stabilized Alexa-568-labeled microtubules (2.5 nM tubulin dimer). Panels show microtubules (a), Ndc80 complex (b), and merge (c). Panel dimensions are 66 by 66 μm. (F) Plot of binding density (v) versus free Ndc80 complex concentration (L). A fit to the Hill model (dashed line) was used to determine the apparent affinity (Kd), Hill coefficient (nH), and lattice occupancy (i, the number of Ndc80 complexes bound per tubulin dimer). (G) Scatchard plot of the same data shown in F, fit to the McGhee and von Hippel model (dashed line) to calculate the Kd, cooperativity parameter (w), and i. For F and G, n = 8–10 replicates per data point, markers are mean ± SEM, and errors on model fit parameters (Kd, nH, w, and i) represent SD.
Ndc80 Complex Directly Stabilizes Disassembling Microtubule Tips and Promotes Microtubule Rescue.
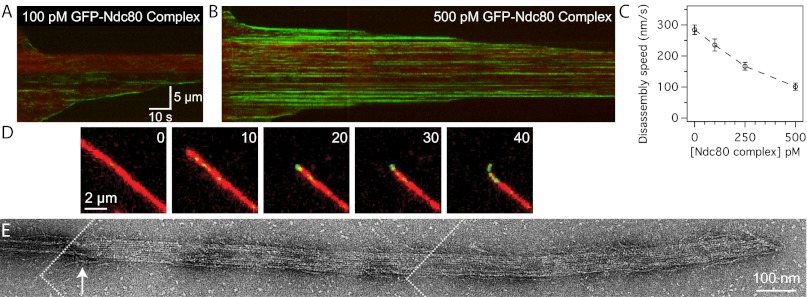
In vivo, kinetochores transmit forces generated by the mitotic spindle to drive chromosome movement (36). This process depends on the ability of microtubule-binding components of the kinetochore to form stable attachments to dynamic microtubule tips. Using TIRF microscopy, we visualized the GFP-tagged Ndc80 complex on disassembling microtubules. In these assays, microtubule disassembly was induced by the removal of free tubulin. The human Ndc80 complex can track with disassembling microtubule tips (Fig. 2A), unlike the budding yeast Ndc80 complex, which requires the Dam1 complex or oligomerization on the surface of beads (23, 24). The human Ndc80 complex also slowed the rate of microtubule disassembly (Fig. 2C). As the concentration of the complex was increased from 0 to 500 pM, microtubule disassembly was slowed from 280 ± 20 nm/s to 100 ± 10 nm/s.
Fig. 2.
The Ndc80 complex slows microtubule disassembly and stabilizes protofilament extensions. Kymographs of disassembling microtubules (red) in the presence of (A) 100 pM or (B) 500 pM GFP-tagged Ndc80 complex (green). Brightness and contrast were adjusted equally in A and B. (C) Mean disassembly speeds ± SEM for microtubules in the presence of increasing concentrations of Ndc80 complex (without Ndc80 complex, n = 80; 100 pM Ndc80 complex, n = 31; 250 pM, n = 29; 500 pM, n = 34). (D) Time-lapse images of a disassembling microtubule (red) in the presence of 500 pM GFP-tagged Ndc80 complex (green) as a curled extension formed at the tip. Inset numbers show elapsed time, in seconds. See Fig. S3B for a gallery of images showing curled extensions. (E) Negative-stain electron micrograph of a disassembling microtubule tip (see SI Materials and Methods) stabilized by the Ndc80 complex. An arrow marks the transition from a closed microtubule to an open sheet. The figure was constructed from three images, the boundaries of which are depicted by dotted white lines.
At 500 pM, bright particles of GFP-tagged Ndc80 complex were observed on microtubules (Fig. 2B), consistent with its cooperative binding behavior in our bulk assays. In some cases, disassembly appeared to stall as the tip reached these particles, and only continued after the Ndc80 complex appeared to detach. This behavior resulted in a step-like appearance in kymographs (Fig. 2B). Furthermore, Alexa-647–labeled tubulin decorated with Ndc80 complex was often seen bending away from the long axis of the microtubule (observed for 66 ± 10% of microtubules) (Fig. 2D, Fig. S3 A and B, and Movie S1). Because these curled extensions can be resolved by light microscopy (116-nm pixels), their curvature is gentler than the tight 20-nm curls seen at bare disassembling tips by cryo-EM in vitro (37). To further investigate tip structure in the presence of Ndc80 complex, we performed a similar disassembly assay and visualized the microtubule tips by negative-stain EM. We observed open protofilament sheets emanating from the tips of microtubules stabilized by Ndc80 complex (Fig. 2E). These sheets were not observed at the tips of microtubules stabilized by taxol or by the Dam1 complex (Fig. S3C) (38). Microtubules exposed to the same conditions in the absence of any stabilizing factor completely disassembled into free tubulin. Although we were unable to distinguish between microtubule plus- and minus-ends in the electron micrographs, curled extensions were observed in the presence of Ndc80 complex at both microtubule ends in the TIRF assay (Fig. S3A). Together, the TIRF and EM assays suggest that the Ndc80 complex slows disassembly by stabilizing protofilament extensions at microtubule tips.
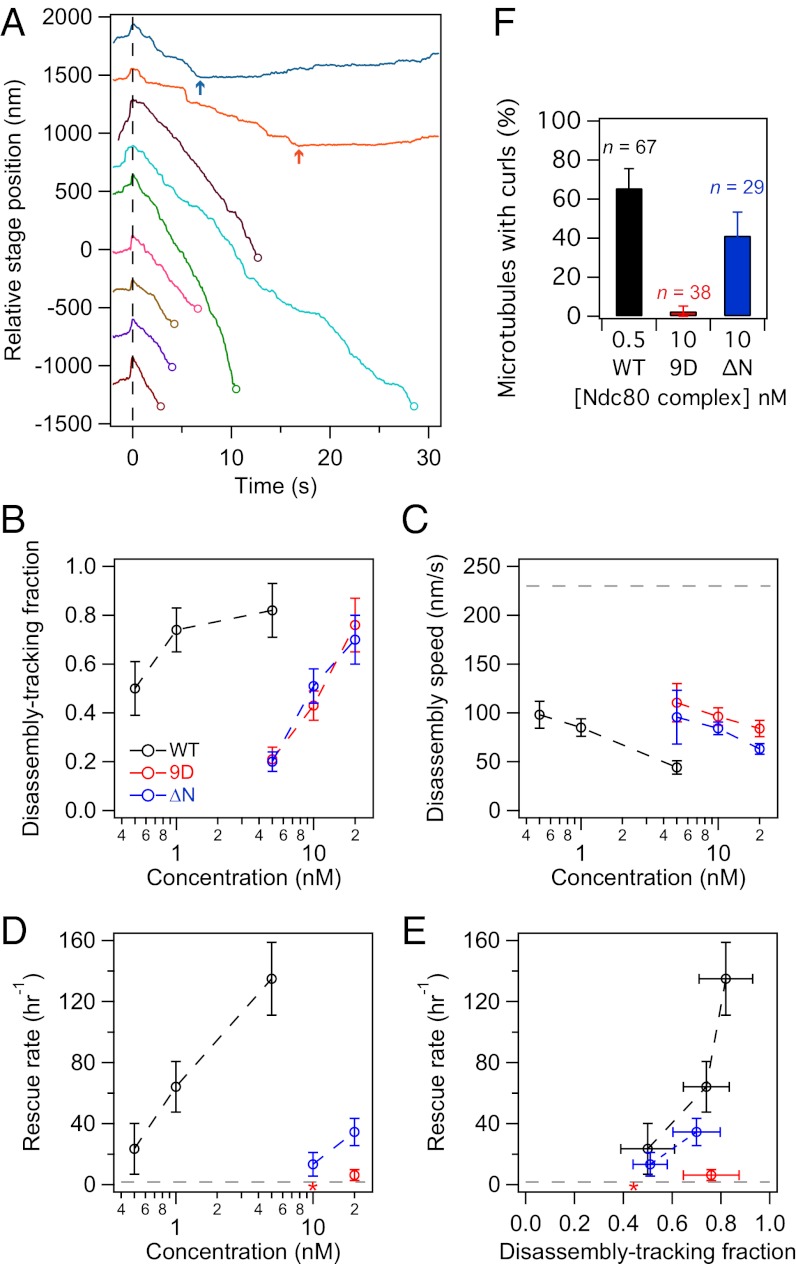
To test how purified Ndc80 complex couples to dynamic microtubule tips under force, we used an optical trap-based bead motility assay (39). By incubating 11 pM beads with 5 nM Ndc80 complex (∼450 complexes per bead), we estimate that up to ∼20 complexes can interact with the microtubule tip based on geometric constraints (23). This number closely approximates the number of Ndc80 complexes per kinetochore microtubule in vivo (40). These beads remained coupled to microtubule tips against 2 pN of tension (Fig. 3 A and B), similar to the forces sustained by kinetochore-microtubule attachments in vivo, which are estimated to be 0.4–8 pN (23, 41, 42). Against the applied force, beads tracked robustly with the tips of disassembling microtubules over an average distance of 970 ± 190 nm (n = 44). Consistent with results from our TIRF-based assays, microtubule disassembly was slowed from 230 ± 14 nm/s (for microtubule tips not coupled to beads and in the absence of force) to 44 ± 7 nm/s by beads coated with Ndc80 complex under 2 pN of force (Fig. 3C). For episodes of disassembly-driven movement against the applied force, about half (53 ± 8%) ended in bead detachment, but a large proportion (40 ± 7%) underwent a microtubule rescue (the remaining events, 7 ± 3%, terminated for other reasons, such as the bead reaching the microtubule seed or nonspecifically adhering to the cover-slip). Strikingly, disassembling microtubule tips coupled to beads coated with the Ndc80 complex rescued ∼70-fold more frequently than bare microtubules (Fig. 3D) (135 ± 24 h−1 compared with 2 ± 1 h−1). Therefore, the Ndc80 complex is an effective tip-coupler that can directly slow microtubule disassembly and promote rescue.
Fig. 3.
Phosphomimetic mutations in the Ndc80 complex inhibit its ability to promote microtubule rescue. (A) Example traces of position vs. time for beads decorated with Ndc80 complex as they tracked microtubule disassembly against ∼2 pN of applied force. Time t = 0 s (dashed vertical line) marks the onset of tracking, when the disassembling microtubule tip began to drive movement of the bead against the force of the trap. Disassembly-driven movement ended when the bead detached (open circles) or when the microtubule rescued (arrows). Traces are offset vertically for visual clarity. (B) The fraction of beads coated with wild-type or mutant Ndc80 complex capable of tracking against ∼2 pN. From the disassembly-tracking events in B, (C) mean microtubule disassembly speeds ± SEM and (D) rescue rates were measured. Without load and in the absence of bead-bound Ndc80 complex, the disassembly rate was 230 ± 14 nm/s (dashed line in C, n = 26) and the rescue rate was 2 ± 1 h−1 (dashed line in D, n = 3 events in 104 min of disassembly). (E) Rescue rate is plotted against the fraction of beads that tracked disassembly against force. (F) Percentage of microtubules for which a curl (Fig. 2D and Fig. S3B) was observed at either tip during disassembly in the TIRF microscopy assay. The n for each data point in B–D is listed in Table S1. Asterisks indicate that no rescues were observed. Unless otherwise noted, all error bars represent uncertainties from counting statistics.
In contrast, in our previous work with the budding yeast Ndc80 complex, we observed little effect on the rate of microtubule rescue (23). Here we analyzed the dataset reported in Powers et al. (23), specifically looking for rescue events. Microtubules rescued at a frequency of 9 ± 5 h−1 while coupled to beads coated with budding yeast Ndc80 complex (n = 4 rescues, ∼100–2,700 complexes per bead, against ∼1 pN of force). This number is close to the rate of rescue for microtubules not coupled to beads (reported above). Therefore, the budding yeast Ndc80 complex, unlike the human complex, appears to have little ability to promote microtubule rescue.
Phosphomimetic Mutations in the Ndc80 Complex Inhibit Its Ability to Influence Microtubule Dynamics.
The Hec1 protein of the Ndc80 complex contains a calponin homology domain that is important for its microtubule binding activity (29, 30). In addition, Hec1 has a disordered N-terminal tail that contributes to the affinity of the complex for microtubules (11, 12, 30). In vivo, the tail is a target for the Aurora B kinase, and mutations that mimic phosphorylation at these sites result in unattached kinetochores (11, 43). Consistent with this observation, Aurora B phosphorylation of a truncated Ndc80 complex reduces its binding to microtubules in vitro (29). On the other hand, mutations that block phosphorylation severely damp kinetochore oscillations in vivo (8). These findings suggest that phosphorylation in the Hec1 tail is required not only for regulation of kinetochore-microtubule attachments, but also for normal kinetochore-microtubule dynamics. Using the optical trap assay, we tested the direct contribution of the tail to microtubule dynamics in vitro. In addition to the wild-type complex, we purified Ndc80 complex with the nine putative Aurora B target sites in the Hec1 tail mutated to aspartic acid to mimic phosphorylation (9D), and Ndc80 complex with the Hec1 tail deleted (ΔN). As a control, we also purified Ndc80 complex with alanine mutations at the Aurora B target sites (9A). Because this construct behaved like the wild-type complex in our TIRF assays (Figs. S3B and S4), it was not further characterized in the optical trap assay.
Beads coated with wild-type, 9D, and ΔN complexes were all able to slow the rate of microtubule disassembly in a concentration-dependent manner when tracking with disassembling tips against ∼2 pN of applied force (Fig. 3 B and C). However, the 9D and ΔN complexes were impaired relative to the wild-type complex; when incubated with 5 nM of Ndc80 complex, 82 ± 11% of wild-type beads tracked with disassembling microtubule tips, but only 21 ± 5% of the 9D beads and 20 ± 4% of the ΔN beads tracked with disassembly (Fig. 3B). Furthermore, 5 nM wild-type beads slowed disassembly to 44 ± 7 nm/s, but 5 nM 9D and 5 nM ΔN beads slowed disassembly to 110 ± 20 and 96 ± 27 nm/s, respectively (Fig. 3C). The ability of the mutant complexes to track with and slow disassembly was recovered to wild-type levels by increasing the density of decoration on beads ∼20-fold (Fig. 3 B and C: compare 0.5 and 1 nM wild-type to 10 and 20 nM mutant complexes, respectively). For example, beads coated with 20 nM 9D or 20 nM ΔN complex tracked with microtubules similarly to 1 nM wild-type beads (9D: 76 ± 11%; ΔN: 70 ± 10%; wild-type: 74 ± 9%). Therefore, increasing the number of mutant complexes on beads compensates for their decreased coupling performance.
When assayed at comparable coupling performance, the wild-type and ΔN complexes promoted microtubule rescue, but the 9D complex did not (Fig. 3 D and E). Deletion of the Hec1 tail reduced the ability of the complex to promote rescue only modestly (∼twofold). In contrast, phosphomimetic mutations in the tail nearly abolished this activity. Using beads coated with 20 nM 9D complex, we observed only three rescue events in 29 min of microtubule disassembly, which is an average rescue frequency similar to that for bare microtubules (6 ± 4 vs. 2 ± 1 h−1). In addition, the ΔN complex but not the 9D complex stabilized curled extensions at disassembling microtubule tips in the TIRF assay (Fig. 3F). Thus, phosphomimetic mutations do not simply negate the activity of the tail, but actively interfere with the ability to modify microtubule tip structure and promote rescue.
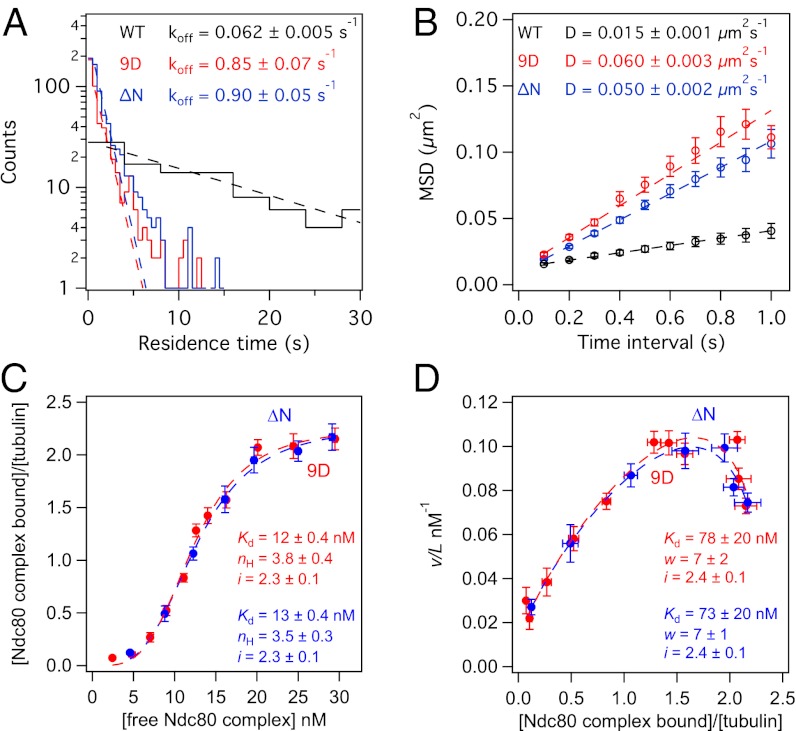
The ΔN and 9D complexes performed similarly in tracking with and slowing microtubule disassembly, suggesting that their disparate effects on microtubule rescue and tip structure are not simply the result of a difference in their microtubule-binding affinities. We quantified binding of GFP-tagged 9D and ΔN complexes directly by single-molecule TIRF microscopy and bulk microtubule binding assays. Unlike the wild-type complex, binding of the mutant complexes was undetectable in standard BRB80 (120 mM K+) buffer conditions, so the assays were performed in BRB40 (60 mM K+) buffer. Single molecules of 9D and ΔN complex exhibited similar dissociation and diffusion rate constants (Fig. 4 A and B) (koff = 0.85 ± 0.07 and 0.90 ± 0.05 s−1, and D = 0.060 ± 0.003 and 0.050 ± 0.002 μm2⋅s−1, respectively). In contrast, wild-type Ndc80 complexes dissociated from microtubules ∼15-times more slowly (koff = 0.062 ± 0.005 s−1) and diffused on the lattice ∼four-times more slowly (D = 0.015 ± 0.001 μm2⋅s−1). It has been previously suggested that the Hec1 tail contributes to microtubule binding by the Ndc80 complex or mediates cooperativity between complexes on microtubules (11, 12, 28). Our results establish that the tail contributes directly to microtubule binding, because deletion of the tail causes individual Ndc80 complexes (in the absence of cooperative binding) to dissociate more quickly from microtubules.
Fig. 4.
The 9D and ΔN Ndc80 complexes exhibit similar binding behavior on microtubules. (A) Histograms of the residence time for single molecules (5 pM complex in solution) of wild-type (black trace, n = 131), 9D (red trace, n = 497), and ΔN (blue trace, n = 705) Ndc80 complex on taxol-stabilized microtubules. Each histogram was fit by a single exponential (dashed lines) to determine the off-rate constant, koff. (B) Plots of MSD vs. time lag for binding events in A. The diffusion constant, D, was measured from linear fits to the data (dashed lines). (C and D) Bulk binding assays of 9D (red traces, n = 4–7 replicates per data point) and ΔN (blue traces, n = 6–7 replicates per data point) Ndc80 complex on taxol-stabilized microtubules. Dashed lines show fits of binding data to (C) Hill and (D) McGhee and von Hippel models. Errors on model fit parameters (Kd, nH, w, and i) represent SD. All markers represent mean ± SEM and all assays were performed in BRB40 buffer (see SI Materials and Methods).
In the bulk binding assay, fits to both the Hill and the McGhee and von Hippel models (Fig. 4 C and D) show that the 9D and ΔN complexes are indistinguishable from one another in their apparent affinities, cooperativity constants, and lattice occupancies (McGhee and von Hippel fit for 9D and ΔN: Kd = 78 ± 20 and 73 ± 20 nM, w = 7 ± 2 and 7 ± 1, i = 2.4 ± 0.1 and 2.4 ± 0.1 per tubulin dimer, respectively). Therefore, phosphomimetic mutations reduce the affinity of the Ndc80 complex for microtubules and impair its ability to promote microtubule rescue. However, these two effects are not strictly coupled; deletion of the Hec1 tail equally reduces the affinity of the complex for microtubules, but is not as detrimental to its ability to modify microtubule tip structure and dynamics. These findings suggest that Aurora B phosphorylation has separable effects on attachment stability and microtubule dynamics at the kinetochore.
Discussion
Ndc80 Complex Directly Modulates Microtubule Dynamics.
The Ndc80 complex is a conserved and essential microtubule-binding component of the kinetochore. Here, we characterized the binding of full-length human Ndc80 complex to microtubules in vitro. The Ndc80 complex bound cooperatively to microtubules with a strong affinity, and directly promoted microtubule rescue. Our in vitro results using unphosphorylated wild-type Ndc80 complex explain observations made in cells. In the absence of Hec1 phosphorylation, we found that the Ndc80 complex antagonizes microtubule disassembly. This effect explains why blocking Hec1 phosphorylation in vivo causes hyper-stabilized K-fibers, and leads to damped sister kinetochore oscillations and severe defects in cell division (8). We believe our findings are unique in representing a demonstration that a core component of the human kinetochore directly modifies microtubule rescue rate in vitro. This ability has been shown previously for a core kinetochore component only once, with the budding yeast Dam1 complex (44), which has no known homolog in higher eukaryotes. Notably, the budding yeast Ndc80 complex does not effectively promote microtubule rescue (23), even though the composition and domain structure of the complex are highly conserved.
Our results also indicate a possible mechanism by which the Ndc80 complex promotes microtubule rescue. In the absence of stabilizing factors, protofilaments at disassembling microtubule tips form tight ∼20-nm curls (37). When microtubules are stabilized by a nonhydrolyzable GTP analog, protofilaments are straighter at disassembling tips (45). We found that the tips of disassembling microtubules in the presence of Ndc80 complex were gently curved (as seen by TIRF microscopy) and formed large protofilament sheets (as seen by EM). These observations suggest that the Ndc80 complex promotes microtubule rescue by stabilizing tip structures with straighter protofilaments. Alushin et al. proposed that the Hec1 calponin homology domain recognizes the interface between tubulin monomers (28) at a putative hinge region (46). Our findings are consistent with this model. Ndc80 complex lacking the Hec1 tail was able to modify microtubule tip structure and promote rescue, indicating that other parts of the complex (outside of the tail) are primarily responsible for this activity. We propose that binding of the Hec1 calponin homology domain at the hinge region between tubulin subunits induces a straighter protofilament conformation that facilitates microtubule rescue.
Aurora B Regulates Microtubule Dynamics Through the Ndc80 Complex.
The Aurora B kinase has an established role in releasing aberrant kinetochore-microtubule attachments (1). Consistent with this model, PtK cells carrying a phosphomimetic mutant Ndc80 (9D) complex have unattached kinetochores (11, 43). We found that the human 9D complex bound to microtubules more weakly relative to the wild-type complex, as determined by three independent in vitro assays. (i) Single molecules of the 9D complex dissociated more quickly (>10-fold) from the microtubule lattice. (ii) In our bulk assays, binding of the 9D complex was undetectable under conditions in which the wild-type complex bound strongly to microtubules. (iii) At equal surface density on beads, the 9D complex was impaired in its ability to track with microtubule disassembly against force. In all three of these assays, the 9D complex behaved similarly to and not worse than Ndc80 complex that lacks the tail domain (ΔN). Therefore, mutations that mimic complete phosphorylation of the Hec1 tail prevent the tail from contributing to microtubule binding.
In vivo observations suggest that in higher eukaryotes, Aurora B does not simply trigger kinetochore-microtubule detachment but additionally regulates microtubule dynamics (8, 10, 47). In PtK cells, syntelic kinetochore-microtubule attachments are not lost immediately following Aurora B activation (10). Instead, reactivation of Aurora B appears to induce disassembly of the kinetochore microtubules, and the kinetochores track with disassembly back to the centrosome, where the attachments are corrected. Our results offer insight into these observations. At higher surface densities on beads (20 nM), the 9D complex tracked robustly with disassembling microtubule tips against force. Based on geometric constraints (23), we estimate ∼80 complexes can interact with the microtubule tip at this surface density. This number is more than the number of Ndc80 complexes per microtubule in vivo (∼20 per microtubule), but fewer than the number of complexes at a single mammalian kinetochore, which binds 20–25 microtubules through more than 400 attachments (40, 48). Relative to the wild-type complex, the 9D and ΔN complexes are similarly impaired in their binding affinity and tracking performance. However, the ΔN complex promotes microtubule rescue, but the 9D complex does not. Thus, a phosphomimetic Hec1 tail interferes with the ability of the Ndc80 complex to modulate microtubule dynamics, possibly by blocking the ability of the calponin homology domain to stabilize a straighter protofilament conformation. Taken together, these in vitro observations explain how phosphorylation relieves microtubule stabilization at syntelic kinetochores to promote K-fiber disassembly, allowing the attached kinetochores to track back to the centrosome.
Here, we show that a conserved core microtubule-binding component of the human kinetochore directly influences microtubule dynamics. In addition, we find that phosphomimetic mutations of essential Aurora B phosphorylation sites in Hec1 not only weaken attachment, but also nearly abolish the ability of the Ndc80 complex to influence dynamics. These effects are separable, and might be independently tunable through phosphorylation of different subsets of target sites in the Hec1 tail. Taken together, our results indicate that microtubule dynamics can be regulated through Aurora B phosphorylation of the Ndc80 complex.
Materials and Methods
Protein Expression and Purification.
The Ndc80 complex was coexpressed from two di-cistronic plasmids encoding Spc25/Spc24-His6 and Hec1/Nuf2 (see SI Materials and Methods) in E. coli BL21 cells (Rosetta; Novagen). Protein expression and purification were carried out as previously described (23).
TIRF Microscopy.
TIRF microscopy was performed on a custom illumination system (49) (see SI Materials and Methods). Taxol-stabilized Alexa-647-labeled microtubules were bound to the cover-slip with “rigor” kinesin (50). GFP-tagged Ndc80 complex was assayed in BRB80 (80 mM Pipes, 120 mM K+, 1 mM MgCl2, and 1 mM EGTA, pH 6.9) or BRB40 (40 mM Pipes, 60 mM K+, 1 mM MgCl2, and 1 mM EGTA, pH 6.9) with 8 mg⋅mL−1 BSA, 10 μM taxol, and an oxygen scavenger system. For dynamic microtubule assays, GMPCPP-stabilized microtubule seeds were bound to the cover-slip using “rigor” kinesin, and Alexa-647-labeled extensions were grown in BRB80 containing 8 mg⋅mL−1 BSA and 1 mM GTP. Microtubule disassembly was triggered by buffer exchange to remove free tubulin and simultaneously introduce GFP-tagged Ndc80 complex in BRB80 with 8 mg⋅mL−1 BSA, 1 mM GTP, and an oxygen scavenger system. See SI Materials and Methods for additional details.
Microtubule Binding Assays.
Microtubule binding assays were performed as previously described (32), with the following modifications: GFP-tagged Ndc80 complex was incubated with taxol-stabilized microtubules in BRB80 with 10 μM taxol and 8% (vol/vol) gel filtration buffer (50 mM Hepes, 200 mM NaCl, pH 7.6), and pelleted through a glycerol cushion onto a cover-slip. The amount of microtubule-bound Ndc80 complex was quantified by fluorescence microscopy. Increasing concentrations of the complex (0–15 nM) were assayed with microtubules (2.5 nM tubulin dimers) to generate a binding curve. Microtubule binding for the 9D and ΔN Ndc80 complexes was undetectable in BRB80, so binding assays were performed with 0–35 nM complex in BRB40. Binding curves were fitted to the Hill (34) and McGhee and von Hippel (35) models in Igor Pro (Wavemetrics) using iterative least-squares fitting. Errors on curve fit parameters (Kd, nH, w, and i) represent the SD estimated by Igor Pro. See SI Materials and Methods for details of the assay.
Electron Microscopy.
Ndc80 complex (50 nM) was incubated with taxol-stabilized microtubules (37 nM tubulin dimers) in BRB80 with 10 μM taxol. For disassembly assays, microtubules were assembled in the absence of taxol and disassembly was induced by dilution into BRB80 containing 25 nM Ndc80 complex. Samples were applied onto carbon-coated copper grids and stained with uranyl formate. Grids were viewed on a transmission electron microscope (Spirit T12; FEI). Additional details are provided in SI Materials and Methods.
Optical Trap Bead Motility Assays.
Anti-His5 antibody-coated polystyrene beads (11 pM) were functionalized by incubation with His6-tagged wild-type (0.5–5 nM) or mutant (5–20 nM) Ndc80 complex. Beads were attached to the tips of disassembling microtubule extensions, which were grown from GMPCPP-stabilized microtubule seeds bound to the cover-slip. An optical trap was used to apply a constant force of ∼2 pN opposite the direction of microtubule disassembly. Assays were performed in BRB80 containing 1.4 mg⋅mL−1 tubulin, 8 mg⋅mL−1 BSA, 1 mM DTT, 250 μg⋅mL−1 glucose oxidase, 30 μg⋅mL−1 catalase, and 4.5 μg⋅mL−1 glucose. Records of bead position over time were generated and analyzed using custom software (Labview and Igor Pro, respectively). These data were used to determine the fraction of beads that tracked with disassembly, and the rates of microtubule disassembly and rescue. Additional details are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Franck, A. Powers, B. Graczyk, and E. Mazanka for helpful discussions. We also thank the Murdock Charitable Trust and the Washington Research Foundation for support of our electron cryomicroscopy facility. This work was supported by National Institutes of Health Grant T32 GM008268 (to N.T.U.); a Natural Sciences and Engineering Research Council of Canada scholarship (to J.F.T.); Searle Scholar Award Grant 06-L-111 (to C.L.A.); Packard Fellowship for Science and Engineering Grant 2006-30521 (to C.L.A.); National Institute of General Medical Sciences Grants R01 GM40506 (to T.N.D.) and R01 GM079373 (to C.L.A.); Public Health Service National Research Science Award 2T32 GM007270 from the National Institute of General Medical Sciences (to B.S.V.); and the Howard Hughes Medical Institute (T.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 15972.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209615109/-/DCSupplemental.
References
- 1.Liu D, Lampson MA. Regulation of kinetochore-microtubule attachments by Aurora B kinase. Biochem Soc Trans. 2009;37:976–980. doi: 10.1042/BST0370976. [DOI] [PubMed] [Google Scholar]
- 2.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 4.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 7.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 8.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 10.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 11.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter AW, et al. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang K, et al. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009;10:857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 16.Manna T, Honnappa S, Steinmetz MO, Wilson L. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 2008;47:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- 17.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: Role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kline-Smith SL, Sandall S, Desai A. Kinetochore-spindle microtubule interactions during mitosis. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei RR, et al. Structure of a central component of the yeast kinetochore: The Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 27.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell. 2011;22:1217–1226. doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 31.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graczyk B, Davis TN. An assay to measure the affinity of proteins for microtubules by quantitative fluorescent microscopy. Anal Biochem. 2011;410:313–315. doi: 10.1016/j.ab.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gestaut DR, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill AV. The possible effects of the aggregation of molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:iv–vii. [Google Scholar]
- 35.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 36.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 37.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: A time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51:242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 42.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller-Reichert T, Chrétien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: Information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate. Proc Natl Acad Sci USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 48.Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 49.Gestaut DR, Cooper J, Asbury CL, Davis TN, Wordeman L. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 2010;95:641–656. doi: 10.1016/S0091-679X(10)95032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.