Abstract
Despite the importance of benzoic acid (BA) as a precursor for a wide array of primary and secondary metabolites, its biosynthesis in plants has not been fully elucidated. BA formation from phenylalanine requires shortening of the C3 side chain by two carbon units, which can occur by a non–β-oxidative route and/or a β-oxidative pathway analogous to the catabolism of fatty acids. Enzymes responsible for the first and last reactions of the core BA β-oxidative pathway (cinnamic acid → cinnamoyl-CoA → 3-hydroxy-3-phenylpropanoyl-CoA → 3-oxo-3-phenylpropanoyl-CoA → BA-CoA) have previously been characterized in petunia, a plant with flowers rich in phenylpropanoid/benzenoid volatile compounds. Using a functional genomics approach, we have identified a petunia gene encoding cinnamoyl-CoA hydratase-dehydrogenase (PhCHD), a bifunctional peroxisomal enzyme responsible for two consecutively occurring unexplored intermediate steps in the core BA β-oxidative pathway. PhCHD spatially, developmentally, and temporally coexpresses with known genes in the BA β-oxidative pathway, and correlates with emission of benzenoid volatiles. Kinetic analysis of recombinant PhCHD revealed it most efficiently converts cinnamoyl-CoA to 3-oxo-3-phenylpropanoyl-CoA, thus forming the substrate for the final step in the pathway. Down-regulation of PhCHD expression in petunia flowers resulted in reduced CHD enzyme activity, as well as decreased formation of BA-CoA, BA and their derived volatiles. Moreover, transgenic lines accumulated the PhCHD substrate cinnamoyl-CoA and the upstream pathway intermediate cinnamic acid. Discovery of PhCHD completes the elucidation of the core BA β-oxidative route in plants, and together with the previously characterized CoA-ligase and thiolase enzymes, provides evidence that the whole pathway occurs in peroxisomes.
Keywords: benzenoid network, multifunctional protein, floral scent
Benzoic acid (BA) and its derivatives are important structural elements in a large number of natural products thus playing many valuable roles in plant metabolism. These compounds not only perform critical functions in plant fitness (e.g., as plant growth regulators, defensive compounds, pollinator attractants) but also are extensively used as preservatives and flavor enhancers, analgesics, antiseptics, chemotherapeutics, and feed stocks for chemical syntheses (1–5). Despite its simple structure and widespread distribution, full understanding of the biochemical pathways leading to BA formation remains incomplete.
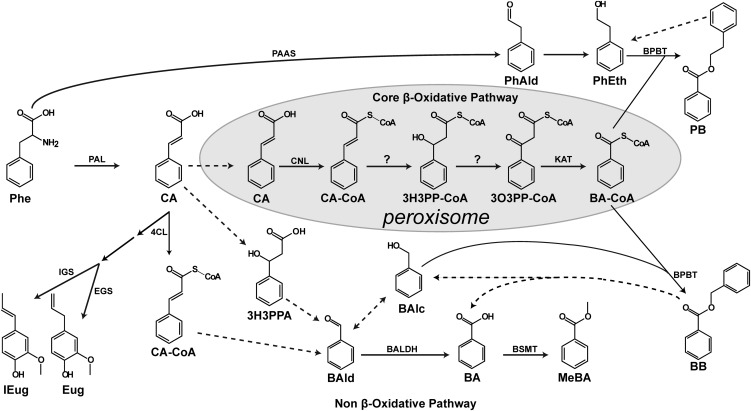
In plants, BA biosynthesis occurs through multiple routes that arise from the phenylpropanoid pathway and begins with the deamination of l-phenylalanine (Phe) to trans-cinnamic acid (CA) by phenylalanine ammonia lyase (PAL) (6–8). Subsequent conversion of CA to BA requires shortening of the C3 side chain by two carbons and is proposed to occur via either a CoA-dependent β-oxidative pathway (9, 10), a CoA-independent non–β-oxidative pathway (11, 12), or a combination of these pathways (13, 14) (Fig. 1). Indeed, in vivo stable-isotope labeling with computer-assisted metabolic flux analysis in petunia flowers has already revealed that both the β-oxidative and non–β-oxidative pathways contribute to the formation of benzenoid compounds in plants (13, 14).
Fig. 1.
The benzoic acid biosynthetic network in plants. Solid arrows show established biochemical reactions, and dashed arrows depict possible steps not yet identified. Stacked arrows show the involvement of multiple enzymatic steps. The CoA-dependent β-oxidative pathway leading to BA-CoA formation is localized in peroxisomes and shown with a gray background. The proposed routes of the non–β-oxidative pathway in cytosol are also depicted. BA-CoA, benzoyl-CoA; BAlc, benzylalcohol; BAld, benzaldehyde; BALDH, benzaldehyde dehydrogenase; BB, benzylbenzoate; BPBT, benzoyl-CoA:benzylalcohol/2-phenylethanol benzoyltransferase; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; CA, cinnamic acid; CA-CoA, cinnamoyl-CoA; 4CL, 4-coumarate-CoA ligase; Eug, eugenol; EGS, eugenol synthase; IEug, isoeugenol; IGS, isoeugenol synthase; 3H3PPA, 3-hydroxy-3-phenylpropionic acid; 3H3PP-CoA, 3-hydroxy-3-phenylpropanoyl-CoA; KAT, 3-ketoacyl-CoA thiolase; MeBA, methylbenzoate; 3O3PP-CoA, 3-oxo-3-phenylpropanoyl-CoA; PAAS, phenylacetaldehyde synthase; PEB, phenylethylbenzoate; PhAld, phenylacetaldehyde; and PhEth, 2-phenylethanol.
The proposed β-oxidative route is analogous to that operating in catabolism of fatty acids and certain branched-chain amino acids in plant peroxisomes (14–16). In this pathway, the first committed step is conversion of CA to its CoA thioester, cinnamoyl-CoA (CA-CoA), catalyzed in petunia by a peroxisomal cinnamate-CoA ligase (PhCNL) (17). Subsequent formation of benzoyl-CoA (BA-CoA) requires three sequential reactions [CA-CoA → 3-hydroxy-3-phenylpropanoyl-CoA (3H3PP-CoA) → 3-oxo-3-phenylpropanoyl-CoA (3O3PP-CoA) → BA-CoA; Fig. 1], the last of which has been described in petunia to occur via a peroxisomal 3-ketoacyl-CoA thiolase (PhKAT1) (18). Gene(s) responsible for the core steps of β-oxidative BA biosynthesis, formation of 3H3PP-CoA and 3O3PP-CoA (Fig. 1), still remain unknown. Biosynthesis of these intermediates requires consecutive enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase activities, both of which could be fulfilled by a single multifunctional protein (MFP) similar to those used in peroxisomal β-oxidation of fatty acids (19).
The flowers of Petunia hybrida cv Mitchell emit high levels of benzenoid volatiles (13, 20), making it an ideal model system for the elucidation of BA biosynthesis. Using a functional genomics approach, we have identified a petunia gene encoding cinnamoyl-CoA hydratase-dehydrogenase (PhCHD), a bifunctional enzyme responsible for the two-step conversion of CA-CoA to 3O3PP-CoA. Transgenic petunia plants with RNAi down-regulation of PhCHD were deficient in BA and BA-CoA while exhibiting four- and fivefold accumulations of CA and CA-CoA, respectively. PhCHD activity is enriched in peroxisomes, and coupling of recombinant PhCHD and PhKAT1 enzymes resulted in formation of BA-CoA from CA-CoA. Together these data revealed involvement of PhCHD in BA biosynthesis and complete elucidation of the core CoA-dependent β-oxidative pathway in plants.
Results
Gene Encoding a Multifunctional Protein Is Highly Expressed in Petunia Flowers.
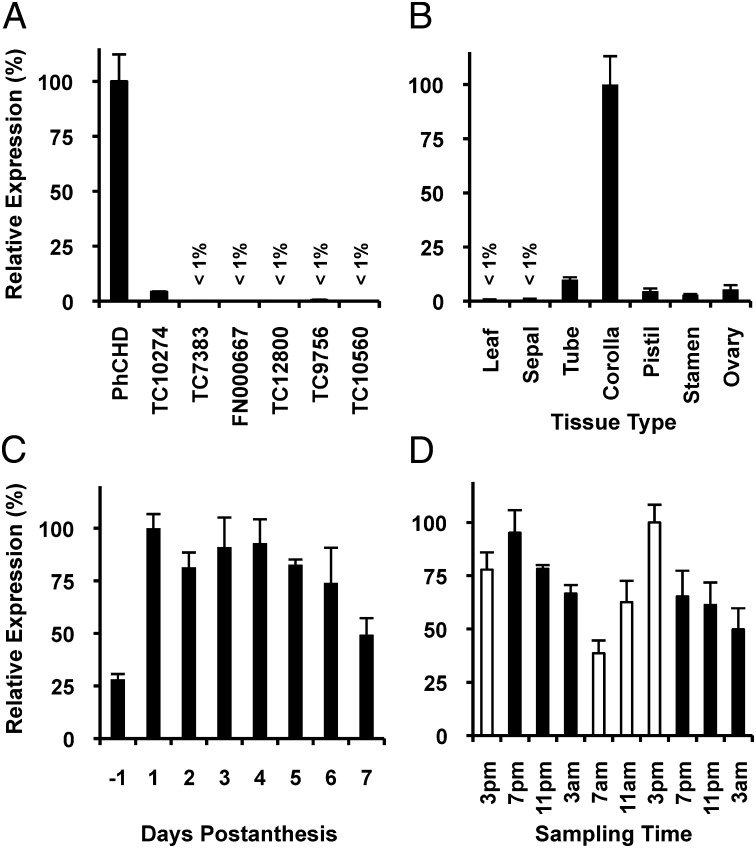
In plants the β-oxidation of Δ2–trans unsaturated short- and long-chain acyl-CoAs require enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase activites fulfilled by a type I MFP, which is encoded by a single fused gene (19). The Arabidopsis genome contains two type I MFPs, AIM1 (At4g29010) and MFP2 (At3g06860), encoding proteins with 58% identity and exhibiting the abovementioned activities (21, 22). Thus, we used these sequences for tblastn searches against a petunia petal-specific EST database (13), the Sol genomics network (www.solgenomics.net), as well as the Gene Indices at Dana Farber Cancer Institute (http://compbio.dfci.harvard.edu/tgi/tgipage.html) to identify an MFP candidate(s) acting on aromatic acyl-CoA intermediates in the BA β-oxidative biosynthetic pathway. Two ESTs (1.1.O05 and SGN-U210479 from the petunia petal-specific EST database and Sol genomics network, respectively), as well as two contigs (TC6988 and TC15042 from the Gene Indices) were found to correspond to a single gene, which based on the work presented below, was designated as PhCHD for Petunia hybrida cinnamoyl-CoA hydratase-dehydrogenase. Searching the Gene Indices uncovered six additional candidates (TC10274, TC7383, FN000667, TC12800, TC9756, and TC10560) highly similar to Arabidopsis AIM1 and MFP2. Because expression of genes involved in biosynthesis of benzenoid volatiles is highest in corolla (13, 17, 18, 23), the scent-producing organ of petunia flowers (13), gene-specific primers were designed for all seven candidates to perform a quantitative RT-PCR (qRT-PCR) screen for those highly expressed in this tissue. Six candidates showed little to no expression in corolla (Fig. 2A), and did not display an expression profile consistent with floral benzenoid production (Fig. S1) (13, 17, 18, 23), and were thus not considered further. PhCHD, however, was highly expressed in the corolla (Fig. 2A) and its tissue expression pattern (Fig. 2B) was consistent with formation of benzenoid volatiles (13, 17, 18, 23). Moreover, PhCHD displayed developmental and rhythmic expression profiles (Fig. 2 C and D) typical for genes involved in scent production (13, 23), including PhCNL (17) and PhKAT1 (18) from the BA β-oxidative pathway. A full-length cDNA corresponding to PhCHD was obtained by 5′-RACE and found to encode a protein of 724 amino acids with a calculated molecular mass of 78.1 kDa. This protein contains one hydratase and two dehydrogenase domains, and is 75 and 58% identical to Arabidopsis AIM1 and MFP2 (21, 22), respectively, 69% identical to MFP from Oryza sativa (24), 65% identical to MFP from Nicotiana tabacum (25), and 60% identical to MFP-b from Cucumis sativus (26).
Fig. 2.
Expression profiles of PhCHD and the six other petunia MFP candidates. (A) Expression of MFP candidates in petunia corollas collected at 3:00 PM on day 2 postanthesis shown relative to the highest expressed candidate, PhCHD. (B) Tissue-specific expression of PhCHD in flowers at 3:00 PM day 2 postanthesis and leaves shown relative to transcript level in corolla. (C) Developmental PhCHD expression profile at 3:00 PM days −1 through 7 postanthesis shown relative to transcript level on day 1. (D) Rhythmic changes in PhCHD expression in corollas of flowers 3:00 PM day 1–3:00 AM day 3 postanthesis during a normal light/dark cycle shown relative to the transcript level at 3:00 PM on day 2. Black and white bars correspond to dark and light periods, respectively. All transcript levels were determined by qRT-PCR either in relation to the reference gene elongation factor 1-α (A) or as absolute amounts based on quantification from a PhCHD DNA standard (B–D). All data are means ± SEM (n = 3–4 biological replicates).
PhCHD Is a Bifunctional Enzyme with Cinnamoyl-CoA Hydratase-Dehydrogenase Activities.
For functional characterization of the isolated PhCHD, its coding region was expressed in Escherchia coli as an inducible fusion protein containing a hexahistidine tag and the recombinant protein was purified using Ni-NTA affinity chromatography followed by gel filtration. Size-exclusion chromatography of recombinant CHD enzyme revealed an apparent molecular weight of 77 kDa, which was nearly identical to the calculated molecular mass (78.1 kDa), indicating that CHD exists as a monomer. Analysis of PhCHD substrate specificity revealed that the enzyme requires NAD+ as a cofactor and is highly specific for CA-CoA and p-coumaroyl-CoA (pCA-CoA), with no detectable activity toward caffeoyl-CoA (CAF-CoA), feruloyl-CoA (FA-CoA), crotonyl-CoA, hexanoyl-CoA, or hexadecanoyl-CoA (Table S1). Moreover, addition of 2 mM Mg2+ increased enzyme activities toward CA-CoA and pCA-CoA by 92%, but did not affect activity toward the other tested substrates. The product formed by PhCHD from CA-CoA was identified by liquid chromatography (LC)-TOF/MS (Fig. S2) as a compound with an exact mass of 912.155 (M-H)− (Fig. S2C), corresponding to the expected product of sequential hydration/dehydrogenation of CA-CoA (3O3PP-CoA; Fig. 1). Omission of NAD+ from the reaction resulted in neither detectable product formation nor 3H3PP-CoA intermediate (Fig. S2B). No activity was observed when NADP+ was supplied as a cofactor instead of NAD+ (Table S1). Coupling of the CHD reaction with recombinant PhKAT1 enzyme (18) in the presence of free CoA-SH resulted in conversion of CA-CoA to BA-CoA as was verified by LC-TOF/MS (Fig. S2 E and F).
Determination of kinetic parameters for recombinant PhCHD revealed its high affinity for CA-CoA and NAD+ with apparent Km values of 89.8 ± 1.8 μM and 13.8 ± 0.7 μM (Table 1), respectively, when incubated together. Whereas Km values for pCA-CoA and NAD+ (when incubated together) were similar (73.7 ± 3.3 μM and 6.1 ± 0.2 μM, respectively), the turnover rate (kcat) with pCA-CoA was more than 10-fold lower than with CA-CoA (0.40 ± 0.01 s−1 and 4.35 ± 0.01 s−1, respectively). Likewise, turnover of NAD+ was nearly 15-fold higher when CA-CoA was used as a substrate compared with pCA-CoA (6.94 ± 0.08 s−1 and 0.24 ± 0.00 s−1, respectively; Table 1).
Table 1.
Kinetic parameters of recombinant PhCHD
| Substrate | Km, μM | Vmax, μmol·min−1·mg−1 | kcat, s−1 | kcat/Km, mM−1·s−1 |
| Cinnamoyl-CoA* | 89.8 ± 1.8 | 3.25 ± 0.01 | 4.35 ± 0.01 | 48.5 ± 0.9 |
| p-coumaroyl-CoA* | 73.7 ± 3.3 | 0.30 ± 0.01 | 0.40 ± 0.01 | 5.5 ± 0.2 |
| NAD+† | 13.8 ± 0.7 | 5.19 ± 0.06 | 6.94 ± 0.08 | 506.2 ± 20.6 |
| NAD+‡ | 6.1 ± 0.2 | 0.18 ± 0.00 | 0.24 ± 0.00 | 39.5 ± 1.4 |
Values are means ± SEM (n = 3).
*[NAD+] 1mM.
†[Cinnamoyl-CoA] 0.4 mM.
‡[p-coumaroyl-CoA] 0.4 mM.
Interestingly, PhCHD was highly sensitive to low levels of CAF-CoA. The presence of 1 μM CAF-CoA in the reaction containing 250 μM CA-CoA (three times Km value) resulted in a 50% loss in activity. No inhibitory effect from FA-CoA was observed. NADH also inhibited PhCHD, so lactate dehydrogenase (with pyruvate as an electron acceptor), was included in all reactions to remove NADH. Characterization revealed that NADH competitively inhibited the PhCHD reaction, with an apparent Ki of 25.3 ± 0.6 μM in the presence of NAD+ (Fig. S3).
PhCHD Activity Is Localized to Peroxisomes, the Site of the β-Oxidative Pathway of BA Biosynthesis.
Due to the presence of the canonical peroxisomal targeting signal 1 ‘SRM’ at its C terminus (27), the protein targeting prediction program PSORT (http://psort.hgc.jp/form.html) identified PhCHD as a peroxisomal protein. To biochemically confirm the predicted localization of PhCHD, peroxisomes were isolated from petunia petals and marker assays were used to assess organellar enrichment. The peroxisomal fraction showed eightfold and 11-fold enrichment in catalase (peroxisomal marker) and PhCHD activities, respectively, compared with crude extract (Table 2), while no enrichment in fumarase activity (mitochondrial marker), alcohol dehydrogenase activity (cytosolic marker) or chlorophyll (plastidial marker) was detected in this fraction.
Table 2.
Enrichement of PhCHD activity in peroxisomes
| Marker assay |
|||||
| Sample | PhCHD* | Catalase† | ADH* | Fumarase† | Chlorophyll‡ |
| Crude extract | 131 ± 11 | 58 ± 2 | 125 ± 13 | 342 ± 34 | 0.16 ± 0.01 |
| Peroxisomes | 1472 ± 111 | 458 ± 54 | <1 | 262 ± 53 | <0.01 |
PhCHD and marker enzyme activities were assayed in crude extracts and percoll-purified peroxisomes from petunia corollas sampled at 8:00 PM on day 2 postanthesis. Catalase, alcohol dehydrogenase (ADH), and fumarase were used as marker enzymes for peroxisomes, cytosol, and mitochondria, respectively, whereas chlorophyll content was used as the plastidic marker. Data are means ± SEM (n = 3).
*pkat·mg−1.
†nkat·mg.
‡ng·g FW−1.
Decrease of CA-CoA Hydratase-Dehydrogenase Activity in Planta Reduces Emission of Benzenoid-Derived Volatiles.
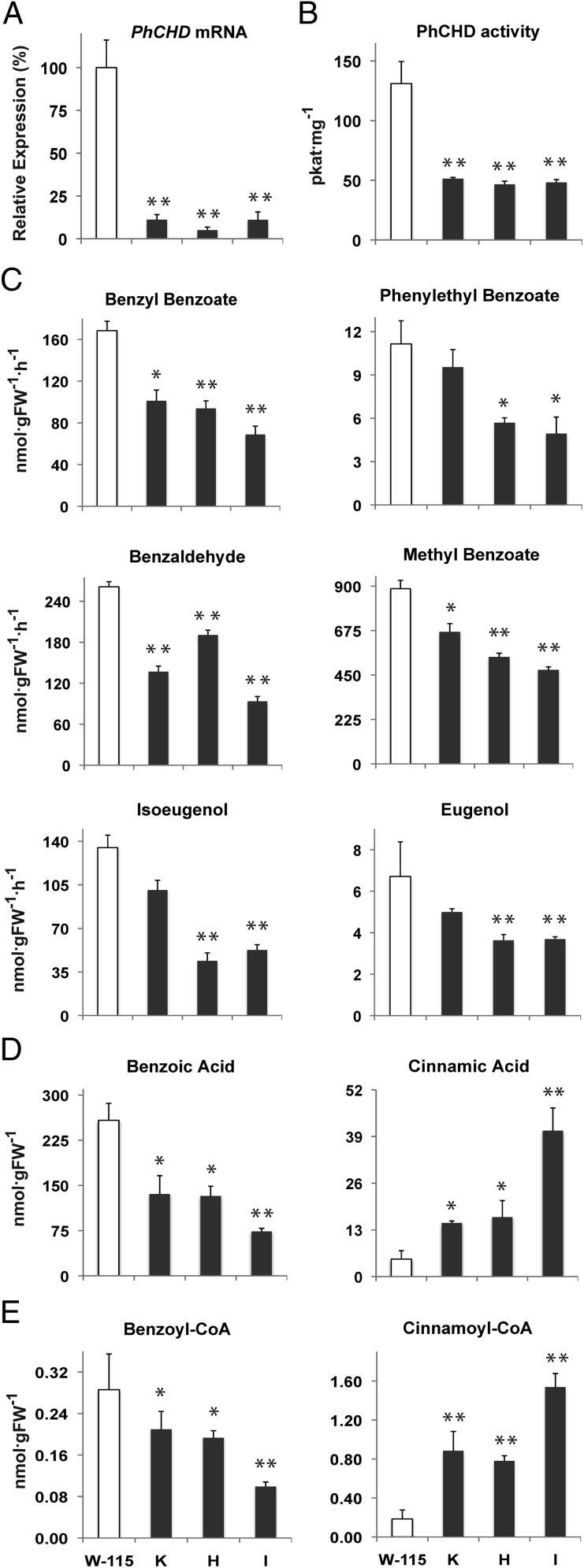
To assess the function of PhCHD in planta, transgenic plants were generated in which PhCHD transcript levels were decreased in flower petals using an RNAi approach under control of a petal-specific promoter (14), designed to eliminate any deleterious effects on plant vitality. The three independently transformed lines that showed the greatest reduction in PhCHD gene expression (85–95%; Fig. 3A) were chosen for further analysis and detailed metabolic profiling. RNAi suppression of PhCHD gene expression resulted in a decrease in PhCHD enzyme activity in crude petal extracts that ranged from 61–64% across the transgenic lines (Fig. 3B). Analysis of volatile compounds collected from transgenic flowers revealed a decline by 48% and 40% (on average relative to controls) in benzylbenzoate and phenylethylbenzoate, respectively (Fig. 3C), both of which directly depend on BA-CoA for their biosynthesis (Fig. 1) (13). Emission of benzaldehyde and methylbenzoate, the latter of which is the result of BA methylation, was also reduced on average by 46 and 37%, respectively, in transgenics compared with controls (Fig. 3C). Interestingly, a 51 and 39% decrease on average was observed in isoeugenol and eugenol emission, respectively (Fig. 3C), whereas no statistically significant changes were detected in emission of benzylalcohol, phenylacetaldehyde and phenylethanol, with exception of a slight reduction in the latter two compounds in line H (Fig. S4A).
Fig. 3.
Effect of PhCHD-RNAi suppression on PhCHD expression and activity, emission of benzenoid/phenylpropanoid compounds, and internal pools of organic acids and CoA esters in corollas of petunia flowers on day 2 postanthesis. (A) PhCHD mRNA levels in tissue collected at 3:00 PM and determined by qRT-PCR. Expression values for transgenic lines are shown as a percentage of PhCHD expression in control petals set at 100%. (B) PhCHD activity in petal crude extracts at 9:00 PM. (C) Floral volatile emission measured from 4:00 to 10:00 PM. Rates are calculated hourly assuming uniform emission over 6 h. (D) Organic acid and (E) CoA-ester internal pools at 9:00 PM. All data are means ± SEM (n = 3 biological replicates). White bars represent wild-type petunia (W-115); black bars correspond to PhCHD-RNAi lines (K, H, and I). *P < 0.05 and **P < 0.01 by analysis of variance (ANOVA). FW, fresh weight.
Analysis of internal pools of hydroxycinnamic acids in transgenic petals revealed (on average) a reduction in free BA (56%) and an increase in free CA (396%; Fig. 3D). Similar effects were observed on the corresponding CoA-esters; the pool of BA-CoA decreased by 42%, whereas that of CA-CoA increased by 479%, (Fig. 3E). No significant changes were observed in internal pools of other hydroxycinnamic acids (p-coumaric, caffeic and ferulic acids), or their corresponding CoA-esters (Fig. S4 B and C).
Discussion
Bifunctional Enzyme Catalyzes Two Intermediate Steps in the BA β-Oxidative Pathway.
In certain bacteria, BA derivatives (i.e., 4-hydroxybenzoic acid (4-HBA), salicylic acid) originate directly from chorismate by action of pyruvate lyases (28, 29), thus bypassing Phe as a precursor. However, genes encoding orthologs of these enzymes are missing in plant genomes. Despite the simple structure and importance of BA for primary and secondary metabolism (1–5) much is still unknown about its biosynthesis in plants. Multiple routes have been proposed for its formation from Phe (9–14, 30), although only a few of the corresponding genes and enzymes have been isolated and characterized (12–14, 17, 18). The most progress on elucidation of the β-oxidative pathway has been done using petunia flowers, which are rich in benzenoid/phenylpropanoid compounds (13). Genes responsible for the first and last reactions within the core of the β-oxidative route, PhCNL and PhKAT1, respectively, (Fig. 1) (17, 18) have been discovered, but those responsible for the two intermediary reactions remain unknown. Here, we identified PhCHD, a petunia gene encoding a bifunctional, peroxisomal enzyme responsible for the two unexplored steps within the BA β-oxidative pathway that convert CA-CoA to 3O3PP-CoA (Fig. 1 and Fig. S2). PhCHD displays a similar expression pattern to PhCNL and PhKAT1, all of which correlate spatially, temporally and developmentally with the emission of benzenoid volatiles (Fig. 2) (17, 18). PhCHD most efficiently uses CA-CoA as substrate (Table 1), which is produced by PhCNL, and together with PhKAT1 provides peroxisomes the capability to synthesize BA-CoA. In addition, the ability of PhCNL and PhCHD to adequately use p-coumaric acid (pCA; 17) and pCA-CoA (Table 1), respectively, may suggest that this pathway also produces 4HBA-CoA in peroxisomes to make 4HBA, which is proposed to proceed from tyrosine through a β-oxidative route analogous to that of BA to produce the ring moiety of ubiquinone (31).
While in mammals β-oxidation of straight-chain fatty acids occurs in mitochondria and peroxisomes, in plants it only takes place in the latter (reviewed in refs. 19, 32). One interesting feature of mammalian β-oxidation is that it appears to be regulated by energy demands in the cell in a redox-mediated fashion through NAD+/NADH levels, via inhibition of 3-hydroxyacyl-CoA dehydrogenase (33). Intriguingly, PhCHD is also inhibited by NADH (Fig. S3). Its inability, however, to use any of the fatty acyl-CoAs tested (Table S1) indicates that it likely does not function in fatty acid breakdown. Thus, any inhibitory affect by NADH is likely not related to energy status, but may instead serve to regulate BA biosynthesis or simply be a vestigial property. The inhibitory impact of CAF-CoA, however, most likely points to an undefined mechanism controlling formation of BA.
Identification of PhCHD adds a unique functional member to the plant MFP family reported to act on an aromatic acyl-CoA substrate. AIM1 shares 75% identity with PhCHD and is one of only two MFPs present in the Arabidopsis genome described to possess enoyl-CoA hydratase activity and the only homolog showing this activity on a substrate (crotonyl-CoA) other than a long-chain acyl-CoA (21). Because AIM1 expression is high in siliques (21), it should be investigated whether AIM1 is capable of using CA-CoA to be involved in BA biosynthesis for benzoyloxyglucosinolate production (34).
CHD Discovery Completes the Elucidation of the Core BA β-Oxidative Pathway.
Down-regulation of PhCHD expression in flowers (>85%) by an RNAi approach resulted in reduction of BA-CoA and BA with simultaneous accumulation of CA-CoA and CA (Fig. 3 D and E) confirming the involvement and position of PhCHD within the BA β-oxidative pathway in planta. Furthermore, PhCHD-RNAi plants emitted lower levels of benzaldehyde, methylbenzoate, benzylbenzoate, and phenylethylbenzoate relative to wild type (Fig. 3C). Because the latter two compounds rely on BA-CoA derived predominantly from the β-oxidative pathway (13, 14), the observed discrepancy between reduction in PhCHD activity (Fig. 3B) and metabolite pool sizes (Fig. 3 C and E) may be explained by a contribution coming from the non–β-oxidative pathway (via benzaldehyde → BA → BA-CoA; Fig. 1) (13). It is possible that a decrease in benzaldehyde emission by 46% (Fig. 3C) in transgenic lines is a result of an increased flux from benzaldehyde to BA and BA-CoA, which partially compensates for the decrease in production of the abovementioned emitted compounds (Fig. 1). Existence of such a flux from the non–β-oxidative pathway to BA-CoA has already been shown to take place in petunia flowers (13). Moreover, down-regulation of PhCHD led to a decrease in isoeugenol and eugenol production (Fig. 3C). Whereas the nature of this decrease is unknown, if flux is indeed increased through the non–β-oxidative pathway toward BA-CoA this may deplete the cytosolic pool of CA available to synthesize these compounds. The total increase in CA detected in PhCHD-RNAi lines (Fig. 3D) is a priori assumed to be sequestered in the peroxisome and not available in the cytosol.
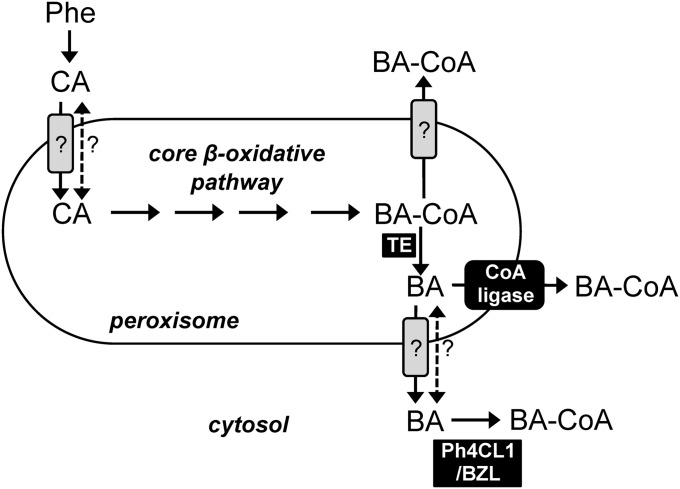
Formation of benzylbenzoate and phenylethylbenzoate occurs in the cytosol (13), but requires BA-CoA largely produced via the peroxisomal β-oxidative pathway (Fig. 1) (13, 17, 18). The open question now is how BA-CoA moves from the peroxisome to the cytosol, because the polar nature of the CoA moiety likely precludes free diffusion across the membrane. It is possible that BA-CoA is directly exported from peroxisomes in a transporter-mediated fashion (scheme in Fig. 4), but to date precedent only exists for fatty acyl-CoA import into this organelle (35–37). Alternatively, peroxisomes may contain thioesterases capable of hydrolyzing BA-CoA to its free acid, BA, which could then be transported and reconverted to its CoA form via different mechanisms (Fig. 4) and used for benzylbenzoate and phenylethylbenzoate biosynthesis. Upon cleavage of the CoA-thioester linkage in peroxisomes, the resulting BA might either freely diffuse across the membrane or be transported and serve as substrate for cytosolic CoA-ligases such as Ph4CL1 (17) and BZL (15), or may be reactivated to BA-CoA by an unknown membrane-associated ligase during export from the peroxisome (Fig. 4). The latter model has already been proposed for passage of free fatty acids across the plasma membrane in cyanobacteria (38) and through the outer plastid envelope in plants (39). The potential for conversion of BA-CoA to BA before transport from peroxisomes is supported by the recent discovery of peroxisomal thioesterases capable of hydrolyzing aromatic acyl-CoA substrates, including BA-CoA, in Arabidopsis (40). Thus, we assayed the same petunia peroxisomal preparation used to confirm subcellular localization of PhCHD activity (Table 2) for BA-CoA thioesterase activity. Indeed, BA-CoA thioesterase activity was enriched (18.1 ± 2.65 compared with 1.60 ± 0.27 pkat mg−1 protein in peroxisomes versus crude extract, respectively) similar to PhCHD (Table 2), suggesting these types of enzymes are not unique to Arabidopsis. Moreover, in humans it has been proposed that peroxisomal CoA-thioesterases serve auxiliary roles in lipid β-oxidation by converting pathway products to transportable forms and maintaining free CoA-SH pools (41). Thus, in addition to investigating the role of the unidentified plant BA-CoA thioesterase in formation of BA for export out of peroxisomes, it should be studied whether the enzyme also has a regulatory role in maintaining organellar CoA-SH levels, and whether it exerts control over the pathway by using other BA β-oxidative pathway CoA intermediates.
Fig. 4.
Possible routes for benzoic acid trafficking out of peroxisomes. Dashed arrow indicates hypothetical diffusion of free protonated acids. Solid arrows with gray boxes indicate putative transporter-mediated steps. Solid arrow with black box indicates a possible membrane-associated CoA ligase coupled to passage of benzoic acid across the peroxisomal membrane. BA, benzoic acid; BA-CoA, benzoyl-CoA; BZL, benzoate:CoA ligase (15); CA, cinnamic acid; Ph4CL1, petunia 4-coumarate:CoA ligase 1 (17); Phe, phenylalanine; and TE, thioesterase.
In conclusion, the discovery of PhCHD provides evidence of a plant MFP acting on aromatic acyl-CoAs. This completes elucidation of the core β-oxidative pathway (CA → CA-CoA → 3H3PP-CoA → 3O3PP-CoA → BA-CoA) for BA biosynthesis in plants, although an auxiliary thioesterase(s) likely exists, which should be explored for regulatory roles in product transport, cofactor recycling, and pathway control. Furthermore, to fully understand the molecular mechanisms connecting the peroxisomal β-oxidative pathway to the benzenoid network in the cytosol, it should also be investigated whether acyl-CoA transporters present in peroxisomes, and/or membrane-associated acyl-CoA ligases contribute to BA metabolism in plants.
Materials and Methods
Chemicals and Reagents.
All hydroxycinnamic acid-CoA esters, CA-CoA, pCA-CoA, CAF-CoA, and FA-CoA, with exception of BA-CoA, were synthesized enzymatically and purified as described previously (42). Crotonoyl-CoA was synthesized and purified as described (43). All other chemicals were from Sigma-Aldrich) unless noted.
Growth Conditions and Generation of PhCHD-RNAi Transgenic Plants.
Petunia hybrida cv. Mitchell plants (W-115; Ball Seed) were grown under standard greenhouse conditions (44) with a light period from 6:00 AM to 9:00 PM. Generation of the PhCHD-RNAi construct was performed as previously described (17) with some modifications (SI Materials and Methods). PhCHD-RNAi transgenic plants were generated via Agrobacterium tumefaciens (strain GV2260 carrying plasmid pLIS-PhCHD-RNAi) transformation using the standard leaf disk transformation method (45).
qRT-PCR.
Sample collection, isolation of RNA and qRT-PCR analyzed relative to the reference gene elongation factor 1-α for PhCHD, TC10274, TC7383, FN000667, TC12800, TC9756, and TC10560 using gene-specific primer pairs (Table S2), were performed as previously described (17). For absolute quantification of PhCHD transcript levels by qRT-PCR see SI Materials and Methods.
Peroxisome Isolation.
Peroxisome isolation from petunia corollas of 2-d-old flowers harvested at 8:00 PM and marker assays to determine integrity and enrichment were done as described previously (17).
Enzyme Assays.
Recombinant PhCHD (SI Materials and Methods describes cloning and purification) activity with aromatic, short, medium, and long-chain acyl-CoAs was determined by measuring consumption of acyl-CoAs at A260 (46) using an HPLC-based assay described in SI Materials and Methods. The standard reaction (100 μL) contained 50 mM Bis⋅Tris propane pH 9.5, 2 mM MgCl2, 1 mM pyruvic acid, 400 μM acyl-CoA, 1 mM NAD+, 2 mM CoA, 2 units of lactate dehydrogenase (Calzyme Laboratories), and 0.15 μg purified PhCHD protein. The reaction was initiated by adding the acyl-CoA substrate and incubated for 60 min at room temperature before termination with 50% (wt/vol) trichloroacetic acid [final concentration 5%]. Ten microliters of the reaction product was analyzed by HPLC-diode array detector (DAD) spectrophotometry as described in SI Materials and Methods. Assays containing all reaction components with boiled protein were used as negative controls. Determination of kinetic parameters for recombinant PhCHD with CA-CoA and pCA-CoA were carried out using 15-min assays also containing 7.5 μg of recombinant PhKAT1 (18). Kinetic data were evaluated by hyperbolic regression analysis (HYPER.EXE, version 1.00, 1992). Triplicate assays were performed for all data points for kinetic analysis. Verification of reaction products was performed by LC TOF/MS (SI Materials and Methods).
Native PhCHD activity in crude extracts (3 μg protein, see SI Materials and Methods for preparation) and peroxisomes (84 ng protein) was determined as described above except using 30- to 60-min assays. Thioesterase assays to measure the hydrolysis of BA-CoA were performed spectrophotometrically using a slightly modified 5,5′-dithiobis-(2-nitroBA) (DTNB) method (40) described in SI Materials and Methods.
Targeted Metabolite Profiling.
Floral volatiles were collected from control and PhCHD-RNAi petunia flowers at the peak of emission, 4:00 PM to 10:00 PM, on day 2 postanthesis in 2-h segments by a closed-loop stripping method and analyzed as previously described (14, 23). To determine the internal pools of organic acids and CoA esters, petal tissue was collected from transgenic and control plants at 9:00 PM on day 2 postanthesis to minimize the effect of rhythmicity. The level of organic acids was determined as described (SI Materials and Methods). Extraction and quantificaiton of CoA esters was performed as described (47).
Supplementary Material
Acknowledgments
We thank Dr. Eran Pichersky for the PhKAT1 construct. This work was supported by Grant MCB-0919987 from the National Science Foundation (to N.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. JX42126).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211001109/-/DCSupplemental.
References
- 1.Dudareva N, Negre F, Nagegowda DA, Orlova I. Plant volatiles: Recent advances and future perspectives. Crit Rev Plant Sci. 2006;25:417–440. [Google Scholar]
- 2.Chipley J. Antimicrobials in Food. 3rd Ed. Boca Raton, FL: CRC; 2005. pp. 11–48. [Google Scholar]
- 3.Quan C, Mok WM, Wang GK. Use-dependent inhibition of Na+ currents by benzocaine homologs. Biophys J. 1996;70:194–201. doi: 10.1016/S0006-3495(96)79563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saïdana D, et al. Studies of the essential oil composition, antibacterial and antifungal activity profiles of Frankenia laevis L. from Tunisia. J Essent Oil Res. 2010;22:349–353. [Google Scholar]
- 5.Veeresham C, Mamatha R, Babu CP, Srisilam K, Kokate CK. Production of taxol and its analogues from cell cultures of Taxus wallichiana. Pharm Biol. 2003;41:426–430. [Google Scholar]
- 6.Yalpani N, Leon J, Lawton MA, Raskin I. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 1993;103:315–321. doi: 10.1104/pp.103.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman P, et al. Salicylic acid in rice: Biosynthesis, conjugation, and possible role. Plant Physiol. 1995;108:633–639. doi: 10.1104/pp.108.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coquoz JL, Buchala A, Metraux JP. The biosynthesis of salicylic acid in potato plants. Plant Physiol. 1998;117:1095–1101. doi: 10.1104/pp.117.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvis AP, Schaaf O, Oldham NJ. 3-hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta. 2000;212:119–126. doi: 10.1007/s004250000377. [DOI] [PubMed] [Google Scholar]
- 10.Hertweck C, Jarvis AP, Xiang L, Moore BS, Oldham NJ. A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid beta-oxidation. ChemBioChem. 2001;2:784–786. doi: 10.1002/1439-7633(20011001)2:10<784::AID-CBIC784>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Abd El-Mawla AM, Beerhues L. Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta. 2002;214:727–733. doi: 10.1007/s004250100657. [DOI] [PubMed] [Google Scholar]
- 12.Long MC, et al. Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. Plant J. 2009;59:256–265. doi: 10.1111/j.1365-313X.2009.03864.x. [DOI] [PubMed] [Google Scholar]
- 13.Boatright J, et al. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 2004;135:1993–2011. doi: 10.1104/pp.104.045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlova I, et al. Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell. 2006;18:3458–3475. doi: 10.1105/tpc.106.046227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beuerle T, Pichersky E. Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys. 2002;400:258–264. doi: 10.1016/S0003-9861(02)00026-7. [DOI] [PubMed] [Google Scholar]
- 16.Wildermuth MC. Variations on a theme: Synthesis and modification of plant benzoic acids. Curr Opin Plant Biol. 2006;9:288–296. doi: 10.1016/j.pbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Klempien A, et al. Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell. 2012;24:2015–2030. doi: 10.1105/tpc.112.097519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Moerkercke A, Schauvinhold I, Pichersky E, Haring MA, Schuurink RC. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 2009;60:292–302. doi: 10.1111/j.1365-313X.2009.03953.x. [DOI] [PubMed] [Google Scholar]
- 19.Graham IA. Seed storage oil mobilization. Annu Rev Plant Biol. 2008;59:115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- 20.Verdonk JC, et al. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry. 2003;62:997–1008. doi: 10.1016/s0031-9422(02)00707-0. [DOI] [PubMed] [Google Scholar]
- 21.Richmond TA, Bleecker AB. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999;11:911–1923. doi: 10.1105/tpc.11.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rylott EL, et al. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. Plant J. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- 23.Maeda H, et al. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell. 2010;22:832–849. doi: 10.1105/tpc.109.073247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuong SDX, Mullen RT, Muench DG. Identification of a rice RNA- and microtubule-binding protein as the multifunctional protein, a peroxisomal enzyme involved in the beta -oxidation of fatty acids. J Biol Chem. 2002;277:2419–2429. doi: 10.1074/jbc.M109510200. [DOI] [PubMed] [Google Scholar]
- 25.Ohya H, Ogata A, Nakamura K, Chung K, Sano H. A stress-responsive multifunctional protein involved in beta-oxidation in tobacco plants. Plant Biotechnol. 2008;25:503–508. [Google Scholar]
- 26.Preisig-Müller R, Gühnemann-Schäfer K, Kindl H. Domains of the tetrafunctional protein acting in glyoxysomal fatty acid beta-oxidation. Demonstration of epimerase and isomerase activities on a peptide lacking hydratase activity. J Biol Chem. 1994;269:20475–20481. [PubMed] [Google Scholar]
- 27.Reumann S, Ma C, Lemke S, Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols BP, Green JM. Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J Bacteriol. 1992;174:5309–5316. doi: 10.1128/jb.174.16.5309-5316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaille C, Kast P, Haas D. Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J Biol Chem. 2002;277:21768–21775. doi: 10.1074/jbc.M202410200. [DOI] [PubMed] [Google Scholar]
- 30.Bjorklund JA, Leete E. Biosynthesis of the benzoyl moiety of cocaine from cinnamic acid via (R)-(+)-3-hydroxy-3-phenylpropanoic acid. Phytochemistry. 1992;31:3883–3887. [Google Scholar]
- 31.Clarke CF. New advances in coenzyme Q biosynthesis. Protoplasma. 2000;213:134–147. [Google Scholar]
- 32.Graham IA, Eastmond PJ. Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res. 2002;41:156–181. doi: 10.1016/s0163-7827(01)00022-4. [DOI] [PubMed] [Google Scholar]
- 33.Eaton S. Control of mitochondrial β-oxidation flux. Prog Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- 34.Kliebenstein DJ, et al. Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J. 2007;51:1062–1076. doi: 10.1111/j.1365-313X.2007.03205.x. [DOI] [PubMed] [Google Scholar]
- 35.Hettema EH, Tabak HF. Transport of fatty acids and metabolites across the peroxisomal membrane. Biochim Biophys Acta. 2000;1486:18–27. doi: 10.1016/s1388-1981(00)00045-7. [DOI] [PubMed] [Google Scholar]
- 36.Verleur N, Hettema EH, van Roermund CWT, Tabak HF, Wanders RJA. Transport of activated fatty acids by the peroxisomal ATP-binding-cassette transporter Pxa2 in a semi-intact yeast cell system. Eur J Biochem. 1997;249:657–661. doi: 10.1111/j.1432-1033.1997.00657.x. [DOI] [PubMed] [Google Scholar]
- 37.Nyathi Y, et al. The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Δ mutant for metabolism of long-chain fatty acids and exhibits fatty acyl-CoA-stimulated ATPase activity. J Biol Chem. 2010;285:29892–29902. doi: 10.1074/jbc.M110.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Berlepsch S, et al. The acyl-acyl carrier protein synthetase from Synechocystis sp. PCC 6803 mediates fatty acid import. Plant Physiol. 2012;159:606–617. doi: 10.1104/pp.112.195263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koo AJK, Ohlrogge JB, Pollard M. On the export of fatty acids from the chloroplast. J Biol Chem. 2004;279:16101–16110. doi: 10.1074/jbc.M311305200. [DOI] [PubMed] [Google Scholar]
- 40.Widhalm JR, et al. Phylloquinone (vitamin K(1) ) biosynthesis in plants: two peroxisomal thioesterases of lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-coa. Plant J. 2012;71:205–215. doi: 10.1111/j.1365-313X.2012.04972.x. [DOI] [PubMed] [Google Scholar]
- 41.Hunt MC, Alexson SEH. Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog Lipid Res. 2008;47:405–421. doi: 10.1016/j.plipres.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Beuerle T, Pichersky E. Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal Biochem. 2002;302:305–312. doi: 10.1006/abio.2001.5574. [DOI] [PubMed] [Google Scholar]
- 43.Chohan SN, Copeland L. Acetoacetyl coenzyme A reductase and polyhydroxybutyrate synthesis in rhizobium (Cicer) sp. Strain CC 1192. Appl Environ Microbiol. 1998;64:2859–2863. doi: 10.1128/aem.64.8.2859-2863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dudareva N, et al. Developmental regulation of methylbenzoate biosynthesis. Plant Cell. 2000;12:949–961. doi: 10.1105/tpc.12.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horsch RB, et al. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 46.Hubbell T, Behnke WD, Woodford JK, Schroeder F. Recombinant liver fatty acid binding protein interacts with fatty acyl-coenzyme A. Biochemistry. 1994;33:3327–3334. doi: 10.1021/bi00177a025. [DOI] [PubMed] [Google Scholar]
- 47.Qualley AV, Cooper BR, Dudareva N. Profiling hydroxycinnamoyl-coenzyme A thioesters: Unlocking the back door of phenylpropanoid metabolism. Anal Biochem. 2012;420:182–184. doi: 10.1016/j.ab.2011.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.