Abstract
Type IB topoisomerases (Top1Bs) relax excessive DNA supercoiling associated with replication and transcription by catalyzing a transient nick in one strand to permit controlled rotation of the DNA about the intact strand. The natural compound camptothecin (CPT) and the cancer chemotherapeutics derived from it, irinotecan and topotecan, are highly specific inhibitors of human nuclear Top1B (nTop1). Previous work on vaccinia Top1B led to an elegant model that describes a straightforward dependence of rotation and religation on the torque caused by supercoiling. Here, we used a single-molecule DNA supercoil relaxation assay to measure the torque dependence of nTop1 and its inhibition by CPT. For comparison, we also examined mitochondrial Top1B and an N-terminal deletion mutant of nTop1. Despite substantial sequence homology in their core domains, nTop1 and mitochondrial Top1B exhibit dramatic differences in sensitivity to torque and CPT, with the N-terminal deletion mutant of nTop1 showing intermediate characteristics. In particular, nTop1 displays nearly torque-independent religation probability, distinguishing it from other Top1B enzymes studied to date. Kinetic modeling reveals a hitherto unobserved torque-independent transition linking the DNA rotation and religation phases of the enzymatic cycle. The parameters of this transition determine the torque sensitivity of religation and the efficiency of CPT binding. This “kinetic clutch” mechanism explains the molecular basis of CPT sensitivity and more generally provides a framework with which to interpret Top1B activity and inhibition.
Keywords: cancer therapy, magnetic tweezers, single-molecule biophysics
Type IB topoisomerase (Top1B) is essential for relaxing DNA supercoils generated during replication, transcription, DNA recombination, and repair (1–4). Due to its prominent roles in DNA metabolism, human Top1B is an important target for cancer therapies (2, 4, 5). Top1B relaxes DNA supercoils by forming a clamp around the DNA and generating a transient nick, which allows rotation of the DNA until the nick is religated (6, 7). This DNA rotation was thought to be hindered by interactions with the protein clamp because the cross-section of rotating DNA at the nick increases twofold during rotation (6, 8, 9). This was confirmed in a pioneering single-molecule study of relaxation by vaccinia Top1B that led to a model of DNA rotation as a random walk along a rugged energy landscape associated with the rotation angle of the 5′ end of the DNA. In this model, the rotation rate is dictated by the escape from multiple energy wells; religation is in kinetic competition with rotation at a specific energy well. Torque on the DNA (i.e., supercoiling) tilts the energy landscape, lowering the rotation energy barriers, thereby increasing the rotation rate and decreasing the probability of religation. As a result, both the rotation rate and the religation probability are torque-dependent to different degrees. This simple yet elegant model describes vaccinia Top1B supercoil relaxation as a function of torque (10). An extension of this model in which the rotation rate is governed by a single energy barrier describes the torque dependence of supercoil relaxation by topoisomerase V (Topo V), a structurally unrelated but mechanistically similar archaeal topoisomerase (11). Although single-molecule measurements of human nuclear Top1B (nTop1) are qualitatively similar to those of vaccinia Top1B and Topo V (4, 10, 12), the torque dependence of the human enzyme has not been investigated in detail. Single-molecule approaches have largely focused on the effects of chemotherapeutic agents on human nTop1, demonstrating that topotecan inhibits religation and hinders supercoil relaxation in a chiral-dependent manner (12).
In this study, we investigated the catalytic activities and the inhibition by camptothecin (CPT) of two human Top1B paralogs: nTop1 and mitochondrial Top1B (Top1mt). Despite extensive sequence similarity (Fig. S1), the two human Top1B enzymes do not functionally complement each other in vivo and differ significantly in their interactions with nuclear DNA and mtDNA (13). These Top1B paralogs afford a unique opportunity to elucidate how subtle differences in domains distal to the active site affect relaxation, catalytic activity, and CPT sensitivity. We included the N-terminal deletion mutant of nTop1 (Top68) in the study to assess the functional role of the N-terminal domain (14–16), which is largely missing from Top1mt (Fig. S1), more directly.
Using a magnetic tweezers-based single-molecule assay (Fig. 1), we measured the catalytic activities of nTop1, Top1mt, and Top68 and the effects of the natural Top1B inhibitor CPT as a function of torque applied to the DNA. Our results show that Top1mt exhibits significant torque dependence, similar to vaccinia Top1B (10). However, nTop1 exhibits less torque dependence; in particular, the religation probability for nTop1 is largely insensitive to torque. Moreover, the torque sensitivity of the three Top1B enzymes is inversely related to the efficacy of CPT inhibition.
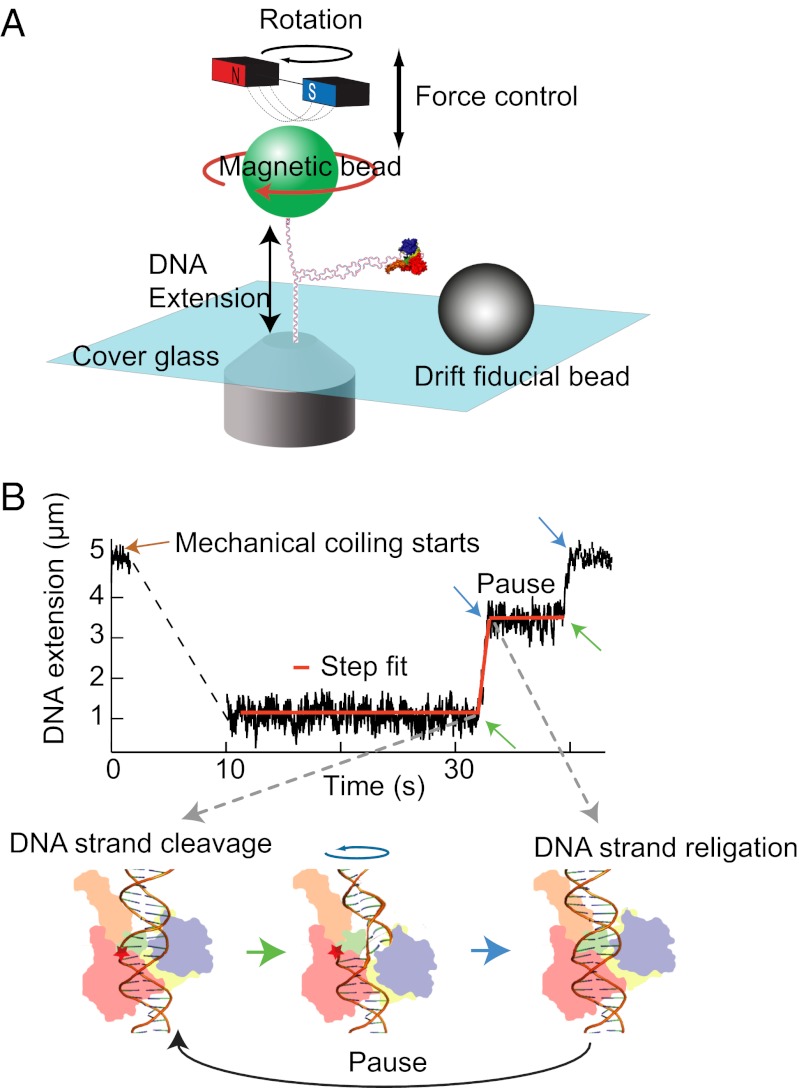
Fig. 1.
Cartoon of experimental setup (not to scale) and an example trace. (A) A DNA molecule is attached between the cover glass of a flow cell and a magnetic bead (green). Force is controlled by moving the magnet assembly above the flow cell. Rotating the magnetic bead by turning the magnets twists the DNA and generates supercoils. (B) DNA extension as a function of time and step-finding fit. DNA extension traces were fitted with a custom written step-finding routine based on a t test algorithm (40). The fitting routine extracts the extension change, duration, and linear velocity for each relaxation event and the duration of pauses between events. These phases of motion correspond to cleavage (green arrow), relaxation, and religation (blue arrow) depicted in the cartoon of Top1B enzyme bound to DNA.
To account for these results, we propose a revised model for relaxation by Top1B enzymes. In the prior model, religation probability is entirely dictated by a free-energy landscape associated with the rotation of the 5′ DNA end within the protein clamp (10, 11). Because our measurements show that religation by nTop1 was weakly dependent on the applied torque, we considered more general reaction schemes that included rate-limiting transitions uncoupled from DNA rotation. The resulting “kinetic clutch” model provides a mechanistic basis for differences in torque and CPT sensitivity among Top1B enzymes.
Results
We used a 23-kb torsionally constrained (“coilable”) DNA molecule attached at one end to a coverslip and at the other to a super paramagnetic bead in a magnetic tweezers instrument (Fig. 1A). Torque on the DNA was controlled by rotating the tethered bead at different applied tensions (Fig. S2) and was calculated using a model developed by Marko and colleagues (11, 17). As the bead is rotated, the torque on the DNA increases until a critical, tension-dependent torque at which the DNA buckles to form plectonemes is reached. Further rotation of the bead extends the length of the plectonemic region, whereas the torque remains fixed at the critical buckling value (Fig. S2). Analyzing only those relaxation events that do not remove all the plectonemes ensures that the measurements were made under constant torque conditions. In the presence of topoisomerases (50–500 pM), DNA supercoils were relaxed, which increased the DNA extension. Extension was converted to the number of turns relaxed per event as described in Fig. S2 (11). Probability density distributions of the relaxation rates were fitted with Gaussian distributions to obtain the mean relaxation rates (Fig. S3).
Human Top1B Exhibits Different Torque-Dependent Relaxation Rates.
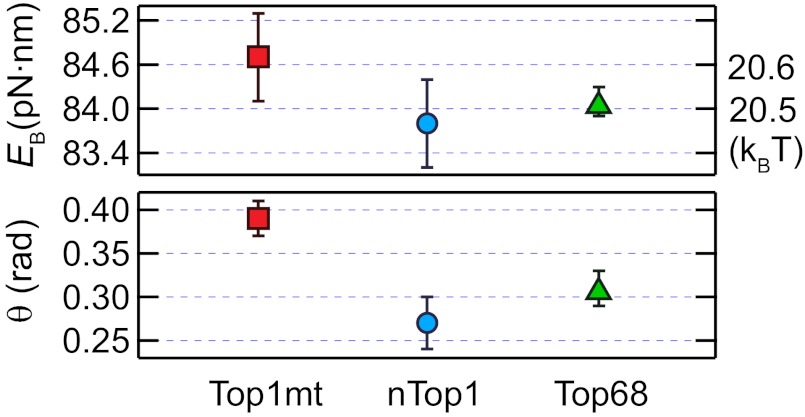
We first measured the relaxation rates for three human Top1B enzymes as a function of torque on the DNA. Consistent with similar measurements of Topo V (11), relaxation rates increased exponentially with increasing positive torque and were fit with a model assuming a single energy barrier associated with the rotation angle of the DNA 5′ end (11). The escape rate, ke, is determined by the height of the energy barrier, EB, the transition angle, θ (rotation angle between the bottom of the energy well and the top of the energy barrier), and the torque, which lowers the energy barrier to rotation in proportion to θ (11, 18) (Fig. S4A). The energy barriers for all three Top1B enzymes were comparable, but the transition angles for Top1mt and Top68 were 40% and 16% larger, respectively, than that for nTop1 (Fig. 2). Measurements of negative supercoil relaxation were limited to low values of torque to avoid complications arising from denaturation of DNA at high negative torque (19). Whereas nTop1 and Top68 relaxed positive and negative supercoils at comparable rates at low torque, Top1mt relaxed positive supercoils ∼1.5-fold faster than negative supercoils, suggesting an asymmetrical energy landscape (Fig. S4B, Inset).
Fig. 2.
Torque dependence of the supercoil relaxation rate. The energy barriers, EB, and transition angles, θ, were obtained from the fits of the torque-dependent relaxation rate (ke) to the barrier-crossing model (10, 11): 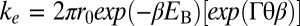 , where r0 is the rate of thermal fluctuations, r0 ∼ (β ηl3)−1 [ where η is the buffer viscosity (∼10−3 Pa⋅s) and l is the radius of DNA (1 nm)], EB is the energy barrier, Γ is the external torque, θ is the transition angle, and β = 1/kBT is the inverse of the thermal energy (SI Materials and Methods). The fitted values were as follows: EB (pN⋅nm): 84.7 ± 0.6 for Top1mt, 83.8 ± 0.6 for nTop1, and 84.1 ± 0.2 for Top68; θ (rad): 0.39 ± 0.02 for Top1mt, 0.27 ± 0.03 for nTop1, and 0.31 ± 0.02 for Top68. All reported errors are SDs.
, where r0 is the rate of thermal fluctuations, r0 ∼ (β ηl3)−1 [ where η is the buffer viscosity (∼10−3 Pa⋅s) and l is the radius of DNA (1 nm)], EB is the energy barrier, Γ is the external torque, θ is the transition angle, and β = 1/kBT is the inverse of the thermal energy (SI Materials and Methods). The fitted values were as follows: EB (pN⋅nm): 84.7 ± 0.6 for Top1mt, 83.8 ± 0.6 for nTop1, and 84.1 ± 0.2 for Top68; θ (rad): 0.39 ± 0.02 for Top1mt, 0.27 ± 0.03 for nTop1, and 0.31 ± 0.02 for Top68. All reported errors are SDs.
nTop1 and Top1mt Display Different Torque-Dependent Religation Probabilities.
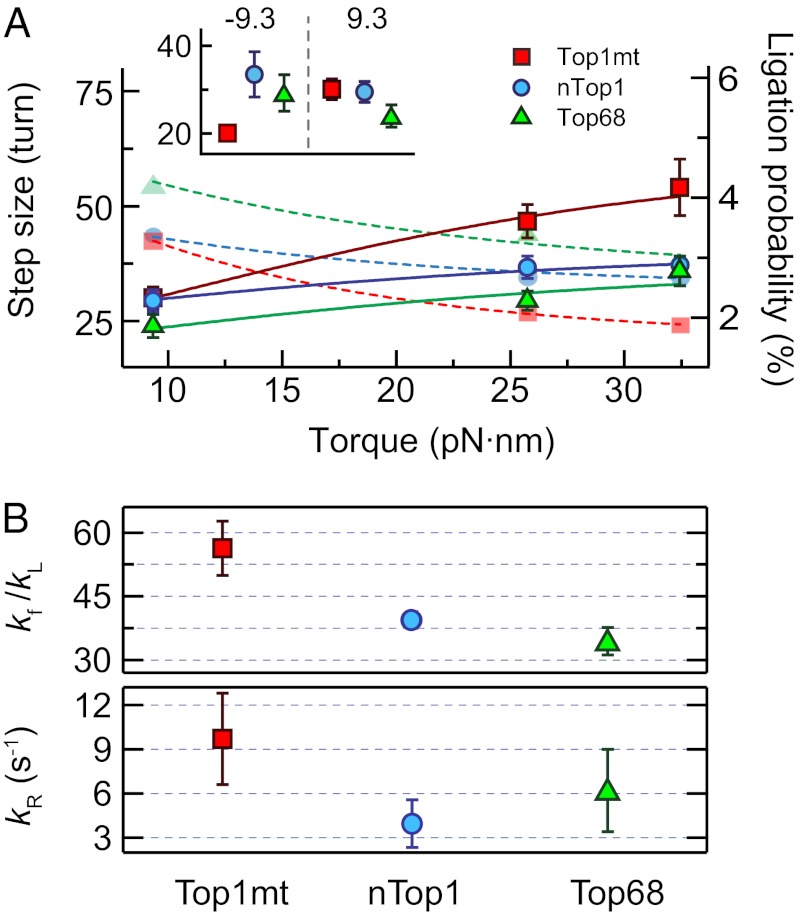
Next, we determined the relaxation step size per enzymatic cycle (from DNA cleavage to religation) as a function of torque (Fig. 3). During supercoil relaxation, the two ends of the DNA can be religated with a certain probability when they come into close proximity in a “religation region.” The number of supercoils relaxed, or step size, is inversely related to the religation probability. Previously, religation probability was modeled as a kinetic competition between religation and escape from the religation energy well in a rotation energy landscape (10). Torque lowers the rotation energy barrier, increasing the escape rate and lowering the religation probability. In our study, we found that the step size of Top1mt increases as a function of torque, similar to vaccinia Top1B (Fig. 3), and both enzymes had comparable transition angles associated with relegation: 0.11 ± 0.02 radians for Top1mt (Fig. S6) and 0.23 ± 0.02 radians for vaccinia Top1B (10). In contrast, the step sizes for Top68 and nTop1 were minimally dependent on torque (Fig. 3), and fits to a rotation energy landscape model resulted in vanishingly small transition angles (0.04 ± 0.01 radians for nTop1 and 0.07 ± 0.03 radians for Top68) (Fig. S6). These small transition angles associated with religation imply that the distance between the energy minimum and the transition state is on the order of an angstrom or less, which we considered unphysical. Alternatively, the weak torque dependence of religation probability for nTop1 suggests that religation occurs from a state that is insensitive to torque.
Fig. 3.
Torque-dependent relaxation step size and religation probability. (A) Relaxation step sizes of Top1mt, nTop1, and Top68 are plotted as a function of torque and fitted with Eq. 1 (solid lines, left axis). The religation probabilities corresponding to the inverse of the relaxation step sizes are shown with fits (dashed lines, right axis). The relaxation step sizes were obtained by fitting the relaxation step size distribution at each torque with a single exponential (Fig. S5). (Inset) Relaxation step sizes of Top1mt, nTop1, and Top68 at positive (9.3 pN⋅nm) and negative (−9.3 pN⋅nm) torques. (B) Ratio kf/kL and kR for three Top1Bs obtained from the fits of the relaxation step sizes as a function of torque to Eq. 1: 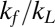 : 56 ± 6 for Top1mt, 39 ± 2 for nTop1, and 35 ± 4 for Top68;
: 56 ± 6 for Top1mt, 39 ± 2 for nTop1, and 35 ± 4 for Top68; 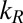 (s−1): 10 ± 3 for Top1mt, 4 ± 2 for nTop1, and 6 ± 3 for Top68. All reported errors are SDs.
(s−1): 10 ± 3 for Top1mt, 4 ± 2 for nTop1, and 6 ± 3 for Top68. All reported errors are SDs.
We developed a revised kinetic scheme, including an energy barrier, along a reaction coordinate orthogonal to DNA rotation (SI Materials and Methods). We hypothesized that once the free end of the DNA rotates back to the global energy minimum along the rotation coordinate, the DNA–protein complex fluctuates between religation-competent (T⋅Dc) and rotation-competent (T⋅Dc*) states with torque-independent rates kf and kR, respectively. The religation-competent state is a torque-independent state in which the ends of the DNA are aligned but unable to rotate due to the relative position of the DNA ends or the conformation of the protein clamp. From this state, the DNA can be religated with rate kL or the complex can enter the rotation-competent state with rate kf. From the rotation-competent state, rotation can proceed with a torque-dependent rate ke (Fig. 2 and SI Materials and Methods) or the enzyme–DNA complex can return to the religation-competent state with rate kR. Within this model, the religation probability per turn is (SI Materials and Methods) as follows:
The mean relaxation step size is 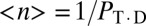 . The transition rates kf and kR, between the religation- and rotation-competent states, provide a mechanistic basis for differences in religation torque sensitivity. As kf or kR decreases, the torque dependence of the religation probability decreases, and eventually vanishes for the extreme case of kR = 0 (Fig. S7). Therefore, the transition rates effectively act as a “clutch,” coupling the torque dependence of rotation (ke) to the religation probability.
. The transition rates kf and kR, between the religation- and rotation-competent states, provide a mechanistic basis for differences in religation torque sensitivity. As kf or kR decreases, the torque dependence of the religation probability decreases, and eventually vanishes for the extreme case of kR = 0 (Fig. S7). Therefore, the transition rates effectively act as a “clutch,” coupling the torque dependence of rotation (ke) to the religation probability.
The mean uncoiling step sizes (<n>) as a function of positive torque for all three enzymes were well-fitted with Eq. 1 (Fig. 3) using the measured relaxation rates (Fig. 2). The fits returned values of kR and the ratio kf/kL. As expected, the values of kR and kf/kL for nTop1 and Top68 were similar, reflecting their minimal sensitivity to torque, whereas the values for Top1mt were larger, consistent with increased torque sensitivity (Fig. 3). Uncoiling step sizes for negative supercoils were measured at a single low value of torque due to complications arising from local melting of the DNA at higher values of negative torque. The relaxation step sizes for nTop1 and Top68 were comparable at equal positive and negative torques (9.3 pN⋅nm), whereas the step size for Top1mt was ∼30% smaller for negative torque in comparison to positive torque (Fig. 3A, Inset).
Religation-Cleavage Equilibrium Is Shifted Toward Religation for Top1mt.
Relaxation events were interrupted by short pauses on the order of a few seconds. We interpret these pauses as states in which Top1B is bound to intact DNA rather than unbinding and rebinding of Top1B. We typically observed a single burst of nearly continuous activity lasting for several hundred seconds, followed by extended periods (greater than 250 s) of no activity, suggesting that activity was due to a single enzyme with low on (kon) and off (koff) rates. Although the binding kinetics of nTop1 have not been measured, the DNA binding kinetics of vaccinia Top1B are slow (kon ∼10−3⋅s−1 and koff ∼10−2⋅s−1 at 50–500 pM enzyme concentration) (20). The pause duration corresponds to the waiting time for DNA rotation to start from a ligated state. Based on our model, we derived an expression for the cleavage rate, kc, using a recursive kinetic calculation (21) (SI Materials and Methods):
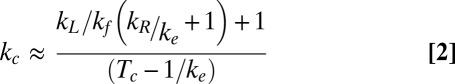 |
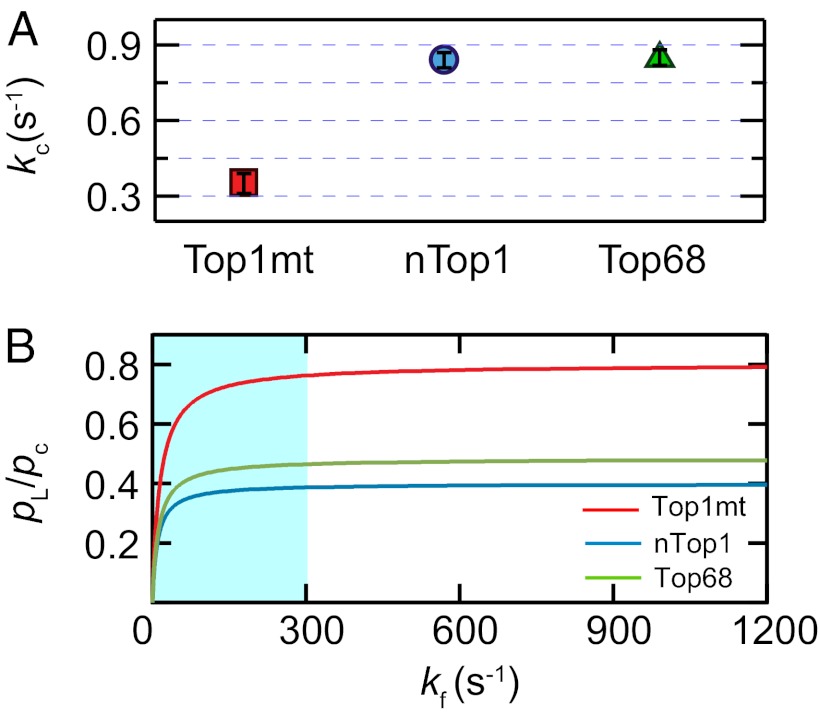
The observed pause duration, Tc, was determined by fitting the histogram of pause durations at each torque with a single exponential (Fig. S8). Using the previously determined values of kR, kf/kL, and ke, we calculated 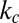 for each enzyme (Fig. 4A). Based on these results, we estimated the equilibrium at zero torque as a function of kf. Because we obtained kf/kL but not kL, we compared the religation-cleavage equilibrium for the three Top1Bs as a function of kf. A lower bound of ∼300 s−1 for kf is imposed by the fact that kf must be significantly larger than the rate-determining step for rotation, ke. As shown in Fig. 4B, the religation-cleavage equilibrium for Top1mt is shifted twofold toward religation compared with nTop1 for all values of kf above the lower bound.
for each enzyme (Fig. 4A). Based on these results, we estimated the equilibrium at zero torque as a function of kf. Because we obtained kf/kL but not kL, we compared the religation-cleavage equilibrium for the three Top1Bs as a function of kf. A lower bound of ∼300 s−1 for kf is imposed by the fact that kf must be significantly larger than the rate-determining step for rotation, ke. As shown in Fig. 4B, the religation-cleavage equilibrium for Top1mt is shifted twofold toward religation compared with nTop1 for all values of kf above the lower bound.
Fig. 4.
Cleavage kinetics. (A) Cleavage rates of the three Top1B enzymes were calculated with Eq. 2 using the mean cleavage time <Tc> averaged over all torques (Fig. S8) and the previously obtained parameters (ke, kR, and kf/kL). The calculated cleavage rates, kc (s−1), were as follows: 0.35 ± 0.04 for Top1mt, 0.83 ± 0.04 for nTop1, and 0.85 ± 0.03 for Top68. All reported errors are SEs. (B) Ratio of the ligation-to-cleavage probabilities at zero torque as a function of kf. Top1mt shows a twofold higher pL/pc at kf ≥ 300 s−1 in comparison to nTop1. Because rotation is the rate-limiting step in the relaxation process, kf is larger than ke for each enzyme. The shaded region corresponds to kf < 300 s−1, the minimal kf consistent with the slowest relaxation rate.
CPT Sensitivity Is Inversely Related to Religation Torque Sensitivity.
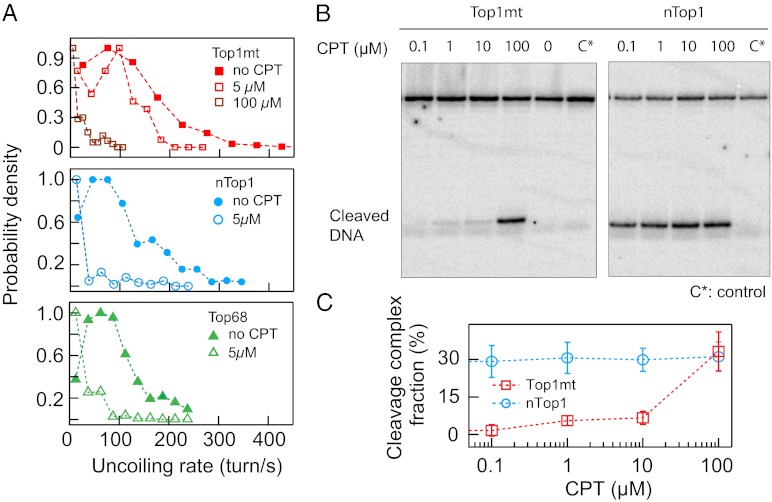
To study inhibition of human Top1B enzymes by CPT, we measured the relaxation rates of positive supercoils in the presence of CPT at a torque of 9.3 pN⋅nm. CPT binding at the Top1B-DNA interface (22) slows down the rotation rate, presumably due to an increase in the rotational energy barrier (12, 18). Consistent with previous single-molecule (12) and ensemble measurements (23), the relaxation rates of nTop1 and Top68 decreased 18-fold and sixfold, respectively, at 5 μM CPT. In contrast, Top1mt did not show significant relaxation rate changes in the presence of 5 μM CPT (Fig. 5A). Top1mt was not completely insensitive to CPT, because the relaxation rate decreased eightfold at 100 μM CPT (Fig. 5A). This differential CPT sensitivity was confirmed with an ensemble assay of the cleavage equilibrium of nTop1 and Top1mt as a function of CPT concentration (Fig. 5 B and C). At CPT concentrations less than 10 μM, nTop1 formed at least sixfold more cleavage complexes than Top1mt, consistent with the single-molecule results at 5 μM CPT (Fig. 5C).
Fig. 5.
CPT sensitivity of human Top1B enzymes relaxing positively supercoiled DNA. (A) Normalized relaxation rate distributions of three Top1B enzymes in the presence and absence of 5 μM CPT and 100 μM CPT for Top1mt. Relaxation rates of nTop1 and Top68 decreased at 5 μM CPT, whereas a comparable decrease in the relaxation rate of Top1mt required 100 μM CPT. (B) Top1B-DNA cleavage complex formation. With nTop1 (Right), cleaved DNA was visible at the lowest CPT concentration (0.1 μM), whereas with Top1mt (Left), cleaved DNA was minimally visible up to 10 μM CPT and became comparable to that observed with nTop1 at 100 μM CPT. C* indicates a control reaction that contains only DNA. (C) Cleavage complex formation as a function of CPT concentration. DNA cleavage was quantified by the intensity in the cleavage band normalized by total DNA (uncleaved and cleaved DNA) corrected by the intensity of the control band.
Discussion
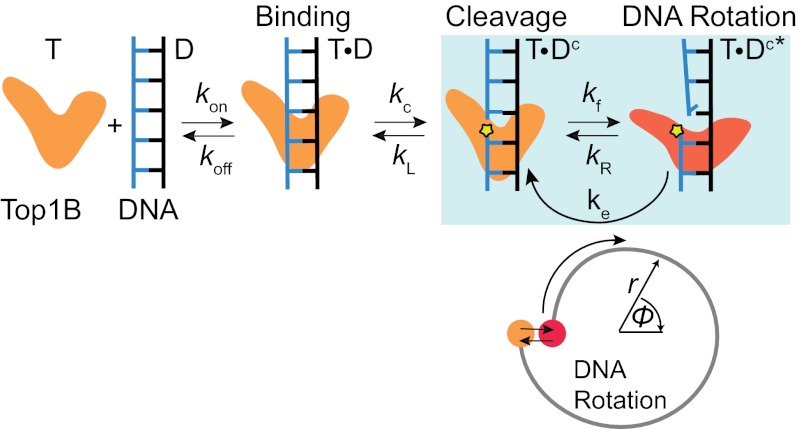
In this study, we compared the human Top1B paralogs nTop1 and Top1mt by measuring their relaxation rate, religation probability, and cleavage rate as a function of torque in the presence and absence of CPT. Despite significant sequence homology, the activities of nTop1 and Top1mt differ significantly, likely reflecting in vivo functional differences. In particular, the religation probability for nTop1 is minimally sensitive to torque in contrast to Top1mt and vaccinia Top1B (10). This finding necessitates revising existing models of Top1B activity. We propose a kinetic clutch model that explains differences in torque sensitivity of the religation probability and CPT sensitivity among Top1B enzymes (Fig. 6).
Fig. 6.
Proposed kinetic clutch model for Top1B. In this scheme, Top1B (T) reversibly binds DNA (D), forming a binary complex (T⋅D) in which the DNA is cleaved at the rate kc. The cleaved complex is initially in a religation-competent state (T⋅Dc) separated from the rotation-competent state (T⋅Dc*) by a torque-independent energy barrier, corresponding to a protein conformational change or a radial DNA distortion (orange and red states). The Top1B–DNA complex fluctuates between the religation-competent (T⋅Dc) and rotation-competent (T⋅Dc*) states at the torque-independent rates kR and kf. From the religation-competent state T⋅Dc, the DNA can be religated at rate kL. DNA rotation corresponds to escape from the T⋅Dc* state at the torque-dependent rate ke. Each rotation of the DNA corresponds to one cycle of T⋅Dc to T⋅Dc* and back to T⋅Dc as denoted by the blue-shaded box. The corresponding reaction coordinates are schematically represented below. The free end of the DNA rotates from the state T⋅Dc* (red dot) but is prevented from further rotation once it enters the state T⋅Dc (orange dot). A reversible radial motion (i.e., crossing an energy barrier orthogonal to rotation) is required to return the complex to the rotation-competent state. Although the model is based on positive supercoil relaxation, we speculate that a similar mechanism holds for negative supercoil relaxation. Based on the “intercalated model,” CPT likely binds Top1B in the religation-competent state (T⋅Dc) in which the two DNA ends are aligned.
The weak torque dependence of religation probability for nTop1 has important implications for the relationship between religation and rotation. In existing models, the state from which religation occurs is on the DNA rotation pathway (4, 10, 11). The resulting torque-dependent religation probability is inconsistent with our nTop1 results (Fig. 3). We propose a model in which, once per rotation, the DNA free end enters a ligation-competent state that is insensitive to torque (Fig. 6). From this state, the DNA can be religated or can reversibly enter a rotation-competent state from which rotation, hindered by a torque-dependent barrier, occurs. The important aspect of this model is that the torque sensitivity of religation depends on the kinetics of the transitions between the ligation and rotation-competent states. This model provides a mechanistic basis for the observed differences in religation torque sensitivity (Fig. 3) and differences in CPT sensitivity (Fig. 5) (see below). The model further postulates the existence of DNA or protein conformational changes that are insensitive to torque (i.e., orthogonal to the angular rotation of the DNA). The present experiments cannot distinguish between these possibilities, but results from recent molecular dynamics (MD) simulations suggest that such orthogonal protein motions may play an important role in the catalytic cycle (18). The kinetic clutch model was developed based on measurements of positive supercoil relaxation due to the limited range of experimentally accessible negative values of torque. We speculate that the same kinetic clutch mechanism applies to the relaxation of negative supercoils. However, the energy landscape for negative supercoil relaxation is not simply the rotational reverse of that for positive supercoil relaxation (Fig. 6). Furthermore, the detailed energy landscape and kinetics are likely different because different domains are presumably affected depending on the direction of rotation (24). This energy landscape asymmetry has been observed in MD simulations (18) and in the religation-cleavage equilibrium (25), and it has been inferred from the large asymmetry in the relaxation rate of negative vs. positive supercoils in the presence of topotecan (12, 18). However, further experimental and computational efforts will be required to elucidate the details of the energy landscape and conformational states for negative supercoil relaxation.
Independent of the mechanistic model developed to explain them, our experimental results demonstrate stark differences in the catalytic activities of nTop1 and Top1mt. The sequence similarity in their core domains suggests that catalytic differences originate from differences in distal domains (Fig. S9). However, the N-terminal domain of nTop1 does not appear to account for these differences, because the activity of Top68 resembles nTop1 more so than Top1mt. Evolutionarily, variations in distal domains may be important for adapting to specific cellular environments and roles (e.g., nuclear DNA metabolism, mtDNA metabolism) without compromising the catalytic core. For example, the N-terminal region of nTop1 has been shown to mediate interactions with nuclear DNA (13). Similarly, the minimal torque dependence of the religation probability (Fig. 3) and high cleavage rate (Fig. 4) of nTop1 may be critical for its biological functions. nTop1 physically interacts with RNA polymerase (RNAP) to remove excessive supercoiling arising during transcription (26). Growing evidence suggests that transcription may be regulated by the supercoiling density of DNA in eukaryotes (27). The weakly torque-dependent relaxation activity of nTop1 could counteract local supercoiling introduced by RNAP at a rate that is independent of the global supercoiling level. In this manner, excessive supercoiling associated with transcription is removed without affecting the global level of supercoiling. Supporting this hypothesis, the aggregate rate of supercoil relaxation by nTop1 is ∼13 supercoils per second, comparable to estimates of ∼10 supercoils per second generated by RNAP in vivo (27). The physical association between RNAP and nTop1 introduces a further complication having to do with the “hand-off” between enzymes (26). The high cleavage rate and bias toward cleavage over religation (Fig. 4) of nTop1 would facilitate this hand-off, allowing nTop1 to remove supercoiling efficiently while minimizing the likelihood of impeding transcription.
In contrast to nTop1, Top1mt behaves similar to vaccinia Top1B and Topo V (10, 11), displaying significant torque dependence of both relaxation rate and religation probability. Top1mt also displays a unique chiral relaxation rate asymmetry (Fig. S4) that may play a role in maintaining the negatively supercoiled status of mtDNA (28). The religation-cleavage equilibrium for Top1mt was shifted approximately threefold toward religation in comparison to nTop1 (Fig. 4B). Biochemical and molecular modeling studies indicate that the linker domain influences cleavage-religation dynamics and equilibrium (14, 29–31). Structural homology modeling and secondary structure prediction (32, 33) suggest that the Top1mt linker domain is less structured and presumably more flexible than the nTop1 linker domain. The resulting lower cleavage rate and higher religation-cleavage equilibrium may be physiologically important for maintaining DNA integrity in mitochondria that have less effective DNA repair systems than nuclei, and may not be able to tolerate high levels of DNA cleavage (34).
The human Top1B paralogs display significant differences in CPT sensitivity (Fig. 5). As previously observed with topotecan (12), the relaxation rate of nTop1 decreased in the presence of CPT. However, Top1mt was less sensitive to CPT, requiring a high concentration to achieve observable effects (Fig. 5). The “intercalated model” postulates that CPT binds between the aligned +1 and −1 nucleosides (35, 36). In our model, this corresponds to the religation-competent state (T⋅Dc) (Fig. 6). Hence, CPT binding should be proportional to the lifetime of the religation state. Our results are consistent with this prediction because the expected escape rate, kf, is larger for Top1mt than for nTop1. Although kf was not directly measured, a lower limit is set by the highest rotation rate (ke) (i.e., kf > 600 s−1 for Top1mt and kf > 300 s−1 for nTop1). These results suggest that CPT sensitivity is inversely related to the torque sensitivity of religation (Eq. 1). Consistent with this interpretation, the torque dependence of religation by vaccinia Top1B is similar to that of Top1mt (10), and both enzymes are largely insensitive to CPT (2).
Overall, our findings demonstrate that subtle changes in distal domains of Top1B profoundly influence the catalytic activity. To account for the striking differences in sensitivity to torque and CPT between Top1mt and nTop1, we propose a kinetic clutch model of religation. These results and the model provide a mechanistic basis with which to interpret Top1B activity and inhibition.
Materials and Methods
Preparation of Human Top1Bs.
Expression and purification of human nTop1 and Top68 from Baculovirus-infected insect cells were performed as previously described (37). Top1mt was expressed and purified in the same manner.
Single-Molecule Experimental Procedures.
A total of 0.3 nM 23-kb DNA was incubated with 32 ng of antidigoxigenin in 50 μL of 1× PBS for 1 h at room temperature. This mixture was introduced into a sample cell coated with a low concentration of stuck beads and incubated overnight at 4 °C. Unbound DNA was washed out by flowing 200 μL of wash buffer (WB) [1× PBS, 0.04% (vol/vol) Tween-20, 0.3% (wt/vol) BSA]. A total of 20 μL of a 20× dilution of streptavidin-coated magnetic beads (My One; Invitrogen) was then introduced in WB, allowed to tether for 1 h, and washed with 1 mL of WB. Once a coilable DNA substrate was found, the chamber was washed with 200 μL of topoisomerase buffer [10 mM Tris (pH 8), 50 mM KCl, 10 mM MgCl2, 0.3% (wt/vol) BSA, 0.04% Tween-20, 0.1 mM EDTA, 5 mM DTT] and topoisomerase was added at a concentration of 50–500 pM in 200 μL of topoisomerase buffer with or without CPT. Topoisomerase activity was measured by tracking the height of the tethered bead at a 100-Hz sample rate. The position of a stuck bead was tracked to correct for sample cell drift actively (38, 39). The data traces were analyzed with a custom written step-finding program (40).
Top1B-DNA Cleavage Assay with CPT.
A Top1B-DNA cleavage assay was performed as previously described (41). The final concentrations of nTop1 and Top1mt were 0.012 mg/mL and 0.03 mg/mL, respectively. The DNA concentration was 40 nM. The CPT concentration was varied from 0.1–100 μM. The DNA substrate was GATTAGGATTGTT∼GTGAAGTATAGTA (top strand, 3′ end-labeled with Cordycepin)/CTAATCCTAACAACACTTCATATCAT (bottom strand). The Top1B cleavage site is indicated by the “∼” in the top-strand sequence. After the cleavage reaction, the oligonucleotide was denatured and run on a denaturing polyacrylamide gel. The gel was imaged using a Phosphor Imager (Molecular Dynamics) and quantified using National Instruments Vision Assistant v. 8.5.
Supplementary Material
Acknowledgments
We thank Dr. Jon Silver, David Levens, and Laura Baranello for suggestions and advice and Hemai Parthasarathy, Tamara Litwin, and Dr. Richard Neuman for editing. This research was supported by the Intramural Research Programs of the National Heart, Lung, and Blood Institute and the National Cancer Institute, Center for Cancer Research, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206480109/-/DCSupplemental.
References
- 1.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 4.Koster DA, Crut A, Shuman S, Bjornsti MA, Dekker NH. Cellular strategies for regulating DNA supercoiling: A single-molecule perspective. Cell. 2010;142:519–530. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart L, Redinbo MR, Qiu X, Hol WGJ, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 7.Wang JC. Untangling the Double Helix. Cold Spring Harbor Laboratory Press, Plainview, NY; 2009. [Google Scholar]
- 8.Schoeffler AJ, Berger JM. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 9.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA topoisomerase I: Evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 10.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 11.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragón A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc Natl Acad Sci USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koster DA, Palle K, Bot ESM, Bjornsti M-A, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 13.Dalla Rosa I, et al. Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 2009;37:6414–6428. doi: 10.1093/nar/gkp708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart L, Ireton GC, Champoux JJ. Reconstitution of human topoisomerase I by fragment complementation. J Mol Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- 15.Champoux JJ. Domains of human topoisomerase I and associated functions. Prog Nucleic Acid Res Mol Biol. 1998;60:111–132. doi: 10.1016/s0079-6603(08)60891-0. [DOI] [PubMed] [Google Scholar]
- 16.Frøhlich RF, Andersen FF, Westergaard O, Andersen AH, Knudsen BR. Regions within the N-terminal domain of human topoisomerase I exert important functions during strand rotation and DNA binding. J Mol Biol. 2004;336:93–103. doi: 10.1016/j.jmb.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 18.Wereszczynski J, Andricioaei I. Free energy calculations reveal rotating-ratchet mechanism for DNA supercoil relaxation by topoisomerase IB and its inhibition. Biophys J. 2010;99:869–878. doi: 10.1016/j.bpj.2010.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuman KC. Single-molecule measurements of DNA topology and topoisomerases. J Biol Chem. 2010;285:18967–18971. doi: 10.1074/jbc.R109.092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA topoisomerase I: Single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry. 1994;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 21.Shaevitz JW, Block SM, Schnitzer MJ. Statistical kinetics of macromolecular dynamics. Biophys J. 2005;89:2277–2285. doi: 10.1529/biophysj.105.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommier Y, Marchand C. Interfacial inhibitors: Targeting macromolecular complexes. Nat Rev Drug Discov. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frøhlich RF, et al. Tryptophane-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sari L, Andricioaei I. Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation. Nucleic Acids Res. 2005;33:6621–6634. doi: 10.1093/nar/gki935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentry AC, Juul S, Veigaard C, Knudsen BR, Osheroff N. The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I. Nucleic Acids Res. 2011;39:1014–1022. doi: 10.1093/nar/gkq822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leppard JB, Champoux JJ. Human DNA topoisomerase I: Relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 27.Baranello L, Levens D, Gupta A, Kouzine F. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochim Biophys Acta. 2012;1819:632–638. doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogenhagen D, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: Introduction of superhelical turns into newly replicated molecules. J Mol Biol. 1978;119:69–81. doi: 10.1016/0022-2836(78)90270-x. [DOI] [PubMed] [Google Scholar]
- 29.Chillemi G, Bruselles A, Fiorani P, Bueno S, Desideri A. The open state of human topoisomerase I as probed by molecular dynamics simulation. Nucleic Acids Res. 2007;35:3032–3038. doi: 10.1093/nar/gkm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chillemi G, Redinbo M, Bruselles A, Desideri A. Role of the linker domain and the 203-214 N-terminal residues in the human topoisomerase I DNA complex dynamics. Biophys J. 2004;87:4087–4097. doi: 10.1529/biophysj.104.044925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiorani P, et al. Evidence of the crucial role of the linker domain on the catalytic activity of human topoisomerase I by experimental and simulative characterization of the Lys681Ala mutant. Nucleic Acids Res. 2009;37:6849–6858. doi: 10.1093/nar/gkp669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 33.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 34.Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging—An update. Exp Gerontol. 2010;45:478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staker BL, et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioanoviciu A, et al. Synthesis and mechanism of action studies of a series of norindenoisoquinoline topoisomerase I poisons reveal an inhibitor with a flipped orientation in the ternary DNA-enzyme-inhibitor complex as determined by X-ray crystallographic analysis. J Med Chem. 2005;48:4803–4814. doi: 10.1021/jm050076b. [DOI] [PubMed] [Google Scholar]
- 37.Laco GS, Pommier Y. Role of a tryptophan anchor in human topoisomerase I structure, function and inhibition. Biochem J. 2008;411:523–530. doi: 10.1042/BJ20071436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seol Y, Neuman KC (2011) Single-molecule measurements of topoisomerase activity with magnetic tweezers. Single Molecule Enzymology, Methods in Molecular Biology, eds Mashanov GI, Batters C (Humana Press, New York), Vol 778, pp 229–241. [DOI] [PubMed]
- 39.Seol Y, Neuman KC. In: Magnetic tweezers for single-molecule manipulation. Single Molecule Enzymology, Methods in Molecular Biology. Peterman EJ, Wuite G, editors. Vol 783. New York: Humana Press; 2011. pp. 265–293. [DOI] [PubMed] [Google Scholar]
- 40.Carter BC, Vershinin M, Gross SP. A comparison of step-detection methods: How well can you do? Biophys J. 2008;94:306–319. doi: 10.1529/biophysj.107.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, et al. Human mitochondrial topoisomerase I. Proc Natl Acad Sci USA. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.